Abstract
This study investigated the combined effects of nitrate (NT) and medium-chain fatty acids (MCFA), including C8, C10, C12, and C14, on methane (CH4) production, rumen fermentation characteristics, and rumen bacteria using a 24 h batch incubation technique. Four types of treatments were used: control (no nitrate, no MCFA), NT (nitrate at 3.65 mM), NT + MCFA (nitrate at 3.65 mM + one of the four MCFA at 500 mg/L), and NT + MCFA/MCFA (nitrate at 3.65 mM + a binary combination of MCFA at 250 and 250 mg/L). All treatments decreased (P < 0.001) methanogenesis (mL/g dry matter incubated) compared with the control, but their efficiency was dependent on the MCFA type. The most efficient CH4 inhibitor was the NT + C10 treatment (− 40%). The combinations containing C10 and C12 had the greatest effect on bacterial alpha and beta diversity and relative microbial abundance (P < 0.001). Next-generation sequencing showed that the family Succinivibrionaceae was favored in treatments with the greatest CH4 inhibition at the expense of Prevotella and Ruminococcaceae. Furthermore, the relative abundance of Archaea decreased (P < 0.05) in the NT + C10 and NT + C10/C12 treatments. These results confirm that the combination of NT with MCFA (C10 and C12 in particular) may effectively reduce CH4 production.
Subject terms: Microbiome, Climate-change mitigation, Animal physiology
Introduction
Globally, the sustainability of agricultural systems relies on efficient livestock production1. Naturally, ruminants lack total nutritional efficiency, as they waste 2–15% of their ingested energy2 by emitting enteric methane (CH4)3. CH4 is now recognized as a major global concern and one of the causes of climate change4. Various feed additives have been explored to effectively mitigate CH4 emissions5, including 3-nitrooxypropanol3 and red seaweed (Asparagopsis spp.)6. However, rumen CH4 is yet to be effectively mitigated in a cost-effective and practical manner for adoption at the farm level5,7,8. Farmers are more likely to adopt a CH4 mitigating option that is particularly economical as well as nutritionally and environmentally beneficial5.
One possible solution for countering fermentation inefficiency is to combine anti-methanogenic inhibitors with complementary modes of action. For example, nitrate (NT) and dietary fat decrease CH4 additively9–11. The effects of NT have been proven10, but the efficacy of lipids depends on their form, concentration, and fatty acid composition12,13. One of the most effective types of lipids for CH4 mitigation are medium-chain fatty acids (MCFA) and polyunsaturated fatty acids (PUFA)13, with MCFA being the most effective14–16. MCFA include caprylic (C8), capric (10), lauric (C12), and myristic acids (C14)16. MCFA have been supplemented in pure forms or in products rich in them (e.g., oils)17; however, it is unclear which MCFA inhibit methanogenesis18. Pure forms of MCFA, as well as their various sources and combinations, affect the rumen differently17,19. For example, C12 and C14 have been extensively evaluated; in addition to being effective individually, their combination has a synergistic effect in reducing CH415,20.
NT and MCFA are effective CH4 inhibitors when used alone; however, their high concentrations have adverse effects on rumen fermentation and feed intake10. Caution must be taken when supplementing NT because of the risk of nitrite poisoning10 and loss of high levels of nitrogen in urine21. Furthermore, high amounts of dietary lipids (8–9%), including MCFA, reduce dry matter intake (DMI), digestibility22,23, and consequently, production efficiency24,25.
NT mainly functions as an electron sink because NT reduction is thermodynamically more favorable than CO2 reduction10. Additionally, NT is toxic to protozoa10, and its intermediate nitrite is toxic to methanogens26,27. MCFA are antibacterial and antiprotozoal agents19,28,29 as they dissociate in bacterial cells17, may cause defaunation30,31, and, therefore, lower H2 supply5,9. Additionally, MCFA can directly inhibit methanogens29,32. The target microorganisms of NT and MCFA appear to overlap, but their effects may be complementary. Mixing low doses of NT and MCFA (and their binary combinations) may decrease rumen methane production without inhibiting fermentation.
Our hypotheses were as follows: (1) different combinations of MCFA and NT would vary in their effectiveness in mitigating CH4 production, and (2) efficient combinations of MCFA and NT capable of lowering methanogenesis without negatively affecting rumen fermentation may be found.
Results
Fermentation parameters
The results of the treatment effects on in vitro total gas (TGP) and CH4 production, apparent dry matter disappearance (aDMd), ammonia-N (NH3-N) concentration, and pH are summarized in Table 1. All treatment combinations of NT + MCFA significantly reduced CH4 production per dry matter incubated (DMi) for 24 h. The extent of reduction varied among the ten combinations tested. Compared with the unsupplemented control, the addition of NT + C10 and NT + C10/C12 led to the highest CH4 reduction, − 40% and − 34%, respectively (Table 1). A significant reduction (P < 0.001) with these two combinations was achieved without any negative effects on fermentation (assessed by net VFA (nVFA) produced) or substrate digestibility (assessed by aDMd; P > 0.05). nVFA production was affected by treatment (P = 0.031); however, specific multiple comparisons after correction using Dunnett’s test did not identify a significant difference between any treatments. The treatment effect on CH4 mitigation (mL/g DMi) then decreased as follows: NT + C8/C10 (− 29.6%) > NT + C10/C14 (− 28.5%) > NT + C8/C12 (− 26.3%) > NT + C8 (− 24.1%) > NT + C12 and NT + C12/C14 (− 23.6%) > NT (− 17.3%) > NT + C14 (− 15.9%). Similarly, other CH4 production parameters, that is, CH4 per percentage of TGP, aDMd, or VFA, significantly decreased (P < 0.001) in a similar manner, with the NT + C10 and NT + C10/C12 treatments being the most effective. NT + C14 was the only combination that reduced (P < 0.05) aDMd (by 6.7%) and the only combination that did not reduce (P < 0.05) TGP.
Table 1.
Effects of MCFA + NT on in vitro gas and methane production, apparent dry matter disappearance (aDMd), ammonia-N concentration, and pH.
| Treatment | TGP† (mL/g DMi§) | Methane | aDMd‡ (g/g) | Net NH3–N (mg/100 mL) | pH | |||
|---|---|---|---|---|---|---|---|---|
| (mL/g DMi) | (% TGP) | (mL/g aDMd) | (mol/mol nVFA¶) | |||||
| Control | 298.7 | 36.5 | 12.22 | 57.6 | 0.2125 | 0.6345 | 13.1 | 5.90 |
| NT | 289.3* | 30.2* | 10.42* | 48.1* | 0.1746 | 0.6313 | 20.7* | 6.06* |
| NT + C8 | 286.6* | 27.7* | 9.64* | 42.4* | 0.1599* | 0.6539 | 20.9* | 6.06* |
| NT + C10 | 263.8* | 21.9* | 8.29* | 34.6* | 0.1498* | 0.6372 | 24.7* | 6.27* |
| NT + C12 | 282.8* | 27.9* | 9.84* | 43.7* | 0.1592* | 0.6405 | 21.0* | 6.13* |
| NT + C14 | 291.7 | 30.7* | 10.51* | 51.9* | 0.1727 | 0.5923* | 21.3* | 6.08* |
| NT + C8/C10 | 277.9* | 25.7* | 9.23* | 39.3* | 0.1825 | 0.6569 | 24.0* | 6.18* |
| NT + C8/C12 | 284.0* | 26.9* | 9.48* | 41.2* | 0.1678* | 0.6565 | 20.9* | 6.13* |
| NT + C8/C14 | 288.8* | 29.1* | 10.06* | 45.6* | 0.1637* | 0.6402 | 22.4* | 6.05* |
| NT + C10/C12 | 271.0* | 24.0* | 8.88* | 38.4* | 0.1478* | 0.6315 | 22.7* | 6.20* |
| NT + C10/C14 | 278.8* | 26.1* | 9.36* | 41.0* | 0.1511* | 0.6383 | 21.2* | 6.15* |
| NT + C12/C14 | 283.1* | 27.9* | 9.85* | 45.2* | 0.1548* | 0.6184 | 21.9* | 6.09* |
| SEM | 2.53 | 0.65 | 0.163 | 1.22 | 0.00484 | 0.00517 | 0.93 | 0.020 |
| P-value | < 0.001 | < 0.001 | < 0.001 | < 0.001 | 0.009 | 0.002 | < 0.001 | < 0.001 |
†Total gas production; ‡apparent dry matter disappearance; §dry matter incubated; ¶net production of volatile fatty acids.
