Abstract
Methicillin resistance in staphylococci is mediated by PBP2a, a penicillin binding protein with low affinity for β-lactam antibiotics. The gene encoding PBP2a, mecA, is transcriptionally regulated in some clinical isolates by mecR1 and mecI, genes divergently transcribed from mecA that encode a signal transducer and repressor, respectively. The biochemical basis of MecI-mediated mecA transcriptional repression was investigated by using purified MecI. In DNase I protection studies, MecI protected a 30-bp palindrome encompassing the predicted mecA −10 and the mecR1 −35 promoter sequences. The larger palindrome contained 15 bp of dyad symmetry within which was a smaller 6-bp palindrome. Electrophoretic mobility shift assays established a requirement for the entire 15-bp half-site for initial repressor binding. Fragments containing the 30-bp palindrome and the entire mecA-mecR1 intergenic region were retarded in gels as multiple discrete bands varying in molecular size, characteristic of cooperative DNA binding. Glutaraldehyde cross-linking confirmed oligomerization of repressor in solution. A naturally occurring MecI mutant (MecI*; D39G) repressed mecA transcription sixfold less well than the wild type in vivo. Although MecI* protected the same target sequences and exhibited similar gel shift patterns to MecI, 5- to 10-fold more protein was required. MecI* exhibited defective oligomerization in solution, suggesting that the MecI amino terminus is important in protein-protein interactions and that protein oligomerization is necessary for optimum repression.
Methicillin resistance in staphylococci is mediated by a novel penicillin binding protein, PBP2a, that has low affinity for β-lactam antibiotics (5). In resistant isolates, PBP2a provides a β-lactam-insensitive transpeptidation activity that is crucial for the synthesis of an intact cell wall (8, 15). PBP2a is encoded by mecA, a gene residing on a 30- to 50-kb DNA fragment (mec DNA [14]) unique to methicillin-resistant isolates that is inserted at a specific chromosomal site. Genetic and molecular analyses of mec DNA have revealed that some isolates contain two additional genes, mecR1 and mecI, located upstream of mecA and divergently transcribed from it (13). Based on structural and functional homologies of mecR1 and mecI to blaR1 and blaI (penI), sequences known to regulate β-lactamase production in Staphylococcus aureus and Bacillus licheniformis, mecI and mecR1 are predicted to encode a repressor and a signal transducer with antirepressor activity, respectively (15). Structure-function and repressor target studies have been performed with the β-lactamase repressors PenI (B. licheniformis [24, 25]) and BlaI (S. aureus [9]). These repressors have 31% predicted amino acid homology to each other and 41 and 60% homology to MecI (15). Both PenI and BlaI bind to two distinct operator regions. One of the PenI operators contains the β-lactamase structural gene (penP) promoter, and the other contains the promoter for the divergently transcribed regulatory gene. The two operators are 154 bp apart. In contrast, the BlaI operators are separated by only 7 bp, and one of the two repressor binding sites contains no predicted promoter sequences. The operators that bind either PenI or BlaI contain regions of dyad symmetry. The amino terminus of each protein has been shown to be essential for operator binding, while the carboxy terminus is needed for dimerization of PenI and is required for optimum binding of BlaI (25). Dimerization of S. aureus BlaI has not been demonstrated. Dimerization of PenI predicts cooperative interactions of proteins that bind at independent operator sites.
The signal transducer/inducer molecule, BlaR, has also been characterized in some detail (10). It is a 601-amino-acid protein containing an extramembrane portion at the carboxy terminus that binds β-lactam antibiotics through a characteristic serine active site, four transmembrane segments, and a cytoplasmic domain with a metal binding motif. The cytoplasmic domain has been predicted to have metallopeptidase activity and to inactivate BlaI by proteolytic cleavage following β-lactam induction. Cleavage of BlaI has been demonstrated in extracts of S. aureus cells following induction (9). Considerable homology exists between S. aureus BlaR1 and MecR1. There is 44% amino acid homology between the β-lactam sensor portion of the two molecules and 30% homology of the signal transduction domains. In addition, there is conservation of structural motifs (10, 15).
Studies such as those described for BlaI and PenI have not been performed with MecI. Allelic replacement mutagenesis studies of mecI have demonstrated its role in tight transcriptional regulation of both mecA and its gene product, PBP2a (20), but there has been no MecI structure-function analysis or identification of repressor binding sites. In this study, we characterized repressor-target interactions of MecI with sequences in the intergenic region between mecA and mecR1. We examined both a purified active repressor protein and a naturally occurring mutant that was defective in mecA transcriptional regulation. Each was from a clinical S. aureus isolate.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The S. aureus and Escherichia coli strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristic and/or genotypea | Comments (reference or source) |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Restriction-deficient mutagenized RN450 | Shuttle plasmid host (16) |
| 450M | Cms Ems Gms Mcr (HE) Tcs Bla− | RN450 transformed with COL mec region DNA (1) |
| N315P | Cms Emr Gms Mcr (HE) Tcs Bla− | Intact mecR1-mecI; clinical isolate from Japan (13) |
| 67-0 | Mcr Emr (HE) Bla− | Clinical isolate from California, 1984 (5) |
| E. coli | ||
| TB1 | recA+lacIqlacZΔM15 | Host for pUC vectors (26) |
| DH5α | F−recA1 hsdR17 φ80dlacZM15Δ(lacZYA-argF) endA1 deoR gyr A96 thi-1 relA supE44 | Recombination-deficient host for pUC derivatives (Gibco) |
| Plasmids | ||
| pUC19 | Apr; 2.7 kb | General-purpose cloning vector (26) |
| pGEX-2T | Apr; 4.8 kb | Vector for the expression of GST-protein fusions (Pharmacia) |
| pSK950 | Spr Tcr Emr ts; 10.4 kb | E. coli-S. aureus shuttle vector for integration of sequences into the S. aureus lipase gene with φL54a att site. The original pCL84 plasmid was modified by addition of pE194 at the PstI site (17). |
| pG0630 or pG0631 | Spr Tcr Emr ts; 16.4 kb | A 2.4-kb fragment from N315P(pG0630) or 67-0(pG063), containing the mecR1-mecI intergenic region with divergent mecR1-mecI and mecA promoters, and the first 50 bases of the mecA coding sequence cloned at EcoRI-BamHI sites of pSK950. A 3.3-kb fragment containing promoterless lacZ was then cloned at the BamHI site to generate a mecA-lacZ transcriptional fusion (this study). |
Abbreviations: Ap, ampicillin; Bla, β-lactamase; Cm, chloramphenicol; Em, erythromycin; Gm, gentamicin; HE, heterotypic; Mc, methicillin; MCS, multiple cloning site; Sp, spectinomycin; Tc, tetracycline; ts, temperature sensitive.
