Abstract
Four genes identified within the late operon of PBSX show characteristics expected of a host cell lysis system; they are xepA, encoding an exported protein; xhlA, encoding a putative membrane-associated protein; xhlB, encoding a putative holin; and xlyA, encoding a putative endolysin. In this work, we have assessed the contribution of each gene to host cell lysis by expressing the four genes in different combinations under the control of their natural promoter located on the chromosome of Bacillus subtilis 168. The results show that xepA is unlikely to be involved in host cell lysis. Expression of both xhlA and xhlB is necessary to effect host cell lysis of B. subtilis. Expression of xhlB (encoding the putative holin) together with xlyA (encoding the endolysin) cannot effect cell lysis, indicating that the PBSX lysis system differs from those identified in the phages of gram-negative bacteria. Since host cell lysis can be achieved when xlyA is inactivated, it is probable that PBSX encodes a second endolysin activity which also uses XhlA and XhlB for export from the cell. The chromosome-based expression system developed in this study to investigate the functions of the PBSX lysis genes should be a valuable tool for the analysis of other host cell lysis systems and for expression and functional analysis of other lethal gene products in gram-positive bacteria.
Host cell lysis upon induction of prophages has been widely studied in a number of gram-negative bacteria (reviewed in reference 28). The best characterized of these systems is lysis of Escherichia coli after induction of phage lambda. Three lysis genes have been identified in lambda: S, R, and Rz. These lysis genes are the first three genes of the late operon, and they are followed by the genes which encode the proteins involved in formation of the phage particle. The function of the Rz gene is unknown, and host cell lysis is not affected by its inactivation (6, 27). The R gene encodes the endolysin, which in the case of lambda, is a transglycosylase (2). The term endolysin is used for a variety of different peptidoglycan-degrading enzymes including lysozyme, transglycosylase, amidase, and endopeptidase (28). A feature of phage endolysins from gram-positive and gram-negative bacteria is that they are synthesized without a signal peptide. Therefore, a second protein is needed to get the enzyme out of the cell so that it can access the extracellular peptidoglycan. In lambda, this protein is encoded by the S gene. The S protein is called a holin since it is thought to function by creating a hole in the cytoplasmic membrane through which the endolysin can pass and access the extracellularly located peptidoglycan (28, 29). When S is expressed in the absence of the R protein, respiration and macromolecular synthesis stop at the normal lysis time; however, no cell lysis is observed (11, 17). Therefore, expression of the S protein is lethal to the cell even though it does not cause cell lysis. The S gene from lambdoid phage 21 (21S) was found to complement mutations in the S gene of lambda, while the lambda S gene was also shown to complement mutations in the 21S gene (5). The two genes display no homology at the amino acid sequence level, but they do share a number of structural characteristics (29): (i) they are small in size (71 and 107 amino acids); (ii) they have two putative membrane-spanning domains separated by a beta-turn linker (the criteria used to designate a protein region as membrane spanning are that it be at least 16 amino acids in length, that it be predominantly hydrophobic in nature, and that it have no net charge); (iii) they have two Met start sites, called a “dual start” motif; and (iv) they have a highly polar, charge-rich C-terminal domain. A number of genes encoding proteins with these characteristics have been identified in phages of both gram-positive and gram-negative bacteria and so have been classed as holins (29). Some putative holins from phages of gram-negative bacteria have been confirmed by mutational and functional analysis to have holin activity in vivo: these holins are encoded by P22 gene 13 (18), phage 21 gene S (5), P2 gene Y (30), and phage P1 gene lydA (21). Three putative holins identified in phages of gram-positive bacteria have been confirmed to function as holins in the lambda phage test system of E. coli: they are encoded by φ29 gene P14 (22, 24), φLC3 gene lysA (3), and φadh gene hol (13). The φ29 holin is lethal when expressed in E. coli and can mediate transit of both the cognate and unrelated (lambda and T7) endolysins to the periplasm. In contrast, the φadh holin can facilitate lysis of E. coli only with heterologous (lambda and φ29) endolysins. It is important to note that the activities of the holins from the phages of gram-positive bacteria have been demonstrated by complementation in the lambda phage system of E. coli. None have been shown to have holin activity in a gram-positive host.
The late operon of PBSX contains genes encoding the structural proteins for phage particle formation and the genes for host cell lysis functions (10, 14, 16, 26). The lysis genes have been located on a 2.6-kb DNA fragment at the distal end of the PBSX late operon (16). Sequence analysis suggests that four open reading frames (ORFs) may be involved in host cell lysis: xepA (ORF1), encoding an exported protein; xhlA (ORF2), encoding a putative membrane-associated protein; xhlB (ORF3), encoding a putative holin; and xlyA (ORF4), encoding a putative endolysin (16). The objective of this work was to investigate the contribution of each of these ORFs to host cell lysis upon PBSX induction. A primary aim was to perform this analysis in Bacillus subtilis, the natural PBSX host, so that the lysis genes would be expressed under the control of their natural promoter and the activity of each protein would be assayed in its natural gram-positive host. The results show that xhlA, xhlB, and xlyA are involved in host cell lysis, whereas xepA is nonessential for lysis. We also show that, in contrast to phages of gram-negative bacteria, expression of the PBSX holin and endolysin alone in their natural host, B. subtilis 168, is not sufficient to effect host cell lysis.
MATERIALS AND METHODS
Plasmid and strain construction.