*Means within a column differ significantly (P < 0.05) from the corresponding control (0 mg/L). SEM, standard error of the mean.
The nVFA (P = 0.031) and the acetate: propionate ratio (P = 0.065) were not altered by any treatment. The effects of NT + MCFAs on VFA production and individual proportions are shown in Table 2. NT + C10 significantly decreased (P < 0.05) the molar proportion of acetate and increased (P < 0.05) the molar proportions of butyrate, valerate, and iso-valerate. Furthermore, butyrate proportions were significantly reduced (P < 0.05) with the following treatments: NT, NT + C14, and NT + C12/C14. Finally, only the NT + C8 and NT + C10/C12 treatments substantially decreased (P < 0.05) the molar proportion of valerate compared to the control.
Table 2.
Impacts of MCFA + NT on in vitro volatile fatty acids production and proportion.
| Treatment | nVFA† (mmol/L) | Molar proportion of VFA (mol/100 mol) | A:P‡ | |||||
|---|---|---|---|---|---|---|---|---|
| Acetate | Propionate | Butyrate | iso-butyrate | Valerate | iso-valerate | |||
| Control | 67.4 | 60.5 | 20.7 | 10.9 | 0.8 | 4.9 | 2.2 | 3.0 |
| NT | 67.9 | 62.9 | 20.0 | 9.5* | 0.9 | 4.5 | 2.2 | 3.2 |
| NT + C8 | 68.1 | 61.1 | 21.6 | 10.3 | 0.9 | 4.0* | 2.1 | 2.9 |
| NT + C10 | 57.8 | 54.9* | 20.1 | 14.4* | 1.3 | 6.7* | 2.6* | 2.8 |
| NT + C12 | 68.5 | 61.2 | 20.3 | 10.0 | 1.3 | 5.1 | 2.2 | 3.1 |
| NT + C14 | 71.1 | 62.1 | 20.3 | 9.5* | 1.1 | 4.7 | 2.3 | 3.1 |
| NT + C8/C10 | 60.0 | 60.1 | 20.0 | 12.0 | 0.4 | 5.2 | 2.3 | 3.1 |
| NT + C8/C12 | 65.4 | 60.1 | 21.4 | 10.6 | 0.7 | 4.9 | 2.2 | 2.8 |
| NT + C8/C14 | 70.1 | 60.5 | 21.5 | 9.9 | 1.7 | 4.3 | 2.2 | 2.8 |
| NT + C10/C12 | 64.0 | 57.3 | 21.8 | 11.6 | 1.2 | 5.8* | 2.3 | 2.7 |
| NT + C10/C14 | 67.9 | 59.4 | 21.4 | 10.5 | 1.5 | 4.9 | 2.1 | 2.8 |
| NT + C12/C14 | 70.9 | 60.7 | 21.3 | 9.5* | 1.5 | 4.9 | 2.1 | 2.8 |
| SEM | 1.65 | 0.40 | 0.33 | 0.31 | 0.12 | 0.19 | 0.08 | 0.05 |
| P-value | 0.031 | < 0.001 | 0.067 | < 0.001 | 0.198 | < 0.001 | 0.013 | 0.065 |
†Net production of volatile fatty acids; ‡acetate:propionate.
*Means within a column differ significantly (P < 0.05) from the corresponding control (0 mg/L). SEM, standard error of the mean.
The pH increased (P < 0.05) in all treatments relative to the control; the highest increase was recorded for the NT + C10. NH3–N concentrations increased (P < 0.05) in all NT + MCFA treatments, with NT + C10 increasing NH3–N concentrations the most.
Microbial community
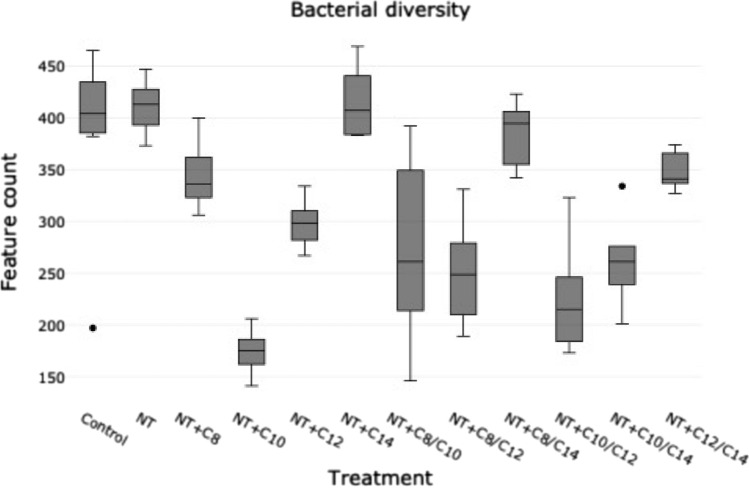
Bacterial diversity varied greatly among treatments and was expressed as ASV counts (Fig. 1) and indices of diversity (Table 3). All indices of alpha diversity significantly (P < 0.05) decreased when the treatments contained C10 or C12, with the NT + C10 treatment having the strongest effect on microbial diversity, richness, and evenness (Figs. 1, 4, Table 3). In contrast, the smallest effect on diversity was observed for treatments containing C8 and C14. Additionally, the NT + C10 and NT + C10/C12 treatments significantly decreased (P < 0.05) the relative abundance of Archaea (Table 4).
Figure 1.
Stack-graphs showing the effect of NT + MCFA on bacterial diversity.
Table 3.
The influence of the NT + MCFA treatments on α-diversity of microbial communities in vitro.
| Treatment | Shannon diversity index | Simpson diversity index | Chao1 richness index | Pielou evenness index |
|---|---|---|---|---|
| Control | 6.954 | 0.955 | 584 | 0.758 |
| NT | 7.101 | 0.965 | 588 | 0.773 |
| NT + C8 | 5.856 | 0.878 | 507 | 0.653 |
| NT + C10 | 3.171* | 0.566* | 259* | 0.396* |
| NT + C12 | 5.125* | 0.802 | 444* | 0.584* |
| NT + C14 | 6.912 | 0.949 | 594 | 0.752 |
| NT + C8/C10 | 4.922* | 0.762* | 437* | 0.560* |
| NT + C8/C12 | 4.271* | 0.703* | 382* | 0.499* |
| NT + C8/C14 | 6.504 | 0.925 | 573 | 0.712 |
| NT + C10/C12 | 3.701* | 0.622* | 336* | 0.441* |
| NT + C10/C14 | 4.524* | 0.731* | 390* | 0.526* |
| NT + C12/C14 | 5.717 | 0.856 | 520 | 0.635 |
| SEM | 0.2287 | 0.0238 | 19.2 | 0.0220 |
| P-value | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 |
*Means within a column differ significantly (P < 0.05) from the corresponding control (0 mg/L). SEM, standard error of the mean.
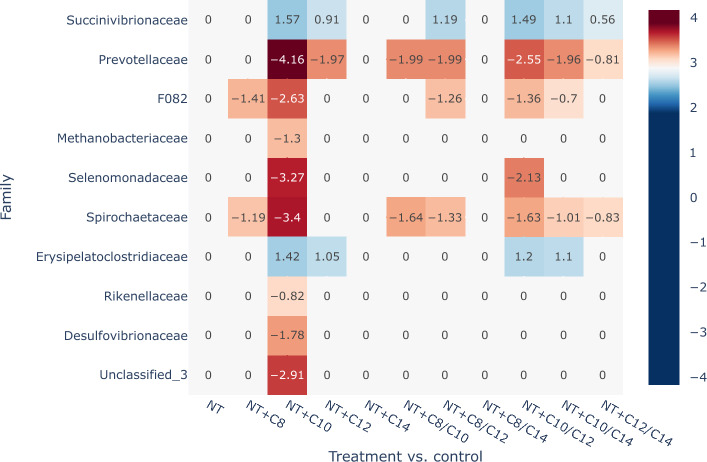
Figure 4.