Materials and media.
Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, Md.) was used for the subculture and maintenance of E. coli and S. aureus strains and transformants. Luria broth (LB) was used for culture of E. coli strains, Trypticase soy broth and brain heart infusion were used for culture of S. aureus, and agar-agar, at a concentration of 1.5%, was added to prepare solid media from these broths; all were purchased from Difco (Detroit, Mich.). The antibiotics and concentrations used were as follows: ampicillin (50 μg/ml) or tetracycline (3 μg/ml) for initial selection of E. coli strains following electroporation; and erythromycin (10 μg/ml) or tetracycline (3 μg/ml) for initial selection of S. aureus strains following electroporation or transduction. All antibiotics and chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). Lysostaphin was obtained from AMBI, Inc. (Tarrytown, N.Y.). Restriction endonucleases and other enzymes were purchased from New England Biolabs (Beverly, Mass.).
Cloning, transformation, and DNA manipulation.
All restriction endonuclease digestions and ligation reactions were performed according to the manufacturers’ directions. Recombinant plasmids were introduced into E. coli and S. aureus by electroporation with a Gene Pulser (Bio-Rad, Richmond, Calif.) at settings of 200 Ω, 25 μF, and 2.5 kV for E. coli and 400 Ω, 25 μF, and 2.5 kV for S. aureus. Electrocompetent cells for E. coli and S. aureus were prepared by the methods described in the Gene Pulsar manual and by Luchanscky et al. (18), respectively. The alkaline lysis procedure described by Hittori and Sakaki (12) was used to screen and to isolate plasmid DNA suitable for restriction digestion, ligation, or electroporation. Isolation of plasmid DNA from S. aureus was accomplished by the hexadecyltrimethylammonium bromide extraction method (21). A Qiagen (Chatsworth, Calif.) Midi-prep kit was used to obtain large quantities of purified plasmid DNA.
Transduction.
S. aureus transducing phage 80α was propagated on S. aureus RN4220 containing plasmids of interest by a modification of the technique of Thompson and Patee (21) as previously described (20). The harvested phage was used to transduce S. aureus.
Determination of β-galactosidase activity.
β-Galactosidase activity was determined as described previously (23). The values were standardized by determining total protein concentration from each lysate analyzed for enzyme activity and expressed as units per milligram of protein.
Northern blot and PBP analyses.
Procedures used for detection of mecA transcript have been described previously (20). PBPs were analyzed by Michael Pucci at the Bristol-Myers Squibb Pharmaceutical Research Institute, using methods previously described (20). Transcript and protein abundance on gels were quantified by scanning densitometry using an AlphaImager 1000 digital imaging system (Alpha Innotech Corp., San Leandro, Calif.).
Purification of MecI and MecI* repressor proteins.
A 372-bp fragment containing the mecI or mecI* coding sequence was isolated by PCR amplification, resolved on a 2% agarose gel, and recovered by electroelution (International Biotechnologies, Inc., New Haven, Conn.) and ethanol precipitation. These fragments were cloned into the expression vector pGEX-2T (Pharmacia LKB Biotechnology, Piscataway, N.J.), and the appropriate translational fusion between the 3′ end of the gene encoding glutathione S-transferase (GST) and the 5′ end of mecI or mecI* was confirmed by DNA sequencing. These plasmids were designated pG0610 (pGEX-2T/mecI) and pG0611 (pGEX-2T/mecI*). An overnight, 20-ml culture of E. coli containing either of these two plasmids was added to 2 liters of LB containing ampicillin at 50 μg per ml, and the cells were grown with vigorous shaking to an A660 of 0.8. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.08 mM to induce transcription from the tac promoter, and the culture was incubated for additional 4 h. The cells were harvested, frozen at −70°C, thawed, and resuspended in 100 ml of cold SET buffer (0.2 M NaCl, 0.001 M Na2EDTA, 0.2 M Tris-HCl [pH 7.4]) containing 10 mg of lysozyme and phenylmethylsulfonyl fluoride at a final concentration of 0.005 M. The cells were incubated in an ice-water bath for 30 min and were lysed by the addition of N-lauroyl sodium sarcosinate, prepared in SET buffer, to a final concentration of 0.1%. The lysed sample was subjected to sonication for 1 min to reduce its viscosity. Triton X-100 was then added to a final concentration of 1%, and the sample, while maintained at 4°C, was shaken intermittently for 30 min. Cellular debris was removed by centrifugation at 14,000 × g for 10 min, and the supernatant was mixed at 4°C for several hours with 2 ml of 50% glutathione-Sepharose beads (Pharmacia LKB Biotechnology). After absorption, the beads were collected by centrifugation at 500 × g for 5 min and washed three times with 100 ml of phosphate-buffered saline (0.01 M Na2HPO4, 0.018 M KH2PO4, 0.14 M NaCl, 0.027 M KCl [pH 7.3]). These beads were resuspended in 2 ml of phosphate-buffered saline containing 400 μg of thrombin and incubated at room temperature for 16 h with gentle shaking. The supernatant, containing the thrombin-cleaved material, was recovered by centrifugation at 500 × g and concentrated by using Centricon-3 concentrator columns (Amicon, Inc., Beverly, Mass.). The purity of the concentrated material was checked by analyzing samples, boiled for 5 min in sodium dodecyl sulfate (SDS) reducing buffer, on an SDS–12% polyacrylamide gel. The conditions for polyacrylamide gel electrophoresis (PAGE), staining, and destaining were as described in the manual for the Mini-PROTEAN II electrophoresis cell (Bio-Rad). The concentration of the purified material was determined by comparing the intensity of the Coomassie blue-stained band with that of a band containing a known quantity of a protein of an equivalent size.