Bacterial strains and plasmids used in this study are described in Table 1. The structure of the PBSX late operon in each of the POP strains is shown diagramatically in Fig. 1. E. coli TP611 (12) was used for cloning fragments containing each of the four ORFs. Four PCR-generated fragments, each encoding one of the four ORFs, were cloned separately into the EcoRV site of the integrating plasmid pDIA5304. The inserts in pDIA5304 were sequenced according to the method of Sanger et al. (20) with a U.S. Biochemical Sequenase sequencing kit to verify that the sequences were those of the wild type. The plasmids containing xepA (ORF1) and xlyA (ORF4) have the fragments inserted such that the ORFs read in the opposite orientation to the α fragment of the lacZ gene on the plasmid, whereas the plasmids containing xhlA (ORF2) and xhlB (ORF3) have the ORFs in the same orientation as the α fragment. The four plasmids obtained (pORF1, pORF2, pORF3, and pORF4) were transformed into L8508 to give strains SK101, SK102, SK103, and SK104 (Fig. 1). Integration occurred by a Campbell-type event. In order to prevent transcription of the late operon progressing beyond the point of the chromosomal insert, the inserted plasmid was amplified by streaking transformants onto plates containing increasing concentrations (from 3 to 30 μg/ml) of chloramphenicol.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TP611 | recBC hsdRM cya610 pcn | 12 |
| B. subtilis | ||
| L8508 | xhi1479 lyt2 | D. Karamata |
| POP0 | L8508 with all ORFs of the PBSX late operon deleted | This work |
| POP4 | L8508 with all ORFs except xlyA of the PBSX late operon deleted | This work |
| POP34 | L8508 with all ORFs except xlyA and xhlB of the PBSX late operon deleted | This work |
| POP234 | L8508 with all ORFs except xlyA, xhlB, and xhlA of the PBSX late operon deleted | This work |
| POP1234 | L8508 with all ORFs except xlyA, xhlB, xhlA, and xepA of the PBSX late operon deleted | This work |
| SK101 | L8508::pORF1, with selection for amplification of integrating plasmid | This work |
| SK102 | L8508::pORF2, with selection for amplification of integrating plasmid | This work |
| SK103 | L8508::pORF3, with selection for amplification of integrating plasmid | This work |
| SK104 | L8508::pORF4, with selection for amplification of integrating plasmid | This work |
| SK105 | POP0::pSK26 | This work |
| SK106 | POP0::pSK27 | This work |
| SK107 | POP0::pSK28 | This work |
| SK108 | POP0::pSK29 | This work |
| SK109 | POP1234::pSK24 | This work |
| SK110 | POP234::pSK24 | This work |
| SK111 | POP34::pSK24 | This work |
| SK112 | POP4::pSK24 | This work |
| Plasmids | ||
| pG+host4 | B. subtilis plasmid with temperature-sensitive replicon and Emr gene | 4 |
| pDIA5304 | Bluescript containing a chloramphenicol resistance gene | 11 |
| pORF1 | xepA-containing fragment cloned into the EcoRV site of pDIA5304 | This work |
| pORF2 | xhlA-containing fragment cloned into the EcoRV site of pDIA5304 | This work |
| pORF3 | xhlB-containing fragment cloned into the EcoRV site of pDIA5304 | This work |
| pORF4 | xlyA-containing fragment cloned into the EcoRV site of pDIA5304 | This work |
| pSK11 | PBSX fragment 38 cloned into the MCSa of pDIA5304 | This work |
| pSK22 | PBSX fragment 615 cloned into the MCS of pDIA5304 | This work |
| pSK24 | Internal 520-bp PstI-SacI fragment from xlyA cloned into pDIA5304 | This work |
| pSK26 | xepA-containing fragment cloned juxtaposed to fragment 38 in pSK11 | This work |
| pSK27 | xhlA-containing fragment cloned juxtaposed to fragment 615 in pSK22 | This work |
| pSK28 | xhlB-containing fragment cloned juxtaposed to fragment 615 in pSK22 | This work |
| pSK29 | xlyA-containing fragment cloned juxtaposed to fragment 38 in pSK11 | This work |
MCS, multiple cloning site.
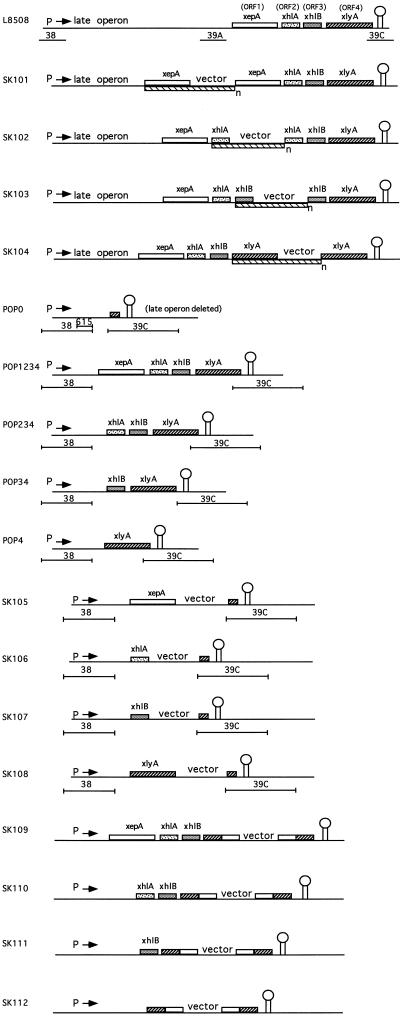
FIG. 1.
Schematic representation of the organization of the PBSX late operon in the strains constructed in this study. The names of the strains are indicated on the left. The promoter of the late operon is indicated by “P,” and the direction of transcription is indicated by an arrow. The term “late operon” represents approximately 24 kb of DNA containing the genes encoding the PBSX head and tail proteins. The four genes in the lysis cluster are indicated by distinctively filled rectangles above each line. The putative terminator of the PBSX late operon is indicated by a stem-loop structure. In cases where DNA is amplified, the extent of the amplification unit is indicated by a rectangle below the line with the suffix “n” indicating an unknown number of copies. The extent and location of four DNA fragments, fragment 38, fragments 39A and 39C, and fragment 615, used to construct some of the strains are indicated by lines below relevant constructs.
