Abstract
While the physical interactions between the Golgi apparatus (Golgi) and lipid droplets (LDs) have been suggested through system-level imaging, the Golgi-LD membrane contact sites (MCSs) remain largely uncharacterized. Here, we show evidence to support the existence of Golgi-LD MCSs in HEK293 cells. We further suggest that vacuolar protein sorting-associated protein 13B (VPS13B) localizes to and promotes the formation of Golgi-LD contacts upon oleic acid (OA) stimulation using 3D high-resolution microscopy. Depletion of VPS13B moderately affects the formation of Golgi-LD contacts upon OA treatment in addition to the fragmentation of the Golgi. Although cellular functions of VPS13B-mediated contacts are still elusive, these findings may provide a new insight into related diseases caused by loss-of-function mutations of VPS13B.
Keywords: Golgi, lipid droplet, endoplasmic reticulum, membrane contact sites, VPS13B
Introduction
Membrane contact sites (MCSs) are microdomains where opposing membranes of two organelles are tethered by protein–protein or protein–lipid interactions (Eisenberg-Bord et al., 2016; Gatta and Levine, 2017; Lees and Reinisch, 2020; Ugur et al., 2020). A growing body of evidence has supported that MCSs are conserved cellular structures with multiple fundamental functions such as organelle inheritance, fusion, and fission, and the transfer of lipids, ions, and other small metabolites (Abrisch et al., 2020; Elbaz and Schuldiner, 2011; Kumar et al., 2018; Lewis et al., 2016; Prinz et al., 2020; Scorrano et al., 2019; Valverde et al., 2019; Wu et al., 2018). The understanding of MCSs has been greatly improved as tethering, effector, and regulatory proteins at MCSs being identified (Eisenberg-Bord et al., 2016; Gatta and Levine, 2017). However, it is clear that only a small portion of the complete cellular puzzle of MCSs has been investigated so far and that, therefore, new MCSs and key players at such MCSs await discovery (Bohnert and Schuldiner, 2018).
Vacuolar protein sorting-associated protein 13B (VPS13B) is a member of bridge-like lipid transfer protein family (BLTP) (Neuman et al., 2022) that is highly conserved in animal species. The human genome contains four VPS13 genes (VPS13A, VPS13B, VPS13C, and VPS13D genes) (Velayos-Baeza et al., 2004). Previous studies have revealed that VPS13A and VPS13C are lipid transporters at endoplasmic reticulum (ER)-associated MCSs including ER-mitochondria/lipid droplet (LD)/plasma membrane (VPS13A) and ER-late endosome/lysosome/LD MCSs (VPS13C) (Guillen-Samander et al., 2022; Kumar et al., 2018; Park et al., 2022). VPS13D has been reported to play roles in mitophagy in Drosophila (Anding et al., 2018; Shen et al., 2021) and localizes to ER-mitochondrial/peroxisomal contacts (Baldwin et al., 2021; Guillen-Samander et al., 2021). Our results further showed that VPS13D plays a regulatory role in ER-mitochondrial MCSs (Du et al., 2021) and facilitates LD remodeling under starvation (Wang et al., 2021). Whether VPS13B is associated with a specific type of MCS remains unclear, though VPS13B has been previously implicated in the maintenance of the Golgi structure (Seifert et al., 2011; Seifert et al., 2015) and in cargo transport from early endosomes to recycling endosomes (Koike and Jahn, 2019).
In the present study, we show evidence to support the existence of Golgi-LD MCSs. We further suggest that VPS13B localizes to the Golgi-LD MCSs under oleic acid (OA)-stimulated conditions. Overexpression of VPS13B promoted the formation of Golgi-LD MCSs, and its depletion affected the formation or the regulation of these contacts. In addition, we show evidence that the ER was often involved in the VPS13B-marked MCSs upon OA stimulation, suggesting that VPS13B may function at ER-Golgi-LD tripartite junctions.
Results and Discussion
A Portion of LD Associates with the Golgi in OA-Stimulated HEK293 Cells
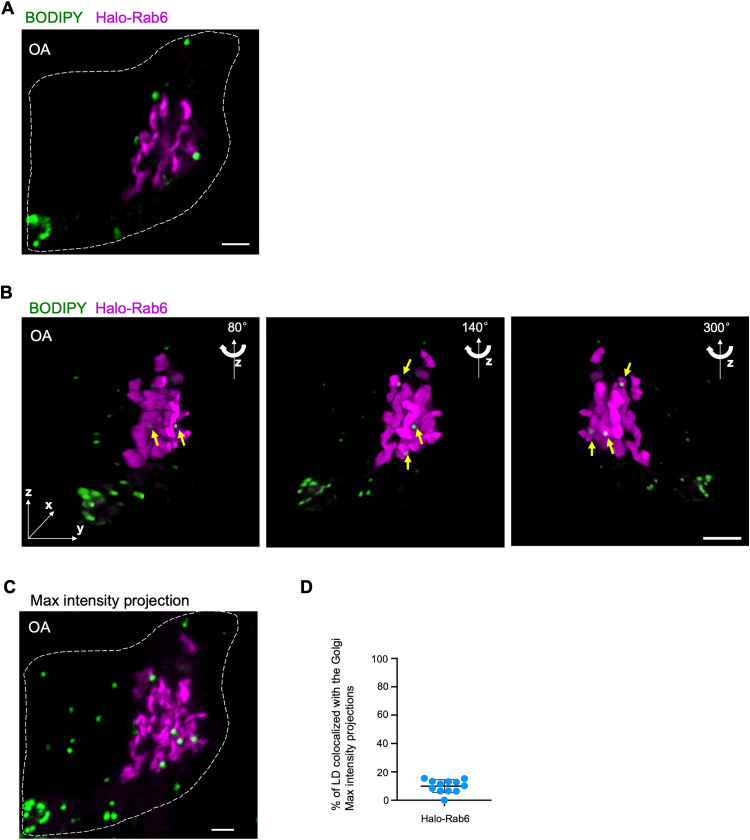
System-level spectral imaging has observed that LDs frequently contacted the Golgi in COS7 cells, suggesting that the Golgi formed MCSs with LDs (Valm et al., 2017). Consistent with the result, we found that a portion of LDs (labeled by BODIPY-558/568) contacted the Golgi (marked by Halo-Rab6A) in HEK293 cells, as revealed by tight associations between these two organelles using high-resolution live-cell airyscan confocal microscopy (Figure 1A). To better visualize LDs, we cultured the cells in medium containing OA, to increase the size and abundance of LDs.