Analysis of Compositions of Microbiomes with Bias Correction (ANCOM-BC) shows positive log fold change (enriched taxa) and negative log fold change (decreased taxa) compared to control. Nonsignificant results (P > 0.05) were replaced by 0. At the family level, the addition of C10 had the most prominent effect on microbiota.
Table 4.
Effects of MCFA + NT on the relative abundances of microbial communities at the kingdom level in vitro.
| Kingdom Treatment |
Bacteria | Archaea |
|---|---|---|
| Control | 97.66 | 2.34 |
| NT | 98.57 | 1.43 |
| NT + C8 | 98.58 | 1.42 |
| NT + C10 | 99.63* | 0.37* |
| NT + C12 | 98.60 | 1.40 |
| NT + C14 | 98.48 | 1.52 |
| NT + C8/C10 | 98.70 | 1.30 |
| NT + C8/C12 | 98.85 | 1.15 |
| NT + C8/C14 | 97.58 | 2.42 |
| NT + C10/C12 | 99.10* | 0.90* |
| NT + C10/C14 | 98.79 | 1.21 |
| NT + C12/C14 | 98.78 | 1.22 |
| SEM | 0.005 | 0.003 |
| P-value | 0.023 | 0.023 |
*Means within a column differ significantly (P < 0.05) from the corresponding control (0 mg/L). SEM, standard error of the mean.
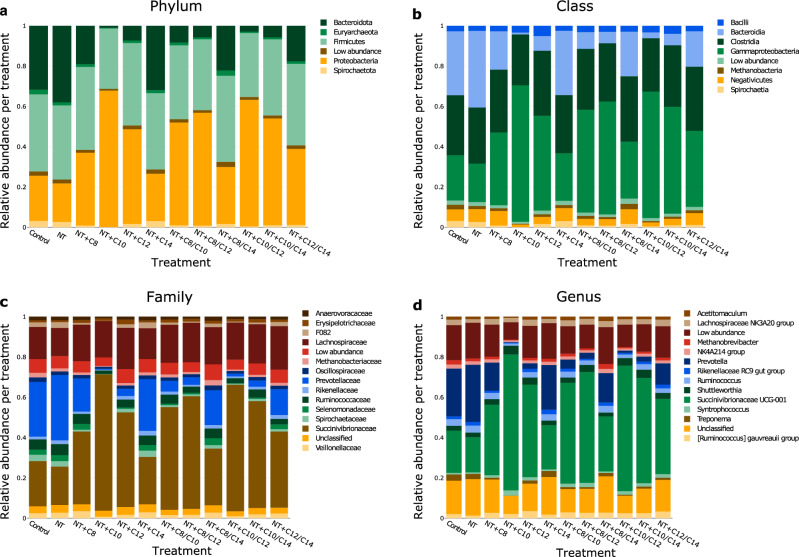
The most abundant phyla (Fig. 2a) were Proteobacteria (relative abundance range 19.5–68.4%), followed by Firmicutes (30.2–43.8%), and Bacteroidota (0.96–37.98%). The dominance of the phylum Proteobacteria was not consistent across all treatments, and the dominance of the respective phyla varied based on the type of MCFA. Proteobacteria thrived in the C10 treatment group at the expense of Bacteroidota. The phylum Bacteroidota showed signs of particular sensitivity to MCFA, as its relative abundance was highest in the NT treatment. Firmicutes, conversely, appear to be favored in treatments containing C8.
Figure 2.
Bar-charts displaying the different effects of NT + MCFA on rumen microbiota at the phylum (a), class (b), family (c), and genus (d) levels.
To gain further insight into the bacterial community, the relative microbial abundances are also shown at the class (Fig. 2b), family (Fig. 2c), and genus (Fig. 2d) levels. The highest relative abundance at the family level was noted for Succinivibrionaceae (25.2–70.7%), followed by Prevotellaceae (0.5–34.5%) and Lachnospiraceae (14.9–23.2%). In general, the family Succinivibrionaceae showed an increasing trend in treatments containing C10 and/or C12. In contrast, the C10 treatment simultaneously decreased the relative abundance of Prevotellaceae.
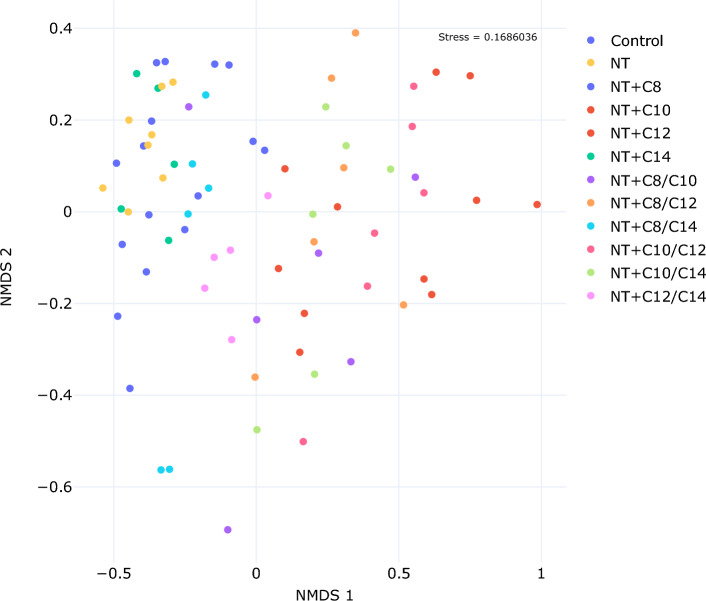
Overall, compared with the control, the treatments containing C10 and C12 had the strongest effect on the relative abundances of bacteria and Archaea. The weakest antimicrobial activity was observed in the C8 and C14 treatments and their respective combinations. Compared to the combination with MCFA, NT alone had little effect on the composition of the microbiota in this experiment. This was confirmed by analysis of Compositions of Microbiomes with Bias Correction (ANCOM-BC) implemented at a family level (Fig. 4) and nonmetric multidimensional scaling (NMDS) analysis used to visualize and assess community composition (Fig. 3).
Figure 3.
A nonmetric multidimensional scaling (NMDS) visualization depicting the microbiome variations at the ASV level in in vitro cultures based on the treatment applied (stress = 0.1686036). The plot clearly illustrates the impact of different treatments on microbiome composition. The treatments involving control, NT, C8, and C14 form a cluster in the upper-left part of the plot, whereas treatments with C10 and C12 are more situated towards the right side.
In the NMDS analysis, the treatments exhibited distinct clustering patterns on the plot. Specifically, treatments comprising control, NT, C8, and C14 formed a cluster located in the upper-left quadrant of the plot (Fig. 3), indicating a high degree of similarity in their microbiome compositions. Conversely, treatments involving C10 and C12 displayed a tendency to be positioned towards the right side of the plot, suggesting differences in microbiome composition compared to the former group.
Discussion
By combining low doses of NT and MCFA, we confirmed our hypotheses that (1) individual combinations varied in their CH4 mitigating efficacy and (2) there was one superior treatment (NT + C10) that decreased methanogenesis the most without negative effects on fermentation. In addition, the microbial community shifted to resemble the rumen microbiota of low-CH4 emitting cattle, with low Ruminococcaceae33,34 (e.g., 4.3%33), and high Succinivibrionaceae35 (e.g., 8.9–9.3%36,37).
