Abstract
RNA-binding proteins (RBPs) regulate essentially every event in the lifetime of an RNA molecule, from its production to its destruction. Whereas much has been learned about RNA sequence specificity and general functions of individual RBPs, the ways in which numerous RBPs instruct a much smaller number of effector molecules, that is, the core engines of RNA processing, as to where, when and how to act remain largely speculative. Here, we survey the known modes of communication between RBPs and their effectors with a particular focus on converging RBP–effector interactions and their roles in reducing the complexity of RNA networks. We discern the emerging unifying principles and discuss their utility in our understanding of RBP function, regulation of biological processes and contribution to human disease.
Introduction
The regulation of gene expression at the RNA level is fundamental to essentially all biological processes. Its complexity presents one of the most formidable challenges in molecular and systems biology1. Human cells must ensure correct processing of each of their more than 20,000 different protein-coding RNAs, many more if one considers alternative isoforms2,3. These transcripts can range in level from fewer than 0.1 copies to thousands of copies per cell4,5. Moreover, each transcript must be processed at the right time, by the right processing molecule and at the right subcellular location, often in a manner that is specific to a particular cell type or conditions in the microenvironment1. There is little room for error, as RNA misprocessing can lead to disrupted cell homeostasis or cell death, as documented experimentally and by cases of human disease3,6,7.
The scale of regulatory complexity began to emerge with the inception of next-generation DNA sequencing in the 2000s8, which allowed RNA processing to be studied from a systems perspective. Since then, a multitude of transcriptome-wide approaches combining high-throughput sequencing with genetic perturbation, biochemistry and mass spectrometry-based approaches have transformed studies of RNA biology. In addition to allowing for the inference of RNA regulatory networks, their principal goal has been to decipher the functions of RNA-binding proteins (RBPs) as the core regulators of RNA processing9–12. Indeed, RNA-centric and RBP-centric systems-wide studies that entail methods based on RNA immunoprecipitation (RIP), crosslinking and immunoprecipitation (CLIP) and its many variants, RNA interactome capture, RNA antisense purification coupled with mass spectrometry (RAP–MS) and other related strategies jointly identified more than 1,500 human RBPs and characterized RNA binding preferences of several hundred of them in vitro and/or in vivo9–12.
Together with loss-of-function analyses and other functional assays, these studies have elucidated some of the basic rules of RNA regulation for numerous RBPs. The above ‘exponential technologies’, however, also created a rift in the field between the rapid cataloguing of protein–RNA interactions, along with their largely correlative functional links, and a lagging understanding of the mechanisms of RBP activities. These activities commonly rely on interactions of RBPs with their effector molecules, that is, proteins or protein complexes that serve as core engines of RNA processing and thus define the activities of many RBPs.
For the purpose of this Review, we designate as ‘effectors’ or ‘effector assemblies’ those molecular entities that operate as executors of RNA processing or otherwise directly affect RNA processing but are not classified as RBPs. We define as RBPs those proteins that bind RNA in a sequence- or structure-specific manner and are not basal or auxiliary components of effector assemblies, to avoid potential confusion when referring to RBP–effector interactions. For instance, TIA1, a sequence-specific RBP, interacts with the spliceosome, an effector, to regulate alternative pre-mRNA splicing13,14. Furthermore, we consider RBP–effector interactions as ‘converging’ if there exists experimental evidence for the capacity of at least two different RBPs in a species to separately connect to the same effector of RNA processing. For example, the RBPs ZFP36 (also known as TTP) and Roquin-1 (also known as RC3H1) can both interact with the effector CCR4–NOT to regulate mRNA stability15–18.
Like RBPs, effectors have crucial roles in essentially all stages of RNA processing; however, unlike RBPs, they and their interactions with RBPs have primarily been studied using reductionist approaches, including X-ray crystallography, in vitro studies of protein–protein interactions (PPIs) and RNA processing, RNA-tethering assays and targeted genetic experiments17,19–22. Studies at the nexus between RBPs and their effectors have also been outpaced by the recent resurgence of interest in the formation of biomolecular condensates, which have begun to shed light on entirely different levels of spatiotemporal control of RNA processing23–26. Together, these developments have propelled our understanding of RNA networks at two discrete levels of the regulatory hierarchy, while leaving behind the intervening level at which RBPs communicate with effectors of RNA processing. This disconnection has arguably hampered our understanding of RBP function as well as the overall operation of RNA networks, which recent studies are now beginning to address.
Here, we focus on the nexus between RBPs and their effectors, with a particular emphasis on its emerging roles in the control of RNA processing, regulation of biological outcomes, and its contribution to human health and disease. We first define the function of the nexus in managing the complexity of modern RNA networks, then discern the unifying features of RBP–effector interactions along with their physiological roles using illustrative examples. We then consider how cellular processes regulate and are regulated at the nexus, followed by a discussion of genetic disorders that affect, and therapeutic opportunities that emerge at, the RBP–effector interface.