EMSA.
The electrophoretic mobility shift assay (EMSA) used was based on the method of Chodosh (6). Six different DNA target fragments were used in this assay. A 210-bp DNA target fragment, containing a 30-bp palindrome and the promoter-operator sequences for mecR1-mecI and mecA, 50 bp of mecA at its 5′ end, and 50 bp of mecR1 at its 3′ end was isolated by PCR amplification. A HindIII site, built into the 3′ end of this fragment, was cleaved with HindIII. The 3′ recessed end thus generated was filled in with Klenow enzyme in the presence of unlabeled dATP, dGTP, dTTP, and [32P]dCTP (NEN/Du Pont Company, Boston, Mass.). Five additional target fragments were generated by annealing oligonucleotides with the sequences shown below to their reverse complements. The first oligonucleotide, 5′ TCGGTAACTACATTTGTAGTGATCCT 3′, contained the 15-base left arm of the 30-base palindrome; a 6-base inverted repeat (underlined) is located within this arm. Second, an oligonucleotide with the sequence 5′ TCGGTAATATTACAAATGTAGTATGATCCT 3′ contained the 15-base right arm of the 30-base palindrome. The third and fourth oligonucleotides, with the sequences 5′ GAGCTCGGTAACTACATGATCCTCTAG 3′ and 5′ GAGCTCGGTAATATTACAAGATCCTCTAG 3′, represented the left and right halves, respectively, of the 6-base inverted repeats that are present in the second oligonucleotide fragment. Finally, an oligonucleotide with the sequence 5′ TCGGTAACTACATTTGTAGTATATTACAAATGTAGTGATCCT 3′ contained the entire 30-base palindrome (underlined). The double-stranded oligonucleotide fragments were labeled at the 5′ hydroxyl termini in a forward exchange reaction using [γ-32P]ATP and T4 polynucleotide kinase. The end-labeled fragments were purified from unincorporated nucleotides and other reaction components by using a QIA quick PCR purification kit (Qiagen, Santa Clarita, Calif.). The DNA binding reaction mixture contained, in a total volume of 30 μl, about 4,000 cpm of labeled target fragment, DNA binding buffer [0.012 M HEPES (pH 7.8), 0.06 M KCl, 8 μg of bovine serum albumin, 1 μg of poly(dI-dC) · poly(dI-dC), 12% glycerol], and various concentrations of MecI or MecI*. After 20 min of incubation at 30°C, the reaction mixture was loaded on a 4% nondenaturing polyacrylamide gel. The gel was subjected to electrophoresis in TGE (0.05 M Tris base, 0.38 M glycine, 0.002 M Na2EDTA [pH 8.5]), chilled to 4°C, at 185 V until the tracking dye reached the bottom of the gel. The gel was dried and autoradiographed.
DNase I footprinting.
The 210-bp DNA target fragment used in EMSA was end labeled by filling in a HindIII-generated recess at its 5′ or 3′ end. About 80,000 cpm of the purified, end-labeled fragment was incubated with various amounts of repressor protein in a total volume of 60 μl. The binding assay was set up as for the EMSA. After 20 min of incubation at 30°C, the reaction mixture was treated with 0.006 U of DNase I. The DNase I treatment and localization of repressor-protected regions were performed according to the procedure described previously (23).
PCR amplification and DNA sequencing.
Oligonucleotide primers with or without added restriction sites were synthesized (Oligos Etc., Wilsonville, Oreg.) for use in amplification of DNA by PCR. The annealing of primer to template and its subsequent amplification by Vent polymerase (New England Biolabs) were carried out under the conditions recommended by the manufacturer. The primers used and the sizes of the resulting PCR fragments were as follows. Amplification of the 2.4-kb mecR1-mecI fragment from N315P and 67-0 plus 56 bp of mecA was performed with the mecA primer 5′ CGCGGATCCCAAACCCGACAACTAC 3′ and the mecI downstream primer 5′ CCGGAATTCGCATATGGATTTCACTGGTG 3′. Amplification of the 442-bp fragment containing the mecI (N315P) or mecI* (67-0) coding sequence and 70 bp of flanking DNA was performed with the forward primer 5′ CGGATCCAGAAATGGAATTAATATAATG 3′ and the reverse primer 5′ CGGAATTCGACTTGATTGTTTCCTC 3′. The primers used in generating these fragments were selected from the published sequence for mecR1-mecI, mecA, and the intervening promoter-operator region. These sequences are accessible in GenBank. The PCR-generated fragments were sequenced directly or were sequenced after cloning into appropriate vectors to confirm that no mutations were introduced into these fragments during the amplification process. DNA sequencing was performed by the Sanger dideoxynucleotide chain termination method using a Sequenase version 2 kit (United States Biochemicals, Cleveland, Ohio) or by using a Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) at the Virginia Commonwealth University Nucleic Acid Synthesis and Analysis Core Facility, using an ABI 377 DNA sequencer.