Figure 1.
A portion of LD associates with the Golgi in OA-stimulated HEK293 cells. (A) Representative airyscan images of a live BODIPY 558/568 (green)-labeled HEK293 cell expressing Halo-Rab6 (magenta). (B) 3D images of the cell in (A) with three different angles (80°, 140°, and 300°) along z-axis were shown. A stack of 26 z-planes with 0.19-μm thickness in each stack was taken. Yellow arrows denoted LDs tightly associated with the Golgi. (C) The maximum intensity projections of 3D reconstructions of the cell in (A) showing Golgi-LD interactions. (D) Quantification of Golgi-LD interactions in BODIPY558/568-labeled cells expressing Halo-Rab6 (n = 12) based on the maximum intensity projections from 3D constructions. Mean ± SD. Scale bar, 10 μm in (A, B, and C).
The Golgi structures remained largely peri-nuclear but occupied a considerable volume in three dimensions (3D). Thereby, we used 3D reconstructions of z-stacks through high-resolution airyscan confocal microscopy to confirm the association between the Golgi and LDs in OA-stimulated HEK293 cells (Figure 1B). A small but significant portion of LDs (∼10%) tightly associated with the Golgi (Figure 1C and D), as revealed by co-localization analysis based on maximum intensity projections of z-stacks.
VPS13B Localizes to Golgi-LD MCSs
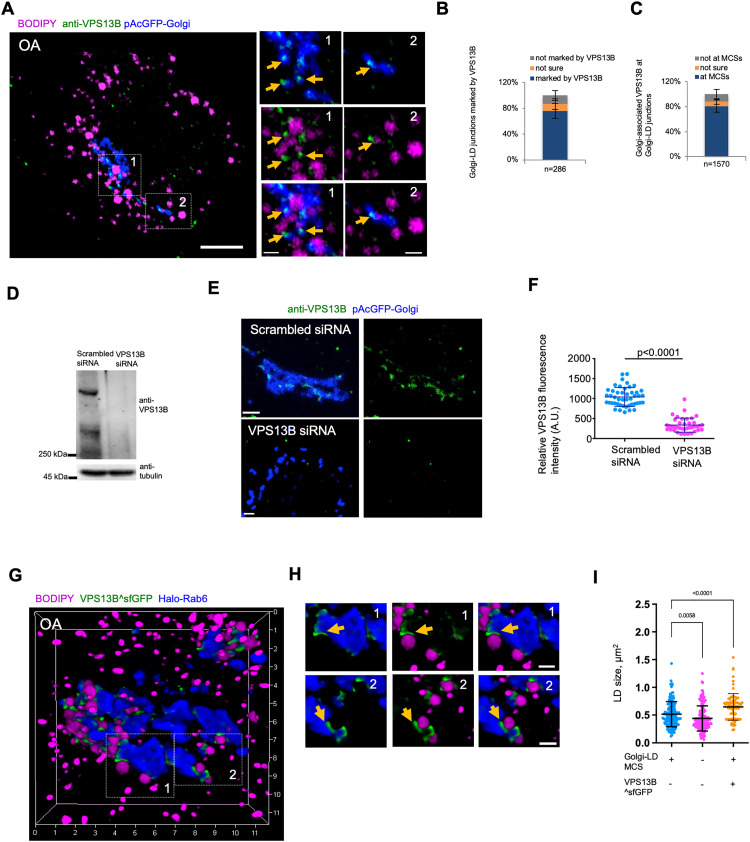
The physical associations between the Golgi and LDs may represent Golgi-LD MCSs. However, the components of such MCSs were unknown. Given that VPS13B is a BLTP associated with the Golgi apparatus (Seifert et al., 2011), we hypothesized that VPS13B may function at Golgi-LDs MCSs. To confirm this hypothesis, we initiated an examination of the cellular localizations of VPS13B within animal cells. We explored the localization of endogenous VPS13B by immunofluorescence staining (IF) in HEK293 cells (Figure 2A). Endogenous VPS13B mainly localized to the Golgi (labeled by Halo-Rab6) (Figure 2A), which is in agreement with previous studies (Koike and Jahn, 2019; Seifert et al., 2015). Interestingly, ∼76% of potential Golgi-LD junctions were marked by VPS13B (Figure 2B), while ∼82% of Golgi-associated VPS13B puncta (or enrichments) were present at Golgi-LD junctions (Figure 2C). The fluorescence of endogenous VPS13B at these junctions was almost completely lost upon small-interfering RNAs (siRNAs)-mediated VPS13B suppression (Figure 2D-F), validating the specificity of the anti-VPS13B antibody in IF.
