Abstract
We used fluorescein-tagged β-lactam antibiotics to visualize penicillin-binding proteins (PBPs) in sporulating cultures of Streptomyces griseus. Six PBPs were identified in membranes prepared from growing and sporulating cultures. The binding activity of an 85-kDa PBP increased fourfold by 10 to 12 h of sporulation, at which time the sporulation septa were formed. Cefoxitin inhibited the interaction of the fluorescein-tagged antibiotics with the 85-kDa PBP and also prevented septum formation during sporulation but not during vegetative growth. The 85-kDa PBP, which was the predominant PBP in membranes of cells that were undergoing septation, preferentially bound fluorescein-6-aminopenicillanic acid (Flu-APA). Fluorescence microscopy showed that the sporulation septa were specifically labeled by Flu-APA; this interaction was blocked by prior exposure of the cells to cefoxitin at a concentration that interfered with septation. We hypothesize that the 85-kDa PBP is involved in septum formation during sporulation of S. griseus.
Sporulation of Streptomyces species is characterized by the formation of chains of uninucleoidal, unicellular spores by growth from multinucleoidal, branched, filamentous cells. Two distinctive features of streptomycete sporulation are its reproductive nature and the requirement for extensive growth during differentiation. Cytological events during sporulation include the growth of specialized filaments, termed aerial (5) or sporogenic (23) hyphae, which are the sites of spore formation; partitioning of copies of the chromosome in these hyphae; massive and relatively synchronous septation to form the spore compartments; and maturation of the spores by thickening of the walls and rounding up of the spore compartments.
The conversion of specialized, reproductive hyphae into chains of spores implicates peptidoglycan synthesis as an important pathway that is likely to respond to temporal and spatial regulatory signals. Penicillin-binding proteins (PBPs) catalyze the terminal steps in the synthesis of peptidoglycan and are targets of inhibition by β-lactam antibiotics. Two major classes of PBPs have been described (10): low-molecular-weight PBPs that have carboxypeptidase and transpeptidase activities in vitro and are typically responsible for cleaving the terminal d-Ala from pentapeptide chains of nascent peptidoglycan, and high-molecular-weight PBPs that form the cross-links between neighboring peptide side chains. Within the latter class are two subgroups. The bifunctional class A PBPs act as both transpeptidases and transglycosylases (10), whereas the class B PBPs contain a transpeptidase domain and at least one additional “module” of controversial function. Ishino and Matsuhashi (17) have suggested that the class B PBPs have transglycosylase activity mediated by the N-terminal domain, but a recent study (12) suggests that the N-terminal domain may act as an intramolecular chaperone to assist in folding the C-terminal transpeptidase module. Most, if not all, of the low-molecular-weight PBPs are not essential for cell growth under standard laboratory conditions (41). Some of the high-molecular-weight PBPs are at least somewhat redundant (32, 40, 42), whereas others appear to be essential (25, 41).
The essential PBPs include those required for septum formation during cell division. In only three bacteria is there convincing evidence of the identity of the septum-specific PBP. In Escherichia coli, FtsI (PBP3) is necessary for septation (37) and may also be required for assembly of the septation site (32). Certain β-lactam antibiotics preferentially inhibit the activity of FtsI (37). C-terminal truncation of PBP2B, the homolog of FtsI in Bacillus subtilis, leads to filamentation; this protein appears to be required for septum formation during vegetative growth (41). Two FtsI homologs have been identified in Pseudomonas aeruginosa. One of these, PBP3, is encoded by a gene that lies within the cell division cluster and is depleted in the stationary phase; the other, PBP3x, is not essential and accumulates during the stationary phase (25, 26).
Although phylogenetic (21) and structural (11, 18, 19, 36) studies have led researchers to propose that streptomycete PBPs may be the progenitors of the serine β-lactamases that inactivate β-lactam antibiotics, little work has been published on the physiological function of PBPs from these bacteria. Early studies of Streptomyces sp. strain R61 demonstrated that β-lactam antibiotics that inhibited transpeptidation in membrane preparations also prevented germination of spores or growth (7). Other researchers have surveyed the PBPs of several Streptomyces species and noted relationships between some PBPs and the growth phase (16, 28). Several PBPs were identified in vegetative cells and mature spores of Streptomyces griseus, but none were correlated with a specific function (1). The physiological role of one streptomycete PBP has been suggested by the work of Paradkar et al. (31), who showed that pcbR, within the cephamycin biosynthetic gene cluster of Streptomyces clavuligerus, could be disrupted in a non-cephamycin-producing mutant but not in the producing parent strain. Disruption of the pcbR gene led to twofold-less resistance to β-lactam antibiotics, and immunological analysis showed that synthesis of PcbR, a PBP of 57 kDa, was increased during the antibiotic production phase. The authors suggested that PcbR was a low-affinity PBP that augmented resistance to β-lactam antibiotics in the β-lactam-producing strain. The role of this PBP in peptidoglycan synthesis remains to be determined.
Because streptomycete sporulation is marked by dramatic changes in cell shape, several PBPs are likely to be required during the life cycle. For example, distinct PBPs (or altered activities of existing PBPs) might be necessary for growth by apical extension (13, 27), branch formation during vegetative growth and at the earliest stage of sporulation to generate sporogenic hyphae, septum formation during growth and differentiation, and spore maturation. Since sporulation and vegetative septa have distinct fates, it is conceivable that different PBPs are responsible for septation at these stages of the life cycle. To begin to understand the roles of PBPs in the Streptomyces life cycle, we have used fluorescence microscopy to visualize PBPs during sporulation and have identified a likely candidate for a PBP involved in septation of S. griseus.
MATERIALS AND METHODS
Media and growth of the organism.
To determine the effect of β-lactam antibiotics on growth and sporulation, cultures were inoculated into sporulation medium (SpM) (20) by dilution (2 × 10−4) from a primary culture grown in SpM for 2 to 5 days at 30°C. At this time the primary culture comprised mature spores. To induce sporulation by phosphate starvation, the primary culture was diluted (10−2 to 10−3) into glucose-ammonia minimal medium containing 1% casein hydrolysate (HyCase SF; Sigma Chemical Co., St. Louis, Mo.) (20) and grown to an absorbance of 3, at which time phosphate was removed from the culture (20). The β-lactam antibiotics were added at the time of transfer to the phosphate-free medium. The number of cells that had developed thick walls, which was used to approximate sporulation efficiency, was measured by determining the viable count of cultures after exposure to ultrasound for 10 min (23).