The complexity of RNA networks
The evolution of progressively more complex RNA networks mandated solutions to the increasingly more challenging regulation and need for flexibility1. Such solutions are built into many modern RNA networks in the form of distinct molecular and systems-level features23,27–29 (Fig. 1). These traits facilitate the regulation of RNA networks by minimizing the need for complexity, including the number of required connections, while also improving network performance and speed of adaptation to the environment.
Fig. 1 |. Traits and organization of RNA networks associated with the management of regulatory complexity.
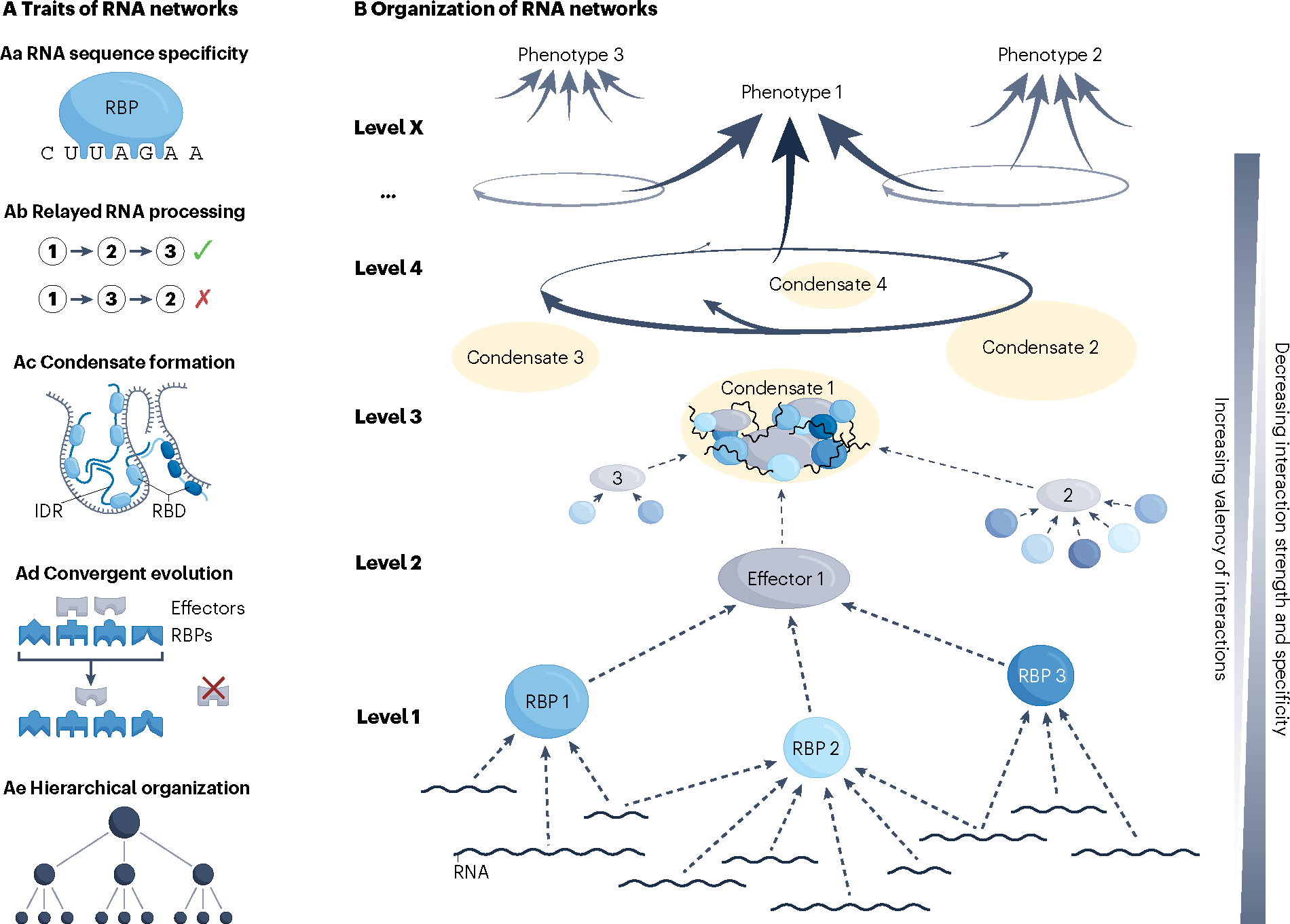
A, Traits of RNA networks. Aa, The recognition of specific RNA sequences and/or structures allows trans-acting factors, including RNA-binding proteins (RBPs) and small RNAs, such as microRNAs (miRNAs), to act upon some but not other transcripts. This trait also allows for synchronized processing of multiple, often functionally related transcripts184. Recognition of specific RNA features defines the most elementary regulatory level of RNA networks that determines which transcripts will be controlled by which trans-acting factors. Different RNA specificities of different RBPs considerably simplify the challenge of coordinated regulation, in addition to allowing for network adaptation through rewiring of RNA targets28,184. Ab, Relayed RNA processing refers to the correct sequence of post-transcriptional processing events. For instance, a typical pipeline would ensure that RNA splicing occurs prior to RNA export and localization to a distal intracellular region, followed by localized translation and that RNA degradation occurs last29. Any other sequence of events could be detrimental to cell homeostasis. General molecular and cellular organization, e.g. association of the splicing machinery with RNA polymerase II (Pol II) and separation of nuclear from cytoplasmic components, respectively, as well as more specific molecular interactions contribute to the correct relay of RNA processing events29,185. Correct and rapid relay of RNA processing events secures directness and energy-efficient regulation of RNA processing. Ac, The formation of ribonucleoprotein (RNP) condensates increases local concentration of RBP–RNA modules along with their effector complexes. RNP condensation is driven primarily by intrinsically disordered regions (IDRs) of different RBP molecules, which multimerize through numerous weak, non-specific interactions, and is aided by transient secondary structures formed within IDRs as well as contributions from the associated RNA23. RNP condensation can stabilize the association of individual RBPs with their recognition motifs on RNA and contribute to correct RNA folding that may be required for processing40,186. In addition, increased local concentration of effector proteins and RNA can