Two plasmids (pSK11 and pSK22) were constructed for the purpose of individually integrating each of the four ORFs into the PBSX locus on the chromosome, so that they could be expressed under the control of the natural PBSX promoter. Plasmid pSK11 was constructed by excising fragment 38 from pWD38 (26) on an EcoRI-BamHI fragment, polishing the ends, and cloning the blunt-ended fragment into XhoI-cut pDIA5304 (the ends of which had been filled in with the Klenow fragment). A recombinant plasmid in which the PBSX late operon promoter on fragment 38 is in the same direction as the α fragment of the lacZ gene was chosen. pSK22 was constructed in a manner similar to pSK11 with the exception that only the last 615 bp of fragment 38 (from PstI to the BamHI site) was cloned into the SpeI site of pDIA5304. In this case, a recombinant plasmid in which the orientation of the 615-bp fragment is opposite to that of the α fragment of the lacZ gene was chosen. Fragments containing xepA and xlyA were excised from pORF1 and pORF4, respectively, on SalI-BamHI fragments and were directionally cloned into pSK11 (SalI and BamHI digested), yielding plasmids pSK26 and pSK29. Similarly, fragments containing xhlA and xhlB were excised from pORF2 and pORF3, respectively, and cloned into BamHI–SalI-cut pSK22 to give plasmids pSK27 and pSK28. Plasmids pSK26, pSK27, pSK28, and pSK29 were then integrated into strain POP0, in which all of the ORFs within the PBSX late operon between fragment 38 and fragment 39C (see below) are deleted. Therefore, in strains SK105, SK106, SK107, and SK108, each of the four ORFs can be expressed alone under the control of the PBSX late operon promoter (Fig. 1).
The POP strains were constructed by the method described by Biswas et al. (4). To generate deletions of chromosomal regions within the PBSX late operon, DNA fragments flanking each end of the region to be deleted were cloned into the conditional integrating plasmid pG+host4 juxtaposed to each other (in the same relative orientation as they are on the chromosome). Five fragment pairs were subcloned in this manner, and each resultant plasmid was used to generate a POP strain having a different region of the chromosome deleted: (i) in strain POP0, the region between fragment 38 and fragment 39C (which overlaps the distal end of xlyA) is deleted; (ii) in POP1234, the region located downstream of fragment 38 and extending to (but not including) xepA is deleted; (iii) in POP234, the region located downstream of fragment 38 and extending to (but not including) xhlA is deleted; (iv) in strain POP34, the region located downstream of fragment 38 and extending to (but not including) xhlB is deleted; and (v) in strain POP4, the region located downstream of fragment 38 and extending to (but not including) xlyA is deleted. Each construct is shown diagramatically in Fig. 1.
Plasmid pSK24 contains an internal 520-bp PstI-SacI fragment from xlyA cloned into pDIA5304. By integrating this plasmid into the chromosome, xlyA was mutagenized in strains POP1234, POP234, POP34, and POP4 to give strains SK109, SK110, SK111, and SK112, respectively (Fig. 1).
The chromosomal rearrangements generated in all strains were verified by PCR with a number of combinations of primers.
Growth curves.
Bacterial cultures were grown in Luria-Bertani (LB) broth at 37°C to an optical density at 550 nm (OD550) of approximately 0.2. Cultures were then moved to a shaking water bath at 48°C to induce PBSX, and further growth was monitored by measuring the turbidity of the culture at OD550 for up to 3 h or until lysis was observed. The time of induction in all experiments was at 1.5 h.
Transformation.
B. subtilis transformation was carried out according to the method of Anagnostopoulos and Spizizen (1), and E. coli transformation was performed as described by Sambrook et al. (19).
General molecular biological methods.
All general molecular biological procedures were performed as detailed by Sambrook et al. (19).
Renaturing gel electrophoresis.
Cells were harvested by centrifugation and prepared for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis as described by Foster (9). The cell extracts were resolved on 15% polyacrylamide gels containing B. subtilis cell walls. The autolytic activities were renatured and visualized according to the method described by Foster (9). The relative mobilities of molecular weight markers were determined on a separate gel, which was stained with Coomassie blue. This gel was prepared identically to and electrophoresed concomitantly with the cell wall-containing gel.
RESULTS
A 2.6-kb fragment of the PBSX late operon encodes sufficient functions to mediate host cell lysis.
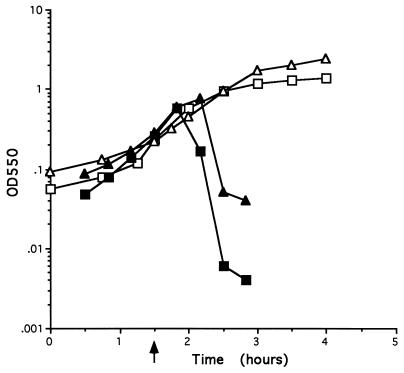
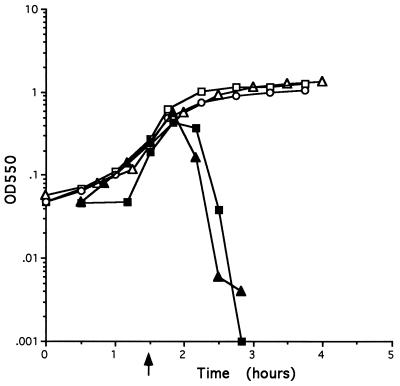
In order to establish experimentally that functions necessary for host cell lysis are contained on the fragment encoding the four ORFs xepA, xhlA, xhlB, and xlyA, strains POP0 and POP1234 were constructed as described in Materials and Methods (Fig. 1). Two noncontiguous regions of chromosome, fragment 38 (which contains the promoter for the PBSX late operon) and fragment 39A, located upstream of the four ORFs (for strain POP1234), or fragment 39C, located downstream of the ORFs (for POP0), were cloned into pG+host4. Each plasmid was integrated into the chromosome, and plasmid excision was subsequently stimulated in the resultant integrants, as previously described (4). In those strains where integration is directed by one of the homologous fragments and excision is directed by the second homologous fragment, the chromosomal regions between the homologous fragments will be deleted. Therefore, in strain POP0 the entire late operon, including the four putative lysis genes, is deleted. In strain POP1234, the late operon is deleted, leaving the four open reading frames intact, juxtaposed to, and under the control of the late operon promoter. The growth curves obtained when PBSX is induced in strains L8508, POP0, and POP1234 are shown in Fig. 2. Cells of strains POP1234 and L8508 lysed after PBSX induction. Lysis of POP1234 cells appears to occur somewhat earlier than lysis of cells of the parental strain L8508. Lysis begins approximately 30 min postinduction and is virtually complete by 60 min. The growth kinetics of POP0 cultures differ from those of POP1234. There is a gradual cessation of growth at 60 to 90 min after PBSX induction in strain POP0 but the cells do not lyse. The growth plateau observed with POP0 cells after PBSX induction occurs much earlier than the natural stationary phase observed with uninduced strain L8508. These results confirm that functions essential for PBSX-induced host cell lysis are contained within the PBSX late operon fragment encoding these four ORFs. They also demonstrate that in the absence of the entire PBSX late operon, induction of PBSX by temperature upshift results in a gradual cessation of growth but the cells do not lyse.
