Abstract
The pilE gene of Neisseria gonorrhoeae is transcribed from a ς70 promoter (pilEp1) with an AT-rich tract extending 65 nucleotides upstream of the −35 box. Within this region is an integration host factor (IHF)-binding core consensus sequence. We have performed a detailed analysis to determine which upstream sequences are required for efficient transcription from pilEp1 in N. gonorrhoeae. Deletion of sequences upstream of the AT-rich tract had no effect on the level of transcription. However, the IHF-binding core consensus sequence and the AT-rich sequence further upstream were both required for enhanced levels of transcription from this promoter in both N. gonorrhoeae and an Escherichia coli strain producing IHF. In addition, an UP-like element positioned between the −35 box and the IHF-binding site was required for maximal transcription. The AT-rich region upstream of the IHF-binding core consensus sequence can also act as an UP-like element when appropriately repositioned upstream of the −35 box.
The production of type 4 pili has been described in over 15 species of gram-negative bacteria, most of which are potential human, animal, or plant pathogens. The presence of type 4 pili has been associated with colonization of eukaryotic cells and twitching motility (41), social gliding motility in Myxococcus xanthus (46), and conjugation associated with plasmid R64 (23).
Despite numerous reports documenting the identification of type 4 pilin gene homologs, only a few studies on the regulation of pilin synthesis have appeared in the literature. In Pseudomonas aeruginosa, the promoter responsible for transcription of the pilin subunit gene (pilA) is ς54 dependent (21), and expression is subject to regulation by a two-component system encoded by pilR and pilS (19). Homologs of pilR and pilS have been identified in M. xanthus in a contiguous cluster with the pilin subunit gene, pilA (46). However, in neither of these systems has the environmental signal detected by the sensor protein, PilS, been determined.
The production of bundle-forming pili (Bfp) by enteropathogenic strains of Escherichia coli is subject to transcriptional regulation involving environmental signals which are potentially relevant to the small intestine (31). However, in contrast to the situation in P. aeruginosa and M. xanthus, the bfpA gene (encoding the pilin subunit) is transcribed from a ς70-dependent promoter with an upstream AT-rich tract. In addition, there is a requirement for an activator protein, BfpT, which is a member of the AraC family of transcriptional activators (42). Transcription of the type 4 pilin subunit gene (tcpA) in Vibrio cholerae is similarly subject to positive regulation by an AraC-type activator protein (ToxT) via a ς70-dependent promoter with an upstream AT-rich region (5). As would be expected, in the absence of the appropriate activator protein, the level of expression of these pilin subunit genes is extremely low when cloned in laboratory strains of E. coli.
The obligate human pathogens Neisseria gonorrhoeae and Neisseria meningitidis are both dependent on the production of type 4 pili for the initial colonization of mucosal surfaces, a prerequisite for subsequent infection (22). The region upstream of the gonococcal pilin subunit gene, pilE, shares features with both the P. aeruginosa pilA and the E. coli bfpA genes. Three promoter consensus sequences have been identified upstream of pilE (11). pilEp1 and pilEp2 are ς70-dependent promoters, and both are functional when the pilE gene is expressed in E. coli. The third promoter, pilEp3, is ς54 dependent and overlaps pilEp1. This promoter is functional in a P. aeruginosa background expressing PilR and PilS (7, 11). However, due to the fact that neither N. gonorrhoeae nor N. meningitidis has an intact rpoN gene, the pilEp3 promoter is nonfunctional in these species (26). In fact, it is clear that in N. gonorrhoeae (at least when grown under standard laboratory conditions), pilE transcription requires only P1 (11).
Interestingly, as was observed in the cases of the bfpA and tcpA promoters, the sequences directly upstream of pilEp1 in both neisserial species are highly AT rich. AT-rich sequences upstream of several E. coli and bacteriophage promoters have been shown to stimulate transcription from those promoters by RNA polymerase (RNAP) in the absence of any other protein factors (14, 32, 33). Such AT-rich sequences, known as UP elements, are generally located within the −40 to −60 region with respect to the transcription start point (TSP) and act as recognition sites for the RNAP α subunit (33). In particular, the C-terminal 85 amino acids of the α subunit (αCTD) make specific protein-DNA interactions with the UP element, resulting in factor-independent transcriptional activation (33). αCTD can also make protein-protein interactions with certain activator proteins in the absence of an UP element, thus enhancing transcription by increasing the efficiency of DNA binding by RNAP (6).
Recently, it was reported that gonococcal integration host factor (IHF) binds to a region upstream of the pilEp1 promoter and is a transcriptional cofactor of pilE (18). This conclusion was based on the observations that purified gonococcal IHF subunits bind to sequences upstream of the pilE promoters and that deletion of 70 bp (from −60 to −130), including both the IHF-binding site and further upstream sequence, resulted in a 10-fold reduction in pilE-specific mRNA. It was suggested that a potential role for IHF in this system was to stabilize a bend in the DNA so as to enhance the protein-protein interaction between RNAP and a putative activator protein.