increase the rate of biochemical reactions, or assist in storage or transport of these molecules23. Upon extensive RNP condensation, a physicochemical phenomenon of liquid–liquid phase separation occurs through which various types of RNA granules, including stress granules, P-bodies, splicing speckles, neuronal granules, and others, are generated. Such granules, also referred to as ‘membraneless organelles’, exist in liquid-like and occasionally solid-like physical states that exhibit distinct physiological roles23,30. Mechanisms that govern the formation and dissolution of RNP condensates include membrane surfaces, molecular chaperones, including nuclear import receptors, RNA helicases, and post-translational modifications of condensate components54,187. Ad, Convergent molecular evolution has an important role at different levels of RNA processing and contributes to the hierarchical structure of RNA networks. At the RBP–RNA level, convergence has been observed in RNA-targeting specificities of RBPs and in the evolutionary adaptation of RNA molecules to a particular mode of post-transcriptional processing, including alternative splicing and mRNA decay28,188,189. Convergent evolution also occurs at the level of RBPs interacting with their effector molecules. Short linear motifs (SLiMs), which are typically located in IDRs of RBPs or adaptor proteins, are specifically recognized by cognate domains of effectors and frequently evolve in a convergent manner93,190. These examples point to a broad potential of convergent evolution to unify the fates of different transcripts by a common regulatory step. Ae, Hierarchical organization is commonly observed in biological networks and is thought to evolve due to the high cost associated with creation and maintenance of network connections27. B, Hierarchically wired networks, including RNA networks, not only exhibit fewer connections, but also adapt faster to the environment and show higher overall performance compared to non-hierarchical networks27. RNA networks show several hierarchical regulatory levels, with control at higher levels having broader effects on RNA processing. RBD, RNA-binding domain.
The associations between RBPs and their effectors have a central role in the regulatory hierarchy of RNA networks by unifying the fates of multiple RBP–RNA modules and serving as a functional bridge to the formation of larger, high-concentration ribonucleoprotein (RNP) assemblies, which confer a higher level of spatiotemporal control23,30 (Fig 1). At the RBP–effector nexus, the effectors often end up being recruited to RNA by RBPs, either directly or indirectly, and act in cis by processing the RBP-bound RNA, although there also exist other modes of RBP-dependent effector engagement with RNA that may have a substantially different impact on RNA processing (Fig. 2).
Fig. 2 |. Modes and dynamics of RBP-dependent effector engagement with RNA.