Microscopy, photography, and image processing.
Samples were prepared for fluorescence microscopy by combining 0.1 ml of culture with 100 μM fluorescein (Flu)-labeled β-lactam in a microcentrifuge tube and incubating the mixture for 20 min at 30°C. In some cases the cells were exposed to a β-lactam antibiotic for 20 min at 30°C prior to two washes with 1.5 ml of 0.85% NaCl (saline) and exposure to the Flu–β-lactam. Then the samples were washed twice with 1.5 ml of saline and suspended in 0.1 ml of saline for microscopy. An identical sample of cells was combined with 5 μl of 1 mg of propidium iodide per ml and held at room temperature for 10 min. Cells were centrifuged and washed twice with saline and then resuspended in 0.1 ml of saline. Samples were viewed with a Zeiss Axioskope microscope equipped for fluorescence and phase-contrast photomicroscopy. To view the Flu label, a Flu isothiocyanate filter set was used, whereas a rhodamine filter set was used for viewing of propidium iodide-treated samples. Images were captured as TIFF files that were subsequently cropped, magnified, and adjusted in brightness and intensity with Corel Photo Paint to resemble the viewed sample as closely as possible. For the dual fluorescent labels, the individual TIFF files were superimposed by using Corel Photo Paint. Each image was imported into Corel Draw for labeling and printed with a Tektronix Phaser 450 dye sublimation printer. Phase-contrast photomicrographs were made by using Kodak TMax 400 film; then the prints were scanned and processed as described above.
For electron microscopy, 1.5 ml of culture was combined with 1.5 ml of 8% glutaraldehyde. The mixture was centrifuged, and the cells were suspended in 1 ml of 4% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.2, and held at 4°C overnight. Cells were processed for thin sectioning as described by Kwak and Kendrick (23). Thin sections were viewed and photographed with a Philips CM12 transmission electron microscope. The prints were scanned and processed as described above.
Preparation of membranes.
All membrane preparation steps were carried out at 4°C. The procedure of Buchanan and Ling (4) was used, with slight modifications. Fifty milliliters of culture was harvested by centrifugation, washed once in 50 mM Tris-HCl (pH 7.5) containing 1 M KCl, suspended in 1 ml of 50 mM Tris-HCl (pH 7.5)–1 mM phenylmethylsulfonyl fluoride, and disrupted by two passages through a French press at 18,000 lb/in2. After centrifugation at 20,000 × g for 30 min, the supernatant was centrifuged at 100,000 × g for 1 h in a Beckman L8-M ultracentrifuge. The supernatant was discarded, the pelleted material was suspended in 3 ml of extraction buffer, and centrifugation was repeated. The material in the pellet was dissolved in extraction buffer to a protein concentration of 10 to 20 mg/ml (8) and either used immediately or stored at −70°C.
For routine treatment with Flu–β-lactam conjugates, 100 μg of protein was combined with at least 50 μM Flu–β-lactam in a final volume of 12 μl. The mixture was incubated for 20 min at 30°C. This was sufficient Flu-6-aminopenicillanic acid (Flu-APA) and reaction time to achieve a concentration between 2.6- and 5-fold greater than the Kd (as calculated from the experimental data shown in Fig. 7) for those PBPs that bound Flu-APA. The reaction was stopped by addition of an equal volume of double-strength sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (24), and the reaction mixture was boiled for 5 min and applied to a sodium dodecyl sulfate-polyacrylamide gel (24). After electrophoresis, the gel was viewed with a Molecular Dynamics Storm imaging device. The relative levels of PBPs were measured by using the quantitation software provided by the manufacturer. This imaging system detects emissions centered at 560 nm and therefore cannot detect very low levels of Flu-tagged PBPs. To confirm that we were able to detect all of the Flu-tagged PBPs by this method, we employed Western immunoblot analysis with anti-Flu antibodies conjugated to alkaline phosphatase and detected proteins bound to Flu-tagged 7-aminocephalosporanic acid (Flu-ACA) or Flu-APA by using a chemiluminescent substrate according to the manufacturer’s instructions (Vistra ECF; Amersham, Buckinghamshire, United Kingdom). This method, which amplified the Flu signal and permitted detection by chemiluminescence at 560 nm, showed no additional PBPs in our sample. Since the direct detection of Flu-tagged PBPs gave better resolution and was more rapid than the Western immunoblot approach, we used the former method for routine analysis.
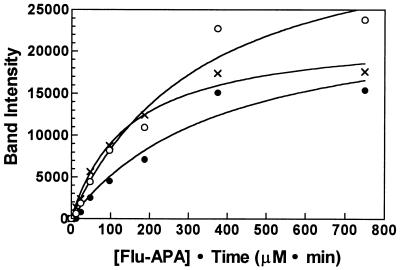
FIG. 7.
Preferential interaction of Flu-APA with the 85-kDa PBP. Increasing concentrations of Flu-APA were incubated for 15 min with a membrane preparation from a culture of S. griseus that had entered stationary phase (A500 = 12) in glucose-ammonia minimal medium supplemented with 1% casein hydrolysate. (Membranes prepared from this culture contained relatively high amounts of the 66- and 140-kDa PBPs, which permitted more accurate quantitation of their levels.) Band intensities were measured as described in Materials and Methods, the background was subtracted, and the results were fit to a hyperbolic curve (one binding site) by using Prism 2.01 (GraphPad, San Diego, Calif.). By this analysis, the relative Kd of the 85-kDa PBP (×) was 2.3- and 2.7-fold lower than that of the 66 (○)- and 140 (•)-kDa PBPs, respectively; the coefficients of determination (r2), which indicate the fit of the data to the equation, were 0.9694 (66-kDa PBP), 0.9892 (85-kDa PBP), and 0.9602 (140-kDa PBP).
To label the cells with Flu-APA prior to denaturing gel electrophoresis, 100 μM Flu-APA was added to an induced culture and incubation was continued for 15 min. The reaction was quenched by adding 1,000-fold excess APA, and the cells were processed as described above.
Synthesis of Flu-ACA and Flu-APA.