Figure 2.
VPS13B localizes to Golgi-LD MCSs. (A) Airyscan images of fixed BODIPY558/568 (magenta)-labeled HEK293 cells expressing pAcGFP-Golgi (blue) in IF with anti-VPS13B antibody (green). Yellow arrows denoted endogenous VPS13B foci adjacent to Golgi-LD junctions. (B) Percentage of Golgi-LD junctions (n = 286 from 20 cells) marked with endogenous VPS13B. Mean ± SD. (C) Percentage of Golgi-associated, endogenous VPS13B puncta or enrichments (n = 1570 from 20 cells) at Golgi-LD junctions. Mean ± SD. (D) Western blots demonstrated the efficiency of siRNA-mediated VPS13B suppression. (E) Confocal images of scrambled (top panel) or VPS13B siRNAs treated (bottom panel) HeLa cells expressing pAcGFP-Golgi (blue) in IF by anti-VPS13B antibody (green). Contrast range of images was set to same level for both cells. (F) Quantification of VPS13B fluorescence intensity in IF of scrambled (n = 46) or VPS13B siRNAs (n = 39) treated cells. Mean ± SD. Two-tailed unpaired Student t-test. (G, H) High-resolution 3D image of a BODIPY558/568 (magenta)-labeled HEK293 cell expressing VPS13B^sfGFP (green) and Halo-Rab6 (blue), with two insets from boxed regions with yellow arrows denoting specific enrichment of VPS13B^sfGFP at Golgi-LD MCSs. (I) Quantification of the size of LDs at the Golgi-LD junctions (n = 157, 24 cells), LDs not at junctions (n = 167, 24 cells) or LDs at VPS13B^sfGFP-marked junctions in cells expressing VPS13B^sfGFP (87, 13 cells). Mean ± SD. Two-tailed unpaired Student t-test. Scale bar, 10 μm in (A) and 2 μm in insets in (A, E, and H).
We ectopically expressed superfolder green fluorescent protein (sfGFP)-tagged VPS13B (NM152564.5 isoform1) (VPS13B^sfGFP), in which sfGFP was internally tagged to VPS13B at a site shown to preserve yeast VPS13 function (Lang et al., 2015), in HEK293 cells. We used HEK293 cells for this purpose as they are well suited for the transfection and expression of the huge VPS13B^sfGFP construct (∼20 K base pairs in size). We further validated the localization of VPS13B^sfGFP at Golgi-LD MCSs via 3D high-resolution imaging. We found that VPS13B^sfGFP was preferentially accumulated at Golgi-LD MCSs with LDs being tightly tethered to the Golgi in response to OA stimulation (Figure 2G). Notably, VPS13B^sfGFP marked an extensive Golgi-LD MCSs (Figure 2H), whereas endogenous VPS13B marked only spot-like contacts between the Golgi and LDs (Figure 2A), suggesting that VPS13B overexpression may result in expansions of these contacts. In addition, we found that the size of LDs at VPS13B-marked contacts appeared to be slightly increased compared with that of those not associating with these contacts, and VPS13B^sfGFP overexpression further increased the size of LDs contacting the Golgi (Figure 2I).
VPS13B Promotes the Formation of Golgi-LD MCSs
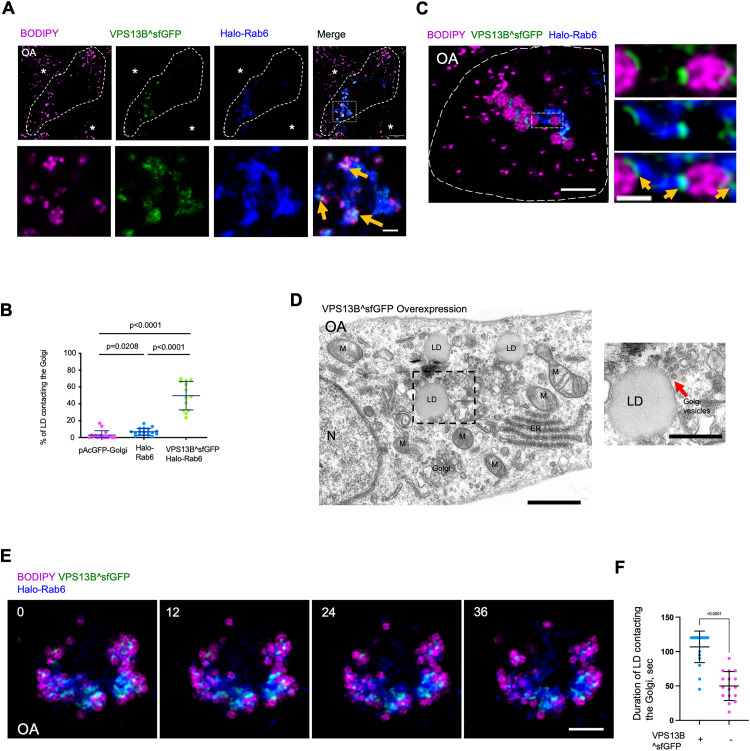
Next, we investigated whether VPS13B promoted the formation of Golgi-LD MCSs by live-cell confocal microscopy. Remarkably, we noted that a substantially higher portion of LDs (∼51%) was closely associated with the Golgi upon expression of VPS13B^sfGFP (Figure 3A and B), compared to either pAcGFP1-Golgi (GFP tagged trans-membrane region of B4GALT1 as a general Golgi marker) or Halo-Rab6 transfected cells, suggesting a potential role of VPS13B in promoting Golgi-LD associations. In addition, high-resolution confocal microscopy suggested that VPS13B^sfGFP appeared to enrich at MCSs between large LDs and the Golgi (Figure 3C).