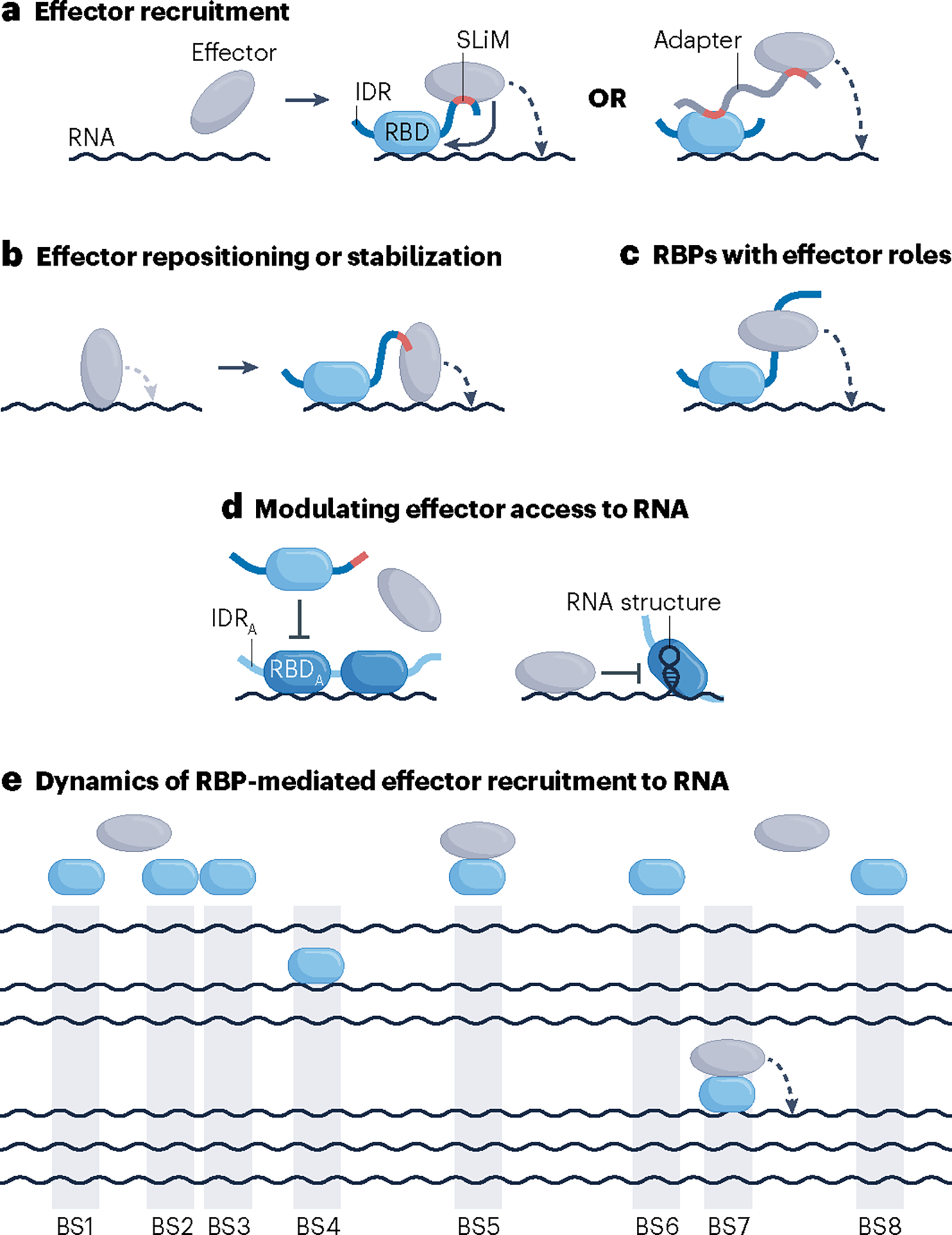
a, RNA-binding proteins (RBPs) (blue shapes) can recruit effectors to target RNA via direct or indirect protein–protein interactions (PPIs) that typically entail a short linear motif (SLiM; red), which typically resides in an intrinsically disordered region (IDR) of the RBP or an adaptor protein, and a structured domain located in the effector. Indirect interactions can involve additional proteins or can be mediated by non-coding RNAs (not shown). Upon recruitment, the effector can exert activity in cis, that is, on the RBP-bound RNA (dashed curved arrow) and occasionally also on the recruiting RBP (solid curved arrow), or in trans, that is, on other molecules (not shown). b, Instead of serving a recruiting role, some RBP–effector interactions may facilitate repositioning or stabilization of a pre-bound effector to modulate its activity137. c,d, Certain activities of RBPs do not entail contacts with effector molecules, either because RBPs themselves operate as effectors, as is the case for RBPs with enzymatic activities11,191–194 (panel c), or because they operate by modulating effector access to RNA, such as heterogeneous nuclear ribonucleoproteins (hnRNPs) in regulation of splicing40 (panel d). e, RBP-mediated recruitment of an effector to RNA is transient and occurs infrequently. Shown is a hypothetical steady-state scenario in which copies of an RBP (blue ovals), the number of which matches the number of RBP-binding sites (BS1–BS8) on RNA (black wavy lines), compete for a limiting number of available effector molecules (grey ovals). Only an RNA–RBP–effector assembly can process RNA in cis (dashed arrow). IDRA, IDR of antagonizing RBP; RBD, RNA-binding domain; RBDA, RBD of antagonizing RBP.
Akin to the capacity of a typical RBP to bind and regulate the fates of different RNA molecules, a typical effector of RNA processing can bind to different RBPs and subdue their bound RNAs to a common processing event. In this manner, effectors handle essentially all common RNA processing events, including the multiple steps of RNA biogenesis, modification, transport, storage, decay and, in the case of coding RNAs, translation.