In this study we have performed a detailed analysis to determine whether upstream sequences, in addition to the IHF-binding site, are required for efficient transcription from pilEp1 in N. gonorrhoeae and E. coli. Results indicate that the IHF-binding core consensus and 5′-proximal AT-rich sequences are associated with significant transcriptional activation of this promoter and that sequences upstream of the AT-rich tract do not further enhance transcription. An UP-like element positioned between the −35 box and the IHF-binding domain was also found to be required for maximal pilEp1 activity. An additional UP-like element was identified upstream of the IHF-binding core consensus sequence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strain used in all cloning experiments was DH5α [F− endA1 thi-1 hsdR17 supE44 relA1 ΔlacU169 (φ80 ΔlacZM15)]. The E. coli strains SØ1718 and SØ1753 (an ihfA::Tn10 derivative of SØ1718), used to assess the role of IHF in pilEp1 transcription, were kindly donated by Per Klemm. The N. gonorrhoeae strain used was MS11-A (36). The pilEp1::cat reporter plasmid used to generate the deletant reporter constructs was pJKD1304, a derivative of pJKD862 with the pilEp2 and pilEp3 promoters inactivated by site-directed mutamutagenesis (11).
Media and culture conditions.
The growth conditions for E. coli and gonococcal strains have been described previously (11). Gonococcal transformations were performed as previously described (3). Chloramphenicol-resistant transformants were selected on GC agar plates containing either 7 or 8 μg of chloramphenicol ml−1, depending on the expected chloramphenicol acetyltransferase (CAT) level.
Recombinant DNA techniques.
DNA manipulations were performed according to standard procedures (34). DNA sequencing and synthesis of oligonucleotide primers were performed as described previously (11). The oligonucleotide primers used to generate the upstream deletion derivatives and fusion derivatives and for site-directed mutagenesis are shown in Table 1. For construction of the deletion and fusion derivatives, DNA fragments with the appropriate pilEp1-containing regions were amplified by PCR, using the appropriate primer in conjunction with M13 reverse primer and with the plasmid pJKD1304 as a template. This required 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C, followed by 1 cycle of 1 min at 94°C, 1 min at 50°C, and 5 min at 72°C in an FTS-1 thermal sequencer (Corbett Research). Site-directed mutagenesis by PCR was performed essentially as described previously (11). The amplified fragments were cloned into pUC18 digested with HincII, and the nucleotide sequences were determined for the regions upstream of cat. Promoter-containing AvaI/AocI fragments were subcloned into pJKD862, replacing the wild-type promoter-containing fragment. The pilEp1::cat cassettes thus generated were subcloned on BamHI fragments into the single BglII site of pJKD1854, a derivative of pJKD1250 (11) containing a 2.9-kb fragment internal to the gonococcal iga gene, with the SphI/BamHI fragment from pJKD1499 (11) as a source of the gonococcal transformation uptake signal. Transformation of N. gonorrhoeae MS11-A with linearized plasmid DNA resulted in recombination of the pilEp1::cat reporter cassettes into the iga gene such that transcription of cat was in the direction opposite to transcription of iga. These recombinant plasmids were also transformed into E. coli strains SØ1718 and SØ1753.
TABLE 1.
Oligonucleotides used in this study for the construction of deleted and mutated pilEp1::cat reporter derivatives
| No. | Construct | Sequence |
|---|---|---|
| 1467 | Δ −111 | 5′ CTTCCGGATCCCACACCCACC 3′ |
| 5828 | Δ −90 | 5′ TACAAAATAAAAACAATTATATAG 3′ |
| 4856 | Δ −82 | 5′ TAAAAACAATTATATAGAG 3′ |
| 4857 | Δ −59 | 5′ CGCATAAAATTTCACCTC 3′ |
| 3997 | Δ −52 | 5′ AACGGATCCAATTTCACCTC 3′ |
| 384 | Δ −37 | 5′ CATAAAATGATCACGAAT 3′ |
| 6161 | IHF mut | 5′ ATAAAAAGTATTATACAGAGATAAACGC 3′ |
| 6162 | IHF mut | 5′ GTTTATCTCTGTATAATACTTTTTATTTTG 3′ |
| 5935 | UP mut | 5′ GGACTGAGGTGAAACTCTATGCGTTTATC 3′ |
| 5936 | UP mut | 5′ CGCATAGAGTTTCACCTCAGTCCATAAAATG 3′ |
| 5817 | Δ −38−77 | 5′ CTAAAAGAAAATACAAAATAAAAACATAAAATGATCACG 3′ |
| 5816 | PL-pilEp1 | 5′ CAAAAAATAAATTCATATAAAAAACATAAAATGATCACG 3′ |
Determination of CAT levels in bacterial cell extracts.
Cell extracts of E. coli and gonococcal strains were prepared by the freeze-thaw method according to the manufacturer’s instructions supplied with the CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim). E. coli cells were harvested from cultures grown for 16 h on L agar plates supplemented with ampicillin (50 mg/μl). Gonococcal cells were harvested from GC agar plates incubated for 20 h at 37°C in the presence of 5% CO2. The determination of CAT levels in these extracts was performed as described previously (11).
RESULTS
pilEp1 transcription is enhanced by upstream sequences in N. gonorrhoeae.