Figure 3.
VPS13B promotes the formation of Golgi-LD MCSs. (A) Representative airyscan confocal images of a BODIPY558/568 (LD marker, magenta)-labeled HEK293 cell expressing VPS13B^sfGFP (green), Halo-Rab6 (blue). Yellow arrows denoted the hyper-tethering of LDs to Golgi upon VPS13B^sfGFP expression. Asterisks represented neighboring cells with normal distribution of LDs. (B) Quantification of Golgi-LD interactions in BODIPY558/568-labeled cells expressing a general Golgi marker pAcGFP-Golgi (n = 16), Halo-Rab6 (n = 18), or co-expressing VPS13B^sfGFP and Halo-Rab6 (n = 12). Mean ± SD. Two-tailed unpaired Student t-test. (C) High-resolution lightning images of a cell as in (A) in medium with OA (500 μM) for 16 hours. Left: whole cell image; right: an inset from a boxed region in whole cell image. Yellow arrows denoted the specific enrichment of VPS13B^sfGFP at Golgi-LD MCSs. (D) Representative electron micrographs showing a cluster of vesicles (red arrows) near LD in HEK293 cells expressing VPS13B^sfGFP in OA medium with one inset from boxed regions on the right. M: mitochondria; N: nucleus; ER: endoplasmic reticulum. (E) Time-lapse images of a BODIPY558/568 (magenta)-labeled HEK293 cell expressing VPS13B^sfGFP (green) and Halo-Rab6 (blue). Time in sec. (F) Duration of Golgi-LD contacts as in (D) or in cells without VPS13B^sfGFP expression by live cell imaging with a time window of 2 min. Time in sec. Mean ± SD. Two-tailed unpaired Student t-test. Scale bar, 10 μm in whole cell image and 2 μm in insets in (A); 2 μm in whole cell image and 0.5 μm in the inset in (C); 1 μm in left panel and 0.5 μm in right panel in (D); 10 μm in (E).
Electron micrographs showed that a cluster of small vesicles were present around LDs in VPS13B overexpressed cells (Figure 3D). Currently, the nature of the vesicle cluster is unclear. Interestingly, SHIP164, a newly identified VPS13-related protein, was localized to a cluster of small vesicles of early endocytic pathway (Hanna et al., 2022). We speculated that the vesicle cluster might be clustered Golgi vesicles possibly from ER-Golgi intermediate compart.
Given the dynamic nature of the Golgi and LDs, we tracked the movement of LDs relative to the Golgi over time in HEK293 cells expressing VPS13B^sfGFP in OA medium. We found that LDs were tightly associated with the Golgi, and these VPS13B^sfGFP-mediated associations could last for more than 2 min by live-cell imaging, which was more stable than those in cells without VPS13B overexpression (Figure 3E and F). Collectively, our findings suggested that VPS13B might facilitate the tethering between the Golgi and LDs under OA-stimulated conditions.
The Depletion of VPS13B Affected the Formation of Golgi-LD Contacts
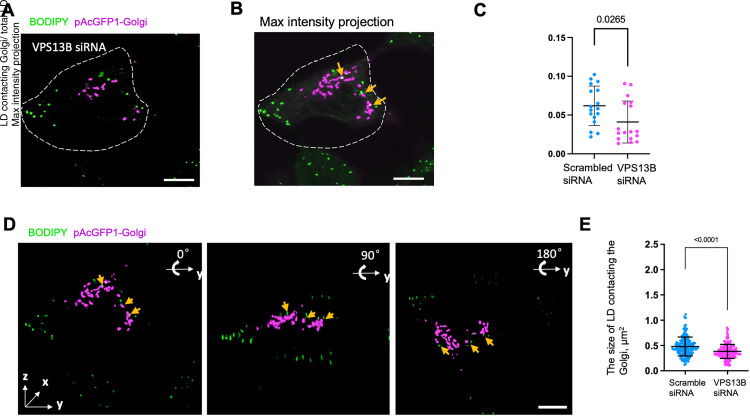
To explore whether VPS13B was required for the formation of Golgi-LD contacts, we examined the effects of siRNA-mediated VPS13B depletion on the Golgi-LD contacts by high-resolution 3D confocal microscopy. In agreement with previous studies (Seifert et al., 2011), VPS13B depletion resulted in Golgi fragmentation (Figure 4A and B and Figure 2E). Further, we quantified the percentage of LDs contacting the Golgi and found that VPS13B depletion reduced the portion of LDs involving in the Golgi contacts, but to a moderate extent (Figure 4C). We speculate that the dispersed Golgi mini stacks in VPS13B-depleted cells increased the probability of contacts with LDs, which might compensate the effect of VPS13B depletion. Notably, we noticed that a portion of LDs appeared to be adjacent to but not physically contacted the Golgi in VPS13B-depleted cells (Figure 4D; yellow arrows), as revealed by 3D reconstruction with three different angles (0°, 90°, and 180°) along with the y-axis. Notably, our results could not exclude that VPS13B depletion indirectly affected the formation or the regulation of Golgi-LD contacts through affecting the Golgi morphology. Of note, we found that the size of LDs at contact with the Golgi was reduced in response to VPS13B siRNA treatment (Figure 4E), suggesting a regulatory role of VPS13B in LD size.
Figure 4.