FIG. 2.
Growth curves of strains after PBSX induction. Strains were grown in LB broth at 37°C to an OD550 of approximately 0.2. PBSX was induced by shifting the cultures to 48°C. Subsequent growth of each strain was monitored by measuring the turbidity at 550 nm. The point of induction is indicated by an arrow. Symbols: ▵, L8508 uninduced; ▴, L8508 induced; □, POP0 induced; ▪, POP1234 induced.
Effect of amplifying each putative lysis gene on the kinetics of cell lysis.
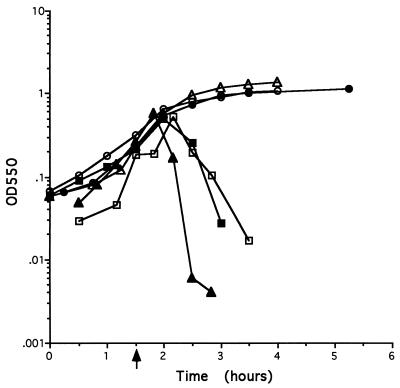
Strains were constructed to test the effect on host cell lysis of amplification of one of the four putative lysis genes coupled with nonexpression of those genes located downstream of the amplified gene. Four plasmids (pORF1 through pORF4), each containing a complete copy of one of the four ORFs, were integrated separately into L8508 (see Materials and Methods and Fig. 1). The integrating plasmid contains a chloramphenicol resistance gene which allows selection of those cells containing multiple copies of the amplification unit. Transcription of the PBSX genes downstream of the site of integration is greatly reduced or abolished in such strains, probably due to terminators located on the vector part of the amplified unit (25a). The late operon construct in each of the resultant strains, SK101, SK102, SK103, and SK104, is shown in Fig. 1, and the growth characteristics of each strain upon thermoinduction of PBSX are shown in Fig. 3. Cultures of L8508 cells continue to grow to an OD of >3.0 in the absence of induction. When PBSX is induced in L8508 cultures, the cells stop growing and cell lysis is virtually complete within 60 min. Upon induction of PBSX in cultures of SK101, the cells continue to grow for up to 60 min, at which time a plateau is reached. No further growth is observed, but, importantly, the cells do not lyse. When PBSX is induced in SK102 cultures, the cells continue to grow for approximately 60 min. Very gradual lysis of the cells is then observed for the subsequent 150 min. When PBSX is induced in cultures of SK103 and SK104, the kinetics of growth and cell lysis are identical to those observed upon induction of the parental strain L8508. These data confirm that the PBSX lysis functions are encoded by the four ORFs. In addition, the data suggest that xepA, xhlA, and xhlB are sufficient for cell lysis under these conditions. However, an aberrant, very slow lysis can be achieved by expression of xepA together with multicopies of xhlA.
FIG. 3.
Growth curves of strains after PBSX induction. Strains were grown in LB broth at 37°C to an OD550 of 0.2. PBSX was induced by shifting the cultures to 48°C. The growth kinetics of each strain were monitored by measuring the turbidity at 550 nm. The point of induction is indicated by an arrow. Symbols: ▵, L8508 uninduced; ▴, L8508 induced; ▪, SK101 induced; □, SK102 induced; ○, SK103 induced; •, SK104 induced.
Individual expression of each putative lysis gene in B. subtilis.
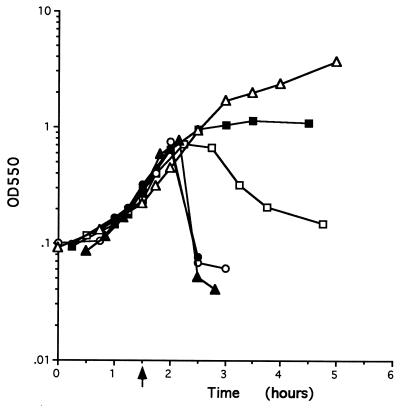
A series of mutants was constructed so that each of the four ORFs could be expressed separately under the control of the natural PBSX promoter in B. subtilis. To achieve this, each complete ORF was cloned into an integrating plasmid which contained either fragment 38, yielding plasmids pSK26 and pSK29, or fragment 615, giving plasmids pSK27 and pSK28 (see Materials and Methods). Each plasmid was then separately integrated into the chromosome of strain POP0 by a Campbell-type event, yielding strains SK105 (expressing xepA only), SK106 (expressing xhlA only), SD107 (expressing xhlB only), and SK108 (expressing xlyA only). The structure of the PBSX late operon in each of these strains is shown in Fig. 1, and their growth kinetics after PBSX induction are shown in Fig. 4. Induction of PBSX in cultures of SK105 and SK107 results in the same growth profile as that seen in cultures of POP0. Cultures of these strains do not lyse but instead enter a premature stationary phase approximately 90 min after PBSX induction. Upon induction of PBSX, cultures of SK106 continue to grow for 60 min with the same kinetics as strains POP0, SK105, and SK107. Thereafter, a decrease in the OD550, which continues for approximately 60 minutes, occurs. However, it is clear that the kinetics of lysis in strain SK106 differ from those seen in the parental strain L8508. Lysis of cells of SK106 occurs more gradually and does not proceed to the same extent as that of strain L8508. The kinetics of lysis of strain SK108 are similar to those of SK106: lysis begins about 60 min after PBSX induction and proceeds more slowly and to a lesser extent than is observed in the parental strain L8508. These data indicate (i) that expression of xepA and xhlB alone are insufficient to effect cell lysis and (ii) that when individually expressed in cells, both xhlA and xlyA can effect cell lysis with aberrant kinetics, being slower and less complete than those seen in the parental L8508 strain.