Flu conjugates of APA and ACA were prepared by the method of Galleni and coworkers (9), as modified by Popham and Setlow (34). When the mixed isomer of Flu was used, the conjugates were purified by thin-layer chromatography on silica gel G (Whatman) in 80% (vol/vol) acetonitrile in water and detected by fluorescence. The regions of the silica gel that contained the fluorescent conjugates, both of which migrated with an Rf of 0.72, were scraped from the plate into 1.5-ml microcentrifuge tubes, combined with 400 μl of water, and centrifuged at 15,000 rpm for 2 min. The supernatant was transferred to a new tube and centrifuged for 5 min. The supernatant was collected, dried in a vacuum centrifuge (Savant, Farmingdale, N.Y.) at room temperature, and dissolved in 50 μl of 50 mM Tris-HCl, pH 7.5. This solution was stored at −20°C in the dark and was stable for at least 1 month. The concentration of conjugate was estimated by measuring its absorbance at 494 nm, assuming that the extinction coefficient of the conjugate was identical to that of Flu (ɛ494 = 71,000 cm−1 M−1).
To confirm their identities, 20 ng of each purified sample was analyzed by negative ion electrospray mass spectrometry with a Perkin-Elmer Sciex API-300 mass spectrometer. The preparation of Flu-APA had an average mass of 688.2 ± 1.7 (expected value of 688.0), and that of Flu-ACA had a mass of 743.0 (expected value of 743.9). The reaction mixture was also tested for antibiotic activity by bioautography of the thin-layer chromatogram. Sufficient soft Luria-Bertani agar containing either E. coli (ATCC 25922) or Staphylococcus aureus (ATCC 29213) to cover the surface of the thin-layer plate was overlaid, and the plate was incubated overnight at 37°C. APA (Rf = 0.30), ACA (Rf <0.05), and Flu-APA did not inhibit either indicator organism, but S. aureus was sensitive to Flu-ACA. No other fluorescent spots inhibited growth of either organism.
RESULTS
Effects of β-lactam antibiotics on growth and sporulation.
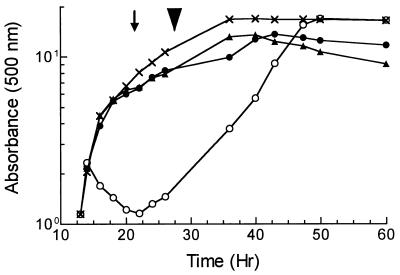
To identify PBPs that were required for morphogenesis of S. griseus, we first surveyed several β-lactam antibiotics for their effects on vegetative growth and sporulation. Cultures of the wild-type strain, S. griseus NRRL B-2682, that were growing exponentially in SpM (20) were exposed to 50 μg of β-lactam antibiotic per ml when they reached an A500 of 1.0 to 1.1. In this medium, the untreated wild-type strain grew with a doubling time of 1.5 to 1.75 h, entered the transition to stationary phase at an A500 of approximately 5 to 6 (Fig. 1), and formed chains of spores 10 h later. Cultures were monitored for growth, cell morphology, and the development of resistance to sonication that occurs when the spore compartments acquire thick walls. This normally happens shortly after the completion of septation (23).
FIG. 1.
Effect of β-lactam antibiotics on growth. S. griseus NRRL B-2682, growing exponentially in SpM (A500 = 1), was subdivided into four cultures and left untreated (×) or exposed to 50 μg of cefoxitin (▴), cefotaxime (•), or penicillin V (○) per ml. The arrow indicates the time at which sporogenic hyphae were evident, and the arrowhead shows when free spores were first visible.
Four penicillins (penicillin V, carbenicillin, benzylpenicillin, and ampicillin) affected the cells during vegetative growth, but the effect was most pronounced with penicillin V. After the addition of penicillin V, the culture underwent an additional doubling before the mycelia formed clumps. Dispersed growth resumed 8 h later (Fig. 1) and culminated in the production of spores to the same extent as the untreated culture.
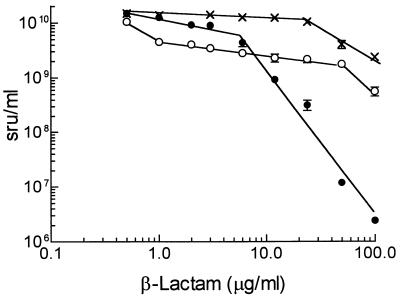
Among nine cephalosporins and their precursor, ACA, seven had no visible effect on vegetative growth or sporulation, whereas cefoxitin, cephalosporin C, and cefotaxime prevented complete sporulation without significantly affecting vegetative growth (Fig. 1). The magnitude of inhibition of spore formation was assessed by measuring the development of sonication-resistant units at concentrations of each drug up to 100 μg/ml. Cefoxitin had the most dramatic effect (Fig. 2), inhibiting complete maturation of the spores at 6 μg/ml, reducing spore formation at 24 μg/ml, and reducing the total viability by 4 orders of magnitude at 100 μg/ml. Cefotaxime inhibited spore formation by 60% at 1 μg/ml; the extent of inhibition increased to 80% at 50 μg/ml. ACA showed little effect until the concentration exceeded 24 μg/ml (Fig. 2). The complex pattern of inhibition by cefoxitin (and perhaps also cefotaxime) suggests that this drug had more than one target in sporulating cells.
FIG. 2.
Dose-response curve for inhibition of sporulation by β-lactam antibiotics. An exponentially growing culture of S. griseus (A500 = 1) in SpM was divided into 3-ml aliquots that were placed in 18- by 150-mm culture tubes containing the indicated concentration of cefoxitin (•), cefotaxime (○), or ACA (×). After incubation for an additional 4 days, the cultures were treated with ultrasound, diluted, and plated for viability. The values (in sonication-resistant units [sru] per ml) are averages ± standard deviations of duplicate experiments, each assayed in duplicate. The concentration of sonication-resistant units for each antibiotic at 0.6 μg/ml was identical to that of an untreated culture.