The Golgi-LD interactions in VPS13B-depleted cells. (A) Representative confocal images of a BODIPY 558/568 (green)-labeled HeLa cell expressing pAcGFP1-Golgi (magenta) upon treatments of VPS13B siRNAs. (B) The maximum intensity projections of 3D reconstructions of the VPS13B-depleted cell in (A) showing possible Golgi-LD interactions. (C) Quantification of Golgi-LD interactions in BODIPY558/568-labeled cells expressing pAcGFP-Golgi in response to either scrambled (n = 17) or VPS13B siRNAs (n = 17). Mean ± SD. Two-tailed unpaired Student t-test. (D) 3D images of the VPS13B-depleted cell in (A) with three different angles (0°, 90°, and 180°) rotating along with y-axis were shown. A stack of 26 z-planes with 0.19-μm thickness in each stack was taken. Yellow arrows denoted LDs adjacent but not physically contacting the Golgi. (E) Quantification the size of LD adjacent to the Golgi in BODIPY558/568-labeled cells in response to either scrambled (n = 17) or VPS13B siRNAs (n = 17). Mean ± SD. Two-tailed unpaired Student t-test. Scale bar, 10 μm in whole cell image and 1 μm in insets in (A); 2 μm in insets in (B–D).
Lack of Stable Association Between VPS13B and VAPs
Previous studies showed that VPS13 proteins, including VPS13A/C/D, tethered the ER via its FFAT or phospho-FFAT motif in its N terminal to the other organelles via a region in its C-terminal, and the organization allowed VPS13 proteins to mediate lipid transfer from the ER to the other organelles (Melia and Reinisch, 2022). Based on a prediction algorithm for the FFAT motif reported in previous studies (Murphy and Levine, 2016; Slee and Levine, 2019), seven putative FFAT motifs were found in the N terminal portion of VPS13B (residues 1-1500) (Figure S1A). However, the N terminal fragment of VPS13B containing these FFAT motifs did not substantially associate with the ER adaptors, such as VAPA (Figure S1B), VAPB (Figure S1C), or MOSPD2 (Figure S1D), in cells without OA stimulation, suggesting that VPS13B may not be able to stably target the ER via FFAT or Phospho-FFAT.
In addition, we asked whether VPS13B overexpression could enhance the interaction between the ER and the Golgi, the latter of which VPS13B localized on. Consistently, expression of VPS13B^sfGFP alone appeared not to result in a strong tethering of the ER to the Golgi, as revealed by the colocalization analysis between these two organelles (Figure S1E and F). Importantly, it should be noted that our results did not exclude the possibility that VPS13B was transiently associated with the ER in the form of ER-Golgi-LD tripartite contacts. Alternatively, VPS13B might be recruited to the ER (or ER subdomains) via unknown adaptors, other than VAPs or MOSPD2.
A growing body of evidence showed that VPS13-containing lipid transporters might be responsible for organelle biogenesis (Melia and Reinisch, 2022). For instance, bridge transporter Atg2 was required for autophagosome biogenesis (Osawa et al., 2019; Valverde et al., 2019). Yeast Vps13 was required for the prospore formation in yeast (Park and Neiman, 2012; Park et al., 2013) and strains lacking the protein failed to sporulate (Enyenihi and Saunders, 2003). Vps13B was essential for acrosome biogenesis during sperm development (Da Costa et al., 2020). Vps13D was suggested to be necessary for peroxisome biogenesis (Baldwin et al., 2021).
Since our results suggested that the ER might be involved in VPS13B-marked MCSs, the mechanisms through which VPS13B recognized the ER membrane remained to be dissected. The following question was whether and how VPS13B transferred lipid across MCSs? The C-terminus of VPS13B is reported to mediate its Golgi localization (Seifert et al., 2011). Therefore, membrane targeting ability of VPS13B appears to be solely attributed to its C-terminus. Since VPS13 proteins (VPS13A/C/D) were bridge-like lipid transporters that anchored the ER on the N-terminus and targeted the other organelles via the C-terminus (Melia and Reinisch, 2022), VPS13B may be distinct from its VPS13 paralogs regarding the lipid transfer mode. Nevertheless, it is possible that VPS13B is transiently associated with the ER via FFAT motifs at its N-terminus; alternatively, there may be an unknown element at the N-terminus of VPS13B that is responsible for targeting VPS13B to the ER.
Furthermore, does VPS13B promote the growth of the Golgi, LDs, or other membrane structures in the ER? These questions are required to be extensively addressed in future. Loss-of-function mutations of VPS13B lead to Cohen disease, a complex syndrome featured by global developmental delay and intellectual disability (Kolehmainen et al., 2004), and the answers to these questions will provide new mechanistic insights on this disease.
Methods and Materials
Plasmids and siRNA Oligonucleotides
Human VPS13B ORF was cloned from HeLa cDNA, and fully sequenced (NM152564.5. isoform1). The internal sfGFP tagged VPS13B (VPS13B^sfGFP) was generated by inserting sfGFP sequence after V1265 of VPS13B in the frame. All truncated VPS13B mutants used in this study were generated by PCR using VPS13B^sfGFP as template and cloned it into mGFP-N1 (addgene 54767) or mGFP-C1 (addgene 54579). pAcGFP-Golgi was bought from Clontech (Cat. No.632464). Halo-Rab6 was generated by inserting Rab6A coding sequence obtained by PCR using HeLa cDNA as template into Halo-C1 vector. mito-BFP was a gift from Gia Voeltz (Addgene 49151). Halo-VAPA/VPAB/Sec61β(Sec61)/MOSPD2 were generated by inserting the coding sequences obtained by PCR from HeLa cDNA as templates into Halo-C1 vector. All of constructs used in this study were generated by using ClonExpress®II One Step Cloning kit (Vazyme, C112-01).