Because a large number of RBPs must converge onto a smaller number of effectors, one might expect different RBPs to use a similar strategy to contact a particular effector. Contacts between RBPs and their effectors must secure specificity to couple the right RBP–RNA modules with the right RNA processing events and be transient enough to enable a rapid reuse of both the RBP and the effector. RBP–effector interactions thus require a compromise between highly specific, high-affinity contacts, such as those that mediate the domain–domain interactions that commonly establish macromolecular complexes31, and weaker, less-specific interactions, such as those implicated in the formation of RNP condensates23.
There is currently little consensus on the nature of interactions between RBPs and their effectors. Consequently, there is limited knowledge about how these interactions enable RBPs to instruct different types of RNA processing, how they may be regulated or how they lead to human disease when misregulated. Below we draw together key aspects of how RBPs directly or indirectly interact with effector assemblies and discuss the importance of these interactions for RNA processing (Figs. 3, 4 and Supplementary Table 1). We review specific examples of RBP–effector interactions that have been characterized at both the molecular and functional levels with an aim to highlight effectors as points of convergence for diverse RBPs and their bound RNA. We gather these findings in a series of vignettes, each specific to a particular effector and its physiological role.
Fig. 3 |. Converging RBP–effector interactions regulating (peri-)nuclear RNA processing.
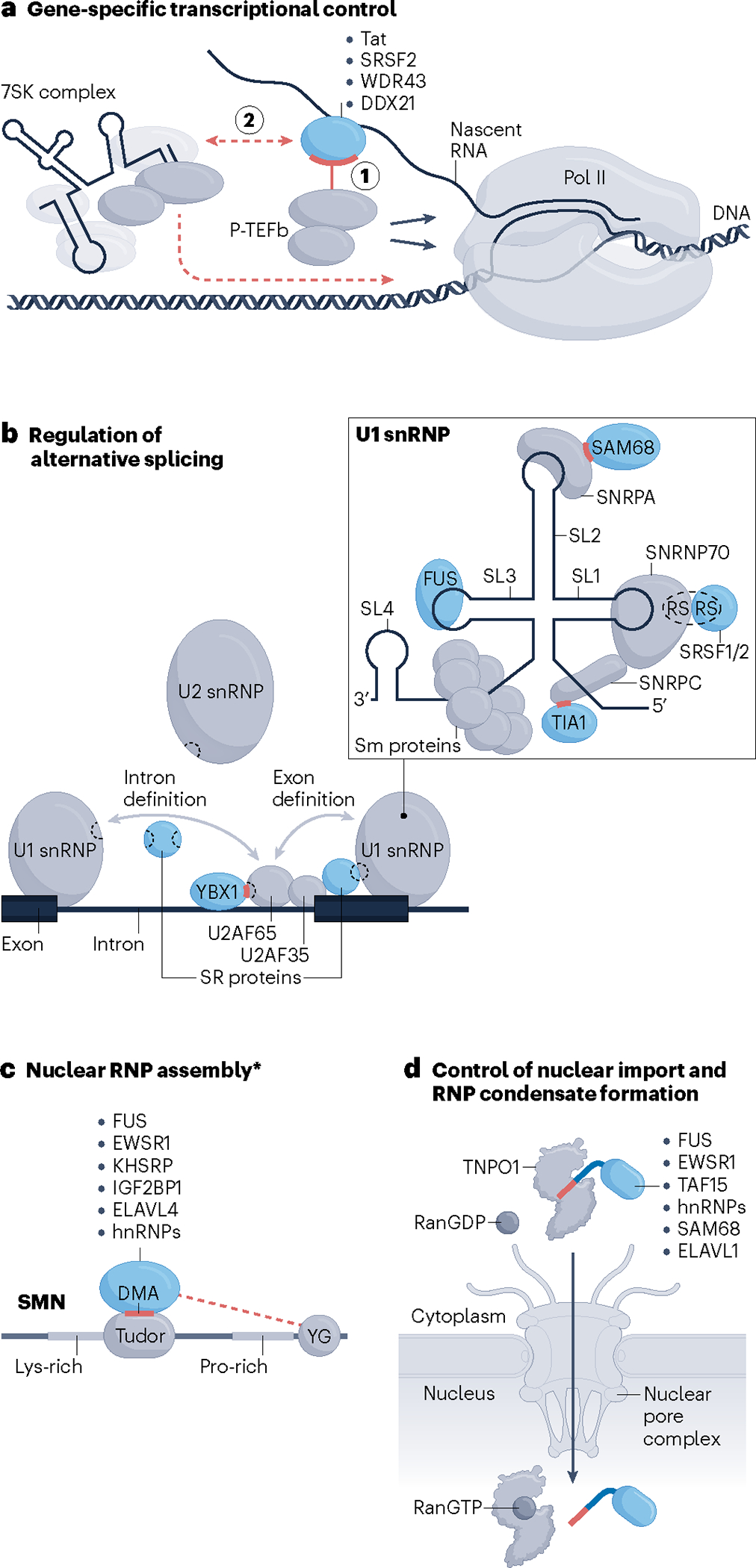
a, RNA-binding protein (RBP)-dependent release of paused RNA polymerase II (Pol II) by positive transcription elongation factor b (P-TEFb). RBPs can stimulate relocation of P-TEFb from a local 7SK complex to the vicinity of Pol II either by recruiting P-TEFb via direct protein–protein interactions (PPIs) while bound to nascent RNA (step 1) or indirectly by associating with or disassociating from the 7SK complex (step 2). b, RBPs directly interact with spliceosomal components, including U1 small nuclear ribonucleoprotein (snRNP), U2 snRNP and U2AF subunits, to promote the early stages of spliceosome assembly. Illustrated is an overview of all interactions (left) along with a zoomed-in view of the U1 snRNP (right). Grey arrows denote intron or exon definition interactions40, several of which are mediated by RS domains (dashed sections of shapes) of SR proteins, such as SRSF1 and SRSF2, and components of the