Glutaraldehyde cross-linking of MecI and MecI*.
MecI or MecI* (3 μg) was incubated with various concentrations of the bifunctional cross-linking reagent glutaraldehyde (J. T. Baker, Phillipsburg, N.J.) in 10 μl of a reaction mixture containing 0.05 M NaH2PO4, 0.05 M NaCl, and 10% glycerol as described by Artsimovich and Howe (2). After incubation for 60 min at 37°C, the reaction was stopped by addition of equal volumes of SDS and reducing buffer. The samples were boiled for 5 min in a water bath and analyzed by SDS-PAGE on a 10% polyacrylamide gel. The gel was stained with Coomassie blue.
RESULTS
Analysis of mecA transcription in S. aureus N315P and 67-0.
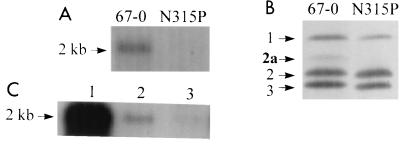
Northern blot analysis of the mecA transcript in clinical S. aureus isolate 67-0 (5) revealed easily detectable transcript, sixfold more abundant than the barely detectable transcript seen in N315P, a clinical isolate previously shown to contain a mecI gene that produced wild-type, active repressor (Fig. 1A and reference 20). The amount of PBP2a in membranes was similarly greater in 67-0 than in N315P (Fig. 1A). The nucleotide sequence of all 67-0 DNA 5′ to the mecA translational start site was determined and found to be identical to that of N315P with the exception of a single nucleotide change at position 117 of the 372-bp mecI coding sequence, resulting in a D-to-G change at position 39 in the predicted MecI amino acid sequence. To confirm that the 67-0 MecI mutation resulted in increased mecA transcription, both 67-0 and N315P regulators were assessed in the same genetic background. Regulatory sequences were PCR amplified by using primers 50 bp into mecA and just 3′ to mecI as previously described (20). The sequences were ligated into pSK950, an E. coli-S. aureus shuttle vector derived from pCL84 (17, 20). This vector contains an E. coli (ColE1) and a temperature-sensitive S. aureus (pE194ts) replication origin; resistance genes for selecting spectinomycin resistance in E. coli (aad3"), tetracycline resistance in both S. aureus and E. coli (tet[K]), and erythromycin resistance in S. aureus (ermC); and the att site for phage L54a. When this plasmid is introduced into an S. aureus strain containing a separate plasmid encoding the φL54a integrase gene (pL150), it integrates into the chromosome at the corresponding att site located within the lipase gene (geh) when the cells are grown at the nonpermissive temperature for plasmid replication (42°C). Following ligation of the mecA regulator and promoter-operator sequences into pSK950, a fragment containing the E. coli lacZ gene preceded by a Bacillus subtilis ribosomal binding site was introduced downstream of the mecA gene remnant, producing a mecA-lacZ transcriptional fusion. The plasmid containing the fusion regulated by the 67-0 mutant sequence was designated pG0631, while the plasmid containing wild-type N315P sequences was designated pG0630 (Table 1). Shuttle plasmids were introduced into the restriction-deficient S. aureus recipient RN4220 by electroporation and then transduced into 450M, a strain isogenic with RN4220 but containing mec sequences from S. aureus COL introduced by transformation (20). The mecA gene in 450M is unregulated due to a naturally occurring deletion of mecI and a portion mecR1 (1). When pG0630 and pG0631 were integrated into the 450M chromosome, they provided single-copy regulation of the mecA-lacZ fusion as well as regulation of the functional mecA gene in trans. The uninduced levels of β-galactosidase activity in 450M/pG0631::geh were sixfold higher than those in 450M/pG0630::geh, confirming the repressor defect conferred by the mecI D39G mutation (Table 2). This difference was further confirmed by Northern blot analysis of mecA transcript in each strain (Fig. 1B). Growth of each strain with 5 μg of the inducer CBAP (2-[2′-carboxyphenyl]benzoyl-β-aminopenicillanic acid) per ml produced a similar-fold increase in β-galactosidase activity for each construct (Table 2), indicating that the mutation had no effect on mecR1-mediated induction. However, because of the higher baseline transcription for 67-0 regulated sequences, induction of 450M/pG0631::geh could achieve 80% of maximum, as determined by β-galactosidase production from a lacZ fusion with mecI deleted, while 450M/pG0630::geh could be induced to only 25% of maximum (data not shown).
FIG. 1.
(A and B) Northern blot analysis of the 2-kb mecA transcript (A) and determination of relative amounts of PBP2a in the cell membranes (B) of S. aureus strains 67-0 and N315P. PBPs are designated by numbers at the left in panel B. (C) Northern blot analysis showing the relative abundance of mecA transcript in 450M (lane 1) and 450M with either of two chromosomally integrated plasmids, pG0631 (lane 2) or pG0630 (lane 3), containing 67-0 or N315P mecR1/mecI sequences, respectively. The images were scanned from their original gels by using an AlphaImager 1000 (Alpha Innotech) and were cropped and labeled by using Canvas 5 graphics software (Deneba, Miami, Fla.). The same systems were used to prepare Fig. 2 and 4 to 6.
TABLE 2.