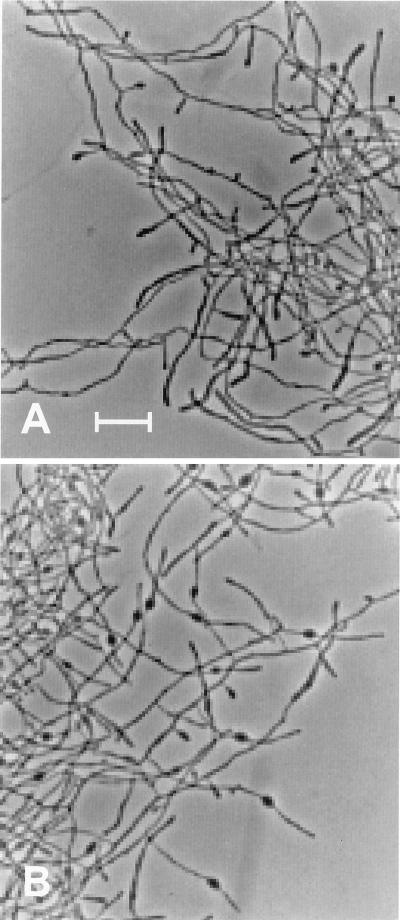
When spores were plated on SpM agar that contained 50 μg of cefoxitin per ml, colonies formed with an efficiency equal to that of spores plated in the absence of the drug and produced aerial hyphae but did not sporulate. Likewise, spores inoculated into liquid SpM initiated growth with equal efficiency in the presence or absence of 24 μg of cefoxitin per ml. At this concentration, cefoxitin altered the morphology of cells only after their entry into the transition phase: the hyphae in cultures treated with cefoxitin from 6 to 50 μg/ml showed bulges, adjacent to the growing tips, that were more prevalent at the higher concentrations of cefoxitin. Only rarely was there more than one bulge per filament. Bulges first appeared in the treated cultures at the same time (20 h) that the control culture was beginning to form sporogenic hyphae. As incubation continued, the distance between the bulge and the tip increased (Fig. 3).
FIG. 3.
Formation of bulges by S. griseus exposed to 50 μg of cefoxitin per ml. A culture of S. griseus that had been grown as described in the legend to Fig. 1 was left untreated (A) or treated with 50 μg of cefoxitin per ml (B), and incubation was continued for 10 h. The swollen tips of branches in panel A are sporogenic hyphae; the bulges are evident in panel B. Bar, 10 μm.
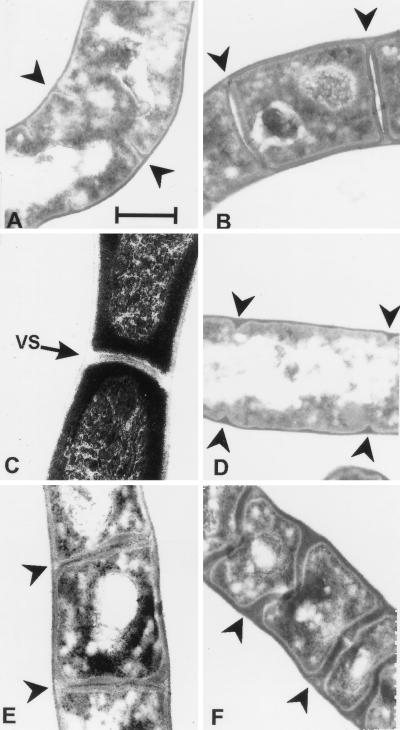
Sporulation was also blocked by cefoxitin and cefotaxime when the drugs were added to cultures of S. griseus that had been induced to sporulate by phosphate starvation. Electron microscopic examination of sporulating cultures that had been exposed to cefoxitin during 12 h of induction confirmed that this antibiotic inhibited septation. At 50 μg/ml, cefoxitin completely prevented progression of septation past the initial stage (Fig. 4D). An electron-dense layer that was continuous with the peptidoglycan in the hyphal wall was evident within the regularly spaced, small septal invaginations. Cefoxitin at 6 μg/ml interfered with but did not prevent septation. The septa in this case were rather diffuse and irregularly shaped, but the spore walls still underwent thickening (Fig. 4E), and the rectangular cells were resistant to ultrasound. The effect of cefoxitin was specific to sporulation since vegetative hyphae contained apparently normal septa (Fig. 4C). Cefotaxime at 12 μg/ml permitted complete septation (Fig. 4F), but the septa were irregularly shaped and the thick-walled spore compartments did not separate into individual cells even after prolonged incubation (4 days) or treatment with ultrasound.
FIG. 4.
Effects of cefoxitin and cefotaxime on septation. Cultures of S. griseus were grown vegetatively (C) or induced to sporulate by phosphate starvation (A, B, and D to F) for 10 or 12 h in the presence or absence of the indicated β-lactam antibiotic. (A) Untreated culture at 10 h of starvation. (B) Untreated culture at 12 h of starvation. (C) Culture treated with 50 μg of cefoxitin per ml in vegetative growth. (D) Culture treated with 50 μg of cefoxitin per ml after phosphate starvation for 12 h. (E) Culture treated with 6 μg of cefoxitin per ml for 12 h. (F) Culture treated with 20 μg of cefotaxime per ml for 12 h. Arrowheads indicate sporulation septa. VS, vegetative septum. Bar, 0.5 μm.
Visualization of binding in situ.