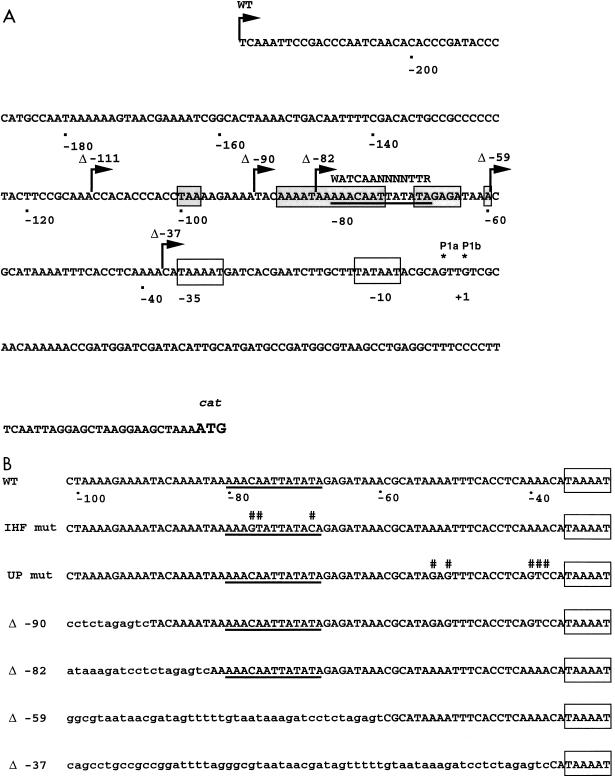
We have shown previously that a pilEp1::cat fusion (in which the P2 −10 and P3 −24 boxes were altered via site-directed mutagenesis), when recombined into the gonococcal chromosome, gave rise to levels of CAT similar to those of a reporter with all three wild-type promoter sequences (11). We were interested in determining whether transcription from pilEp1 was dependent on additional sequences upstream of the poorly conserved −35 box (TAAAAT). As shown in Fig. 1A, the sequence upstream of the pilEp1 TSP contains an 80% AT-rich tract extending from −30 to −100, within which a region protected by IHF has been mapped via DNase I footprinting (18). This protected region was originally proposed to contain two putative 13-bp IHF-binding core consensus sequences (domains 1 and 2) which partially overlap and are on opposite sides of the DNA helix. However, the sequence of domain 2 is closest to the E. coli consensus, WATCAANNNNTTR (9), and the DNase 1 footprint (18) is consistent with IHF binding preferentially to domain 2. Consequently, this is the sequence which is designated the putative IHF-binding core consensus sequence in this study, as shown in Fig. 1A. Deletions were generated from a pilEp1::cat fusion to remove the sequence upstream of the AT-rich tract (Δ −111), the AT-rich sequence upstream of the primary IHF-protected domain (Δ −90), the sequence upstream of the putative IHF-binding core consensus sequence (Δ −82), all of the sequence protected by IHF (Δ −59), and additional sequence between this region and the poorly conserved pilEp1 −35 box (Δ −37). Three base substitutions were also introduced at conserved positions within the putative IHF-binding core consensus sequence by site-directed mutagenesis (Fig. 1B). These substitutions were expected to have a severe effect on IHF binding, based on previous studies (14, 17), and the resulting construct was designated IHF mut. The new upstream regions generated by cloning the appropriate fragments into pUC18, and subsequently into pJKD1854, are shown in Fig. 1B.
FIG. 1.
Construction of deletant and mutant derivatives of a pilEp1::cat reporter cassette. (A) Nucleotide sequence of the region containing the pilEp1 promoter (open boxes) fused to the cat gene in the wild-type (WT) reporter cassette. The nucleotides contained within the shaded boxes are protected by IHF (18), with the solid underline indicating the putative 13-bp core consensus sequence previously designated domain 2 (18). The E. coli core consensus sequence (9) is shown above for comparison. Bent arrows indicate the sequences contained within each of the deletant derivatives (Δ −111, Δ −90, Δ −82, Δ −59, and Δ −37). Asterisks indicate the two TSPs (Pla and P1b) associated with pilEp1 (11). The large ATG defines the start codon of cat. (B) The sequences from −30 to −101 for the WT pilEp1::cat reporter, the mutated reporter derivatives (IHF mut and UP mut), and each of the deletant reporter constructs (Δ −90, Δ −82, Δ −59, and Δ −37) following subcloning into the unique BglII site within the iga gene fragment in pJKD1854. Nucleotides represented in lowercase letters are vector- or iga-derived. The putative IHF-binding core consensus sequences are underlined. #, a nucleotide substitution generated by site-directed mutagenesis. The −35 regions are boxed.
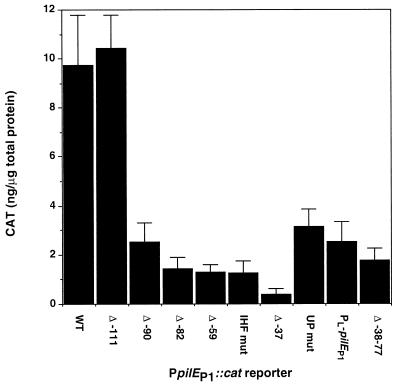
The wild-type, deletant, and mutated reporter cassettes were recombined into the chromosomal iga gene of N. gonorrhoeae MS11-A. Cell extracts were prepared from the recombinant gonococcal strains, and CAT levels were determined. The results shown in Fig. 2 clearly indicated that deletion of the region upstream of the AT-rich tract (Δ −111) had no significant effect on pilEp1 transcription as measured by CAT levels. However, deletion of a further 21 nucleotides (Δ −90) was associated with a fourfold reduction in transcriptional activity. Deletion of the entire AT-rich region upstream of the 13-bp core consensus sequence (Δ −82) resulted in a level of transcription similar to that obtained when the entire IHF-binding domain was deleted (Δ −59) or mutated (IHF mut). A further threefold reduction in CAT levels was observed upon replacement of the AT-rich sequence upstream of nucleotide −37 with a vector- or iga-derived sequence. A possible explanation for this observation is that the sequence between −37 and −59 functions as an UP element, enhancing the basal level of transcription from this promoter. Although no consensus sequence has yet been determined for an UP element in E. coli, the αCTD is thought to interact with DNA as a twofold symmetric dimer, recognizing two distinct regions within the UP element (12). Furthermore, DNA flexibility is thought to be crucial for the role of an UP element (28). Comparison of the −37 to −60 region upstream of pilEp1 with several well-characterized UP elements (data not shown) suggested that one or both of the two poly(A) tracts centered at −39.5 and −52.5, which were disrupted in Δ −37 (Fig. 1B), may be important for transcriptional enhancement. Consequently, five base substitutions were introduced by site-directed mutagenesis (at positions −38, −39, −40, −51, and −53) to specifically disrupt the poly(A) tracts (Fig. 1B). The activity of pilEp1 with this mutant upstream region substituted in the absence of the IHF-binding domain was comparable with that of Δ −37 (data not shown). In the presence of the IHF-binding domain (UP mut), promoter activity was shown to be threefold lower than that of the wild-type reporter (Fig. 2), confirming that the sequence located between the IHF-binding domain and the pilEp1 −35 box is important for promoter activity in N. gonorrhoeae.
