Abstract
The catalase gene, katA, of the sepiolid squid symbiont Vibrio fischeri has been cloned and sequenced. The predicted amino acid sequence of KatA has a high degree of similarity to the recently defined group III catalases, including those found in Haemophilus influenzae, Bacteroides fragilis, and Proteus mirabilis. Upstream of the predicted start codon of katA is a sequence that closely matches the consensus sequence for promoters regulated in Escherichia coli by the alternative sigma factor encoded by rpoS. Further, the level of expression of the cloned katA gene in an E. coli rpoS mutant is much lower than in wild-type E. coli. Catalase activity is induced three- to fourfold both as growing V. fischeri cells approach stationary phase and upon the addition of a small amount of hydrogen peroxide during logarithmic growth. The catalase activity was localized in the periplasm of wild-type V. fischeri cells, where its role could be to detoxify hydrogen peroxide coming from the external environment. No significant catalase activity could be detected in a katA null mutant strain, demonstrating that KatA is the predominately expressed catalase in V. fischeri and indicating that V. fischeri carries only a single catalase gene. The catalase mutant was defective in its ability to competitively colonize the light organs of juvenile squids in coinoculation experiments with the parent strain, suggesting that the catalase enzyme plays an important role in the symbiosis between V. fischeri and its squid host.
The luminous marine bacterium Vibrio fischeri occupies a unique niche in nature: it is the only bacterial species found within the symbiotic light-emitting organ of the Hawaiian squid, Euprymna scolopes (7). Interestingly, not all strains of V. fischeri are equally capable of establishing and maintaining a colonization of the light organ (24, 31), suggesting that the symbiosis-competent strains express special factors required for growth in the squid. Such factors could include an important adhesin, enzymes for metabolizing a specific host-derived nutrient, or defenses that protect against a host-produced stress.
When E. scolopes juveniles hatch, their light organs are devoid of bacteria (47), but they rapidly become colonized if competent V. fischeri cells are present in the surrounding seawater. Within 24 h of the initiation of the association, a few infecting bacteria have multiplied to fully colonize the juvenile light organ, which can contain approximately 106 cells (36). These cells, however, have undergone dramatic morphological alterations, such as the loss of their flagella and a decrease in cell volume (36). These and other changes demonstrate that V. fischeri cells recognize and respond to the particular conditions present inside the light organ, perhaps including environmental stresses that result from their association with the eukaryotic host. While the oxidative conditions inside the light organ are only now being analyzed, host mRNA that encodes a halide peroxidase is abundant (43, 48). It has been suggested that this peroxidase, which converts hydrogen peroxide into toxic hypohalous acids (e.g., hypochlorous acid), could serve as a host defense mechanism, like the human myeloperoxidase to which it is related (48). If this hypothesis is correct, oxidative stress may be a significant condition that V. fischeri cells encounter in the light organ environment.
One strategy that bacteria use to combat the oxidative stress resulting from exposure to hydrogen peroxide is to produce the enzyme catalase, which decomposes this reactive oxygen species into water and oxygen (26). The addition of hydrogen peroxide to a colony of V. fischeri cells results in a vigorous bubbling, a reaction which suggests that this organism does indeed possess an active catalase. In this report we describe (i) the cloning of the V. fischeri gene encoding this activity, which we have called katA, and the analysis of its predicted protein sequence; (ii) the expression pattern of the katA product during growth of the cells in culture; and (iii) the construction of a katA mutant and investigation of its ability to colonize juveniles of E. scolopes squid.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Wild-type V. fischeri strain ES114 (7) is a natural isolate from the squid E. scolopes. Strain ESR1 (20) is a rifampin-resistant derivative of ES114 and is the parent strain used in the construction of the catalase mutant. The following Escherichia coli strains were also used: DH5α, UM2 (katE katG [27]), MC1000 (parental strain for JV1012 [11]), and JV1012 (rpoS [44]).
LB (12) broth was used for the growth of E. coli strains. V. fischeri strains were grown either in LBS medium (16), which contains 1% tryptone, 0.5% yeast extract, 2% NaCl, and 0.3% glycerol in 50 mM Tris-HCl (pH 7.5), or in SWT medium (7), which contains 0.5% tryptone, 0.3% yeast extract, and 0.3% glycerol in 70% seawater. Agar was added to a concentration of 1.5% for solid media. Antibiotics were added when appropriate to the following final concentrations: ampicillin, 100 μg/ml; erythromycin, 150 μg/ml for E. coli and 5 μg/ml for V. fischeri.
Plasmid construction.