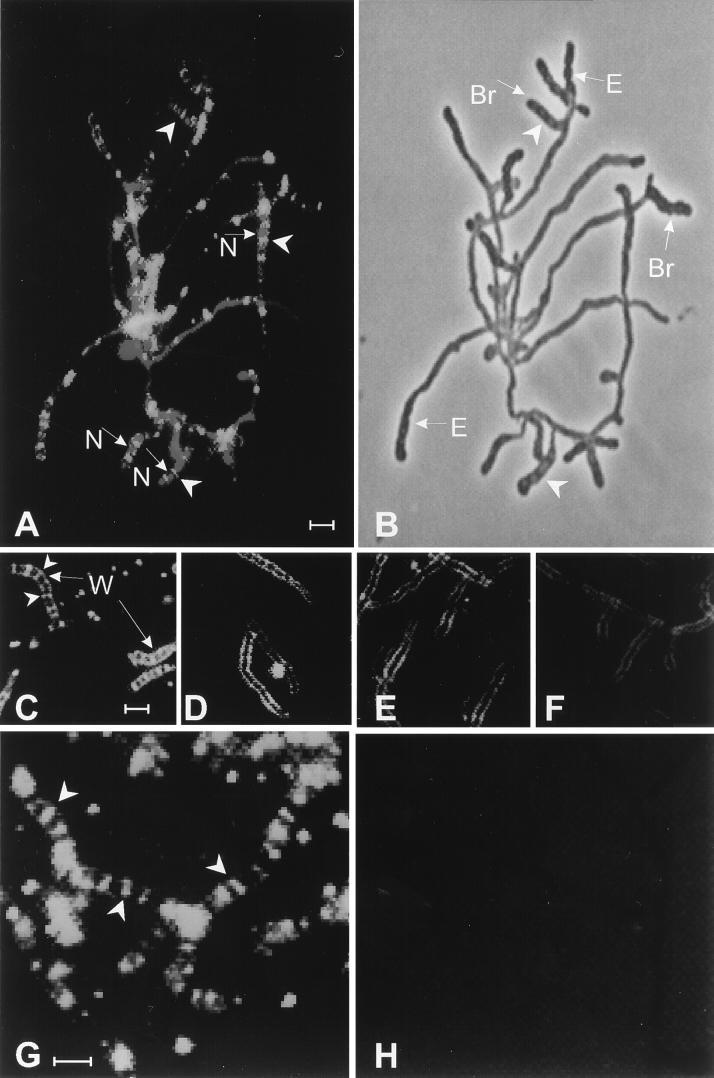
Because massive, synchronous septation occurs in the sporogenic hyphae at 10 h of sporulation of S. griseus when induced by phosphate starvation (Fig. 4) (23), we reasoned that the PBP(s) required for septation may be present at a relatively high concentration in the sporogenic hyphae at this time. We examined sporulating cultures by fluorescence microscopy to determine whether the Flu-conjugated β-lactam antibiotics Flu-APA and Flu-ACA could be visualized upon interaction with their target PBPs. During vegetative growth there was substantial autofluorescence, but no localization of either Flu-APA or Flu-ACA was evident. Early in sporulation, neither Flu–β-lactam bound noticeably to the cells. At 10 h of sporulation, Flu-APA bound to sites distributed at regular intervals through the sporogenic hyphae (data not shown). Most of these zones of fluorescence did not completely transect individual sporogenic hyphae. Since electron microscopic observations had shown that the majority of the septa were still forming at this time (Fig. 4), these zones may correspond to nascent septa. Flu-tagged “ladders” were evident at 12 h (Fig. 5A and G), shortly after the completion of septation (23). The “rungs” of these ladders lay between the nucleoids, consistent with their localization to the sporulation septa (Fig. 5A). Inspection of the same sample by phase-contrast microscopy showed the sporulation septa as thin, dark bands at intervals within the swollen sporogenic hyphae (Fig. 5B); these bands coincided with the zones stained by Flu-APA. This sample also showed that spore-bearing hyphae formed not only by growth from new branches (23) but also by extension from existing hyphal tips (Fig. 5B). By examining this and other samples, we estimated that hyphal regions undergoing septation comprised no more than 25% of the biomass at 12 h of induction. Although Flu-APA localized only at septation sites, Flu-ACA bound to both the sites of septation and the walls of the sporogenic hyphae (Fig. 5C). When cells were treated with 6 μg of cefoxitin per ml during induction, subsequent binding of Flu-APA and Flu-ACA to the septa was blocked (cf. Fig. 5C and D and Fig. 5G and H). A higher concentration of cefoxitin was required to prevent binding of Flu-ACA to the hyphal walls (cf. Fig. 5C to F).
FIG. 5.
Visualization of PBPs by fluorescence microscopy. Cells of S. griseus that had been induced to sporulate for 12 h in the presence or absence of cefoxitin were exposed to Flu-conjugated β-lactams. (A) Flu-APA and propidium iodide. (B) Phase-contrast micrograph of the sample shown in panel A. (C) Flu-ACA. (D) Cefoxitin (6 μg/ml), then Flu-ACA. (E) Cefoxitin (20 μg/ml), then Flu-ACA. (F) Cefoxitin (60 μg/ml), then Flu-ACA. (G) Flu-APA. (H) Cefoxitin (6 μg/ml), then Flu-APA. Arrowheads in panels A, C, and G point to examples of Flu-tagged “ladders” in sporogenic hyphae. Arrowheads in panel B point to sporulation septa. The arrows marked “N” in panel A point to nucleoids stained with propidium iodide; in panel B, those marked “Br” point to sporogenic hyphae that emerged from new branches and those marked “E” point to sporogenic hyphae that formed by extension of preexisting vegetative hyphae. The arrows marked “W” in panel C point to Flu–ACA-labeled hyphal walls. Bar, 2 μm. The color version of this figure can be viewed at http://www.biosci.ohio-state.edu/~kek/html/j_bacteriol_1998.html.
Identification of PBPs in membrane preparations.
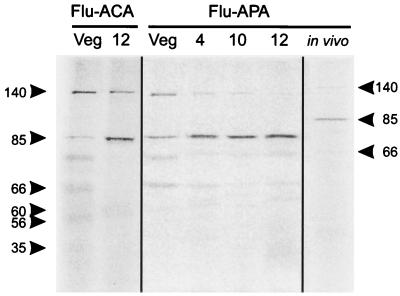
The formation of abundant septa at 10 h of sporulation of S. griseus would be expected to require an increase in the activity of septum-specific PBPs. Moreover, the specificity of Flu-APA for the sporulation septa suggested that biochemical studies with this reagent might permit us to identify a septum-specific PBP. Incubation of Flu-APA with membrane preparations from vegetative and sporulating cultures of S. griseus revealed three PBPs, all of relatively high molecular weights. The largest of these (140 kDa) decreased at least threefold in intensity, ultimately to a barely detectable level, during the first 12 h of sporulation (Fig. 6). An 85-kDa PBP that was present during vegetative growth increased 1.5-fold during the first 4 h of sporulation, at which time the sporogenic hyphae had begun to form (23). By 10 h, the 85-kDa PBP signal had increased threefold, coincident with the onset of septum formation. At 12 h, when complete septa were evident, the amount of signal was approximately fourfold greater than that during vegetative growth (Fig. 6). The third PBP (66 kDa) was present at a low level during growth and sporulation. In addition to these three high-molecular-weight PBPs, three lower-molecular-weight PBPs were faintly evident when membranes were treated with Flu-ACA. One (60 kDa) was detectable only in membranes from sporulating cells (Fig. 6). We classified these proteins as PBPs because the bands were absent from membranes that were not exposed to Flu-ACA (in contrast to 75- and 30-kDa autofluorescent proteins), they disappeared entirely when the membranes were incubated with 50 μg of cefoxitin per ml followed by Flu-ACA, and they were also detectable by immunoblot with anti-Flu antibodies.
FIG. 6.
Identification of PBPs after denaturing polyacrylamide gel electrophoresis. PBPs were labeled with Flu-ACA or Flu-APA in membranes prepared from vegetative hyphae (Veg) or cells induced to sporulate for 4, 10, or 12 h prior to separation of proteins by denaturing gel electrophoresis. Cells induced to sporulate for 10 h were treated with 100 μM Flu-APA, and then membranes were prepared and proteins were separated as described above. Arrowheads point to PBPs of the indicated molecular masses (in kilodaltons). In the absence of treatment with either Flu–β-lactam, membrane preparations showed two inherently fluorescent proteins of 75 and 30 kDa.