Determination of β-galactosidase activity from mecA-lacZ transcriptional fusions
| Strain | β-Galactosidase activitya in the presence of CBAP at:
|
Fold increase in β-galactosidase activityb | |
|---|---|---|---|
| 0 μg/ml | 5 μg/ml | ||
| 450M/pG0630::geh | 496.2 ± 105.9 | 1,822.4 ± 312.6 | 3.6 |
| 450M/pG0631::geh | 2,934.8 ± 622.6 | 13,618.4 ± 1,759.1 | 4.6 |
Miller units per milligram of protein per hour; mean of five experiments ± standard deviation.
Calculated by dividing the activity units obtained in the presence of CBAP by that in the absence of CBAP.
Purification of MecI and MecI*.
The mecI genes from N315P and 67-0 were PCR amplified by using primers that amplified a 442-bp fragment containing the 372-bp mecI coding sequence, 17 bp 5′ to the translational start site, and 53 bp 3′ to the stop codon. The fragments were ligated into the pGEX-2T expression vector, adding bases to ensure conservation of the GST reading frame. Following isolation of fusion protein by using glutathione-Sepharose beads, the repressor proteins were cleaved from GST with thrombin and the purity and size of cleaved protein were checked by SDS-PAGE. The mutant protein was designated MecI*. The cleaved protein was of the expected size for each repressor (14 kDa). The amounts of cleaved MecI and MecI* used in experiments described below were equalized by visual inspection and by scanning densitometry of the 14-kDa protein band on gels and confirmed to vary by no more than 5%.
DNase I protection assay.
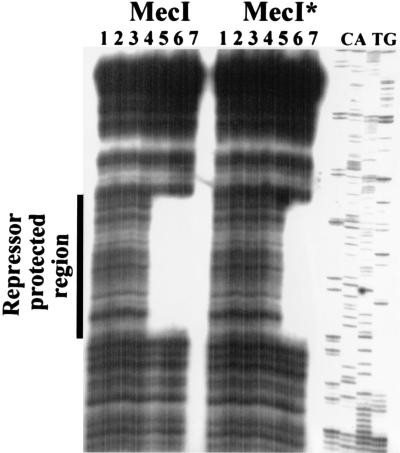
First, a 210-bp fragment, containing 56 bp of mecA at one end, the entire 99-bp intergenic region, and 55 bp of mecR1 at the other end, was generated by PCR and digested with DNase I following incubation with either MecI or MecI*. Fragments were end labeled by cleaving one end with HindIII and filling in the 3′ recess with Klenow fragment in the presence of [α-32P]dCTP. Either strand was labeled by including a HindIII site in either the mecA or mecR1 primer. Concentrations of purified repressor of 1 to 1,000 ng were assessed for the ability to protect the fragment. As shown in Fig. 2, both MecI and MecI* protected the same DNA sequences, but while protection was achieved with 0.15 μg of MecI, 0.4 μg of MecI* was required to produce the same result. Sequences protected on opposite strands were staggered by four bases at the mecR1 and two bases at the mecA end. The 43-bp protected sequence contained a 30-bp palindrome with 15 bp of dyad symmetry. Within each 15-bp half-site was another area of dyad symmetry with 6 bp in each arm. This palindrome included the predicted −10 mecA and −35 mecR1 promoter sequences. Protected sequences are indicated in Fig. 3. Control DNA, consisting of a 267-bp fragment from the trs region of staphylococcal plasmid pG01 (see below), was also incubated with purified repressor protein, and no DNase protection was demonstrated (data not shown).
FIG. 2.
DNase I footprint of the 210-bp fragment containing the region between the mecA and mecR1 translational start sites. MecI and MecI* protected regions are shown. Lanes 1 through 7 contain 0, 0.001, 0.01, 0.05, 0.15, 0.4, and 1 μg of MecI or MecI*, respectively. The repressor-protected region is marked on the left. C, A, T, and G are the labeled nucleotides of the sequencing ladder generated from the 210-bp fragment by using a primer complementary to its 5′ end. The sequence is read from bottom (5′ end) to the top (3′ end).
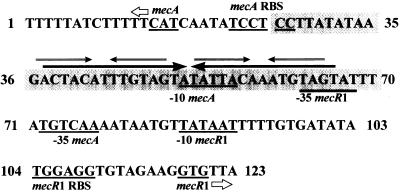
FIG. 3.
DNA sequence of the portion of the 210-bp fragment showing the maximum number of nucleotides (shaded) protected on both DNA strands by MecI and MecI*. The protection begins at nucleotide 26 and ends at nucleotide 70. The area protected on each DNA strand is staggered by two bases at the end closest to the mecA translational start site and four bases at the end closest to the mecR1 start site (data not shown). The 30-bp palindrome, present in the protected region, is identified with long arrows. Two 6-bp inverted repeats, present within the larger palindrome, are identified with smaller arrows. The mecA and mecR1 translational start sites and ribosomal binding sites (RBS) are identified and underlined. The predicted −10 and −35 promoter regions for mecA and mecR1, as also described in reference 14, are designated and underlined.
EMSA.