A library of EcoRI-digested V. fischeri chromosomal DNA cloned into pBluescript KS (Stratagene Inc., La Jolla, Calif.) was used to transform E. coli UM2 (katE katG) cells. Ampicillin-resistant isolates were screened for the presence of a functional catalase clone by visual identification of bubbles when hydrogen peroxide was dropped on the individual colonies. Several such hydrogen peroxide-decomposing colonies were obtained, and the plasmids isolated from these strains all appeared to carry an EcoRI insert of about 4.4 kb in size. One of these plasmids, pLP2 (Fig. 1A), was saved for further study.
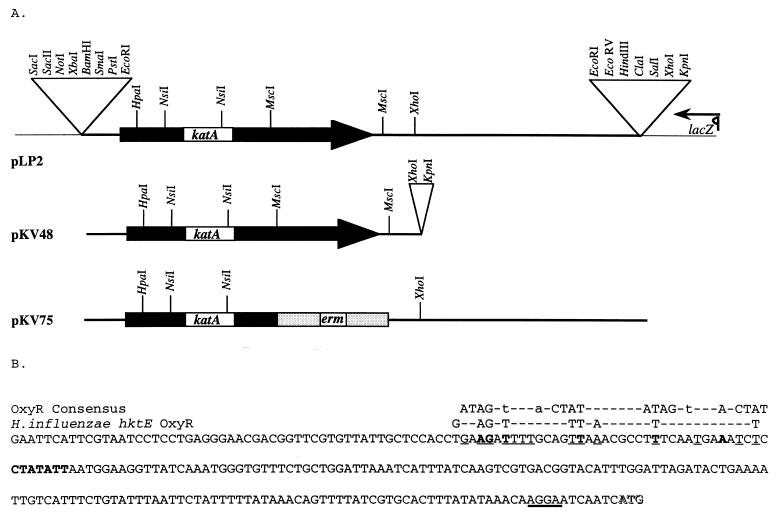
FIG. 1.
(A) Partial maps of plasmids used in this study. Plasmids pLP2, pKV48, and pKV75 are derivatives of pBluescript KS (Stratagene), a portion of which, including the multiple cloning site, is denoted by the thin line. The V. fischeri catalase gene, katA, is indicated by the black box, and the erythromycin resistance gene, erm, is indicated by the gray box. Transcription of the lacZ gene is in the direction of the arrow. (B) Nucleotide sequence upstream of the V. fischeri katA gene from the EcoRI site to the putative ATG start codon. Nucleotides in the putative ribosome binding site upstream of the gene are underlined in bold, and a potential −10 promoter sequence is in boldface. Nucleotides that match the proposed OxyR consensus sequence (41, 42) are in boldface and underlined, while those that match the region upstream of the H. influenzae hktE gene, where an OxyR binding site has been proposed (6), are underlined. The proposed OxyR motifs are displayed above. The putative ATG start site is shown in shadowbox letters.
Plasmid pKV48 (Fig. 1A) was derived from pLP2 by the deletion of a 2.2-kb XhoI fragment. The following constructs (not shown) were derived from pKV48: pKV52, which was deleted for a 0.3-kb fragment of DNA from SmaI in the multiple cloning site to HpaI; pKV53, which was deleted for the 0.75-kb fragment of DNA between the two MscI sites; and pKV54, which carries a 0.4-kb deletion of DNA between the two NsiI sites.
Plasmid pKV75 (Fig. 1A) was derived from pLP2 as follows. pLP2 was digested with MscI, and the resulting DNA fragments were purified by gel electrophoresis. The larger fragment was extracted from the gel and purified by using a Geneclean kit (Bio101, Inc., Vista, Calif.). The 1.1-kb fragment of DNA carrying the erythromycin resistance gene was purified in a similar manner after SmaI and EcoRV digestion of pKV25 (a derivative of pUC19 [50] in which the erythromycin resistance gene from pLS3 [45] was cloned at the SalI and PstI sites). The two purified fragments were ligated by using T4 DNA ligase (Promega, Madison, Wis.), and the resulting mix was used to transform E. coli DH5α cells made competent by treatment with calcium chloride. Ampicillin- and erythromycin-resistant clones were further examined by restriction analysis to identify a clone in which the erythromycin resistance marker had replaced the C-terminal portion of katA, resulting in pKV75 (Fig. 1A).
Catalase activity measurements.
Extracts of V. fischeri cells were made as follows. Cells were pelleted by a 2-min room temperature microcentrifugation and then resuspended in 0.5 ml of an extract buffer containing 5 mM potassium phosphate (pH 7.0), 5 mM EDTA, and 10% glycerol and stored on ice. The cells were lysed by a gentle sonication (four to five rounds of four to five short pulses) in a Vibra-cell microtip sonicator (Sonics and Materials, Inc., Danbury, Conn.) set at an amplitude of 50 U. Cell debris was removed by microcentrifugation at 4°C for 10 min, and the resulting extracts were kept on ice until assayed. Catalase activity was measured by using a quantitative spectrophotometric assay for the decomposition of hydrogen peroxide (4) and reported as units/milligram of protein in the extract, where 1 U equals 1 μmol of hydrogen peroxide decomposed per min. Soluble protein concentrations were determined by the method of Lowry et al. (28).