Combinations of NT and various MCFA differed in their CH4 suppressing effect, confirming previous findings that CH4 mitigation is dependent on the MCFA type19. Overall, our results suggested that the most effective combinations were those containing C10. The most effective treatment was NT + C10 (− 40%; mL/g DMi). This is consistent with the anti-methanogenic effect of C10 reported by Goel et al. (2009), who showed a dose-dependent effect of C1017. In their study, C10 at 400 and 600 mg/L of incubation medium decreased CH4 production by 44% and 88%, respectively, but also inhibited overall fermentation, as suggested by decreased VFA production17. In contrast, Dohme et al.18 reported no effect of C10 (approximately 600 mg/L) on methanogenesis using a RUSITEC, an in vitro rumen simulation technique. Previous in vitro studies suggested that CH4 is reduced by about 5% for each mmol of NT38,39. However, in our study, it was only about 3.3% per mmol of NT, indicating an incomplete reduction of NT. Latham et al. (2016)40 previously reported that the mitigation potential of NT may be reduced due to its partial metabolization to nitrous oxide either via dissimilatory NT reduction or via incomplete denitrification.
The most efficient binary combination of MCFA in the present study was NT + C10/C12. When expressed as CH4 production per VFA, this treatment inhibited CH4 production to the greatest extent. Consistent with this, Desbois and Smith29 reported that MCFA with 10 or 12 carbons is the most biologically active and that the antimicrobial activity of MCFA decreases with any change in carbon chain length. C12 is one of the most frequently examined MCFAs19. In previous studies, C12 has decreased CH4 production in the rumen by up to 89% in vitro18,41 and by as much as 76% in vivo42,43.
The fact that binary combinations of MCFA might be effective anti-methanogenic additives was demonstrated by Soliva et al.36 In their study, C12 and C14 exhibited synergistic CH4 suppression effects. Their various proportions decreased methanogenesis by 50–96%, and the extent of inhibition increased with increasing amounts of C12 in the mixture. However, in our study, NT + C12/C14 decreased methanogenesis by much less in comparison with the study of Soliva et al.36 The lower efficiency of our binary combination may be attributed to the higher concentration used in their study (1000 mg/L vs. 500 mg/L in our study). Another reason could be the C12/C14 ratio. CH4 production was not as inhibited in our study as it was in the study of Soliva et al.36, where CH4 production progressively decreased with an increasing proportion of C12 in the mixture. We assessed a 1:1 ratio, whereas the most effective ratio reported by Soliva et al.36 was 2:1 or higher.
In general, the treatments that did not contain C10 or C12 were less effective. Namely, NT with C8, C14, and their binary combination decreased methanogenesis only by 16–24%. In line with this, C14 has been reported to have low efficiency14,18,20, and the low potential for CH4 inhibition by C8 is in agreement with the findings of Ajisaka et al.44.
In general, individual MCFA with NT had little effect on nVFA production or individual VFA proportions. This is in line with the findings of Yanza et al. (2020), who showed that VFA concentrations were not significantly decreased by MCFA in vitro but only in vivo. Furthermore, in our study, sole NT treatment had no effect on nVFA, supporting the results of other in vitro studies with NT38,45. The P-value in the analysis of variance was statistically significant (P = 0.031); however, Dunnett´s post-hoc analysis failed to identify significant differences between the treatments (P > 0.05). Nevertheless, the numerical differences showed the NT + C10 treatment decreased nVFA the most (− 14.2%). Previously, similar doses of C10 (400–600 mg/L) decreased total VFA production by 17–23%17,18. This difference could be due to the NT in our treatments, which might have mitigated the inhibitory effects of MCFA on nVFA production, as NT has previously increased VFA concentrations46,47. Conversely, treatments C8 and C14 had the lowest effect on rumen fermentation, as indicated by their weak effect on CH4 inhibition. In line with the generally low inhibitory effect on CH4 production, the effects of the C8 and C14 treatments on nVFA production were negligible.
The molar proportions of the individual VFA were most prominently affected by the NT + C10 treatment. These effects are consistent with the strong anti-methanogenic and antimicrobial activities of C1029. The NT + C10 treatment decreased acetate production. Acetate formation, an H2 releasing pathway, usually decreases at higher H2 concentrations, which occurs when methanogenesis is inhibited48,49. High H2 concentrations favor H2 sinks such as propionate, butyrate, and valerate48–50. We did not measure the levels of H2, however, this theory was confirmed by an increase in butyrate concentrations, which, as demonstrated in a fermentation balance experiment, may provide 14% of the H2 sinks in the rumen51. Propionate is a more common H2 sink than butyrate51; however, its concentration did not increase. This could be because propionate-producing bacteria were inhibited. Indeed, the propionate-producing families Prevotellaceae, Veillonellaceae, and Selenomonadaceae52,53 were reduced, but at the expense of Succinivibrionaceae, which produce succinate, a precursor to propionate52. As a result, another explanation could be that the bacteria producing propionate from succinate were inhibited. Furthermore, the added NT, a favorable H2 sink49, was available and could have consumed the free H2 required for propionate production54. Acetate is normally produced by cellulolytic microorganisms along with H250,55. The decrease in acetate might be due to C10 inhibiting cellulolytic microorganisms, such as the family Ruminococcaeae56, which was inhibited in the NT + C10 treatment along with acetate production.
The effects of MCFA on digestibility are type- and dose-dependent19,31,57. Higher doses of MCFA typically decrease nutrient digestibility both in vivo and in vitro19,31,57. For example, in in vitro continuous culture, C8, C10, and C12 at 5% DM (approximately 600 mg/L) reduced NDF digestion by 2.4, 6.0, and 8.7%, respectively18. This reduction may be due to the absorption of fatty acids (FA) on feed particles, limiting the access of enzymes and microbes, and/or FA may be directly absorbed by fiber-degrading microbes (protozoa or cellulolytic bacteria), to which they are toxic19,24.
Nevertheless, in the present study, digestibility was not influenced by MCFA, presumably because of their low dosages. The only exception was the NT + C14 treatment (− 6.7%), which had no significant effect on rumen fermentation or nVFA, and the proportion of microorganisms was very similar to that of the control. This result is similar to that of Dohme et al. (2001), who reported the lowest organic matter degradability in C14 (− 7.4%). NT alone (3.65 mM) did not decrease digestibility in our study. Previously, 5 mM NT did not decrease digestibility as well27. However, higher doses of NT may be toxic to cellulolytic bacteria and decrease digestibility in vitro27.
All treatments, including NT alone, increased pH in our study. This increase may have been due to the addition of NT, which is reduced to ammonia in the rumen. These results are consistent with those of Zhou et al.26, who reported an increase in pH at higher NT concentrations. In our study, the NT + C10 treatment increased the pH to a maximum of 6.27, which is in line with the numerically lower nVFA levels in this treatment. Similarly, Dohme et al.18 reported the highest pH when supplemented with C10. However, all values of ruminal pH remained within the physiological range (5.5–7.554,55).
Various combinations of MCFA with NT added to ruminal fluid significantly affected the richness and diversity of bacterial populations to different degrees (Table 3, Fig. 1). Currently, treatments containing C10 and/or C12 significantly decreased the alpha diversity indices (Table 3), consistent with their effects on other ruminal parameters in our study and their strong antibacterial activity reported previously17,18,29. Burdick et al.31 used a mixture of MCFA (C8, C10, and C12) and, contrary to our results, did not report any changes in alpha diversity. This might be due to the low concentrations used in their study, although there was a reported tendency to reduce the bacterial richness. Previous studies on MCFA either did not investigate ruminal microbiota17,32 or used less sophisticated (chamber counting method) and insufficiently specific methods18.
The dominant bacterial phyla in the current study were Firmicutes, Bacteroidota, and Proteobacteria; this is consistent with the findings of previous studies47,52,58. The treatments (particularly those containing C10 and C12) that decreased CH4 production also increased the relative abundance of Proteobacteria (Fig. 2a). Proteobacteria predominantly belonged to the family Succinivibrionaceae (Fig. 2c). Hydrogen plays a central role in CH4 production59,60. The amount of H2 in the rumen can be influenced by the abundance of H2-producing and H2-consuming bacteria associated with CH4 emissions61. Previously, low-CH4-emitting ruminants and tammar wallabies were associated with the H2-consuming family Succinivibrionaceae34,35,62. Succinivibrionaceae utilize H2 to generate succinate (a precursor to propionate) and, therefore, can reduce CH4 emissions52. Treatment with NT + C10 increased the relative abundance of Succinivibrionaceae the most (70.7%) (Fig. 2c). In contrast, Succinivibrionaceae were the least abundant in the NT + C14 treatment (25.2%). NT + C10 and NT + C14 treatments decreased CH4 production the most and least, respectively.