FIG. 4.
Growth curves of strains after PBSX induction. Strains were grown in LB broth at 37°C to an OD550 of 0.2, and PBSX was induced by shifting the cultures to 48°C. The point of induction is indicated by an arrow. Symbols: ▵, POP0; ▴, POP1234; ▪, SK105; □, SK106; •, SK107; ○, SK108.
Expression of combinations of the four putative cell lysis genes.
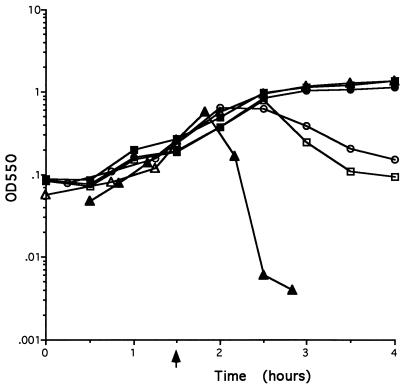
A series of strains were constructed to assess the contribution of different combinations of the four putative lysis genes to host cell lysis. In the first set of strains, the lysis genes in strain POP1234 were sequentially deleted in the order of xepA, xhlA, xhlB, and xlyA to give strains POP234, POP34, POP4, and POP0, respectively (Fig. 1). The strategy used to generate this series of mutant strains was the same as that used for the generation of POP0 and POP1234 (4). An important consequence of this construction method is that each gene is present in single copy and expressed from the PBSX late operon promoter. The growth curves for these strains are presented together with control growth curves for POP0 and POP1234 in Fig. 5. The results show that strain POP234 lyses approximately 30 to 60 min after PBSX induction with kinetics virtually indistinguishable from those of strain POP1234. In contrast, the strains expressing xhlB and xlyA alone (POP34) or xlyA only (POP4) do not lyse upon PBSX induction. Instead, both strains enter a growth plateau approximately 60 min after PBSX induction, displaying growth kinetics similar to those observed with strain POP0 after PBSX induction (Fig. 2). These data demonstrate that xepA is not essential for host cell lysis. It is also clear that in this PBSX system, cell lysis can be effected neither by expression of xhlB and xlyA (holin and endolysin) together nor by expression of xlyA (endolysin) alone. Therefore, XhlA is an essential component for PBSX-mediated host cell lysis.
FIG. 5.
Growth curves of strains after PBSX induction. Strains were grown in LB broth at 37°C to an OD550 of 0.2, and PBSX induction was effected by shifting the cultures to 48°C. The point of induction is indicated by an arrow. Symbols: ▵, POP0; ▴, POP1234; ▪, POP234; □, POP34; ○, POP4.
A second series of mutant strains were generated to assess the ability of combinations of the lysis genes to effect host cell lysis upon PBSX induction. The integrating plasmid pSK24, which contains an internal 520-bp fragment from xlyA (ORF4) was integrated into the chromosomes of strains POP1234, POP234, POP34, and POP4. The lysis genes that are functional in the resultant integrant strains are SK109 (xepA, xhlA, and xhlB), SK110 (xhlA and xhlB), SK111 (xhlB), and SK112 (Fig. 1). It is important to note that each gene is present in single copy and is expressed from the natural PBSX control system in these strains. The growth curves (Fig. 6) show that cells of strains SK109 and SK110 both lyse approximately 90 min after PBSX induction. Lysis in these strains therefore is delayed by approximately 30 min compared to the lysis kinetics of strains carrying the wild-type prophage. When PBSX was induced in strains SK111 (expressing only xhlB) and SK112, a gradual cessation of growth was observed but the cells did not lyse, a pattern similar to that observed with POP0. These data confirm that xepA is not essential for cell lysis. They also demonstrate that cell lysis can be effected when xhlA and xhlB are expressed alone and that that XhlA is an essential component of the PBSX-encoded host cell lysis system.
FIG. 6.
Growth curves of strains after PBSX induction. Strains were grown in LB broth at 37°C to an OD550 of 0.2, and PBSX was induced by shifting the cultures to 48°C. The point of induction is indicated by an arrow. Symbols: ▵, POP0; ▴, POP1234; ▪, SK109; □, SK110; •, SK111; ○, SK112.
Can XlyA autolysin activity be detected in strain POP34 after thermoinduction of PBSX?