spliceosome. RBPs other than SR proteins, including YBX1, SAM68 and TIA1, use short stretches of their intrinsically disordered regions (IDRs) (red dashes) to contact the indicated spliceosomal proteins. FUS recognizes the stem–loop (SL) region 3 (SL3) of the U1 small nuclear RNA (snRNA). Sm proteins are seven core spliceosomal proteins that make up a stable ring-like structure. c, Dimethylated arginines (DMAs) in the RGG/RG-rich regions of the indicated RBPs are recognized by the aromatic cage within the Tudor domain of survival motor neuron protein (SMN). Dashed red line denotes additional IDR-mediated interactions of some of the listed RBPs with the YG box domain (YG). The asterisk indicates that the same RBP–SMN interactions might also participate in processes other than RNP assembly in the nucleus43. For clarity, RBP-bound RNA is not drawn. d, RBPs that use a proline-tyrosine-rich nuclear localization sequence (PY-NLS) short linear motif (SLiM) (red dash) to interact with transportin 1 (TNPO1) for their nuclear import. The disaggregase activity of TNPO1 is not indicated (Fig. 4e). Drawings of multiple RBPs binding to the same effector molecule in individual panels solely illustrate that different RBPs can bind to a particular effector; they do not imply simultaneous interactions of multiple RBPs with different segments of the same effector or competition between different RBPs for binding to a particular region of an effector molecule.
Fig. 4 |. Converging RBP–effector interactions regulating cytoplasmic RNA processing.
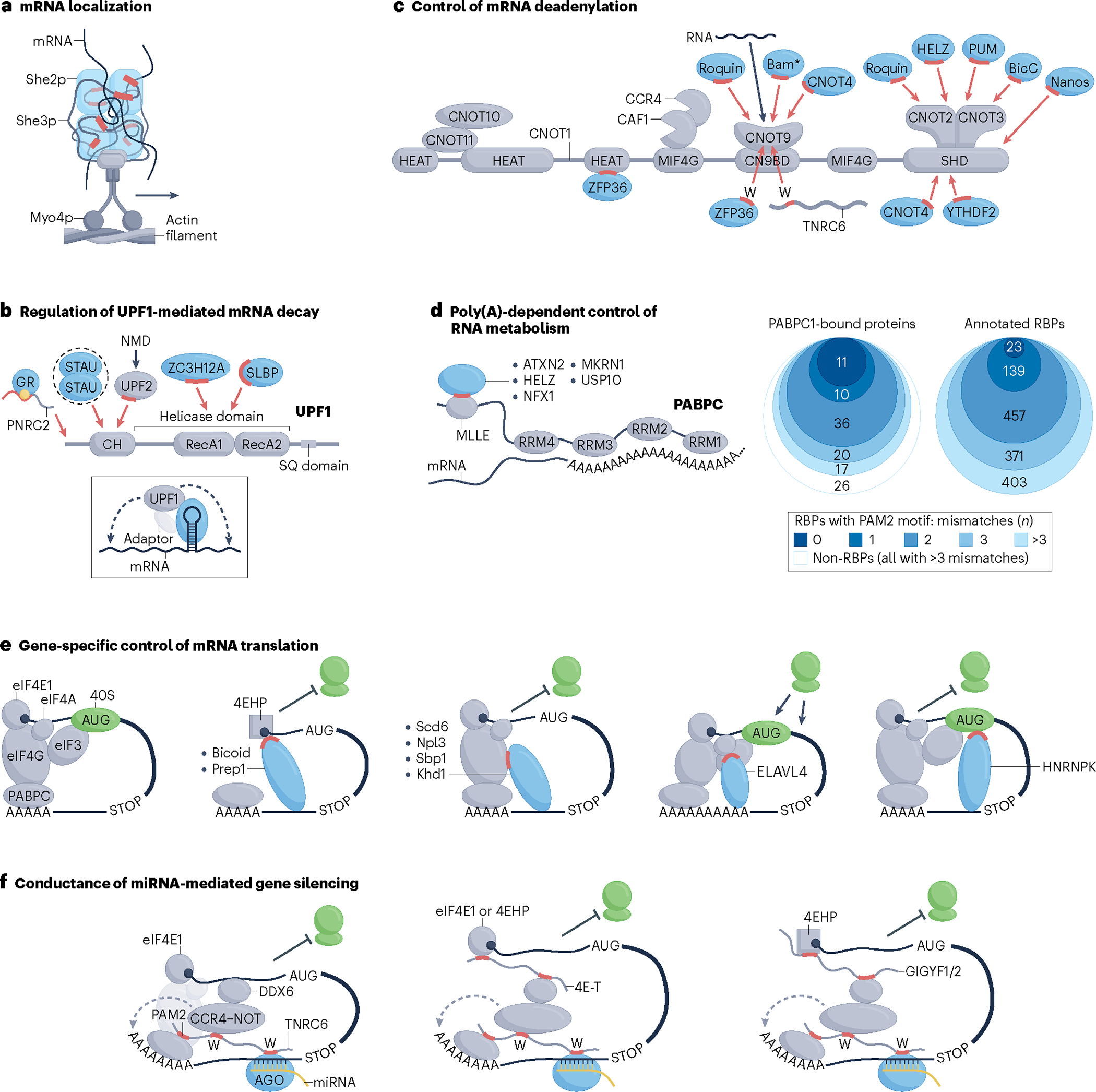
a, Myo4p motor protein-mediated transport of She2p/She3p RNA-binding protein (RBP)-bound mRNA along an actin filament, a process required for asymmetrical mRNA localization in budding yeast. Red segments indicate intrinsically disordered regions (IDRs) of She3p in contact with She2p and mRNA. b, Interactions of RBPs with the RNA helicase UPF1 implicated in regulation of mRNA decay. Red highlights represent IDR segments that form key protein–protein interactions (PPIs) required for activation of specific RBP-mediated mRNA decay pathways. Yellow dot denotes glucocorticoid, the ligand required for efficient association of the glucocorticoid receptor (GR) with the PNRC2 adaptor. Dashed oval indicates dimerization of Staufen (STAU) proteins. Red arrows point to approximate (domain-resolution) sites of contact on UPF1. Drawing in the inset illustrates a generalized mode of RBP-stimulated UPF1-mediated mRNA decay. Dashed arrows denote indirect stimulation in cis of exo- or endo-ribonucleolytic cleavage or translational repression. c, Schematic