The affinity of Flu-APA for the 85-kDa PBP was also evident after labeling the cells at 10 h of sporulation and examination of the PBPs in membrane samples (Fig. 6). The pattern of labeling was similar to that seen in vitro, with the 85-kDa PBP predominating under these conditions. Titration of the PBPs by the addition in vitro of increasing concentrations of Flu-APA confirmed that the 85-kDa PBP preferentially bound Flu-APA, followed by the 66- and 140-kDa PBPs (Fig. 7). Binding of the 85-kDa PBP to Flu-APA in vivo was inhibited approximately 90% by 6 μg of cefoxitin per ml (data not shown).
DISCUSSION
The technique of detection of PBPs by coupling β-lactam antibiotics to Flu was developed by Galleni and colleagues (9). We have applied this technique to the visualization of streptomycete PBPs in situ. The success of this method may reflect the relative abundance of certain PBPs during sporulation, when the demand for septation proteins and enzymes required for spore wall thickening is expected to be high. Since the stoichiometry between a PBP and its bound Flu–β-lactam is 1, no signal amplification occurs in this system. The visualization of the signal most likely depends on an abundant or highly localized target protein. This method is therefore probably less sensitive than immunofluorescence microscopy, which detected FtsI at an estimated 100 molecules per cell when the protein localized to the midcell (39).
It is noteworthy that Flu-ACA revealed several more PBPs than did Flu-APA. Although we do not know whether Flu-ACA reveals the full complement of PBPs expressed in S. griseus under the conditions of this study, our results demonstrate the importance of using more than one labeled β-lactam compound to detect PBPs.
Our previous studies (23) showed that sporogenic hyphae developed by growth from newly formed branches after induction of sporulation. Here we have extended this observation, noting that the sporogenic hyphae also grew from existing tips of vegetative hyphae. It is not known whether the same occurs during the formation of aerial hyphae during sporulation on a solid surface. The sporogenic hyphae that formed by extension of vegetative hyphae were longer and, upon subdivision, generated more spores than those that formed at new branch points. At 12 h of induction, two subpopulations of cells existed in this culture: old vegetative hyphae and newly formed sporogenic hyphae, approximately 50% of which were undergoing septation.
The inhibitory effect of cefoxitin on sporulation of S. griseus was complex. Electron micrographs showed that septum formation progressed through the earliest stage but no further in the presence of 50 μg of cefoxitin per ml. Phase-contrast microscopy demonstrated that this effect was caused by cefoxitin in excess of 20 μg/ml; lower concentrations of cefoxitin resulted in complete but deformed septa, perhaps because the target PBP was incompletely inhibited. The presence of a layer of peptidoglycan within the small septal ingrowth evident in the culture treated with 50 μg of cefoxitin per ml indicated that the inhibited PBP was not necessary to effect the change in direction of peptidoglycan growth. The peptidoglycan within these invaginations is therefore presumably synthesized either by a mechanism that does not require the transpeptidase activity of the septum-specific PBP or by an additional PBP that is relatively insensitive to cefoxitin. This result is consistent with radiolabeling studies with E. coli that suggested that FtsI is responsible for the centripetal extension of peptidoglycan during septation but not the initiation of septation (29, 40).
Comparison of the in vivo and in vitro effects of cefoxitin showed that a concentration of 6 μg/ml was sufficient to abrogate fluorescence at the septum but not the hyphal walls. This concentration resulted in greater than 90% inhibition of the 85-kDa PBP and visibly altered the ultrastructure of the sporulation septa. Even at 6 μg of cefoxitin per ml, however, the spores still had thick walls sufficient to withstand sonication. The absence of a strict correlation between the concentration of cefoxitin required to block binding of Flu-APA and that required for a morphological effect is probably a consequence of the rather insensitive methods (electron microscopy and development of resistance to sonication) that are available to assess structural changes during sporulation.
We cannot yet explain the bulges that formed after exposure to cefoxitin during growth. These bulges resembled those described by several researchers for E. coli cells in which the activity of FtsL (14) or FtsI (the latter in conjunction with PBP2 [2], RodA [3], or soluble lytic transglycosylase [38]) was inhibited. In all of these cases the formation of bulges correlated with the inhibition of septation. Although comparisons with the results for E. coli would suggest that the bulges in S. griseus marked sites of aborted septation, phase-contrast microscopic examination suggested that this was not the case; bands resembling septa were evident, sometimes traversing the bulge but more commonly immediately proximal or distal to the bulge. Moreover, the bulges formed at the growing tips, which are unlikely to be undergoing vegetative septation (22). Bulges always formed during the transition between vegetative growth and sporulation in cultures that were growing in sporulation medium, even though cefoxitin had been present since inoculation. This observation suggests that there is something unique about the hyphal tips at this stage of the life cycle. Since the control culture was beginning to form sporogenic hyphae at the time that the bulges first appeared, it is conceivable that these bulges mark the junctions between the preexisting vegetative hyphae and the nascent sporogenic hyphae.
We might reasonably expect that the presence of a β-lactam antibiotic that targets a septum-specific PBP would slow vegetative growth, yet this was not observed. Cultures grew with essentially identical rates (μ = 0.4 ± 0.07 h−1) at concentrations of cefoxitin as high as 24 μg/ml. Microscopic observations confirmed that septa were present in vegetative hyphae of a culture growing in the presence of 50 μg of cefoxitin per ml. These results suggest that the septum-specific PBP that is the target of cefoxitin during sporulation either is not accessible to cefoxitin or is not required for septation during vegetative growth. This observation supports the possibility that distinct PBPs are responsible for septation during vegetative growth and sporulation.
The simplest hypothesis to explain our results is that the 85-kDa PBP is involved in septation during sporulation of S. griseus. This PBP showed the greatest affinity for Flu-APA in vitro and was prominent in membranes from sporulating cells that had been exposed to Flu-APA in vivo. Flu-APA bound specifically to the sites of sporulation septation, and this binding was inhibited by cefoxitin at a concentration that interfered with septation. Like the septum-specific PBPs in other bacteria (10, 25, 41), the 85-kDa PBP has a high molecular weight. Nevertheless, there may be additional PBPs involved in septation. For example, the electron micrographs suggest that a cefoxitin-insensitive peptidoglycan-synthesizing enzyme deposits the material within the invaginations that mark the earliest stage of septum formation.