Subcellular localization of the catalase protein.
Periplasmic proteins from V. fischeri were selectively released using a chloroform permeabilization treatment (2) as follows. Cells from 1 ml of a culture of stationary-phase V. fischeri grown in SWT were pelleted in a microcentrifuge, the supernatant was discarded, and the cells were resuspended in the residual fluid. Chloroform (20 μl) was added, and the cells were vortexed briefly. After a 10-min exposure, 200 μl of sterile 70% seawater was added, and the cell suspension was gently mixed, followed by a 2-min centrifugation at 4°C to pellet the cells. The upper 125 μl of supernatant were removed to a fresh tube, and aliquots were assayed both for catalase activity and for the activity of luciferase (30), a cytoplasmically located enzyme.
Construction of the catalase mutant.
A catalase mutant of V. fischeri was constructed by marker exchange using pKV75, a plasmid in which about 750 nucleotides between the two MscI sites in the katA gene are replaced by the gene for erythromycin resistance (Fig. 1A). This plasmid was introduced sequentially into a dam mutant strain of E. coli, and then into V. fischeri strain ESR1, by electroporation (45). Erythromycin-resistant colonies of V. fischeri were purified by several passages on SWT agar, and each of these was tested for the inability to decompose hydrogen peroxide. Several colonies that produced no bubbles upon the addition of hydrogen peroxide were designated as presumptive catalase mutants. Southern blot analysis of one such strain (KV433) and its parent (ESR1), using the 32P-radiolabeled katA gene as a probe, revealed that the internal MscI restriction fragment that is present in ESR1 is missing in KV433. The predicted insertion was further confirmed when the probe hybridized to an EcoRI fragment of KV433 DNA that was slightly larger than the hybridizing fragment of the parent strain (data not shown).
Colonization of the juvenile squid light organ.
To determine the effect of the katA mutation on colonization of juvenile E. scolopes, animals were inoculated with either KV433 or ESR1. The development of the symbiotic infection was monitored by using the onset of luminescence of the squids as an indicator, as previously described (35). After 45 and 72 h postinoculation, individual squids were rinsed in sterile seawater and homogenized. Dilutions of the homogenates were spread on SWT agar, and the V. fischeri colonies that arose were counted to estimate the extent of colonization. Mixed culture experiments were performed with a 1:1 ratio of KV433 to ESR1 cells. The juvenile squid in these experiments were exposed for a short amount of time (3 to 3.5 h) with a low inoculum of cells (about 600 to 800 CFU/ml of seawater) and then removed and placed in symbiont-free seawater for the remainder of the experiment. Squid homogenates were diluted, and aliquots were spread on SWT agar plates and erythromycin-containing LBS plates to determine the level of colonization by KV433 (erythromycin resistant) and ESR1 (erythromycin sensitive).
Sequence analysis.
The V. fischeri katA gene was sequenced by using a dye terminator cycle sequencing system at the Biotechnology-Molecular Biology Instrumentation Facility of the University of Hawaii, Manoa. Subclones of the pLP2 plasmid were constructed (not shown), and sequencing of both strands was performed with primers complementary to the vector DNA. A BLAST search (1, 18) was used to compare the V. fischeri KatA deduced amino acid sequence to those of catalases from other organisms.
Nucleotide sequence accession number.
The GenBank nucleotide accession number for the V. fischeri katA gene sequence is AF011784.
RESULTS
Isolation of the katA gene of V. fischeri.
A plasmid, pLP2 (Fig. 1A), was identified from a plasmid library carrying V. fischeri genomic DNA based on its ability to complement a catalase-negative (katE katG double mutant) strain of E. coli. Restriction digests and subcloning experiments localized the catalase activity to within a 2.2-kb region of the insert DNA (pKV48 [Fig. 1A]) that retained the ability to complement the E. coli mutant. A deletion of DNA between either the two MscI sites or the two NsiI sites, however, resulted in a loss of catalase activity. In addition, a 0.3-kb deletion of DNA between the EcoRI and HpaI sites also resulted in a loss of catalase activity (not shown). Taken together, these data suggested that the V. fischeri catalase gene extends most of the length of the DNA located between the EcoRI and XhoI restriction sites in pLP2 (Fig. 1A).
Sequence analysis of the V. fischeri katA gene.