Oligonucleotides for human VPS13B siRNAs were synthesized by RiboBIO (Guangzhou, CN) against target sequence 5′-GACCTTACTTGTCATAATA-3′ (siRNA#1); 5′-GCCTATGTTTATTCGTATA-3′ (siRNA#2); As a control, scrambled siRNA 5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′ (RiboBIO) was used.
Antibodies and Reagents
Anti-Tubulin (100109-MM05 T; Sinobiological) was used at 1:1000 dilutions for Western blot. Anti-VPS13B (HPA043865; Sigma), anti-GM130 (11308-1-AP; Proteintech) antibodies were used 1:100 for IF. The following reagents were used in this study: OA (O1008; sigma); BODIPY 558/568 (ThermoFisher, D3835), and antibiotics such as G418 (10131027) and puromycin (A1113803) were obtained from Thermofisher. All EM reagents were purchased from Electron Microscopy Sciences.
Cell Culture and Transfection
Human embryonic kidney 293 cells (ThermoFisher, R70507) were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin. All of the cell lines used in this study are free of mycoplasma contamination.
For transfection, cells were seeded at 4 × 105 cells per well in a six-well dish 16 h before transfection. Plasmid transfections were performed in OPTI-MEM (Invitrogen) with 2 µl Lipofectamine 2000 per well for 6 h, followed by trypsinization and replating onto glass-bottom confocal dishes at 3.5 × 105 cells per well. Cells were imaged in live-cell medium (DMEM with 10% FBS and 20 mM Hepes no antibiotics) 16–24 h after transfection. For all transfection experiments in this study, the following amounts of DNA were used per 3.5 cm well (individually or combined for cotransfection): 1000 ng for VPS13B^sfGFP; 500 ng for truncated VPS13B mutations; 500 ng for pAcGFP-Golgi; 50 ng for Halo-Rab6 or Rab6 mutants; and 500 ng for mito-BFP.
BODIPY 558/568 Staining in Live Cell
Cells were washed once with PBS and were changed to complete medium containing 3 μM BODIPY558/568 and incubated at 37°C for the indicated period of time. Cells were washed with PBS three times and changed to imaging medium (DMEM supplemented with 10% FBS and 20 mM Hepes without phenol red) prior to imaging.
Halo Staining in Live Cell
Cells were incubated with complete medium with 5 nM Janilia Fluo® 646 HaloTag® Ligand for 30 min. Cells were washed three times with complete medium to remove extra ligands, followed by incubation for another 30 min. Medium was replaced with imaging medium to remove unconjugated Halo ligands that have diffused out of the cells prior to imaging.
Live Imaging by Confocal Microscopy
Cells were grown on glass-bottom confocal dishes. Confocal dishes were loaded to a laser scanning confocal microscope (LSM780, Zeiss, Germany) equipped with multiple excitation lasers (405 nm, 458 nm, 488 nm, 514 nm, 561 nm, and 633 nm) and spectral fluorescence GaAsP array detector. Cells were imaged with the 63× 1.4-NA iPlan-Apochromat 63× oil objective using the 405-nm laser for BFP, 488-nm for GFP, 561-nm for mStrawberry, OFP, tagRFP, or mCherry. Cells were imaged in live cell chamber supplied with 5% CO2 at 37°C.
High-Resolution Airyscan Live Cell Imaging
Cells on confocal dishes were loaded to a laser scanning confocal microscope (LSM900, Zeiss, Germany) with Airyscan2 in equipped with ≥ 32 GaAsP-PMT array detector, 63×/1.4 oil objective (working distance 190 μm), four lasers (405 nm 5 mW, 488 nm 5 mW, 561 nm 5 mW, and 640 nm 5 mW) with corresponding filters. Cells were imaged with 405-nm laser for BFP, 488-nm for GFP, 561-nm for OFP, tagRFP, or mCherry and 640-nm for Janilia Fluo® 646 HaloTag® Ligand.
Live Cell Imaging by Leica SP8 Equipped with Lightning Super-Resolution Module
Cells on confocal dishes were loaded to Leica SP8 equipped with lightning super-resolution module equipped with HC PL APO CS2 100×/1.4 oil objective, four lasers (415 nm, 499 nm, 567 nm, and 662 nm) with corresponding filters. Cells were imaged in optiMEM with Hepes buffer in live cell chamber supplied with 5% CO2 at 37°C.
Immunofluorescence Staining
Cells were fixed with 4% PFA (paraformaldehyde, Sigma) in PBS for 10 min at room temperature. After washing with PBS three times, cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min on ice. Cells were then washed three times with PBS, blocked with 0.5% BSA in PBS for 1 h, incubated with primary antibodies in diluted blocking buffer overnight, and washed with PBS three times. Secondary antibodies were applied for 1 h at room temperature. After washing with PBS three times, samples were mounted on Vectashield (H-1000; Vector Laboratories).