At this time we cannot determine whether the 85-kDa PBP band represents one or multiple PBP species. The intensity of fluorescence of this protein band increased three- to fourfold during the first 12 h of sporulation; this may be a consequence of either the enhanced production of a single PBP during sporulation or the production of one 85-kDa PBP during vegetative growth and the subsequent production of a different, sporulation-specific 85-kDa PBP. The good fit of our data to the single-site hyperbolic equation for binding of Flu-APA to the 85-kDa PBP is consistent with a single protein species, yet we cannot readily explain the presence of this PBP during vegetative growth if it is required only during sporulation. The absence of localized sites of fluorescence in hyphae during vegetative growth and early in morphogenesis may be attributable to interference from autofluorescence, the relative rarity and asynchrony of vegetative septation, the absence of the target PBP in sufficient quantity, or the presence of the target PBP(s) in diffuse, unlocalized regions. In light of the results of Pogliano et al. (32), who suggested that FtsI is required not only for synthesis of peptidoglycan at the septum but also for assembly of the septation sites, one possibility is that the 85-kDa PBP is required for synthesis of septa during sporulation as well as placement or assembly of the septation sites during vegetative growth. Cefoxitin may inhibit the former function but not the latter.
Why should a relatively small (three- to fourfold) increase in the binding of the 85-kDa PBP be sufficient to permit visualization in situ? We suggest that the increased activity of this PBP may be confined to the sporogenic hyphae. Since the regions of the sporogenic hyphae that are undergoing septation at 12 h of induction comprise roughly 20 to 25% of the biomass of this culture, the amount of 85-kDa PBP in this location would actually have increased 12- to 20-fold. Once the structural gene for the 85-kDa PBP is isolated, we will be able to determine whether its expression or activity is spatially regulated, as our hypothesis would predict.
We consider it unlikely that a different PBP, undetectable by our methods, is solely responsible for septum formation, because the visible interaction between the sporulation septa and Flu-APA suggests that the reactive PBP(s) should be relatively abundant and readily bind Flu-APA in vivo and in vitro. At the time of septation, the 85-kDa PBP was the predominant PBP labeled by Flu-APA in vivo. We cannot yet rule out the possibility that the 66-kDa PBP is also involved in septation, but its very low level at the time when sporogenic hyphae were undergoing septation and its somewhat lower affinity for Flu-APA argue against this possibility. Despite the physiological evidence suggesting that the 85-kDa PBP is involved in septation during sporulation, the definitive test of our hypothesis requires disruption of its structural gene. Current work is directed toward this goal.
In keeping with the various morphologies assumed by streptomycetes during their complex life cycle, the synthesis of peptidoglycan in these organisms appears to be complex, differing in fundamental ways in vegetative growth, upon entry into the transition phase, and during sporulation. The nature of submerged sporulation by S. griseus dictates that any β-lactam antibiotic that inhibited only septation during spore formation would exert a relatively small effect on the development of sonication-resistant structures. This is because in our system the sporogenic hyphae typically subdivide into chains of 5 to 20 spores. Related work in our laboratory indicates that the septum near the base of each sporogenic hypha and the thick spore walls form even when sporulation septation is prevented (6, 15). From these preliminary results and the observations presented in this paper, we would expect that the absence of sporulation septation would result in a decrease in sonication-resistant units of approximately 1 order of magnitude. This effect is clearly illustrated by our results with cefotaxime. The spore compartments formed but did not separate in cultures exposed to cefotaxime, even after treatment with ultrasound. Because the septa in this case were deformed, it is likely that a change in the structure of peptidoglycan in these septa led to a form that was refractory to subsequent hydrolysis. This proposed action is consistent with the results of the dose response experiment, which showed that cefotaxime at 50 μg/ml reduced the proportion of sonication-resistant units to approximately 20% of that of the untreated culture. Microscopic observations showed that the sonication-resistant units in this culture corresponded to spore chains, comprising 5 to 10 spore compartments, that failed to separate into individual spores.
To our knowledge, the 140-kDa PBP is considerably larger than others that have been identified in unicellular gram-positive and gram-negative bacteria. The largest reported to date appears to be PBP1a of Caulobacter crescentus (30), which has a molecular mass of 130 kDa on the basis of its migration on a denaturing polyacrylamide gel. The next largest is PBP1 of B. subtilis, which is approximately 100 kDa on the basis of its amino acid sequence but migrates on a denaturing polyacrylamide gel as if it were about 110 kDa (33). Like S. griseus, other streptomycetes also contain PBPs having apparent molecular masses in excess of 120 kDa (16, 28). The dramatic decrease in the 140-kDa signal during the early stages of sporulation implies not only that this PBP is not involved in spore septation or maturation in S. griseus but also that the structure of the peptidoglycan differs in young sporogenic and vegetative hyphae. Sporogenic hyphae, which begin to form from rapidly growing tips during the first 2 to 4 h of sporulation and complete elongation by approximately 10 h, do not branch as they elongate prior to septation (23). In contrast, vegetative hyphae form branches, generating new growing tips and thereby maintaining exponential growth (13, 27, 35). It is therefore tempting to speculate that the 140-kDa PBP is required for branch formation. This function, necessary for growth of actinomycetes and perhaps also prosthecate bacteria, might require a PBP that would have no homolog in most unicellular bacteria.
ACKNOWLEDGMENTS
This research was supported by grants from the National Science Foundation (MCB-9210743 and MCB-9632468).
We thank F. R. Tabita for use of the chemiluminescence imager, Scott Taylor and Kathy Wolken for assistance with fluorescence and electron microscopy, and Tim Vojt for assistance with image processing. David Popham, John Reeve, and Julie Schwedock provided helpful comments.
REFERENCES
- 1.Barabás J, Barabás G, Szabó I, Veemhuis M, Harder W. Penicillin-binding proteins of protoplast and sporoplast membranes of Streptomyces griseus strains. Arch Microbiol. 1988;150:105–108. [Google Scholar]
- 2.Begg K J, Donachie W D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg K J, Spratt B G, Donachie W D. Interaction between membrane proteins PBP3 and RodA is required for normal cell shape and division in Escherichia coli. J Bacteriol. 1986;167:1004–1008. doi: 10.1128/jb.167.3.1004-1008.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan C E, Ling M-L. Isolation and sequence analysis of dacB, which encodes a sporulation-specific penicillin-binding protein in Bacillus subtilis. J Bacteriol. 1992;174:1717–1725. doi: 10.1128/jb.174.6.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 6.Dharmatilake, A. J. Unpublished results.