Both strands of a 1,913-base region of pLP2, including the presumed location of the catalase-encoding gene, were sequenced (Fig. 1B and data not shown). An open reading frame 482 codons in length, which could potentially encode a protein with a molecular mass of 54,830 Da, was located in this region. A potential ribosome binding site (38) (AGGA) was found 9 bases upstream of the putative start codon (Fig. 1B). Further upstream, at position −161 relative to the presumed translational start, is a sequence (CTATAAT) that is a reasonable match to the consensus −10 sequence recognized by E. coli RNA polymerase carrying the housekeeping sigma subunit, ς70 (21). However, there was no such match for the corresponding −35 region that would be expected for a ς70-regulated promoter. Promoters recognized by the alternative sigma factor ςS (encoded by rpoS), a key positive regulator of genes induced by stress and at stationary phase in many bacteria (22), have a −10 consensus sequence similar to ς70 promoters but often lack identifiable −35 regions (17, 49). The sequence features upstream of the katA gene suggest that it could be regulated by such an alternative sigma factor.
The putative promoter region of the katA gene may also contain a binding site for OxyR, a conserved activator of transcription of catalases and other genes required for defense against oxidative stress (40). Although this potential binding site matches only poorly the proposed E. coli OxyR binding site consensus sequence (Fig. 1B) (41, 42), there is a higher similarity of the V. fischeri upstream sequence to the nucleotides in a proposed OxyR binding site upstream of the catalase gene in Haemophilus influenzae (Fig. 1B) (6). A hypothetical V. fischeri OxyR protein might, therefore, regulate the activation of katA in response to environmental cues.
The V. fischeri catalase sequence shows high identities to catalases of many other bacteria, including H. influenzae (78% [6]), Bacteroides fragilis (74% [34]), Proteus mirabilis (71% [10]), Bordetella pertussis (73%), and Bacillus subtilis (54%). In addition, katA has significant similarity to human (60%), bovine (59%), and other mammalian catalases. These similarities suggest that the V. fischeri katA gene is a “typical monofunctional” catalase as defined by Loewen (26) and, specifically, that it can be classified with the very recently identified group III phylogenetic class of bacterial catalases described by Klotz et al. (23). The amino acid sequence of the V. fischeri catalase also shows a lower sequence identity (46%) to the E. coli catalase, HPII (46), that is induced at stationary phase.
Regulation of expression of the catalase enzyme in response to entry into stationary phase and to oxidative stress.
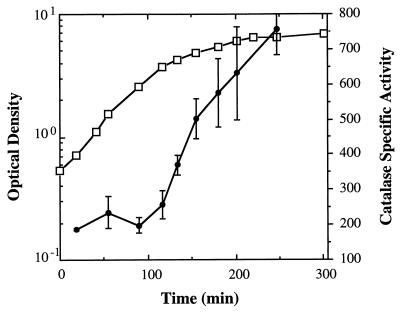
Catalase activity in cell extracts of wild-type V. fischeri strain ES114 was measured at different times during its growth in a rich medium. While significant catalase activity could be detected during all phases of growth, the amount of catalase was induced more than threefold in cells that were beginning to enter stationary phase (Fig. 2). Similar results were obtained for V. fischeri ESR1 (not shown). The level of activity of the V. fischeri catalase enzyme in extracts of cells in stationary phase, approximately 800 U/mg of protein, is about 10-fold higher than that observed in extracts of an overnight culture of E. coli (data not shown) and may reflect different levels of catalase expression in the two organisms.
FIG. 2.
Catalase specific activity during the growth of V. fischeri ES114. The average optical density of the cultures is indicated by open squares; catalase specific activity, reported in units/milligram of protein, is designated by black circles. The data are averages of activity assays performed on extracts of triplicate cultures. The error bars are equal to 1 standard deviation.
The induction of catalase activity in V. fischeri cells entering stationary phase suggested that the gene product of an rpoS homolog might be involved in the regulation of katA. Because V. fischeri rpoS mutants have not yet been isolated or constructed, this hypothesis was addressed by an alternative method. The katA-encoding plasmid pKV48 (Fig. 1A) was introduced into both an E. coli rpoS mutant and its wild-type parent. When catalase specific activity was assayed in cell extracts after overnight growth, high levels were observed in the wild-type strain carrying pKV48 (Table 1). These levels were over 15-fold higher than the activity of the endogenous E. coli catalases observed in a control strain transformed with the vector alone. In contrast, when pKV48 was present in the rpoS mutant, this high level of catalase activity was not observed (Table 1). These data suggest that in E. coli, transcription of the katA gene may be directed almost exclusively by RNA polymerase containing ςS. This finding, taken together with the temporal pattern of catalase expression in V. fischeri, lends support to the model that the katA gene is transcribed under the control of an rpoS homolog in V. fischeri.
TABLE 1.