Electron Microscopy
Oleic acid–stimulated HEK293 cells ectopically expressing VPS13B^sfGFP were fixed with 2.5% glutaraldehyde in 0.1 M Phosphate buffer, pH7.4 for 2 h at room temperature. After washing three times with 0.1 M phosphate buffer, cells were scraped and collected with 0.1 M phosphate buffer followed by centrifugation at 3000 rpm. The pellet was resuspended in PBS (0.1 M), and centrifuged at 3000 rpm for 10 min. This step was repeated three times. The samples were post-fixed with pre-cold 1% OsO4 in 0.1 M Phosphate buffer for 2-3 h at 4°C, followed by rinsing with PBS 3 times (3 × 20 min). The samples were dehydrated in graded ethanol (50%, 70%, 85%, 90%, 95%, and 2 × 100%) with 15 min for each condition. The penetrations were performed in an order of acetone-epoxy (2:1); acetone-epoxy (1:1); epoxy. Each round of penetration was performed at 37°C for 12 h. The samples were embedded in epoxy resin using standard protocols. Sections parallel to the cellular monolayer were obtained using a Leica EM UC7 with a thickness of 60–100 nm and examined under HT7800/HT7700. Golgi apparatus, the ER, mitochondria, and LDs were identified based on their respective morphology and were traced by hand.
Image Analysis
All image analysis and processing were performed using ImageJ (National Institutes of Health). MCSs were automatically identified by colocalization plugin with overlapping pixels representing potential MCSs. LD or Golgi number was measured manually with the assistance of an ImageJ plugin Cell Counter.
Statistical Analysis
All statistical analyses and p-value determinations were performed in GraphPad Prism6. All the error bars represent mean ± SD. To determine p-values, ordinary one-way ANOVA with Tukey's multiple comparisons test was performed among multiple groups, and a two-tailed unpaired Student t-test was performed between two groups.
Supplemental Material
Video 1: 3D reconstructions of Golgi-LD interactions were rotating along z-axis in HEK293 cell cultured in OA medium expressing VPS13BsfGFP (green) and Halo-Rab6 (Golgi, blue), stained with BODIPY 558/568 (LD, magenta). A stack of 31 z-planes with 0.2μm thickness in each stack was taken.
Video 2: Dynamics of Golgi-LD interactions in HEK293 cell expressing VPS13BsfGFP (green) and Halo-Rab6 (Golgi, blue), stained with BODIPY 558/568 (LD, magenta). Time in sec. Scar bar: 2 μm.
Supplemental material, sj-docx-3-ctc-10.1177_25152564231195718 for A Possible Role of VPS13B in the Formation of Golgi-Lipid Droplet Contacts Associating with the ER by Yuanjiao Du, Xuewen Hu, Weiping Chang, Lin Deng, Wei-Ke Ji and Juan Xiong in Contact
Acknowledgements
Majority of the data reported in the manuscript was shown in a study as a BioRxiv preprint (2020.12.16.423147v1).
Author Contributions: Y. Du, X. Hu, W. Chang, L. Deng, W. Ji, and J. Xiong conceived the project and designed the experiments. Y. Du, X. Hu, W. Ji, and J. Xiong performed the experiments. Y. Du, W. Ji, and J. Xiong analyzed and interpreted the data. W. Ji prepared the manuscript with inputs and approval from all authors.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Data and Materials Availability: All the data and relevant materials, including reagents and primers, that supports the findings of this study are available from the corresponding author upon reasonable request.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: W. Ji was supported by National Natural Science Foundation of China (32122025; 91854109). J. Xiong was supported by National Natural Science Foundation of China (81901166).
ORCID iDs: Weiping Chang https://orcid.org/0000-0002-2547-8030
Wei-Ke Ji https://orcid.org/0000-0003-1788-0879
Supplemental Material: Supplemental material for this article is available online.
References
- Abrisch RG, Gumbin SC, Wisniewski BT, Lackner LL, Voeltz GK. (2020). Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J Cell Biol 219. 10.1083/jcb.201911122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding AL, Wang C, Chang TK, Sliter DA, Powers CM, Hofmann K, Youle RJ, Baehrecke EH. (2018). Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr Biol 28, 287–295.e6. 10.1016/j.cub.2017.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin HA, Wang C, Kanfer G, Shah HV, Velayos-Baeza A, Dulovic-Mahlow M, Bruggemann N, Anding A, Baehrecke EH, Maric D, et al. (2021). VPS13D promotes peroxisome biogenesis. J Cell Biol 220. 10.1083/jcb.202001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M, Schuldiner M. (2018). Stepping outside the comfort zone of membrane contact site research. Nat Rev Mol Cell Biol 19, 483–484. 10.1038/s41580-018-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa R, Bordessoules M, Guilleman M, Carmignac V, Lhussiez V, Courot H, Bataille A, Chlemaire A, Bruno C, Fauque P, et al. (2020). Vps13b is required for acrosome biogenesis through functions in Golgi dynamic and membrane trafficking. Cell Mol Life Sci 77, 511–529. 10.1007/s00018-019-03192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wang J, Xiong J, Fang N, Ji WK. (2021). VPS13D Interacts with VCP/p97 and negatively regulates endoplasmic reticulum-mitochondria interactions. Mol Biol Cell 32, 1474–1486. 10.1091/mbc.E21-03-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M. (2016). A tether is a tether is a tether: Tethering at membrane contact sites. Dev Cell 39, 395–409. 10.1016/j.devcel.2016.10.022. [DOI] [PubMed] [Google Scholar]