- 7.Dusart J, Leyh-Bouille M, Ghuysen J M. The peptidoglycan crosslinking enzyme system in Streptomyces strains R61, K15 and rimosus. Kinetic coefficients involved in the interactions of the membrane-bound transpeptidase with peptide substrates and β-lactam antibiotics. Eur J Biochem. 1977;81:33–44. doi: 10.1111/j.1432-1033.1977.tb11924.x. [DOI] [PubMed] [Google Scholar]
- 8.Ehresmann B, Imbault P, Weil J H. Spectrophotometric determination of protein concentration in cell extracts containing tRNA’s and rRNA’s. Anal Biochem. 1973;54:454–463. doi: 10.1016/0003-2697(73)90374-6. [DOI] [PubMed] [Google Scholar]
- 9.Galleni M, Lakaye B, Lepage S, Jamin M, Thamm I, Joris B, Frère J-M. A new, highly sensitive method for the detection and quantification of penicillin-binding proteins. Biochem J. 1993;291:19–21. doi: 10.1042/bj2910019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghuysen J-M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 11.Ghuysen J-M. Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 12.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J-M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module, and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray D I, Gooday G W, Prosser J I. Apical hyphal extension in Streptomyces coelicolor A3(2) J Gen Microbiol. 1990;136:1077–1084. doi: 10.1099/00221287-136-6-1077. [DOI] [PubMed] [Google Scholar]
- 14.Guzman L-M, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 15.Hao, J. Unpublished results.
- 16.Horikawa S, Nakazawa H, Ogawara H. Penicillin-binding proteins in Streptomyces cacaoi and Streptomyces clavuligerus. J Antibiot. 1980;33:1363–1368. doi: 10.7164/antibiotics.33.1363. [DOI] [PubMed] [Google Scholar]
- 17.Ishino F, Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981;101:905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- 18.Joris B, Ghuysen J M, Dive G, Renard A, Dideberg O, Charlier P, Frere J M, Kelly J A, Boyington J C, Moews P C, Knox J R. The active-site serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988;250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly J A, Dideberg O, Charlier P, Wery J P, Libert M, Moews P C, Knox J R, Duez C, Fraipont C, Joris B, Dusart J, Frere J M, Ghuysen J M. On the origin of bacterial resistance to penicillin: comparison of a β-lactamase and a penicillin target. Science. 1986;231:1429–1431. doi: 10.1126/science.3082007. [DOI] [PubMed] [Google Scholar]
- 20.Kendrick K E, Ensign J C. Sporulation of Streptomyces griseus in submerged culture. J Bacteriol. 1983;155:357–366. doi: 10.1128/jb.155.1.357-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby R. Evolutionary origin of the Class A and Class C β-lactamases. J Mol Evol. 1992;34:345–350. doi: 10.1007/BF00160242. [DOI] [PubMed] [Google Scholar]
- 22.Kretschmer S. Septation behaviour of the apical cell in Streptomyces granaticolor. J Basic Microbiol. 1989;29:587–595. [Google Scholar]
- 23.Kwak J, Kendrick K E. Bald mutants of Streptomyces griseus that prematurely undergo key events of sporulation. J Bacteriol. 1996;178:4643–4650. doi: 10.1128/jb.178.15.4643-4650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Liao X, Hancock R E W. Cloning and characterization of the Pseudomonas aeruginosa pbpB gene encoding penillin-binding protein 3. Antimicrob Agents Chemother. 1995;39:1871–1874. doi: 10.1128/aac.39.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao X, Hancock R E W. Identification of a penicillin-binding protein 3 homolog, PBP3x, in Pseudomonas aeruginosa: gene cloning and growth phase-dependent expression. J Bacteriol. 1997;179:1490–1496. doi: 10.1128/jb.179.5.1490-1496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miguélez E M, Martín C, Manzanal M B, Hardisson C. Growth and morphogenesis in Streptomyces. FEMS Microbiol Lett. 1992;100:351–360. doi: 10.1111/j.1574-6968.1992.tb14063.x. [DOI] [PubMed] [Google Scholar]
- 28.Nakazawa H, Horikawa S, Ogawara H. Penicillin-binding proteins in Streptomyces strains. J Antibiot. 1981;34:1070–1072. doi: 10.7164/antibiotics.34.1070. [DOI] [PubMed] [Google Scholar]
- 29.Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 30.Nathan P, Newton A. Identification of two new cell division genes that affect a high-molecular-weight penicillin-binding protein in Caulobacter crescentus. J Bacteriol. 1988;170:2319–2327. doi: 10.1128/jb.170.5.2319-2327.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paradkar A S, Aidoo K A, Wong A, Jensen S E. Molecular analysis of a β-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J Bacteriol. 1996;178:6266–6274. doi: 10.1128/jb.178.21.6266-6274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogliano J, Pogliano K, Weiss D S, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham D L, Setlow P. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J Bacteriol. 1995;177:326–335. doi: 10.1128/jb.177.2.326-335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popham D L, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichl U, Buschulte T K, Gilles E D. Study of the early growth and branching of Streptomyces tendae by means of an image processing system. J Microsc. 1990;158:55–62. doi: 10.1111/j.1365-2818.1990.tb02976.x. [DOI] [PubMed] [Google Scholar]
- 36.Samraoui B, Sutton B J, Todd R J, Artymiukm P J, Waley S G, Phillips D C. Tertiary structural similarity between a class A β-lactamase and a penicillin-sensitive d-alanyl carboxypeptidase-transpeptidase. Nature. 1986;320:378–380. doi: 10.1038/320378a0. [DOI] [PubMed] [Google Scholar]
- 37.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Templin M F, Edwards D H, Höltje J-V. Inhibition of an autolysin together with PBP3 causes the formation of bulges: identification of the soluble lytic transglycosylase in E. coli as the specific target of bulgecin. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 241–244. [Google Scholar]
- 39.Weiss D S, Pogliano K, Carson M, Guzman L-M, Fraipont C, Nguyen-Distèche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 40.Wientjes F, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanouri A, Daniel R A, Errington J, Buchanan C E. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J Bacteriol. 1993;175:7604–7616. doi: 10.1128/jb.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yousif S Y, Broome-Smith J K, Spratt B G. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]