Catalase specific activities of E. coli strains carrying the V. fischeri katA gene and of V. fischeri cells challenged with hydrogen peroxide
| Expt | Avg catalase sp act ± 1 SD (U/mg of protein)a |
|---|---|
| 1b | |
| E. coli MC1000/pKV48 | 756 ± 210 |
| E. coli MC1000/pBS | 23 ± 12 |
| E. coli JV1012/pKV48 | 64 ± 5 |
| E. coli JV1012/pBS | 38 ± 8 |
| 2c | |
| −H2O2 | 197 ± 34 |
| +H2O2 | 709 ± 32 |
Average of activity assays performed on extracts of triplicate cultures.
Stationary-phase cultures of E. coli MC1000 and its isogenic rpoS mutant derivative JV1012, carrying either plasmid pKV48, which encodes the V. fischeri katA gene, or the control vector pBluescript (pBS) were assayed for catalase activity.
Exponentially growing cultures of V. fischeri ES114 were exposed to 60 μM hydrogen peroxide for 30 min and then assayed for the ability to decompose hydrogen peroxide. Extracts were made from cells either exposed or not exposed to hydrogen peroxide.
Catalases from a number of other organisms, including the highly similar group III enzyme from H. influenzae, are induced upon exposure to low levels of hydrogen peroxide. In E. coli, however, the catalase (HPI) that is induced in response to this reactive oxygen species is distinct from the catalase (HPII) that is induced at stationary phase; the former requires OxyR for induction, while the latter is dependent on ςS. We therefore asked whether wild-type V. fischeri would induce the expression of its catalase in response to the presence of hydrogen peroxide. Exposing a culture of V. fischeri ES114 cells to 60 μM hydrogen peroxide for 30 min early in logarithmic growth resulted in a 3.5-fold increase in the level of catalase specific activity (Table 1), e.g., up to the level seen in stationary phase cells (Fig. 2). Thus, V. fischeri cells can respond to the presence of hydrogen peroxide in their external environment by inducing the expression of catalase activity. This induction most likely occurs at the transcriptional level and may be dependent on OxyR acting at the putative OxyR binding site described above (Fig. 1B).
Subcellular localization of the catalase protein.
Although many of the well-characterized bacterial catalases are typically present in the cytoplasm, some, such as the KatA catalase from B. subtilis (8, 29), are exported. Because the V. fischeri enzyme has a high degree of identity with the B. subtilis catalase, and because a secreted catalase might be of particular value if the light organ environment were a significant source of reactive oxygen species, we investigated whether the catalase protein from V. fischeri was exported. While there was no evidence of catalase activity in the cell-free supernatant from an overnight culture of V. fischeri, permeabilization of cells with chloroform to specifically release periplasmic proteins yielded 100% of the catalase activity but less than 5% of the cytoplasmic luciferase activity. We conclude that the catalase enzyme is localized in the periplasm of V. fischeri.
Characterization of a V. fischeri katA mutant.
To determine whether V. fischeri has multiple catalase genes and to investigate the physiological role of KatA, we constructed a katA null mutant of V. fischeri in which the C-terminal portion of the gene was replaced by an erythromycin resistance marker. While high levels of catalase activity were present in the parent strain after overnight growth in a rich medium, no significant activity (<3% of wild-type activity) was detectable in the katA mutant at any time in the growth cycle. Thus, V. fischeri cells apparently synthesize one catalase, and this enzyme is responsible for the high specific activity and efficient decomposition of hydrogen peroxide displayed by wild-type cells.
The susceptibility of the katA mutant and its parent to external hydrogen peroxide was assayed to determine whether KatA has a physiological role in protecting V. fischeri from oxidative stress. When the cells were grown to early stationary phase and exposed to 20 mM hydrogen peroxide, the catalase mutant was readily killed (<0.001% survival), whereas its parent survived well (>50%). In spite of this greatly enhanced sensitivity, there was no detectable difference between the aerobic growth rates or yields of the katA mutant and its parent in a rich medium.
Survival of the katA mutant during colonization of the squid light organ.
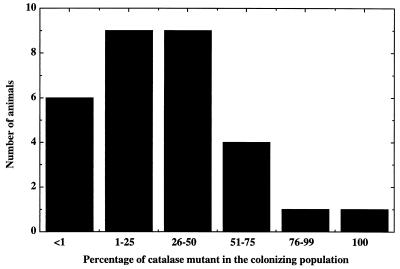
To determine whether the ability of KatA to protect against hydrogen peroxide killing in culture enhanced the survival of V. fischeri in the light organ environment, the effectiveness of colonization by the katA mutant was compared to that of its parent. The course of colonization by these two strains was monitored by observing the luminescence levels of infected juvenile E. scolopes squid. When presented by itself, the katA mutant was not defective in initiating a colonization of the light organ of E. scolopes, and at both 2 and 3 days postinoculation, the extent of colonization (as determined by CFU per light organ) was indistinguishable between the two strains (data not shown). However, the katA mutant was clearly defective in its ability to compete in a mixed infection with its catalase-positive parent strain (Fig. 3). When the two strains were mixed in equal ratios and used to coinoculate juveniles of E. scolopes, the ratios of the two strains in the colonized animal at 17 h postinoculation were skewed in favor of the wild-type parent. This bias was observed in each of the several times the experiment was performed. These data suggest that although a catalase mutant is able to establish a symbiotic colonization, it carries a deficiency that decreases the overall fitness of the strain. The presence of a functional catalase enzyme is likely therefore to be critical to the ability of V. fischeri to achieve a successful colonization in nature.