of the CCR4–NOT complex with indicated protein subunits and domains of the largest, CNOT1 subunit. Blue shapes indicate regulatory RBPs that bind to CCR4–NOT whereas red and black arrows point to sites of contact of RBPs and RNA, respectively. Red dash denotes short linear motif (SLiM)-containing IDR segments and letters W indicate tryptophan residues that form PPIs with CCR4–NOT. The asterisk indicates that the RNA-binding capacity of Bam is currently uncertain. The grey wavy line represents TNRC6 proteins that can serve as adaptors to connect RBPs with CCR4–NOT. RBP-bound mRNA has been omitted for clarity. d, RBPs that use a PAM2 or a PAM2-like SLiM (red dash) to interact with the MLLE domain of cytoplasmic poly(A)-binding protein (PABPC) bound to a poly(A) RNA sequence (left). Numbers of human PABPC1-bound proteins or annotated human RBPs that contain a PAM2 or a PAM2-like SLiM shown in proportional Venn diagrams. Total pools of proteins in each group were defined previously11,88 and are listed in Supplementary Table 3. The two PAM2 motifs, LIG_PAM2_1 and LIG_PAM2_2, annotated in the ELM database were considered as canonical (no mismatching residues; darkest shade of blue). Proteins that harbour motifs with one, two or three residues that deviate from either of the canonical PAM2 motifs are indicated in progressively lighter shades of blue (right). Note that the PAM2-like SLiM of makorin 1 (MKRN1) contains three residues that mismatch LIG_PAM2_1. None of the 26 PABPC1-bound non-RBPs harbours any PAM2 or PAM2-like motifs (that is, those with zero to three mismatches). e, Direct RBP–effector interactions that regulate translation initiation. Red segments of mRNA-bound RBPs denote IDRs that interact with different initiation factors or the 40S ribosomal subunit, as indicated. f, Cooperation of RBP–adaptor–effector conduits in miRNA–AGO-mediated gene silencing. Several co-associated molecules and processes have been omitted for clarity. Blunt or sharp arrows towards or away from the ribosomes (green shapes) indicate repressive or activating net effect on translation, respectively. Drawings of multiple RBPs binding to the same effector molecule in panels b–d solely illustrate that different RBPs can bind to a particular effector; they do not imply simultaneous interactions of multiple RBPs with different segments of the same effector or competition between different RBPs for binding to a particular region of an effector molecule. NMD, nonsense-mediated decay; RRM, RNA recognition motif.
The nexus in and around the nucleus
RBPs and positive transcription elongation factor b.