FIG. 2.
Effect of upstream sequences on pilEp1 transcription in N. gonorrhoeae. CAT levels were measured in cell extracts prepared as described in Materials and Methods from gonococcal reporter strains containing pilEp1::cat cassettes with wild-type (WT) or substituted upstream sequences (explained in the legends to Fig. 1 and 4). The results shown are averages of at least four separate experiments, and error bars represent 1 standard deviation.
An intact ihfA gene is required for maximal transcription from pilEp1 in E. coli.
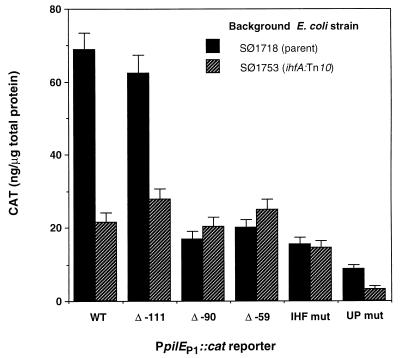
Our results are consistent with those of a previous study (18), where a 10-fold reduction in pilE-specific mRNA was observed when a 70-nucleotide deletion was made upstream of nucleotide −59. On the basis that this deletion removed the IHF-binding domain, it was concluded that IHF was required for optimal pilE transcription. Despite the fact that the gonococcal ihfA and ihfB genes (encoding the IHF α and β subunits) have been cloned, it has not been possible to isolate a mutant with a mutation in either of these genes (18a). Therefore, it was necessary to evaluate the role of IHF in the enhancement of pilEp1 transcription in an E. coli background. To this end, pJKD1854-derived plasmids containing the wild-type pilEp1::cat, deletant derivatives Δ −111, Δ −90, and Δ −59, and the IHF mut and UP mut reporters were transformed into the E. coli strains SØ1718 and SØ1753 (an ihfA::Tn10 derivative of SØ1718). Cell extracts were prepared and assayed for CAT. The results (Fig. 3) indicated that the levels of pilEp1 transcription obtained in the ihfA mutant background were three- to fourfold lower for the wild-type and Δ −111 reporters than the levels measured in the wild-type E. coli background. However, when the IHF-binding domain upstream of pilEp1 was partially deleted (Δ −90) or completely deleted (Δ −59), or when the core consensus sequence was mutated (IHF mut), this difference was negated. It has been well documented that certain IHF-binding core consensus sequences require additional 5′-proximal bases, with a high AT content, for efficient binding of IHF to occur (17). The fact that CAT levels obtained for the Δ −90 reporter were significantly reduced relative to those of the wild-type and Δ −111 reporters in both N. gonorrhoeae and E. coli indicated that the additional AT-rich sequence upstream of the core consensus sequence is absolutely required for IHF to bind and/or enhance transcription from pilEp1. The observation that deletion of sequences upstream of the IHF-protected domain (Δ −111) similarly had no effect on pilEp1 transcription in either N. gonorrhoeae or E. coli indicated that the role of IHF in the enhancement of transcription from this promoter is unlikely to be associated with the binding of a neisseria-specific activator protein to sequences further upstream.
FIG. 3.
Effect of an ihfA mutation on pilEp1 transcription in E. coli. CAT levels were measured in cell extracts prepared as described in Materials and Methods from E. coli SØ1718 (parent strain) and SØ1753 (ihfA::Tn10) transformed with pJKD1854-derived plasmids containing pilEp1::cat cassettes with and without upstream deletions or mutations (Fig. 1). The results shown are averages of at least four separate experiments, and error bars represent 1 standard deviation.
It also appears from the data presented in Fig. 3 that the UP-like element directly upstream of the pilEp1 −35 box plays a significant role in the transcriptional enhancement of this promoter in an E. coli background. In fact, mutation of the UP-like element (UP mut) resulted in an eightfold reduction in promoter activity in the ihfA+ background and a similar reduction in the ihfA mutant. This effect is apparently greater than that observed in N. gonorrhoeae, where mutation of the UP-like element resulted in only a threefold reduction in promoter activity. However, in a gonococcal background, deletion of the IHF-binding domain had a more deleterious effect on pilEp1 transcription (eightfold reduction) than that observed in E. coli (three- to fourfold reduction). These observed differences in the relative roles of the IHF-binding site and UP-like element may be a reflection of the fact that, in E. coli, the reporters were present on multicopy plasmids, while they were integrated into the chromosome as single copies in N. gonorrhoeae, potentially resulting in differences in the levels of DNA supercoiling. However, in both cases the two elements appear to be functionally independent.