FIG. 3.
Colonization of juveniles of the squid E. scolopes by the katA mutant and its parent. Juveniles of E. scolopes were exposed to a 1:1 mixture of the katA mutant and its wild-type parent. The percentage of the katA mutant strain in the population of V. fischeri cells colonizing the light organ was determined for each animal at 17 h postinoculation.
DISCUSSION
Despite the fact that all animals typically have long-term, cooperative associations with microbial symbionts, little is known about such benign interactions between bacteria and their animal hosts (13). We believe that the lessons learned from the study of one specific host-symbiont pair, the squid E. scolopes and the bioluminescent bacterium V. fischeri, will advance our understanding of the mechanisms by which many diverse bacteria, including pathogens, interact with their hosts. In this paper, we report the characterization of a stationary-phase catalase that is important in protecting V. fischeri from oxidative stress and that is required for the organism to successfully compete against other bacteria for the symbiotic colonization of juvenile squid in coinoculation experiments.
Our characterization of the V. fischeri katA gene and its product yielded a number of interesting results: (i) wild-type cells express a high catalase specific activity that is further induced upon stationary phase growth in culture, (ii) the presence of low levels of hydrogen peroxide in the culture medium induces log-phase cells to express a similarly high level of catalase specific activity, and (iii) the enzyme appears to be entirely localized to the periplasm. These and other data suggested that V. fischeri cells have developed a powerful system of protection against the potential oxidative stress posed by hydrogen peroxide in their environment.
The light organ of E. scolopes has an abundance of mRNA encoding a squid halide peroxidase activity that catalyzes the conversion of hydrogen peroxide into the bactericidal compound hypochlorous acid (43, 48). This enzyme is similar to the mammalian defense protein, myeloperoxidase, and although the role of the halide peroxidase in the light organ has not yet been demonstrated, these data support a hypothesis in which oxidatively stressful conditions exist in the light organ environment (48). This hypothesis, in light of our results regarding the activity and location of the catalase enzyme, can be extended to predict that the catalase enzyme would be a requirement for successful symbiotic colonization by V. fischeri.
Curiously, when presented as the only bacterial strain in the inoculation, a catalase mutant displayed no observable defect in the ability to colonize juveniles of E. scolopes. However, when competition assays were performed to determine whether the catalase mutant could compete in mixed culture against its catalase-positive parent strain, it was found that the catalase mutant was deficient in its ability to compete for colonization. Thus, in the natural environment of the ocean, a catalase mutation would confer a serious fitness defect upon a strain competing against wild-type bacteria co-occurring in the seawater. This type of phenotype, in which a mutant that is competent for colonization when presented alone but that is unable to successfully compete when present with a wild-type parent strain, has been reported in the Rhizobium-root nodule association as well (3, 19). It is likely that such subtle, competition-dependent phenotypes, while easily missed in the laboratory, may have dramatic consequences for bacteria existing in the natural environment.
Our data confirm a requirement for the bacterium to protect itself from oxidative stress in the light organ but suggest that the bacterium does not rely solely on the catalase enzyme for protection from stress factors in the light organ environment. Similarly, it is known from work performed with E. coli that the rpoS gene is important for the resistance of this organism to hypochlorous acid, although interestingly, the catalase genes katE and katG are not the major contributors to this resistance (15). Other contributors may include the superoxide-induced gene regulators, SoxRS, which are also activated by hypochlorous acid (14). Similarly, cholera toxin, produced by Vibrio cholerae, is an ADP-ribosyltransferase enzyme that has been shown to inhibit the superoxide (and ultimately hydrogen peroxide)-generating respiratory burst (37). Two ADP-ribosylating activities have recently been found in V. fischeri (32, 33), and these exported enzymes may also be involved in reducing the oxidative stress environment of the light organ. Clearly, many avenues exist for bacteria to combat oxidative stress, and the production of catalase is just one of these.