Within the phylum Bacteroidota, treatments with greater inhibition of methanogenesis decreased the relative abundance of the genus Prevotella (Fig. 2d). The genus Prevotella utilizes H2 and produces propionate61,63,64; in a study with Colombian buffalos, a higher abundance of Prevotella was associated with lower CH4 emissions65. However, our findings are not consistent with this, as the genus Prevotella was less abundant in treatments resulting in lower CH4 production. This lower relative abundance of Prevotella could be explained by H2 availability in our study. Theoretically, the low H2 availability may have been due to the H2 being used for the reduction of supplemented NT to ammonia63,66. Furthermore, H2 could have been utilized by the family Succinivibrionaceae, and therefore outcompeted the Prevotella genus. This shift in relative abundance from Prevotella to Succinivibrionaceae has been previously noted67,68. Furthermore, the relative abundance of Prevotella is decreased by supplementation with NT69. However, the majority of the previous studies on NT have reported an increase in the relative abundance of Prevotella12,64,70,71, because Prevotella is associated with nitrate metabolism71.
In the phylum Firmicutes, treatments with the greatest anti-methanogenic potential (NT + C10 and NT + C10/C12) decreased the relative abundance of the family Ruminococcaceae (Fig. 2c). The Ruminococcaeae belong to the H2-producing bacteria72 and are present in higher abundance in high CH4-emitting rumens33,34. This finding is consistent with our results. However, this family plays a significant role in fiber metabolism, and its reduction may cause a decrease in fiber digestion73. Unfortunately, we did not measure fiber digestion, and aDMd was not negatively affected.
A decrease in methanogenesis was also observed in the methanogenic population. The relative abundance of Archaea, the sole producers of CH4 in the rumen60, was decreased by NT + C10 and NT + C10/C12 treatments by 84.2% and 45.7%, respectively (Table 4). Dohme et al.18 and Burdick et al.31 reported no change in the methanogen population when supplemented with MCFA (C8, C10, and C12). A meta-analysis showed that the Archaea population diminished quadratically only under in vitro conditions with increasing doses of MCFA19. Furthermore, NT supplementation decreases methanogenesis and consistently reduces the abundance of methanogenic Archaea60,69. Notably, it has been reported that instead of the overall abundance of methanogens, the community structure of methanogens60 and differential gene expression of methanogenesis pathways are the decisive factors in ruminal methanogenesis74.
Conclusion
This in vitro study showed that the effects of NT and MCFA combinations depend on the type of MCFA. The tested treatments have the potential to lower CH4 production without negatively affecting ruminal fermentation. The most effective CH4 inhibitors were combinations of NT and C10/C12. These combinations also had the greatest impact on the ruminal microbiota, as reflected in the changes in bacterial diversity and shifts in the relative abundances of bacteria and Archaea. The reported results from our 24 h batch incubations should be verified for their long-term effects in vitro and efficiency in vivo.
Material and methods
Ethical compliance
Procedures with animals were conducted in accordance with Czech legislation (Act No. 246/1992 Coll., on the protection of animals against cruelty) and applicable European guidelines and regulations (Directive 2010/63/EU, on the protection of animals used for scientific purposes) for experimentation with animals. The experimental protocol was approved by the Animal Ethics Committee of the Institute of Animal Science (Prague, Czech Republic). This study was conducted in accordance with ARRIVE guidelines to ensure an appropriate animal care. The donor cows were housed at the experimental farm of the Institute of Animal Science (Netluky, Prague, Czech Republic).
Treatments and experimental design
A 24 h batch incubation was used to evaluate the combined effect of NT and MCFA on CH4 production and rumen fermentation. The treatments were: control (no NT, no MCFA); NT (nitrate at 3.65 mM), NT + MCFA (nitrate at 3.65 mM + one of four MCFA (C8, C10, C12, and C14) at 500 mg/L), and NT + MCFA/MCFA (nitrate at 3.65 mM + binary combination of MCFA (C8/C10, C8/C12, C8/C14, C10/C12, C10/C14, and C12/C14) at 250 and 250 mg/L). Sodium nitrate was used as the nitrogen source. Three runs were performed over two weeks. In each run, four bottles were assigned to the control (no NT, no MCFA) to obtain a robust average value, three bottles for the NT and blank (no substrate), and two bottles for each of the ten treatments (NT + MCFA and NT + MCFA/MCFA). The average values of the bottles for each treatment in each run were used as experimental replicates.
Animals, diets, and substrate
Samples of rumen content were manually collected through a rumen cannula (internal diameter, 10 cm) from different sites in the rumen 3 h after morning feeding. Two early lactation Holstein cows (584 ± 20 kg body weight; 32 ± 2 kg of milk/d) were used as donors. Cows were fed a mixed ration consisting (DM basis) of corn silage (337 g/kg), legume-cereal silage (58 g/kg), alfalfa silage (53 g/kg), high-moisture corn silage (50 g/kg), brewer grain (37 g/kg), wheat straw (18 g/kg), liquid supplement (50:50 mixture of molasses and glycerol; 83 g/kg), and concentrate mixture (364 g/kg). The diet was provided twice daily (0400 and 1600) ad libitum.
Rumen content samples were immediately transported to the laboratory in thermal flasks. The interval between sampling and the next sample processing was 20–30 min. In the laboratory, rumen content was strained under continuous CO2 flushing through a stainless-steel sieve (250 μm; Retsch, Haan, Germany) to obtain rumen fluid. Rumen fluids from the two donor cows were pooled in equal proportions.
The experimental substrates consisted of (on a DM basis) corn silage (300 g/kg), alfalfa silage (300 g/kg), and barley (400 g/kg). The dried feeds were ground and passed through a 1-mm screen. The chemical composition per kilogram of substrate (DM basis) was as follows: organic matter, 951 g/kg; crude protein, 154 g/kg; ether extract, 25 g/kg; starch, 306 g/kg; neutral detergent fiber (NDF), 354 g/kg; and acid detergent fiber (ADF), 193 g/kg.
In vitro incubations
The in vitro incubations were conducted over 24 h in 120-mL serum bottles. The dried ground substrate (200 mg) was weighed into sterile CO2-flushed serum bottles the day before incubation. The culture fluid (20 mL) was dispensed into each serum bottle using a bottle-top dispenser (Calibrex 530 Salutae, Socorex, Switzerland) under a stream of CO2. The culture was prepared by mixing composite rumen fluid with a medium (1:3, v/v) as described previously75. The resulting mixture was immediately gassed with CO2 at 39 °C for 10 min before being added to the bottles. Sodium nitrate and MCFA were supplied to bottles by adding 200 µL of filter-sterilized distilled water and ethanol stock solutions, respectively, to reach the desired concentration in 20 mL of culture fluid. Equivalent amounts of water and ethanol were added to the control and blank bottles to compensate for the possible effects of solvents on fermentation. The initial headspace gas phase in all incubations was CO2. All serum bottles were sealed and placed in a temperature-controlled water bath (SW 22; Julabo, Germany) at 39 °C with a shaking frequency of 90 rpm for 24 h.
Sampling and chemical analyses
Total volume of gas produced was estimated from the headspace gas pressure using Boyle’s law76. Headspace gas pressure was measured using a manometer (Traceable; Fisher Scientific, Pittsburgh, PA, USA) after 24 h of incubation. The headspace gas was then sampled by displacement into a tube (5 mL) prefilled with distilled water by inserting a 23 gauge needle through the stopper of the bottle. The CH4 concentration in the headspace gas was measured using gas chromatography77.
The concentration of each VFA in the cultures was examined using gas chromatography77. VFA concentrations were calculated as the difference between concentrations in the fluid sample after 24 h of incubation and initial concentrations and were therefore reported as nVFA produced. The ammonia-N concentrations in the cultures were determined using the indophenol method78. The aDMd was determined gravimetrically by calculating the difference between the weight of the incubated substrate and the dry weight of the fermentation residue, normalized to the residue weight in the blank77.