The AT-rich sequence upstream of the IHF-binding core consensus sequence is capable of enhancing pilEp1 transcription.
The AT-rich sequence 5′ to the IHF-binding core consensus sequence upstream of pilEp1 is reminiscent of a sequence similarly located upstream of the bacteriophage λ early PL promoters (13, 14), as shown in Fig. 4. Transcription of λ PL1 is activated by IHF, in the presence of an intact αCTD (13). In contrast to pilEp1, the DNA between the −35 box and the IHF-binding site upstream of λ PL1 is not particularly AT rich, and a role for this region as an UP element has not been reported. However, an αCTD-binding site has been identified upstream of the IHF core consensus sequence, within the IHF-protected region. This region has been shown to function as an UP element for the λ PL2 promoter (in the absence of IHF) and, when placed directly upstream, of λ PL1 or Plac (14). The role of this UP element, when located at the normal position (−79 to −102 with respect to the PL1 TSP), in the IHF-mediated activation of PL1 has not been confirmed. However, it has been suggested that the change in DNA conformation induced by IHF binding could increase the affinity of RNAP for PL1 through direct contact between αCTD and this UP element (14).
FIG. 4.
Comparison of the sequence upstream of pilEp1 with the sequence upstream of the λ PL1 promoter (14). The −35 regions of pilEp1 and λ PL1 are contained within the open boxes. The −10 and −35 sequences associated with λ PL2 are in italics and underlined, with the TSP denoted by the bent arrow. The nucleotides contained within the hatched box have been shown to function as an UP element through the binding of the α subunit of RNAP. Asterisks denote identical nucleotides shared between the pilEp1 and λ PL sequences within this region. IHF-binding core consensus sequences are underlined in boldface. Nucleotides below the double-headed arrow were deleted in the construction of the Δ −38−77 and PL-pilEp1 reporter derivatives, resulting in the relocation of either the gonococcal putative UP element or the λ PL UP element directly upstream of nucleotide −37.
Comparison of the sequences directly upstream of the pilEp1 and λ PL1 IHF-binding core consensus sequences reveals that 17 of the 24 nucleotides are identical (Fig. 4). Based on this similarity, a pilEp1::cat reporter was constructed in which the sequence from −78 to −101 was fused by PCR, directly upstream of nucleotide −37, thus relocating the putative UP element immediately upstream of the pilEp1 −35 box (Fig. 4). The effect of creating this fusion was the same as that of deleting the sequence from nucleotides −38 to −77, including the IHF-binding core consensus sequence. The pilEp1::cat reporter thus generated was designated Δ −38−77. Similarly, the 21-nucleotide λ PL UP element was fused upstream of pilEp1 (PL-pilEp1), locating the sequence at the same position relative to the pilEp1 −35 box as it is located upstream of λ PL2 (14). The results of CAT enzyme-linked immunosorbent assays performed on gonococcal strains containing these reporters are shown in Fig. 2. It can be seen from this data that the λ PL UP element can enhance transcription from pilEp1 approximately sevenfold with respect to the basal level associated with Δ −37. The Δ −38−77 construct gave rise to levels of CAT comparable with those of Δ −82, Δ −59, and IHF mut constructs (each of which contain the wild-type −37 to −59 sequence), indicating that the sequence from −78 to −101 upstream of pilEp1 is also able to enhance transcription from that promoter when relocated directly upstream of the −35 box, but to a lesser extent than the λ PL UP element.
DISCUSSION
Previous work in this laboratory has clearly shown that, of the three promoter consensus sequences identified upstream of the pilE gene, the only one which is functional in N. gonorrhoeae during growth in vitro is the ς70-dependent promoter pilEp1 (11). Disruption of the pilEp1 −10 box via site-directed mutagenesis resulted in insignificant levels of transcription from the pilE upstream region. A gonococcal Pn strain (with a deletion encompassing the promoter and 5′ end of the chromosomal pilE gene) containing pilE on a multicopy plasmid, with only pilEp2 and pilEp3 intact, did not express pili. On the other hand, a similar strain with pilEp1 as the only intact promoter was highly piliated (our unpublished observations). Perhaps in an ancestor of the pathogenic neisseriae, transcription of the pilE gene was ς54 dependent, but it has since evolved to become ς70 dependent. It is unclear whether this was a consequence of the deletion in rpoN (25) and/or the gonococcal pilR and pilS homologs (8) or whether these occurred later. It is intriguing that both the ς54 promoter upstream of pilE and a sequence capable of functioning as an activator-binding site have been conserved in all gonococcal strains examined to date, and in some meningococcal strains, despite the fact that they are apparently no longer functional. However, this could be merely an indication of a relatively recent evolutionary event.
Prior to the demonstration that the pathogenic neisseriae contain an inactive rpoN gene, studies on the regulation of pilE were performed with the aim of identifying genes encoding putative transcriptional regulators (38). Using a pilEp::cat transcriptional fusion as a reporter, two divergently transcribed genes, designated pilA and pilB (not homologs of the P. aeruginosa pilA and pilB genes), were cloned in E. coli (38). PilA and PilB were reported to function as a two-component regulatory system controlling the transcription of pilE via the ς54 promoter (38–40), despite the fact that significant amino acid similarity was identified between PilA and FtsY (39). In E. coli, FtsY has been shown to be the functional homolog of the eukaryotic “docking protein” (26), the receptor for the signal recognition particle. The signal recognition particle is essential for the biogenesis of a subset of inner membrane proteins (37, 43). PilB, on the other hand, has recently been shown to function as a peptide methionine sulfoxide reductase (45). It is intriguing that purified PilA protein has been shown to bind to a DNA fragment containing the pilE promoter. Gel mobility shift assays performed on fragments containing the pilE promoters identified several regions necessary for this binding (2). In particular, a fragment lacking the sequence from −125 to −183 was no longer able to bind PilA. However, in this study, deletion of a sequence upstream of nucleotide −111 had no effect on transcription from pilEp1, suggesting that this region does not contain an essential activator-binding site, and thus that PilA is not a transcriptional activator of pilE. Indeed, much of the data accumulated in relation to the putative role of PilA in gonococcal piliation and viability (24, 39, 40) can be explained on the basis that this protein is the gonococcal homolog of FtsY and therefore more likely to be involved in regulation at a posttranscriptional level.