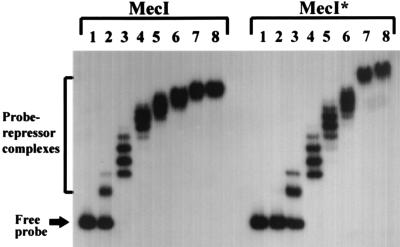
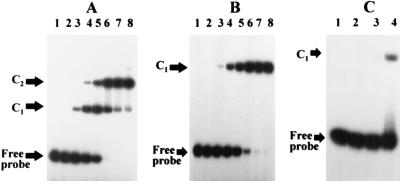
Fragments containing the entire mecA-mecR1 intergenic region and various components of the binding site itself were end labeled with [γ-32P]ATP, incubated with a range of MecI or MecI* protein concentrations, and electrophoresed through a nondenaturing 4% polyacrylamide gel in order to assess patterns of fragment migration caused by protein-DNA and protein-protein interactions. To confirm the specificity of repressor binding, protein was incubated with unlabeled specific and nonspecific target DNA before incubation with labeled DNA as follows. Each reaction mixture included poly(dI-dC) · poly(dI-dC) with an average length of 300 bp, as described in Materials and Methods. In addition, a 500-fold excess of either the specific 210-bp target used in the DNase I protection assay or nonspecific DNA was included in a competition assay. The nonspecific DNA was a 267-bp fragment amplified from the trs region of plasmid pG01, a region containing genes for conjugative transfer (19). This fragment (bases 2072 to 2338) included the 3′ end of trsC and the 5′ end of trsD with an intervening promoter region but contained no sequences that exhibited dyad symmetry. Either MecI or MecI* produced mobility-shifted complexes in the presence of excess unlabeled nonspecific DNA, while an excess of unlabeled specific target DNA completely abolished the altered mobility of the labeled 210-bp promoter-operator fragment (data not shown). The mobility shift of the 210-bp fragment is shown in Fig. 4. At increasing protein concentrations, multiple bands were seen. The patterns were similar for MecI and MecI*, but 5- to 10-fold-greater concentrations of mutant protein were required to produce the same shift as for the wild type. At concentrations of 10 μg of MecI and 50 μg of MecI*, there were multiple discrete retarded complexes of increasing size. To assess the precise sequence required for repressor binding, oligonucleotides that contained the specific target sequence flanked by nonspecific nucleotides were synthesized. The fragments were as follows: the entire 30-bp palindrome in a 42-bp oligonucleotide, each 15-bp half-site in a 26- and a 30-bp oligonucleotide, and two 6-bp half-sites of the internal palindrome within a 27- and a 29-bp oligonucleotide. Increasing concentrations of MecI and MecI* were incubated with each fragment. The results of this mobility shift assay with MecI are shown in Fig. 5. The fragment containing the 15-bp half-site was shifted into a single band of identical mobility at all concentrations tested. The gel shifts were identical for each 15-bp half-site, indicating equal protein-target affinity for each site. The band shift was seen first at a concentration of 10 ng of protein and was completely converted from the unbound to the bound form at 500 ng. When the 6-bp half-site of the internal palindrome was evaluated, no shift was seen until 500 ng of protein was used, and at 1,000 ng only about 30% of the free target had been converted to the bound form. Finally, when the complete 30-bp palindrome was used as a target, discrete bands differing in molecular size were seen at increasing protein concentrations. The same gel shift patterns were seen with MecI* as with MecI, but 5- 10-fold-higher concentrations of the mutant were required to produce the same amount of bound target as for the wild type. Taken together, these results suggest that the 15-bp half-site containing internal dyad symmetry is required for optimum repressor binding; the 6-bp internal half-site is not sufficient. The presence of additional retarded bands is characteristic of cooperative DNA binding.
FIG. 4.
EMSA showing the binding of MecI and MecI* to the 210-bp fragment labeled at the 5′ end with [α-32P]dCTP. Lanes 1 through 8 contain 0, 0.001, 0.01, 0.05, 0.15, 0.4, 0.7, and 1 μg of MecI or MecI* and 4,000 cpm of the labeled fragment (probe), respectively. After incubation under appropriate conditions as described in Materials and Methods, the samples were analyzed on a 4% nondenaturing polyacrylamide gel. Positions of free and repressor-bound probe are indicated by arrows.
FIG. 5.
EMSA showing the binding of MecI to a 42-bp synthetic oligonucleotide containing, as shown in Fig. 4, the 30-bp palindrome (A), a 26-bp oligonucleotide containing the 15-bp left arm of the 30-bp palindrome (B), and a 27-bp oligonucleotide containing the 6-bp inverted repeat found in the left arm of the palindrome (C). These oligonucleotides were labeled at the 5′ end with [γ-32P]ATP. Labeled oligonucleotides (4,000 cpm) were incubated with 0, 0.001, 0.01, 0.05, 0.1, 0.25, 0.5, and 1 μg (lanes 1 through 8, respectively, in panels A and B) and 0, 0.1, 0.5, and 1 μg (lanes 1 through 4, respectively, in panel C) of MecI. After incubation under appropriate conditions as described in Materials and Methods, the samples were analyzed on a 10% nondenaturing polyacrylamide gel. Positions of free and repressor-bound (C1 and C2) oligonucleotides are indicated by arrows.
Cross-linking of repressor protein in solution.
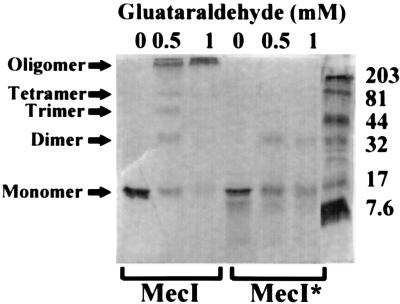
Because of the suggestion from mobility shift experiments that protein-protein interactions were occurring, either MecI or MecI* was incubated with the bifunctional cross-linking reagent glutaraldehyde in order to demonstrate oligomerization of the repressor proteins in solution. As shown in Fig. 6, MecI and MecI* were present as monomers in the absence of cross-linking reagent. However, in the presence of 0.5 mM glutaraldehyde, MecI formed a mixture of dimers, trimers, tetramers, and higher-order forms. At 1 mM, all of the MecI was present as a large oligomeric complex. In contrast, at these two concentrations, MecI* produced predominately dimers, with a significant proportion of the protein still existing in monomeric form. At 10 mM glutaraldehyde, MecI* had still not been converted to the single oligomeric form seen with MecI (data not shown).