DNA extraction, polymerase chain reaction (PCR), and amplicon sequence variant (ASV) analysis
DNA (0.5 mL) was extracted from each homogenized sample of the incubation fluid using the repeated bead beating and column purification methods77 with modifications. The DNA yield and quality of each sample were quantified using an ND-1000 UV spectrophotometer (NanoDrop Technologies, Witec AG, Littau, Switzerland). The extracted DNA was kept at − 20 °C prior to PCR. The V4 region of the 16S rRNA gene was amplified using the primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′)79. The PCR mixture (20 μL) was prepared using the GoTaq G2 DNA polymerase (Promega, USA); briefly, it consisted of 4 μL 5 × GoTaq buffer, 1.6 μL of magnesium chloride, 1 μL of T4 bacteriophage Gene 32 product, 0.4 μL of each dNTP, 0.1 μL of each primer, 0.1 μL GoTaq Polymerase, 1 μL of template DNA, and 11.1 μL of PCR water.
The PCR reaction was performed with the following steps: initial denaturation at 95 °C for 45 s, annealing at 50 °C for 30 s, elongation at 68 °C for 30 s and a final 5 min extension at 68 °C; the reaction was put to hold at 4 °C. The number of cycles was adjusted separately for each sample to minimize chimeric sequence formation. The resulting PCR products were verified using agarose gel (1%) electrophoresis with staining of the gels with SYBR™ Green I Nucleic Acid Gel Stain (Thermo Fisher, USA). The banding patterns were documented using the Gel Doc XR + System (BioRad, USA). All PCRs were performed in triplicate and pooled.
PCR products were sequenced using the MiniSeq platform (Illumina, USA). The resulting amplicons were analyzed using the DADA2 pipeline (Callahan et al., 2017) and SILVA database (v128)80. After normalizing the data to the lowest sample depth (32 000 sequences/sample), relative bacterial abundance was plotted at the phylum (a), class (b), family (c), and genus (d) levels (Fig. 2).
We employed the 'vegan' package in R to visualize and assess community compositional differences (beta diversity) through NMDS (Fig. 3) analysis using the metaMDS function81. The ordination patterns were acceptable, as the stress values of the two-dimensional NMDS analysis were below 0.2082 (stress = 0.1686036). NMDS stress values are reported after 100 tries and the best solution was repeated 13 times. Next, we utilized ANCOM-BC (version 2.2.2), implemented through the 'ANCOMBC' package83, to identify variations in taxa abundance between the groups at the family level (Fig. 4). We applied the Holm-Bonferroni method to adjust for multiple testing and to decrease the likelihood of type I error84. Nonsignificant results (P > 0.05) were replaced by 0.
Statistical analysis
The main effect of treatment was analyzed using the PROC MIXED procedure in SAS (SAS Enterprise Guide 6.1, SAS Institute Inc., Cary, NC, USA) according to a randomized complete block design. Runs (n = 3) were the blocking factors. Prior to the statistical analysis, technical replicates (parallel bottles for each treatment) were averaged per run. The model was:
where Yij is the dependent variable, µ is the overall mean, Ti is the fixed effect of the treatment (i = 12 levels; control, nitrate, and 10 combinations of nitrate with MCFA), Rj is the random effect of the run (j = 1, 2, and 3), and eij is the residual error. Treatment means were compared with the control using Dunnett’s adjustment, and differences between each treatment and the control were considered significant at P < 0.05.
Acknowledgements
This study was supported by the Ministry of Agriculture of the Czech Republic (grant number NAZV QK23020011). Computational resources were provided by the e-INFRA CZ project (ID:90254), supported by the Ministry of Education, Youth and Sports of the Czech Republic.
Author contributions
M.J. designed the research. M.J., M.V., A.V., Y.T., P.H., and D.T. collected the samples. M.J., K.J., and M.V. led the laboratory work and were assisted by A.Š. M.J., M.V., and A.Š. analyzed the data. M.V. and M.J. wrote the initial draft of the manuscript and all authors participated in its revision.
Data availability
The datasets utilized and/or analyzed in the present study can be obtained from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cabezas-Garcia EH, Gordon AW, Mulligan FJ, Ferris CP. Revisiting the relationships between fat-to-protein ratio in milk and energy balance in dairy cows of different parities, and at different stages of lactation. Animals. 2021;11:3256. doi: 10.3390/ani11113256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saeed, M. I., Sabow, A. B., Muhamad, A. A. & Hermiz, H. N. Comparative growth performance and carcass traits of calves of different cattle breeds fed pasture and concentration. Tikrit J. Agric. Sci.22, 44–50 (2021).
- 3.Alvarez-Hess P, et al. Effect of combining wheat grain with nitrate, fat or 3-nitrooxypropanol on in vitro methane production. Anim. Feed Sci. Technol. 2019;256:114237. doi: 10.1016/j.anifeedsci.2019.114237. [DOI] [Google Scholar]
- 4.Hadipour A, Mohit A, Darmani Kuhi H, Hashemzadeh F. Recent nutritional advances to mitigate methane emission in cattle: A review. Iran. J. Appl. Anim. Sci. 2021;11:1–14. [Google Scholar]
- 5.Patra AK. Animal Feed Science and Nutrition-Production, Health and Environment. IntechOpen; 2022. Introductory chapter: Animal feed science and nutrition-production, health and environment. [Google Scholar]
- 6.Stefenoni H, et al. Effects of the macroalga Asparagopsis taxiformis and oregano leaves on methane emission, rumen fermentation, and lactational performance of dairy cows. J. Dairy Sci. 2021;104:4157–4173. doi: 10.3168/jds.2020-19686. [DOI] [PubMed] [Google Scholar]
- 7.Soliva CR, Amelchanka SL, Duval SM, Kreuzer M. Ruminal methane inhibition potential of various pure compounds in comparison with garlic oil as determined with a rumen simulation technique (Rusitec) Br. J. Nutr. 2011;106:114–122. doi: 10.1017/S0007114510005684. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Álvaro M, et al. Bovine host genome acts on rumen microbiome function linked to methane emissions. Commun. Biol. 2022;5:1–16. doi: 10.1038/s42003-022-03293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyader J, et al. Additive methane-mitigating effect between linseed oil and nitrate fed to cattle. J. Anim. Sci. 2015;93:3564–3577. doi: 10.2527/jas.2014-8196. [DOI] [PubMed] [Google Scholar]
- 10.Patra A, Park T, Kim M, Yu Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017;8:1–18. doi: 10.1186/s40104-017-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villar ML, Hegarty RS, Nolan JV, Godwin IR, McPhee M. The effect of dietary nitrate and canola oil alone or in combination on fermentation, digesta kinetics and methane emissions from cattle. Anim. Feed Sci. Technol. 2020;259:114294. doi: 10.1016/j.anifeedsci.2019.114294. [DOI] [Google Scholar]
- 12.Patra AK, Yu Z. Effective reduction of enteric methane production by a combination of nitrate and saponin without adverse effect on feed degradability, fermentation, or bacterial and archaeal communities of the rumen. Bioresour. Technol. 2013;148:352–360. doi: 10.1016/j.biortech.2013.08.140. [DOI] [PubMed] [Google Scholar]
- 13.Fouts JQ, Honan MC, Roque BM, Tricarico JM, Kebreab E. Enteric methane mitigation interventions. Transl. Anim. Sci. 2022;6:txac041. doi: 10.1093/tas/txac041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odongo NE, Or-Rashid MM, Kebreab E, France J, McBride BW. Effect of supplementing myristic acid in dairy cow rations on ruminal methanogenesis and fatty acid profile in milk. J. Dairy Sci. 2007;90:1851–1858. doi: 10.3168/jds.2006-541. [DOI] [PubMed] [Google Scholar]
- 15.Broucek J. Options to methane production abatement in ruminants: A review. J. Anim. Plant Sci. 2018;28:348–364. [Google Scholar]
- 16.Honan, M., Feng, X., Tricarico, J. & Kebreab, E. Feed additives as a strategic approach to reduce enteric methane production in cattle: Modes of action, effectiveness and safety. Anim. Prod. Sci. 62, 1303–1317 (2021).