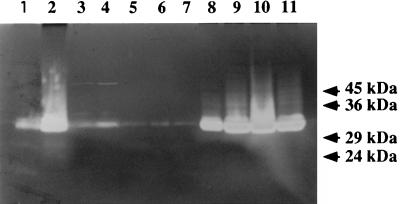
An underlying assumption of the experiments previously described is that xhlA, xhlB, and xlyA are expressed upon PBSX induction despite deletion of most of the PBSX late operon (yielding POP1234) and despite deletion and/or amplification of individual genes within the lysis cluster (yielding the remaining SK and POP strains). The autolysin, N-acetylmuramoyl-l-alanine amidase, is the only gene product of the lysis gene cluster for which an assay is readily available. The xlyA gene is in a distal position (relative to the PBSX promoter) in the lysis cluster. Therefore, if PBSX-encoded autolysin activity can be detected in the POP strains, thereby demonstrating xlyA expression, it is likely that in those strains where xhlA, xhlB, or both genes are positioned between xlyA and the promoter, they also will be expressed. Cell extracts were prepared according to the method described by Foster (9) from the parental strain L8508 and from strains POP0 and POP34, both before and after thermoinduction of PBSX. The extracts were resolved on SDS-polyacrylamide gels containing 0.1% (wt/vol) B. subtilis cell walls, and autolysin activity was detected after enzyme renaturation followed by staining with methylene blue (9). Therefore, this assay directly detects production of a functional PBSX autolysin. A representative gel is shown in Fig. 7. Lanes 1 and 2 show the autolysin profiles of the parental strain L8508 at the point of temperature upshift and at 30 min after thermoinduction at 48°C, respectively. There is a very significant increase in autolytic activity after induction, which migrates with an Mr of approximately 32,000. The size of this activity correlates with the Mr of XlyA estimated by Foster (10) and verified by sequencing of the gene (16). This increase in activity is a result of temperature upshift and is not growth phase dependent, since cells grown at 37°C do not show this increased activity at any stage of the growth cycle (data not shown). The autolytic profile observed in POP0 cell extracts (in this strain the entire PBSX late operon, including the lysis gene cluster, is deleted [Fig. 1]) is shown at the point of temperature upshift (Fig. 7, lane 3) and at 120 min after induction at 48°C (lane 4). It is clear that the increased autolytic activity of Mr ∼32,000 is no longer observed in this strain even after this prolonged growth period at the elevated temperature (under these conditions, the parental strain L8508 lyses at approximately 60 min after induction). This shows that the increased autolysin activity observed in L8508 is encoded within the late operon of PBSX. When extracts of POP34 are examined for autolytic activity at 37°C, it is clear that only low levels of activity are present throughout the growth cycle (lane 5, extracts prepared from cells at an OD550 of 0.29; lane 6, extracts prepared from cells at an OD550 of 2.91 [Fig. 7]). The autolytic activity profile of strain POP34 at the point of temperature upshift and at 30, 60, 90, and 120 min postinduction can be seen in Fig. 7, lanes 7 to 11, respectively. It is clear that there is a very dramatic increase in autolytic activity of Mr 32,000 at 30 min postinduction and which persists throughout the remainder of the experiment. Since the only difference between POP0 and POP34 is the presence of the xhlB and xlyA genes, it is clear that the induced autolytic activity of the latter strain must be encoded by xlyA. Therefore, in POP34, the xlyA gene is expressed at 30 min postinduction and the N-acetylmuramoyl-l-alanine amidase activity remains active in the cell for a further 90 min, during which time the turbidity of the medium rises from an OD550 of 0.8 to an OD550 of 1.5. No cell lysis of POP34 is observed at any time under these conditions, in contrast to the parental and POP234 strains, which lyse within 60 min of thermoinduction. These data show that the autolysin activity of the late operon lysis gene cluster is expressed and is active in POP34 cells after thermoinduction of PBSX.
FIG. 7.
Detection of autolysin activity by renaturing SDS gel electrophoresis. Cell extracts were resolved on SDS–15% polyacrylamide gels containing 0.1% (wt/vol) B. subtilis cell walls. The autolysin activity was renatured as described in the text, and gels were stained with methylene blue and destained in water. Cell extract equivalent to 1 ml of cells at an OD550 of 0.9 was loaded into each well. The mobility of molecular weight markers was determined from a separate gel (without added cell walls) identically prepared and concomitantly electrophoresed with the gel shown. Lane 1, L8508 at the point of 37°C-to-48°C upshift; lane 2, L8508 at 30 min postthermoinduction; lane 3, POP0 at the point of 37°C-to-48°C upshift; lane 4, POP0 at 120 min postthermoinduction; lane 5, POP34 before shifting to 48°C; lane 6, POP34 retained at 37°C for 120 min while the other half of the culture was shifted to 48°C; lane 7, POP34 at the point of 37°C-to-48°C upshift; lane 8, POP34 at 30 min after thermoinduction; lane 9, POP34 at 60 min after thermoinduction; lane 10, POP34 at 90 min after thermoinduction; lane 11, POP34 at 120 min after thermoinduction.
DISCUSSION
The objective of this study was to examine the contribution of xepA, xhlA, xhlB, and xlyA to host cell lysis upon induction of PBSX. A summary of the lysis genes expressed in each strain and of the host cell lysis phenotypes is presented in Table 2. Three distinct growth profiles are evident after PBSX induction: (i) gradual cessation of growth with no cell lysis, (ii) aberrantly slow cell lysis, and (iii) normal saltatory cell lysis. The results show that expression of xhlA, xhlB, and xlyA is sufficient to effect saltatory host cell lysis upon induction of PBSX. The xepA gene does not appear to be involved in cell lysis. Strain POP234, in which the PBSX late operon contains only three genes (xhlA, xhlB, and xlyA), displays growth and lysis kinetics identical to those observed upon induction of the wild-type PBSX phage. In addition, expression of xepA does not appear to affect cell growth or lysis of strains SK101 and SK105 (even when it is amplified [strain SK101]). The assumption that xepA was involved in cell lysis was based on the observation that a homologous gene is found upstream of cwlA, an endolysin (amidase) from B. subtilis which shows homology to xlyA (8, 15). However, we have since established that cwlA is located within an operon of the skin element, which is highly homologous to the late operon of PBSX both in terms of gene organization and the identity percentages between homologous gene pairs (14, 23). This discovery makes it likely that xepA in fact encodes a phage protein which is not involved in host cell lysis.
TABLE 2.