FIG. 6.
SDS–10% polyacrylamide gel showing the relative oligomerization of MecI and MecI* upon treatment with 0.5 or 1 mM glutaraldehyde. Relative positions of monomeric and higher-order species formed after incubation with the cross-linking agent are indicated by the arrows. Positions of protein molecular mass standards are shown at the right in kilodaltons.
DISCUSSION
In this study, we have shown that the purified MecI repressor protein binds to a single 30-bp target sequence that includes both the mecA −10 and mecR1 −35 promoter sequences. This interaction provides tight repression of mecA transcription and PBP2a production. We have previously shown that MecI inactivation leads to a 5-fold increase in mecA transcription and a 40-fold increase in the amount of PBP2a in membranes (20). Because of the involvement of predicted mecR1 promoter sequences in the repressor binding site, autoregulation is presumed to occur, but this has not been demonstrated. The 30-bp target sequence contains 15 bp of almost perfect dyad symmetry. The entire 15-bp half-sequence is necessary for optimum MecI binding, and the two half-sites bind with equal affinity. The presence of more than one initial binding site should promote cooperative DNA binding if protein oligomerization occurs. We demonstrated the oligomerization of MecI in solution by using the bifunctional cross-linking agent glutaraldehyde. Furthermore, mobility shift of the 30-bp target, containing the two binding half-sites, produced discrete bands of different mobilities. Similarly, the 210-bp target yielded multiple bands of different mobilities at increased MecI concentrations. In contrast, the 15-bp half-site alone produced a single band at all protein concentrations. Although we have not presented direct evidence that protein complexes form on DNA, these data are consistent with cooperative DNA binding occurring as a result of dimers binding to the palindrome half-sites followed by oligomerization due to further protein-protein interactions. There is also some evidence that the cognate PenI and BlaI repressors bind in a similar manner. While oligomerization of S. aureus BlaI has not been demonstrated directly, different-sized bands are seen in mobility shift assays (9) and PenI has been shown to form dimers (24). It is interesting that one of the two BlaI operator sites involves no promoter sequences. It is tempting to speculate that this target exists solely to provide cooperative DNA interactions with protein bound to the other site.
In contrast to the single site for MecI binding, the BlaI target consists of two sites, each with 9 bp of dyad symmetry and each having equal affinity for repressor. The centers of symmetry for the two bla sites are 31 bp apart, while they are only 16 bp apart for mec. The presence of different spacings in the promoter-operator regions of the bla and mec may have functional implications. The 16-bp spacing between the centers of symmetry of the mec target sequences allows one and one-half helical turns, putting protein bound to each sequence on opposite sides of the DNA helix. Thus, interactions may lead to torsion or twist of the helix. In contrast, 30-bp spacing puts protein bound to the two bla targets on the same surface of the helix, three turns apart. Interactions between protein bound to these sites may produce DNA conformations different from those seen for mec. The extent to which the different target arrangements and protein sequences determine differences in the regulation and induction of MecI versus BlaI remains to be elucidated.
Analysis of the mutant protein, MecI*, further established the importance of protein oligomerization for optimum binding. The mutation at position 39 of the predicted protein sequence is in the turn portion of a putative amino terminal helix-turn-helix motif as established by protein analysis programs (7) and by comparison with similar motifs (15). Several investigators have established the importance of the amino termini of PenI and BlaI in DNA binding by making deletions and analyzing mutations in this area (9, 25). In each case of amino terminus alterations, DNA binding was markedly altered. In contrast, MecI* bound to target in a manner identical to MecI, footprinting the same sequence and binding to each 15-base half-site with equal affinity. However, 5- to 10-fold more MecI*, as determined by protein titration in EMSA, was required to produce the same pattern seen with MecI. The mutant protein was clearly deficient in repressor function in vivo, as shown by a sixfold-higher production of β-galactosidase from mutant repressor gene-lacZ fusions than from wild-type gene fusions and by Northern blot analysis showing a reduced ability of MecI* to repress mecA transcription. A possible explanation for the functional alteration of the MecI* protein was shown in glutaraldehyde cross-linking studies. Approximately 10-fold-higher concentrations of glutaraldehyde were required to produce multimers in cross-linking experiments with MecI* than with MecI and the predominant cross-linked form of MecI* was a dimer, whereas MecI produced all forms of multimer at the lowest concentration of glutaraldehyde that resulted in any cross-linking. Most proteins that bind DNA by means of a helix-turn-helix motif form dimers in order to achieve binding to palindromic operator sequences (3, 11). However, it has also been shown for the lambda cI repressor that optimum cooperative DNA binding does not occur unless dimerization leads to the formation of higher-order oligomers (4). Studies with PenI and BlaI have shown that the carboxy terminus of the protein is required for dimerization (9, 25). A deletion of the carboxy-terminal 23 amino acids of BlaI produced alterations such that the same sequences were protected in DNase I footprints but much higher quantities of deleted protein were required to achieve the same footprint (9). Thus, just as for MecI*, mutations that affected protein-protein interactions altered repressor-target binding without changing the binding site. MecI* may not provide maximal repression if its quantity cannot be increased sufficiently to meet demands imposed by deficient oligomerization.