- 17.Goel G, et al. Effects of capric acid on rumen methanogenesis and biohydrogenation of linoleic and α-linolenic acid. Animal. 2009;3:810–816. doi: 10.1017/S1751731109004352. [DOI] [PubMed] [Google Scholar]
- 18.Dohme F, Machmüller A, Wasserfallen A, Kreuzer M. Ruminal methanogenesis as influenced by individual fatty acids supplemented to complete ruminant diets. Lett. Appl. Microbiol. 2001;32:47–51. doi: 10.1046/j.1472-765x.2001.00863.x. [DOI] [PubMed] [Google Scholar]
- 19.Yanza, Y. R. et al. The effects of dietary medium‐chain fatty acids on ruminal methanogenesis and fermentation in vitro and in vivo: A meta‐analysis. J. Anim. Physiol. Anim. Nutr. 105, 874–889 (2020). [DOI] [PubMed]
- 20.Soliva CR, Meile L, Cieślak A, Kreuzer M, Machmüller A. Rumen simulation technique study on the interactions of dietary lauric and myristic acid supplementation in suppressing ruminal methanogenesis. Br. J. Nutr. 2004;92:689–700. doi: 10.1079/BJN20041250. [DOI] [PubMed] [Google Scholar]
- 21.Berça AS, et al. Animal Feed Science and Nutrition-Production, Health and Environment. IntechOpen; 2021. Advances in Pasture management and animal nutrition to optimize beef cattle production in grazing systems. [Google Scholar]
- 22.Ibrahim NA, et al. Effects of vegetable oil supplementation on rumen fermentation and microbial population in ruminant: A review. Trop. Anim. Health Prod. 2021;53:1–11. doi: 10.1007/s11250-021-02863-4. [DOI] [PubMed] [Google Scholar]
- 23.Zain, M., Ningrat, R. W. S., Suryani, H. & Jamarun, N. Effect of various feed additives on the methane emissions from beef cattle based on an ammoniated palm frond feeds. in Animal Feed Science and Nutrition-Production, Health and Environment (IntechOpen, 2021).
- 24.Shi L, et al. Moderate coconut oil supplement ameliorates growth performance and ruminal fermentation in hainan black goat kids. Front. Vet. Sci. 2020;7:622259. doi: 10.3389/fvets.2020.622259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Temesgen T, Ayeneshet B, Aman Y. Nutritional mitigation of enteric methane gas emission from livestock sector: A Review. Forage Res. 2021;47(2):139–146. [Google Scholar]
- 26.Zhou Z, Yu Z, Meng Q. Effects of nitrate on methane production, fermentation, and microbial populations in in vitro ruminal cultures. Bioresour. Technol. 2012;103:173–179. doi: 10.1016/j.biortech.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Patra AK, Yu Z. Effects of adaptation of in vitro rumen culture to garlic oil, nitrate, and saponin and their combinations on methanogenesis, fermentation, and abundances and diversity of microbial populations. Front. Microbiol. 2015;6:1434. doi: 10.3389/fmicb.2015.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hristov AN, et al. Effect of lauric acid and coconut oil on ruminal fermentation, digestion, ammonia losses from manure, and milk fatty acid composition in lactating cows. J. Dairy Sci. 2009;92:5561–5582. doi: 10.3168/jds.2009-2383. [DOI] [PubMed] [Google Scholar]
- 29.Desbois AP, Smith VJ. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010;85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- 30.Machmüller A, Kreuzer M. Methane suppression by coconut oil and associated effects on nutrient and energy balance in sheep. Can. J. Anim. Sci. 1999;79:65–72. doi: 10.4141/A98-079. [DOI] [Google Scholar]
- 31.Burdick M, Zhou M, Guan L, Oba M. Effects of medium-chain fatty acid supplementation on performance and rumen fermentation of lactating Holstein dairy cows. Animal. 2022;16:100491. doi: 10.1016/j.animal.2022.100491. [DOI] [PubMed] [Google Scholar]
- 32.Machmüller A. Medium-chain fatty acids and their potential to reduce methanogenesis in domestic ruminants. Agric. Ecosyst. Environ. 2006;112:107–114. doi: 10.1016/j.agee.2005.08.010. [DOI] [Google Scholar]
- 33.Kittelmann S, et al. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS ONE. 2014;9:e103171. doi: 10.1371/journal.pone.0103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira AM, de Lurdes Nunes Enes Dapkevicius M, Borba AES. Alternative pathways for hydrogen sink originated from the ruminal fermentation of carbohydrates: Which microorganisms are involved in lowering methane emission? Anim. Microbiome. 2022;4:5. doi: 10.1186/s42523-021-00153-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizrahi I, Wallace RJ, Moraïs S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021;19:553–566. doi: 10.1038/s41579-021-00543-6. [DOI] [PubMed] [Google Scholar]
- 36.Stepanchenko N, et al. Microbial composition, rumen fermentation parameters, enteric methane emissions, and lactational performance of phenotypically high and low methane-emitting dairy cows. J. Dairy Sci. 2023;106:6146–6170. doi: 10.3168/jds.2022-23190. [DOI] [PubMed] [Google Scholar]
- 37.Wallace RJ, et al. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics. 2015;16:1–14. doi: 10.1186/s12864-015-2032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patra AK, Yu Z. Combinations of nitrate, saponin, and sulfate additively reduce methane production by rumen cultures in vitro while not adversely affecting feed digestion, fermentation or microbial communities. Bioresour. Technol. 2014;155:129–135. doi: 10.1016/j.biortech.2013.12.099. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z, Yu Z, Meng Q. Effects of nitrate on methane production, fermentation, and microbial populations in in vitro ruminal cultures. Bioresour. Technol. 2012;103:173–179. doi: 10.1016/j.biortech.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 40.Latham EA, Anderson RC, Pinchak WE, Nisbet DJ. Insights on alterations to the rumen ecosystem by nitrate and nitrocompounds. Front. Microbiol. 2016;7:228. doi: 10.3389/fmicb.2016.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soliva C, Hindrichsen I, Meile L, Kreuzer M, Machmüller A. Effects of mixtures of lauric and myristic acid on rumen methanogens and methanogenesis in vitro. Lett. Appl. Microbiol. 2003;37:35–39. doi: 10.1046/j.1472-765X.2003.01343.x. [DOI] [PubMed] [Google Scholar]
- 42.Machmüller A, Soliva CR, Kreuzer M. Methane-suppressing effect of myristic acid in sheep as affected by dietary calcium and forage proportion. Br. J. Nutr. 2003;90:529–540. doi: 10.1079/BJN2003932. [DOI] [PubMed] [Google Scholar]
- 43.Božic AK, et al. Effects of the methane-inhibitors nitrate, nitroethane, lauric acid, Lauricidin® and the Hawaiian marine algae Chaetoceros on ruminal fermentation in vitro☆. Bioresour. Technol. 2009;100:4017–4025. doi: 10.1016/j.biortech.2008.12.061. [DOI] [PubMed] [Google Scholar]
- 44.Ajisaka N, et al. Effects of medium-chain fatty acid-cyclodextrin complexes on ruminal methane production in vitro. Anim. Sci. J. 2002;73:479–484. doi: 10.1046/j.1344-3941.2002.00066.x. [DOI] [Google Scholar]
- 45.Wu H, Meng Q, Zhou Z, Yu Z. Ferric citrate, nitrate, saponin and their combinations affect in vitro ruminal fermentation, production of sulphide and methane and abundance of select microbial populations. J. Appl. Microbiol. 2019;127:150–158. doi: 10.1111/jam.14286. [DOI] [PubMed] [Google Scholar]
- 46.Nolan JV, Hegarty RS, Hegarty J, Godwin IR, Woodgate R. Effects of dietary nitrate on fermentation, methane production and digesta kinetics in sheep. Anim. Prod. Sci. 2010;50:801. doi: 10.1071/AN09211. [DOI] [Google Scholar]