Summary of expressed genes and lysis phenotypes for each strain
| Strain | Expression ofa:
|
Phenotype | |||
|---|---|---|---|---|---|
| xepA | xhlA | xhlB | xlyA | ||
| L8508 | + | + | + | + | Lysis |
| SK101 | (+)n | − | − | − | No lysis |
| SK102 | + | (+)n | − | − | Lysis (slow) |
| SK103 | + | + | (+)n | − | Lysis |
| SK104 | + | + | + | (+)n | Lysis |
| SK105 | + | − | − | − | No lysis |
| SK106 | − | + | − | − | Lysis |
| SK107 | − | − | + | − | No lysis |
| SK108 | − | − | − | + | Lysis (slow) |
| POP0 | − | − | − | − | No lysis |
| POP1234 | + | + | + | + | Lysis |
| POP234 | − | + | + | + | Lysis |
| POP34 | − | − | + | + | No lysis |
| POP4 | − | − | − | + | No lysis |
| SK109 | + | + | + | − | Lysis |
| SK110 | − | + | + | − | Lysis |
| SK111 | − | − | + | − | No lysis |
| SK112 | − | − | − | − | No lysis |
Symbols: +, expressed; (+)n, amplified; −, not expressed.
XhlA is the most intriguing of the four proteins. Even when expressed alone (in SK106), it is capable of causing host cell lysis, although in this case lysis occurs more slowly and to a lesser extent than in induction of wild-type PBSX. In addition, expression of xepA and xhlA (SK102) results in a pattern of lysis identical to that observed when xhlA alone is expressed (SK106). It should be noted, however, that amplification of xhlA is possible in both strains even though amplification was only selected for in strain SK102. Therefore, we cannot conclude with certainty that expression from a single copy of xhlA can effect this slow host cell lysis.
Expression of xhlB in single copy (strain SK111) cannot effect cell lysis. In fact, the growth kinetics of this strain after PBSX induction are virtually indistinguishable from the strains which have no functional lysis gene (POP0 and SK112). However, cell lysis occurs when xhlA and xhlB are expressed together in single copy (strain SK110), albeit delayed by 30 min. This supports the view that xhlA plays an essential role in cell lysis. XhlA is a small protein with a hydrophilic amino terminus and a putative transmembrane helix located in the carboxy terminus of the protein. This suggests that XhlA is membrane associated, perhaps forming a complex with XhlB to effect host cell lysis.
The endolysin encoded by xlyA, when expressed in single copy, results in very slow and aberrant lysis of the cell population (SK108). However, to achieve wild-type saltatory cell lysis, xlyA must be expressed in conjunction with xhlA and xhlB. An interesting observation from the lysis patterns observed (Table 2) is that cells of four strains (SK103, SK106, SK109, and SK110) lyse upon induction of PBSX even though the endolysin-encoding xlyA gene is not present. A probable explanation for these data is that induction of PBSX results in expression of a second endolysin activity which also requires the activities of XhlA and XhlB to access the peptidoglycan and effect cell lysis. None of the genes encoded in the 28 kb of PBSX sequenced in this laboratory (14), other than XlyA, are homologous to an endolysin. However, a second PBSX-encoded lytic activity, XlyB, has been reported recently; it may function to effect the observed cell lysis in those strains lacking xlyA (7). This finding confirms previous reports of the existence of a second PBSX-encoded autolytic activity (10, 16, 25).
The endolysin encoded by xlyA appears to lack (as judged by visual sequence inspection) a signal peptide sequence for transport across the membrane, a feature also observed for other endolysins found in phages of both gram-negative and gram-positive hosts. Therefore, it is not surprising that XlyA requires other proteins for transport from the cytoplasm. The results presented demonstrate that the activities of both XhlA and XhlB are required to transport the endolysin from the cell. In strains where only XhlB and XlyA are expressed (POP34), no cell lysis is observed up to 120 min after thermoinduction. Under these conditions, the parental strain L8508 lyses within 60 min of induction. We have demonstrated that xlyA is expressed in POP34 by showing that these cells contain a B. subtilis cell wall-degrading activity with the following properties: (i) it is approximately 32 kDa in size, consistent with the molecular mass of XlyA determined by sequence analysis; (ii) it migrates on SDS-polyacrylamide gels to the same extent as a thermoinducible autolysin activity seen in the parental strain L8508; (iii) the activity is observed only after thermoinduction; and (iv) the activity can be thermoinduced in strain POP34, which contains xlyA, but not in strain POP0, where xlyA is absent. While we have demonstrated expression of xlyA, we have not shown that xhlB is expressed in strain POP34. There is no readily available assay for holin activity. However, since xhlB is positioned on the promoter-proximal side of xlyA in this strain, we assume that it is also expressed in POP34. The assumption that xhlB is expressed in POP34 (which does not lyse) is supported by the finding that placing xhlA upstream of xhlB and xlyA (strain POP234) results in saltatory cell lysis with kinetics similar to those of the parental strain. This suggests that the expression kinetics and levels of the lysis genes in POP234 are very similar to those of the parental strain. XhlA is likely to be a membrane-associated protein (but is probably not a holin), whereas XhlB displays the characteristics of holins. Our results, therefore, suggest that host cell lysis mediated by this PBSX system differs from the systems encoded by the phages of gram-negative bacteria. In these cases, the holin protein alone appears to be sufficient to transport the endolysin out of the cell. Although holin genes have been identified in phages of gram-positive bacteria and have been shown to function in E. coli (P14 from φ29 [22], lysA from φLC3 [3], and hol from φadh [13]), their functioning in gram-positive bacteria has never been established. Gram-negative and gram-positive bacterial cell walls differ structurally and may require different functions to allow access of the endolysin to the peptidoglycan outside the cytoplasmic membrane. Since expression of both xhlA and xhlB is required to achieve host cell lysis in B. subtilis, it is possible that XhlA and XhlB act together to provide the function(s) which can be provided by holins alone in gram-negative bacteria.