- Elbaz Y, Schuldiner M. (2011). Staying in touch: The molecular era of organelle contact sites. Trends Biochem Sci 36, 616–623. 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Enyenihi AH, Saunders WS. (2003). Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163, 47–54. 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Levine TP. (2017). Piecing together the patchwork of contact sites. Trends Cell Biol 27, 214–229. 10.1016/j.tcb.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Guillen-Samander A, Leonzino M, Hanna MG, Tang N, Shen H, De Camilli P. (2021). VPS13D bridges the ER to mitochondria and peroxisomes via Miro. J Cell Biol 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen-Samander A, Wu Y, Pineda SS, Garcia FJ, Eisen JN, Leonzino M, Ugur B, Kellis M, Heiman M, De Camilli P. (2022). A partnership between the lipid scramblase XK and the lipid transfer protein VPS13A at the plasma membrane. Proc Natl Acad Sci U S A 119, e2205425119. 10.1073/pnas.2205425119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna MG, Suen PH, Wu Y, Reinisch KM, De Camilli P. (2022). SHIP164 is a chorein motif lipid transfer protein that controls endosome-Golgi membrane traffic. J Cell Biol 221. 10.1083/jcb.202111018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Jahn R. (2019). SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat Commun 10, 1608. 10.1038/s41467-019-09617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolehmainen J, Wilkinson R, Lehesjoki AE, Chandler K, Kivitie-Kallio S, Clayton-Smith J, Traskelin AL, Waris L, Saarinen A, Khan J, et al. (2004). Delineation of Cohen syndrome following a large-scale genotype-phenotype screen. Am J Hum Genet 75, 122–127. 10.1086/422197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, Wheeler H, Reinisch KM, De Camilli P. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217, 3625–3639. 10.1083/jcb.201807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AB, John Peter AT, Walter P, Kornmann B. (2015). ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol 210, 883–890. 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Reinisch KM. (2020). Inter-organelle lipid transfer: A channel model for Vps13 and chorein-N motif proteins. Curr Opin Cell Biol 65, 66–71. 10.1016/j.ceb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SC, Uchiyama LF, Nunnari J. (2016). ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549. 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia TJ, Reinisch KM. (2022). A possible role for VPS13-family proteins in bulk lipid transfer, membrane expansion and organelle biogenesis. J Cell Sci 135. 10.1242/jcs.259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Levine TP. (2016). VAP, a Versatile access point for the endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta 1861, 952–961. 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Neuman SD, Levine TP, Bashirullah A. (2022). A novel superfamily of bridge-like lipid transfer proteins. Trends Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, Ohsumi Y, Noda NN. (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol 26, 281–288. 10.1038/s41594-019-0203-4. [DOI] [PubMed] [Google Scholar]
- Park JS, Hu Y, Hollingsworth NM, Miltenberger-Miltenyi G, Neiman AM. (2022). Interaction between VPS13A and the XK scramblase is important for VPS13A function in humans. J Cell Sci. 10.1242/jcs.260227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Neiman AM. (2012). VPS13 Regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J Cell Sci 125, 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Okumura Y, Tachikawa H, Neiman AM. (2013). SPO71 Encodes a developmental stage-specific partner for Vps13 in Saccharomyces cerevisiae. Eukaryot Cell 12, 1530–1537. 10.1128/EC.00239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Toulmay A, Balla T. (2020). The functional universe of membrane contact sites. Nat Rev Mol Cell Biol 21, 7–24. 10.1038/s41580-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnoczky G, Kornmann B, Lackner LL, Levine TP, Pellegrini L, Reinisch K, et al. 2019). Coming together to define membrane contact sites. Nat Commun 10, 1287. 10.1038/s41467-019-09253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert W, Kuhnisch J, Maritzen T, Horn D, Haucke V, Hennies HC. (2011). Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J Biol Chem 286, 37665–37675. 10.1074/jbc.M111.267971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert W, Kuhnisch J, Maritzen T, Lommatzsch S, Hennies HC, Bachmann S, Horn D, Haucke V. (2015). Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J Biol Chem 290, 3349–3358. 10.1074/jbc.M114.608174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JL, Fortier TM, Wang R, Baehrecke EH. (2021). Vps13D functions in a Pink1-dependent and Parkin-independent mitophagy pathway. J Cell Biol 220. 10.1083/jcb.202104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee JA, Levine TP. (2019). Systematic prediction of FFAT motifs across eukaryote proteomes identifies nucleolar and eisosome proteins with the predicted capacity to form bridges to the endoplasmic reticulum. Contact (Thousand Oaks) 2, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur B, Hancock-Cerutti W, Leonzino M, De Camilli P. (2020). Role of VPS13, a protein with similarity to ATG2, in physiology and disease. Curr Opin Genet Dev 65, 61–68. 10.1016/j.gde.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J. (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ. (2019). ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798. 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco AP. (2004). Analysis of the human VPS13 gene family. Genomics 84, 536–549. 10.1016/j.ygeno.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Fang N, Xiong J, Du Y, Cao Y, Ji WK. (2021). An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D-TSG101 interactions. Nat Commun 12, 1252. 10.1038/s41467-021-21525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Carvalho P, Voeltz GK. (2018). Here, there, and everywhere: The importance of ER membrane contact sites. Science 361. 10.1126/science.aan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1: 3D reconstructions of Golgi-LD interactions were rotating along z-axis in HEK293 cell cultured in OA medium expressing VPS13BsfGFP (green) and Halo-Rab6 (Golgi, blue), stained with BODIPY 558/568 (LD, magenta). A stack of 31 z-planes with 0.2μm thickness in each stack was taken.
Video 2: Dynamics of Golgi-LD interactions in HEK293 cell expressing VPS13BsfGFP (green) and Halo-Rab6 (Golgi, blue), stained with BODIPY 558/568 (LD, magenta). Time in sec. Scar bar: 2 μm.
Supplemental material, sj-docx-3-ctc-10.1177_25152564231195718 for A Possible Role of VPS13B in the Formation of Golgi-Lipid Droplet Contacts Associating with the ER by Yuanjiao Du, Xuewen Hu, Weiping Chang, Lin Deng, Wei-Ke Ji and Juan Xiong in Contact