- 47.Zhao L, et al. Effects of nitrate addition on rumen fermentation, bacterial biodiversity and abundance. Asian–Australas. J. Anim. Sci. 2015;28:1433. doi: 10.5713/ajas.15.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guyader J, Ungerfeld EM, Beauchemin KA. Redirection of metabolic hydrogen by inhibiting methanogenesis in the rumen simulation technique (RUSITEC) Front. Microbiol. 2017;8:393. doi: 10.3389/fmicb.2017.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ungerfeld EM. Metabolic hydrogen flows in rumen fermentation: Principles and possibilities of interventions. Front. Microbiol. 2020;11:589. doi: 10.3389/fmicb.2020.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang LZ, Zhou ML, Wang JW, Wu D, Yan T. The effect of dietary replacement of ordinary rice with red yeast rice on nutrient utilization, enteric methane emission and rumen archaeal diversity in goats. PLoS ONE. 2016;11:e0160198. doi: 10.1371/journal.pone.0160198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolin MJ. A theoretical rumen fermentation balance. J. Dairy Sci. 1960;43:1452–1459. doi: 10.3168/jds.S0022-0302(60)90348-9. [DOI] [Google Scholar]
- 52.Li F, et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome. 2019;7:92. doi: 10.1186/s40168-019-0699-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuste S, Amanzougarene Z, de la Fuente G, de Vega A, Fondevila M. Rumen protozoal dynamics during the transition from milk/grass to high-concentrate based diet in beef calves as affected by the addition of tannins or medium-chain fatty acids. Anim. Feed Sci. Technol. 2019;257:114273. doi: 10.1016/j.anifeedsci.2019.114273. [DOI] [Google Scholar]
- 54.Zhang D-F, Yang H-J. Combination effects of nitrocompounds, pyromellitic diimide, and 2-bromoethanesulfonate on in vitro ruminal methane production and fermentation of a grain-rich feed. J. Agric. Food Chem. 2012;60:364–371. doi: 10.1021/jf203716v. [DOI] [PubMed] [Google Scholar]
- 55.Mitsumori M, Sun W. Control of rumen microbial fermentation for mitigating methane emissions from the rumen. Asian–Australas. J. Anim. Sci/ 2008;21:144–154. doi: 10.5713/ajas.2008.r01. [DOI] [Google Scholar]
- 56.Zhang XM, et al. Combined effects of 3-nitrooxypropanol and canola oil supplementation on methane emissions, rumen fermentation and biohydrogenation, and total tract digestibility in beef cattle. J. Anim. Sci. 2021;99:skab081. doi: 10.1093/jas/skab081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollmann M, et al. Enteric methane emissions and lactational performance of Holstein cows fed different concentrations of coconut oil. J. Dairy Sci. 2012;95:2602–2615. doi: 10.3168/jds.2011-4896. [DOI] [PubMed] [Google Scholar]
- 58.Rossi CAS, Grossi S, Dell’Anno M, Compiani R, Rossi L. Effect of a blend of essential oils, bioflavonoids and tannins on in vitro methane production and in vivo production efficiency in dairy cows. Animals. 2022;12:728. doi: 10.3390/ani12060728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janssen PH. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 2010;160:1–22. doi: 10.1016/j.anifeedsci.2010.07.002. [DOI] [Google Scholar]
- 60.Wang K, Xiong B, Zhao X. Could propionate formation be used to reduce enteric methane emission in ruminants? Sci. Total Environ. 2023;855:158867. doi: 10.1016/j.scitotenv.2022.158867. [DOI] [PubMed] [Google Scholar]
- 61.Ahmed E, et al. Impacts of mootral on methane production, rumen fermentation, and microbial community in an in vitro study. Front. Vet. Sci. 2021;7:623817. doi: 10.3389/fvets.2020.623817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pope P, et al. Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science. 2011;333:646–648. doi: 10.1126/science.1205760. [DOI] [PubMed] [Google Scholar]
- 63.Hassan F, et al. Effect of methionine supplementation on rumen microbiota, fermentation, and amino acid metabolism in in vitro cultures containing nitrate. Microorganisms. 2021;9:1717. doi: 10.3390/microorganisms9081717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Y, et al. Effect of sodium nitrate and cysteamine on in vitro ruminal fermentation, amino acid metabolism and microbiota in buffalo. Microorganisms. 2022;10:2038. doi: 10.3390/microorganisms10102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aguilar-Marin SB, Betancur-Murillo CL, Isaza GA, Mesa H, Jovel J. Lower methane emissions were associated with higher abundance of ruminal Prevotella in a cohort of Colombian buffalos. BMC Microbiol. 2020;20:364. doi: 10.1186/s12866-020-02037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Zijderveld SM, et al. Effects of a combination of feed additives on methane production, diet digestibility, and animal performance in lactating dairy cows. J. Dairy Sci. 2011;94:1445–1454. doi: 10.3168/jds.2010-3635. [DOI] [PubMed] [Google Scholar]
- 67.Salami SA, et al. Characterization of the ruminal fermentation and microbiome in lambs supplemented with hydrolysable and condensed tannins. FEMS Microbiol. Ecol. 2018 doi: 10.1093/femsec/fiy061. [DOI] [PubMed] [Google Scholar]
- 68.Ran T, et al. Effects of brewers’ spent grain protein hydrolysates on gas production, ruminal fermentation characteristics, microbial protein synthesis and microbial community in an artificial rumen fed a high grain diet. J. Anim. Sci. Biotechnol. 2021;12:1–14. doi: 10.1186/s40104-020-00531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Veneman JB, et al. Does dietary mitigation of enteric methane production affect rumen function and animal productivity in dairy cows? PLoS ONE. 2015;10:e0140282. doi: 10.1371/journal.pone.0140282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang R, et al. Nitrate improves ammonia incorporation into rumen microbial protein in lactating dairy cows fed a low-protein diet. J. Dairy Sci. 2018;101:9789–9799. doi: 10.3168/jds.2018-14904. [DOI] [PubMed] [Google Scholar]
- 71.Xie F, et al. Sodium nitrate has no detrimental effect on milk fatty acid profile and rumen bacterial population in water buffaloes. AMB Express. 2022;12:1–17. doi: 10.1186/s13568-022-01350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang C, Rooke JA, Cabeza I, Wallace RJ. Nitrate and inhibition of ruminal methanogenesis: Microbial ecology, obstacles, and opportunities for lowering methane emissions from ruminant livestock. Front. Microbiol. 2016;7:132. doi: 10.3389/fmicb.2016.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patra AK, Yu Z. Effects of essential oils on methane production and fermentation by, and abundance and diversity of, rumen microbial populations. Appl. Environ. Microbiol. 2012;78:4271–4280. doi: 10.1128/AEM.00309-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beauchemin KA, Ungerfeld EM, Eckard RJ, Wang M. Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal. 2020;14:s2–s16. doi: 10.1017/S1751731119003100. [DOI] [PubMed] [Google Scholar]
- 75.Menke K, et al. The estimation of the digestibility and metabolizable energy content of ruminant feedingstuffs from the gas production when they are incubated with rumen liquor in vitro. J. Agric. Sci. 1979;93:217–222. doi: 10.1017/S0021859600086305. [DOI] [Google Scholar]
- 76.Romero-Pérez A, Beauchemin KA. Estimating gas volume from headspace pressure in a batch culture system. Can. J. Anim. Sci. 2018;98:593–596. doi: 10.1139/cjas-2017-0100. [DOI] [Google Scholar]
- 77.Joch M, et al. In vitro and in vivo potential of a blend of essential oil compounds to improve rumen fermentation and performance of dairy cows. Anim. Feed Sci. Technol. 2019;251:176–186. doi: 10.1016/j.anifeedsci.2019.03.009. [DOI] [Google Scholar]
- 78.Weatherburn M. Phenol-hypochlorite reaction for determination of ammonia. Analyt. Chem. 1967;39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 79.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oksanen, J. et al. vegan: Community Ecology Package. (2022).
- 82.Yang W, et al. Succession of bacterioplankton community in intensive shrimp (Litopenaeus vannamei) aquaculture systems. Aquaculture. 2018;497:200–213. doi: 10.1016/j.aquaculture.2018.07.053. [DOI] [Google Scholar]
- 83.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets utilized and/or analyzed in the present study can be obtained from the corresponding author upon reasonable request.