We chose to manipulate the PBSX lysis genes on the B. subtilis chromosome in order to examine their function. Although more cumbersome and time consuming than cloning the genes on plasmids, this strategy has two very important advantages. (i) It allows the functions of these proteins to be examined in their natural host. This is an important feature, since the functions of holins isolated from phages of gram-positive bacteria have not been previously examined in their natural hosts (28, 29). (ii) Expression of the genes encoding the lysis functions is directed by the homologous PBSX control and expression system. Therefore, upon PBSX induction the level of each gene product in the cell is the same as that produced when the wild-type prophage is induced. Expression of the late operon is very tightly regulated because of the lethal consequences of PBSX induction. Therefore, the chromosome-based system developed in this study to investigate the functions of the PBSX lysis genes should be a valuable tool for the analysis of other host cell lysis systems and also for expression and functional analysis of other lethal gene products in gram-positive bacteria.
ACKNOWLEDGMENTS
We thank D. Karamata for the gift of strain L8508 and Simon Foster for the gift of bacterial cell walls.
This work was supported by EU grant BIOT-CT91-0268 and by Forbairt grant SC/95/124.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienkowska-Szewczyk K, Lipinska B, Taylor A. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol Gen Genet. 1981;184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- 3.Birkeland N-K. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage phi LC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. doi: 10.1139/m94-104. [DOI] [PubMed] [Google Scholar]
- 4.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonovich M T, Young R. Dual start motif in two lambdoid S genes unrelated to lambda S. J Bacteriol. 1991;173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens S, Eppler K, Parr R, Poteete A R. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology. 1989;171:588–598. doi: 10.1016/0042-6822(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 7.Dasilva E, Longchamp P S, Karamata D. 9th International Conference on Bacilli. Lausanne, Switzerland, 15 to 19 July 1997. 1997. Identification of XlyB, the second lytic enzyme of the prophage PBSX, abstr. B25. [Google Scholar]
- 8.Foster S J. Cloning, expression, sequence analysis and biochemical characterization of an autolytic amidase of Bacillus subtilis 168 trpC2. J Gen Microbiol. 1991;137:1987–1998. doi: 10.1099/00221287-137-8-1987. [DOI] [PubMed] [Google Scholar]
- 9.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster S J. Analysis of Bacillus subtilis 168 prophage-associated lytic enzymes: identification and characterization of CWLA-related prophage proteins. J Gen Microbiol. 1993;139:3177–3184. doi: 10.1099/00221287-139-12-3177. [DOI] [PubMed] [Google Scholar]
- 11.Garrett J R, Young R. Lethal action of bacteriophage λ S gene. J Virol. 1982;44:886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser P, Kunst F, Arnaud M, Coudart M-P, Gonzales W, Hullo M-F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweizer J, Vertes A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97 kilobase region from 325° to 333°. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 13.Henrich B, Binishofer B, Bläsi U. Primary structure and functional analysis of the lysis genes of Lactobacillus gasseri bacteriophage φadh. J Bacteriol. 1995;177:723–732. doi: 10.1128/jb.177.3.723-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogh S, O’Reilly M, Nolan N, Devine K M. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology. 1996;142:2031–2040. doi: 10.1099/13500872-142-8-2031. [DOI] [PubMed] [Google Scholar]
- 15.Kuroda A, Sekiguchi J. Cloning, sequencing and genetic mapping of a Bacillus subtilis cell wall hydrolase gene. J Gen Microbiol. 1990;136:2209–2216. doi: 10.1099/00221287-136-11-2209. [DOI] [PubMed] [Google Scholar]
- 16.Longchamp P F, Mauel C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 17.Reader R W, Siminovitch L. Lysis defective mutants of bacteriophage lambda: on the role of the S function in lysis. Virology. 1971;43:623–637. doi: 10.1016/0042-6822(71)90287-x. [DOI] [PubMed] [Google Scholar]
- 18.Rennell D, Poteete A R. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and lambda genes. Virology. 1985;143:280–289. doi: 10.1016/0042-6822(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt C, Velleman M, Arber W. Three functions of bacteriophage P1 involved in cell lysis. J Bacteriol. 1996;178:1099–1104. doi: 10.1128/jb.178.4.1099-1104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiner M, Lubitz W, Bläsi U. The missing link in phage lysis of gram-positive bacteria: gene 14 of Bacillus subtilis phage φ29 encodes the functional homolog of lambda S protein. J Bacteriol. 1993;175:1038–1042. doi: 10.1128/jb.175.4.1038-1042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takemura K, Mizuno M, Sato T, Takeuchi M, Kobayashi Y. Rearrangement during sporulation in Bacillus subtilis. Microbiology. 1995;141:323–327. doi: 10.1099/13500872-141-2-323. [DOI] [PubMed] [Google Scholar]
- 24.Tedin K, Resch A, Steiner M, Blasi U. Dual translational start motif evolutionarily conserved in the holin gene of Bacillus subtilis phage phi29. Virology. 1995;206:479–484. doi: 10.1016/s0042-6822(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 25.Ward B, Curtis C A M, Taylor C, Buxton R S. Purification and characterization of two phage-induced lytic enzymes of Bacillus subtilis 168: an N-acetylmuramoyl-l-alanine amidase and an N-acetylmuramidase. J Gen Microbiol. 1982;128:1171–1178. doi: 10.1099/00221287-128-6-1171. [DOI] [PubMed] [Google Scholar]
- 25a.Wood, H. Unpublished data.
- 26.Wood H E, Dawson M T, Devine K M, McConnell D J. Characterization of PBSX, a defective prophage of Bacillus subtilis. J Bacteriol. 1990;172:2667–2674. doi: 10.1128/jb.172.5.2667-2674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young R, Way J, Way S, Yin J, Syvanen M. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J Mol Biol. 1979;132:307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- 28.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 30.Ziermann R, Bartlett B, Calendar R, Christie G E. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J Bacteriol. 1994;176:4974–4984. doi: 10.1128/jb.176.16.4974-4984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]