The role of IHF in the transcriptional enhancement of pilE may originally have been architectural, i.e., in the bending of the DNA between the ς54-dependent promoter and the activator-binding site so as to enhance the protein-protein interactions between the RNAP and the bound activator. Several examples of such an indirect role for IHF in the activation of ς54 promoters in E. coli and other bacterial species have been documented (16). There are also examples of IHF playing an architectural role in the activation of a ς70 promoter in E. coli (35) and an AlgU-dependent promoter in P. aeruginosa (10).
The results presented in this study suggest that IHF plays a direct role in the transcriptional activation of the pilEp1 promoter, i.e., no additional activators appear to be required. To our knowledge, this is the first example of such a role for IHF to be described in a bacterial species other than E. coli. The importance of the AT-rich region, 5′ to the IHF-binding core consensus sequence, for the pilEp1 transcription-activating role of IHF was also demonstrated in this study. Only a subset of E. coli IHF-binding sites have such additional domains (15). However, placement of an AT-rich element upstream of a core consensus element can significantly increase the affinity of that element for IHF-binding (17).
The introduction of mutations at nucleotides −90, −93, and −98 within the IHF-protected region upstream of λ PL resulted in a threefold decrease in IHF binding (14). We observed that replacement of the two poly(A) tracts immediately 5′ to nucleotide −90, upstream of pilEp1, with vector-derived sequence resulted in a fourfold reduction in P1 transcriptional activity. One possible explanation for this result is that the conformation and flexibility of the DNA in the vicinity of the IHF core consensus sequence plays an important role in determining the efficiency of IHF binding (15). Further experiments are required to determine the likelihood of this explanation.
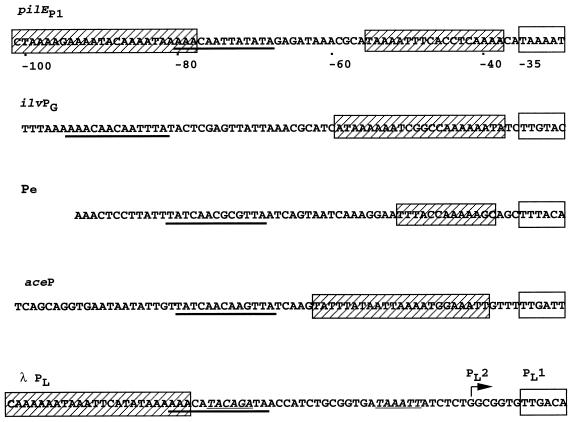
In addition to the domain bound by IHF, the AT-rich region upstream of pilEp1 has been shown to contain two UP-like elements. The first of these, positioned within the −37 to −59 region, enhances pilEp1 transcription three- to fourfold in N. gonorrhoeae, independently of the IHF-mediated activation. Similar elements have been described directly upstream of several E. coli and bacteriophage promoters, including ones which are directly activated by IHF. Figure 5 shows an alignment between the sequence upstream of pilEp1 and the equivalent sequences upstream of several promoters which have been shown to be directly activated by IHF. In the case of the E. coli ilvGMEDA operon, it was suggested that the formation of an IHF-DNA nucleoprotein structure caused a conformational change in the DNA helix at the promoter and that this enhanced transcription initiation via increased open complex formation in a manner which was sensitive to DNA supercoiling (30). In addition to the contribution of the IHF-induced bend to transcriptional activation, the AT-rich sequence centered at nucleotide −50 significantly enhanced ilvpG transcriptional levels through the formation of an intrinsic DNA bend (29). The transcriptional activation properties of the IHF-binding domain and the region associated with the intrinsic bend were functionally independent in the case of the ilvpG promoter (30). On the other hand, IHF-mediated activation of the early promoter of bacteriophage Mu (Pe) is thought to be associated with improved binding of αCTD to an UP element located between −39 and −51 (44). An αCTD-binding UP element has likewise been identified between the IHF-binding site and the promoter region upstream of the E. coli acetate operon, aceBAK (28). However, the functional independence or interaction of the two elements was not reported in this study. It appears, on the basis of the alignments presented in Fig. 5, that pilEp1 shares the AT-rich sequence within the −40 to −60 region, characteristic of the Pe, acep, and ilvpG promoters. Clearly this region enhances transcription from pilEp1, but the mechanism involved has yet to be confirmed. The most likely explanations, based on the sequence similarities, are that the region functions (i) as a binding site for αCTD (or some other factor) or (ii) by introducing an intrinsic DNA bend upstream of the promoter, which in turn enhances binding of RNAP or alters the kinetics of open complex formation. The observation that the level of activation associated with the presence of the IHF-binding domain (eightfold in N. gonorrhoeae and three- to fourfold in E. coli) was the same in the presence and absence of the wild-type UP-like element indicated that the two regions are functionally independent.