Due to the periplasmic location of the catalase enzyme in V. fischeri, and data regarding its activity in culture, one might predict that in mixed-culture experiments, the wild-type strain could protect the catalase mutant strain by removing hydrogen peroxide in the general vicinity of both of the cell types. Our colonization data suggest that this is not the case. What then is the mechanism underlying the catalase mutant’s competitive defect even though, when not forced to compete with the wild type, it can colonize and persist normally for several days? We envision two hypotheses that address this question, both of which arise from the population dynamics that characterize the daily pattern of symbiont growth within the squid light organ. Each morning about 90% of the symbiont cells are expelled from the light organ crypts (25), and the remaining 10% go on to fully recolonize the crypt spaces by nightfall. In our first hypothesis, as a result of its greater susceptibility to a constant, low level of oxidative stress during this period of regrowth, the catalase mutant would experience a higher level of attrition than the wild-type parent, and its relative abundance in the recovering population could ultimately decrease. Our second hypothesis is based on recent evidence suggesting that the host peroxidase may be most highly expressed during a specific period each day (39). Thus, the catalase mutant may experience an episodic exposure to oxidative stress that diminishes its numbers relative to the wild type. Subsequent proliferation of the surviving symbionts in a mixed-infection organ would thereby result in a predominance of wild type over the catalase mutant. Under either of these scenarios, the catalase mutant, when occupying the light organ by itself, could ultimately recolonize to the same level as the wild type; however, we would predict that this recolonization should take somewhat longer as a result of either the mutant’s diminished growth rate or its lower abundance due to its reduced resistance to an ambient oxidative stress.
An understanding of these events will require a description of the kinetics of recolonization (i.e., how rapidly the population multiplies and over what portion of the day). Unfortunately, these kinetics remain experimentally difficult to determine due to the wide variation in colonization levels among individual animals (36). In addition, it is not known whether the bacteria achieve a stationary phase of growth for any significant period of time in the light organ, thus making it difficult to assign a significance to the stationary-phase regulation of katA. We expect that further studies of the pattern of growth and recolonization by the catalase mutant after the expulsion event, as well as studies of the expression pattern of this stationary-phase-regulated catalase gene in the light organ, will begin to answer some of these questions.
The ability of a cell to use catalase to protect itself from host-imposed oxidative stress has been shown not to be a requirement for tissue colonization by several species of bacterial pathogens (5, 9). However, in these studies, mixed-inoculation experiments were not performed. A thorough investigation of mutant phenotypes may require the examination of strains presented to the host in mixtures, a situation that not only provides an experimental test of subtle phenotypes but also more closely mimics the natural environment. Studies such as this one, investigating the symbiotic association between a marine bacterium and its squid host, may thus provide insight into other naturally occurring microbe-animal interactions.
ACKNOWLEDGMENTS
Many thanks are due to Joerg Graf, who constructed the V. fischeri DNA library from which pLP2 was isolated, to Lornie Phillips II, who identified the katA-containing clone, and to Jonathan Visick for strains and protocols. We also thank A. Small and M. McFall-Ngai for communicating data prior to publication.
This work was funded by National Science Foundation grant IBN96-01155 to M. McFall-Ngai and E.G.R. K.L.V. was funded by National Institutes of Health Research Service Award 1F32GM174724-01A1.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ames G F-L, Prody C, Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo R S, Robleto E A, Handelsman J. A hydrophobic mutant of Rhizobium etli altered in nodulation competitiveness and growth in the rhizosphere. Appl Environ Microbiol. 1994;60:1430–1436. doi: 10.1128/aem.60.5.1430-1436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 5.Bishai W R, Howard N S, Winkelstein J A, Smith H O. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect Immun. 1994;62:4855–4860. doi: 10.1128/iai.62.11.4855-4860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishai W R, Smith H O, Barcak G J. A peroxide/ascorbate-inducible catalase from Haemophilus influenzae is homologous to the Escherichia coli katE gene product. J Bacteriol. 1994;176:2914–2921. doi: 10.1128/jb.176.10.2914-2921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boettcher K J, Ruby E G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bol D K, Yasbin R E. The isolation, cloning and identification of a vegetative catalase gene from Bacillus subtilis. Gene. 1991;109:31–37. doi: 10.1016/0378-1119(91)90585-y. [DOI] [PubMed] [Google Scholar]