The mutation in MecI* did not significantly affect its induction, as determined by the fold increase in β-galactosidase from a mecA-lacZ fusion in the presence of a strong inducer (CBAP), in comparison with the MecI-repressed fusion. Thus, if proteolytic cleavage of MecI is the mechanism of derepression, as suggested previously (9, 15), the mutation does not affect the cleavage site and this degree of deficiency in oligomerization does not impair repressor-protease interactions. However, because baseline repression is impaired, greater induction of mecA transcription can be achieved, a result that may affect phenotypic expression of resistance when methicillin-resistant staphylococci are exposed to inducing concentrations of β-lactam antibiotics.
The results of this study establish a requirement for MecI repressor oligomerization and cooperative DNA binding for optimal function. In addition, the amino terminus of MecI, while it likely has a major role in initial DNA binding, as shown for the cognate repressors PenI and BlaI, also has some role in protein-protein interactions. Although the carboxy terminus probably mediates dimerization, the formation of higher-order oligomers may involve other parts of the molecule.
ACKNOWLEDGMENTS
This work was supported in part by USPHS grant R37 AI35705 from the National Institute of Allergy and Infectious Diseases.
We are indebted to Michael J. Pucci, Bristol-Myers Squibb Research Institute, for performing the penicillin binding protein analysis.
REFERENCES
- 1.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artsimovitch I, Howe M M. Transcription activation by the bacteriophage Mu Mor protein: analysis of promoter mutations in Pm identifies a new region required for promoter function. Nucleic Acids Res. 1996;24:450–457. doi: 10.1093/nar/24.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burz D S, Beckett D, Benson N, Ackers G K. Self-assembly of bacteriophage λ cI repressor: effects of single-site mutations. Biochemistry. 1994;33:8399–8405. doi: 10.1021/bi00194a003. [DOI] [PubMed] [Google Scholar]
- 4.Burz D S, Ackers G K. Cooperativity mutants of bacteriophage λ cI repressor: temperature dependence of self-assembly. Biochemistry. 1996;35:3341–3350. doi: 10.1021/bi952055x. [DOI] [PubMed] [Google Scholar]
- 5.Chambers H F, Hartman B, Tomasz A. Increased amounts of a novel penicillin-binding protein in methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985;76:325–331. doi: 10.1172/JCI111965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chodosh L A. Mobility shift DNA-binding assay using gel electrophoresis. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Smith J A, Struhl K, editors. Current protocols in molecular biology. Unit 12.2.1, Suppl. 13. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. pp. 12.0.1–12.4.16. [Google Scholar]
- 7.Chou P Y, Fasman G D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 8.Gaisferd C, Reynolds P E. Methicillin resistance in Staphylococcus epidermidis. Relationship between the additional penicillin-binding protein and an attachment transpeptidase. Eur J Biochem. 1989;185:211–218. doi: 10.1111/j.1432-1033.1989.tb15104.x. [DOI] [PubMed] [Google Scholar]
- 9.Gregory P D, Lewis R A, Curnock S P, Dyke K G H. Studies of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. Mol Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardt K, Joris B, Lepage A, Brasseur R, Lampen J O, Frére J-M, Fink A L, Ghuysen J-M. The penicillin sensory transducer, BlaR, involved in the inducibility of β-lactamase synthesis in Bacillus licheniformis is embedded in the plasma membrane via a four-α-helix bundle. Mol Microbiol. 1997;23:935–944. doi: 10.1046/j.1365-2958.1997.2761642.x. [DOI] [PubMed] [Google Scholar]
- 11.Harrison S C, Aggarwal A K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 12.Hattori M, Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986;152:232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence of the regulator region of mecA gene in methicillin resistant Staphylococcus aureus. FEBS Lett. 1992;298:133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 15.Joris B, Hardt K, Ghuysen J-M. Induction of β-lactamase and low-affinity penicillin binding protein 2′ synthesis in gram-positive bacteria. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science; 1994. pp. 505–515. [Google Scholar]
- 16.Kreiswirth B N, Lofdahl S, Betley M J, O’Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock exotoxin structural gene is not detectably transmitted by prophage. Nature (London) 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 17.Lee C Y, Buranen S L, Ye Z. Construction of a single-copy integration vector for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 18.Luchansky T B, Muriana P M, Klaenhammer T R. Bio-Rad technical bulletin no. 1350. Richmond, Calif: Bio-Rad Laboratories; 1988. Electrotransformation of gram-positive bacteria; pp. 1–3. [Google Scholar]
- 19.Morton T M, Eaton D M, Johnston J L, Archer G L. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pG01. J Bacteriol. 1993;175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson N E, Pattee P A. Genetic transformation in Staphylococcus aureus: demonstration of a competence-conferring factor of bacteriophage 80α lysates. J Bacteriol. 1981;148:294–300. doi: 10.1128/jb.148.1.294-300.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townsend D E, Ashdown N, Bolton J, Grubb W B. The use of cetyltrimethylammonium bromide for the rapid isolation from Staphylococcus aureus of relaxable and nonrelaxed plasmid DNA suitable for in vitro manipulation. Lett Appl Microbiol. 1985;1:87–94. [Google Scholar]
- 23.Sharma V K, Johnston J L, Morton T M, Archer G L. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pG01. J Bacteriol. 1994;176:3445–3454. doi: 10.1128/jb.176.12.3445-3454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittman V, Wong H C. Regulation of the penicillinase genes of Bacillus licheniformis: interaction of the pen repressor with its operators. J Bacteriol. 1988;170:3206–3212. doi: 10.1128/jb.170.7.3206-3212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wittman V, Lin H C, Wong H C. Functional domains of the penicillinase repressor of Bacillus licheniformis. J Bacteriol. 1993;175:7383–7390. doi: 10.1128/jb.175.22.7383-7390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C J, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]