FIG. 5.
Comparison of the sequence upstream of and including the pilEp1 −35 box with the corresponding sequences of a series of IHF-activated promoters described in E. coli and bacteriophages. ilvpG is located upstream of the E. coli ilvGMEDA operon (30), Pe is an early promoter of phage Mu (44), acep is located upstream of the operon encoding the enzymes for acetate utilization in E. coli (28), and λ PL is an early promoter of phage λ (13, 14) containing the PL2 promoter (italicized and underlined) in addition to the PL1 −35 box. The IHF-binding core consensus sequences are underlined in boldface, the −35 regions are defined by open boxes, and sequences shown to be associated with transcriptional enhancement, or confirmed UP elements, are contained within hatched boxes.
The second region capable of enhancing pilEp1 transcription is positioned between −78 and −101, upstream of the IHF-binding core consensus sequence but within the IHF-protected domain. This element shares 17 identical nucleotides out of 24 with a similarly placed UP element within the λ PL upstream region.
It remains to be directly demonstrated that one or both gonococcal UP-like elements enhance transcription through binding to the RNAP α subunit. Extensive analysis of the E. coli RNAP α subunit (4, 12) have suggested that interaction of the CTD with the rrnBp1 UP element involves dimerization and binding to two distinct regions (centered at −42 and −52) within the UP element. Recent evidence suggests that in the presence of an activator (e.g., cyclic AMP receptor protein [CRP]) bound upstream of a promoter, the two α subunits are capable of binding different DNA sites as monomers (27). It was shown in this study that the β′-associated α subunit was capable of contacting a site as far upstream as −96 in the presence of a CRP dimer bound at −74.5 with respect to the TSP. It is intriguing that the locations of these contact and binding sites are remarkably similar to the locations of the centers of the pilEp1 upstream UP-like element and IHF-binding core consensus sequence.
The nucleotide sequence of the gonococcal rpoA gene (encoding the α subunit of RNAP) was identified following a BLAST search (1) of the genomic sequence data for strain FA1090, released on the University of Oklahoma N. gonorrhoeae Genome Database (32a). This sequence was translated, and an alignment was generated between the deduced amino acid sequence and the amino acid sequence of the E. coli α subunit (20). It was clear from this alignment that the amino acid sequence of the C-terminal end of the gonococcal α subunit is very similar to the equivalent region of the E. coli protein, with 60 of 85 amino acids identical (data not shown). Seven amino acids within two domains of the E. coli αCTD have been shown to be crucial for DNA binding to the rrnB UP element (12). All of them are conserved in the gonococcal α subunit (data not shown). Thus, it is likely that similar interactions occur between gonococcal UP elements and αCTD.
Work is currently under way in our laboratory to determine the relative roles and interactions of IHF, the UP-like elements, and the α subunit of RNAP in the transcriptional activation of the gonococcal pilE gene. The potential role of DNA supercoiling in the regulation of this important gene is also under investigation.
ACKNOWLEDGMENTS
We thank Per Klemm for kindly providing bacterial strains. We are also grateful to Jim Pittard and Ji Yang for helpful discussions during the preparation of the manuscript.
This work was supported by a project grant from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–415. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arvidson C G, So M. Interaction of the Neisseria gonorrhoeae PilA protein with the pilE promoter involves multiple sites on the DNA. J Bacteriol. 1995;177:2497–2504. doi: 10.1128/jb.177.9.2497-2504.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black C G, Fyfe J A M, Davies J K. A promoter associated with the neisserial repeat (NR) can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J Bacteriol. 1995;177:1952–1958. doi: 10.1128/jb.177.8.1952-1958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatter E E, Ross W, Tang H, Gourse R L, Ebright R H. Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994;78:889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 5.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 6.Busby S, Ebright R H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 7.Carrick C S, Fyfe J A M, Davies J K. The normally silent ς54 promoters upstream of the pilE genes of both Neisseria gonorrhoeae and Neisseria meningitidis are functional when transferred to Pseudomonas aeruginosa. Gene. 1997;198:89–97. doi: 10.1016/s0378-1119(97)00297-7. [DOI] [PubMed] [Google Scholar]
- 8.Carrick, C. S., J. A. M. Fyfe, and J. K. Davies. Unpublished data.
- 9.Craig N L, Nash H A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 10.Delic-Atree I, Toussaint B, Froger A, Willison J C, Vignais P M. Isolation of an IHF-deficient mutant of a Pseudomonas aeruginosa mucoid isolate and evaluation of the role of IHF in algD gene expression. Microbiology. 1996;142:2785–2793. doi: 10.1099/13500872-142-10-2785. [DOI] [PubMed] [Google Scholar]
- 11.Fyfe J A M, Carrick C S, Davies J K. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a ς70 promoter during growth in vitro. J Bacteriol. 1995;177:3781–3787. doi: 10.1128/jb.177.13.3781-3787.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaal T, Ross W, Blatter E E, Tang H, Jia X, Krishnan V V, Assa-Munt N, Ebright R H. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding architecture. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 13.Giladi H, Igarashi K, Ishihama A, Oppenheim A B. Stimulation of the phage λ pL promoter by integration host factor requires the carboxy terminus of the α-subunit of RNA polymerase. J Mol Biol. 1992;227:985–990. doi: 10.1016/0022-2836(92)90514-k. [DOI] [PubMed] [Google Scholar]
- 14.Giladi H, Murakami K, Ishihama A, Oppenheim A B. Identification of an UP element within the IHF binding site at the PL1-PL2 tandem promoter of bacteriophage lambda. J Mol Biol. 1996;260:484–491. doi: 10.1006/jmbi.1996.0416. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich J A, Schwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goosen N, van de Putte P. The regulation of transcription by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 17.Hales L M, Gumport R I, Gardner J F. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J Bacteriol. 1994;176:2999–3006. doi: 10.1128/jb.176.10.2999-3006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill S A, Samuels D S, Carlson J H, Wilson J, Hogan D, Lubke L, Belland R J. Integration host factor is a transcriptional cofactor of pilE in Neisseria gonorrhoeae. Mol Microbiol. 1997;23:649–656. doi: 10.1046/j.1365-2958.1997.2321612.x. [DOI] [PubMed] [Google Scholar]
- 18a.Hill, S. A. Personal communication.