- 9.Buchmeier N A, Libby S J, Xu Y, Loewen P C, Switala J, Guiney D G, Fang F C. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Invest. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buzy A, Bracchi V, Sterjiades R, Chroboczek J, Thibault P, Gagnon J, Jouve H M, Hudry-Clergeon G. Complete amino acid sequence of Proteus mirabilis PR catalase. Occurrence of a methionine sulfone in the close proximity of the active site. J Protein Chem. 1995;14:59–72. doi: 10.1007/BF01888363. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Douglas A E. Symbiotic interactions. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 14.Dukan S, Dadon S, Smulski D R, Belkin S. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl Environ Microbiol. 1996;62:4003–4008. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dukan S, Touati D. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol. 1996;178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunlap P V. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J Bacteriol. 1989;171:1199–1202. doi: 10.1128/jb.171.2.1199-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinosa-Urgel M, Chamizo C, Tormo A. A consensus structure for ςS-dependent promoters. Mol Microbiol. 1996;21:657–659. doi: 10.1111/j.1365-2958.1996.tb02573.x. [DOI] [PubMed] [Google Scholar]
- 18.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1996;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 19.Gordon D M, Ryder M H, Heinrich K, Murphy P J. An experimental test of the rhizopine concept in Rhizobium meliloti. Appl Environ Microbiol. 1996;62:3991–3996. doi: 10.1128/aem.62.11.3991-3996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengee-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss R I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 23.Klotz M G, Klassen G R, Loewen P C. Phylogenetic relationships among prokaryotic and eukaryotic catalases. Mol Biol Evol. 1997;14:951–958. doi: 10.1093/oxfordjournals.molbev.a025838. [DOI] [PubMed] [Google Scholar]
- 24.Lee K-H, Ruby E G. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J Bacteriol. 1994;176:1985–1991. doi: 10.1128/jb.176.7.1985-1991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K-H, Ruby E G. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loewen P C. Bacterial catalases. In: Scandalios J G, editor. Oxidative stress and the molecular biology of antioxidant defenses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 273–308. [Google Scholar]
- 27.Loewen P C, Triggs B L, George C S, Hrabauchuk B E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J Bacteriol. 1985;162:661–667. doi: 10.1128/jb.162.2.661-667.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 29.Naclerio G, Baccigalupi L, Caruso C, De Felice M, Ricca E. Bacillus subtilis vegetative catalase is an extracellular enzyme. Appl Environ Microbiol. 1995;61:4471–4473. doi: 10.1128/aem.61.12.4471-4473.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nealson K H. Isolation, identification, and manipulation of luminous bacteria. Methods Enzymol. 1978;57:153–166. [Google Scholar]
- 31.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. Unpublished data.
- 32.Reich K A, Biegel T, Schoolnik G K. The light organ symbiont Vibrio fischeri possesses two distinct secreted ADP-ribosyltransferases. J Bacteriol. 1997;179:1591–1597. doi: 10.1128/jb.179.5.1591-1597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich K A, Schoolnik G K. Halovibrin, secreted from the light organ symbiont, Vibrio fischeri, is a member of a new class of ADP-ribosyltransferases. J Bacteriol. 1996;178:209–215. doi: 10.1128/jb.178.1.209-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha E R, Smith C J. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1995;177:3111–3119. doi: 10.1128/jb.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruby E G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 36.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 37.Seifert R, Schultz G. The superoxide-forming NADPH oxidase of phagocytes. Berlin, Germany: Springer-Verlag; 1991. [PubMed] [Google Scholar]
- 38.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small, A., and M. McFall-Ngai. Personal communication.
- 40.Tao K, Makino K, Yonei S, Nakata A, Shinagawa H. Molecular cloning and nucleotide sequencing of oxyR, the positive regulatory gene of a regulon for an adaptive response to oxidative stress in Escherichia coli: homologies between OxyR protein and a family of bacterial activator proteins. Mol Gen Genet. 1989;218:371–376. doi: 10.1007/BF00332397. [DOI] [PubMed] [Google Scholar]
- 41.Tartaglia L A, Storz G, Ames B N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 42.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: A mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 43.Tomarev S I, Zinovieva R D, Weis V M, Chepelinsky A B, Piatigorsky J, McFall-Ngai M J. Abundant mRNAs in the squid light organ encode proteins with a high similarity to mammalian peroxidases. Gene. 1993;132:219–226. doi: 10.1016/0378-1119(93)90199-d. [DOI] [PubMed] [Google Scholar]
- 44.Visick J E, Clarke S. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J Bacteriol. 1997;179:4158–4163. doi: 10.1128/jb.179.13.4158-4163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick K L, Ruby E G. Construction and symbiotic competence of a luxA-deletion mutant of Vibrio fischeri. Gene. 1996;175:89–94. doi: 10.1016/0378-1119(96)00129-1. [DOI] [PubMed] [Google Scholar]
- 46.von Ossowski I, Mulvey M R, Leco P A, Borys A, Loewen P C. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J Bacteriol. 1991;173:514–520. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei S L, Young R E. Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar Biol. 1989;103:541–546. [Google Scholar]
- 48.Weis V M, Small A L, McFall-Ngai M J. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wise A, Brems R, Ramakrishnan V, Villarejo M. Sequences in the −35 region of Escherichia coli rpoS-dependent genes promote transcription by EςS. J Bacteriol. 1996;178:2785–2793. doi: 10.1128/jb.178.10.2785-2793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 cloning vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]