- 19.Hobbs M, Collie E S R, Free P D, Livingston S P, Mattick J S. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi K, Fujita N, Ishihama A. Sequence analysis of two temperature-sensitive mutations in the alpha subunit gene (rpoA) of Escherichia coli RNA polymerase. Nucleic Acids Res. 1990;18:5945–5948. doi: 10.1093/nar/18.20.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimoto K S, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellogg D S, Jr, Peacock W L, Jr, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S-R, Komano T. The plasmid R64 thin pilus identified as a Type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larribe M, Taha M-K, Topilko A, Marchal C. Control of Neisseria gonorrhoeae pilin gene expression by environmental factors: involvement of the pilA/pilB regulatory genes. Microbiology. 1997;143:1757–1764. doi: 10.1099/00221287-143-5-1757. [DOI] [PubMed] [Google Scholar]
- 25.Laskos L, Dillard J P, Seifert H S, Fyfe J A M, Davies J K. The pathogenic neisseriae contain an inactive rpoN gene and do not utilize the pilE ς54 promoter. Gene. 1998;208:95–102. doi: 10.1016/s0378-1119(97)00664-1. [DOI] [PubMed] [Google Scholar]
- 26.Luirink J, ten Hagen-Jongman C M, van der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami K, Owens J T, Belyaeva T A, Meares C F, Busby S J W, Ishihama A. Positioning of two alpha subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc Natl Acad Sci USA. 1997;94:11274–11278. doi: 10.1073/pnas.94.21.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negre D, Bonod-Bidaud C, Oudot C, Prost J F, Kolb A, Ishihama A, Cozzone A J, Cortay J C. DNA flexibility of the UP element is a major determinant for transcriptional activation at the Escherichia coli acetate promoter. Nucleic Acids Res. 1997;15:713–718. doi: 10.1093/nar/25.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagel J M, Winkelman J W, Adams C W, Hatfield G W. DNA topology-mediated regulation of transcription initiation from the tandem promoters of the ilvGMEDA operon of Escherichia coli. J Mol Biol. 1992;224:919–935. doi: 10.1016/0022-2836(92)90460-2. [DOI] [PubMed] [Google Scholar]
- 30.Parekh B S, Hatfield G W. Transcriptional activation by protein-induced DNA bending: evidence for a DNA structural transmission model. Proc Natl Acad Sci USA. 1996;93:1173–1177. doi: 10.1073/pnas.93.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 32.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Gourse R L. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 32a.Roe, B. A., S. Clifton, and D. W. Dwyer.http://www.genome.ou.edu.
- 33.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schroder I, Darie S, Gunsalus R P. Activation of the Escherichia coli nitrate reductase (narGHJI) operon by NarL and Fnr requires integration host factor. J Biol Chem. 1993;268:771–774. [PubMed] [Google Scholar]
- 36.Segal E, Billyard E, So M, Storzbach S, Meyer T F. Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell. 1985;40:293–300. doi: 10.1016/0092-8674(85)90143-6. [DOI] [PubMed] [Google Scholar]
- 37.Seluanov A, Bibi E. FtsY, the procaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 38.Taha M-K, So M, Seifert H S, Billyard E, Marchal C. Pilin expression in Neisseria gonorrhoeae is under both positive and negative transcriptional control. EMBO J. 1988;7:4367–4378. doi: 10.1002/j.1460-2075.1988.tb03335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taha M-K, Dupuy B, Saurin W, So M, Marchal C. Control of pilus expression in Neisseria gonorrhoeae as an original system in the family of two-component regulators. Mol Microbiol. 1991;5:137–148. doi: 10.1111/j.1365-2958.1991.tb01834.x. [DOI] [PubMed] [Google Scholar]
- 40.Taha M-K, Giorgini D. Phosphorylation and functional analysis of PilA, a protein involved in the transcriptional regulation of the pilin gene in Neisseria gonorrhoeae. Mol Microbiol. 1995;15:667–677. doi: 10.1111/j.1365-2958.1995.tb02376.x. [DOI] [PubMed] [Google Scholar]
- 41.Tennent J M, Mattick J S. Type 4 fimbriae. In: Klemm P, editor. Fimbriae. Adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press Inc.; 1994. pp. 127–170. [Google Scholar]
- 42.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 43.Ulbrandt N D, Newitt J A, Bernstein H D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 44.van Ulsen P, Hillebrand M, Kainz M, Collard R, Zulianello L, van de Putte P, Gourse R L, Goosen N. Function of the C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase in basal expression and integration host factor-mediated activation of the early promoter of bacteriophage Mu. J Bacteriol. 1997;179:530–537. doi: 10.1128/jb.179.2.530-537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wizemann T M, Moskovitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure H R. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]