Research of RBPs has traditionally focused on their post-transcriptional activities; however, it is becoming increasingly clear that RBPs also have a role in regulating gene transcription itself32. One notable example involving converging RBP–effector interactions is transcriptional control of paused genes. Promoter-proximal RNA polymerase II (Pol II) pausing has emerged as a widespread mechanism of transcriptional regulation, affecting approximately 30% of metazoan genes and enabling rapid transcriptional responses to activation signals33. Release of paused Pol II into productive transcriptional elongation can be triggered by the kinase activity of the multiprotein complex positive transcription elongation factor b (P-TEFb) upon its delivery to paused genes34 (Fig. 3a). A viral RBP, called Tat, and a few cellular RBPs have been found to stimulate transcription of specific genes by recruiting P-TEFb to paused Pol II while bound to nascent RNA in a sequence- and/or structure-specific manner34–38. It is presently unclear whether the cellular RBPs recruit P-TEFb via direct PPIs, akin to the viral Tat protein, or indirectly increase the pool of locally available P-TEFb by liberating it from the promoter-proximal 7SK RNP complex (Fig. 3a).
RBPs and the spliceosome.
A large proportion of multi-exon genes in higher eukaryotes (more than 95% in humans) undergoes alternative splicing, a process that is crucial for generating proteomic diversity and is thought to sustain speciation and phenotypic complexity39. Sequence-specific RBPs regulate alternative splicing by modulating the activity of the spliceosome in the vicinity of their targeted loci on pre-mRNA2. A notable type of RBP is SR proteins, which stimulate splice site selection by locally stabilizing the ‘early’ components of the spliceosome through PPIs between arginine- and serine-rich intrinsically disordered regions (IDRs), also called RS domains, present in both the SR proteins and the spliceosome21,40 (Figs. 2b,3b, Table 1 and Supplementary Table 2). Such bonding is strengthened by the observed phosphorylation of the RS domains41, which sensitizes alternative splicing to signal transduction pathways and presents an important therapeutic opportunity, as discussed below. Regulatory strategies that rely on spliceosome stabilization are also employed by other RBPs with characterized interactions that use short stretches of IDRs to contact various protein subunits of the early spliceosomal components (Fig. 3b and Table 1). FUS, the splicing activity of which relies on an interaction with spliceosomal RNA rather than a protein subunit, represents an apparent exception to this rule42.
Table 1 |.
Interface between RBPs and their effectors
| Effector | Effector proteina | Effector contact regionb | RBPa | RBP contact regionb | Kd (μM)c | Process regulated by the RBP-effector interaction |
|---|---|---|---|---|---|---|
| Spliceosome | SNRNP70 (U1-70K)d | RS domain (IDR) | SRSF1/2 (SC35) | RS domain (IDR) | Pre-mRNA splicing | |
| SNRPA (U1A)d | RRM1 domain (SD) | SAM68 (KHDRBS1) | Y-rich (YY) region (IDR) | |||
| SNRPC (U1C)d | N terminus (SD) | TIA1 | Q-rich domain (IDR) | |||
| U1 snRNAd | SL3 of U1 snRNA | FUS | RRM domain (SD) | |||
| U2AF65 | RS domain (IDR) | YBX1 | Basic/acidic repeats (IDR) | |||
| SMN | SMN | Tudor domain (SD) | ELAVL4 (HuD) | NR | mRNA transport, translation | |
| IGF2BP1 (ZBP1) | KH domain region | |||||
| KHSRP (FUBP2) | NR (methylated Arg) (IDR) | mRNA stability | ||||
| FUS | RGG box (IDR) and RRM (SD) | Pre-mRNA splicing | ||||
| Tudor domain (SD) and YG box (SD) | HNRNPR | RGG box (IDR) | mRNA transport, pre-mRNA splicing | |||
| SYNCRIP | RGG box (IDR) | |||||
| EWSR1 | RG domain (IDR) | NR | ||||
| YG box (SD) | FMRP | Internal segment (IDR) | mRNA transport, translation | |||
| NR | HNRNPU | RGG box (IDR) | NR | |||
| NIRs | TNPO1 | HEAT repeats (SD) | HNRNPA1 | PY-NLS SLiM (IDR)e | 0.042 | Nuclear import, RBP chaperoning/disaggregation |
| HNRNPA2B1 | ||||||
| EWSR1 | ||||||
| TAF15 | ||||||
| FUS | 0.0095 | |||||
| HNRNPF | Nuclear import | |||||
| HNRNPM | 0.01 | |||||
| HNRNPD | 0.0032 | |||||
| HNRNPDL | 1 | |||||
| SAM68 (KHDRBS1) | ||||||
| ELAVL1 (HUR) | ||||||
| KAP104(yeast) | Nab2p(yeast) | 0.037 | ||||
| Hrp1p(yeast) | 0.032 | |||||
| Kapβ1/Impα | HEAT/Arm repeats (SD) | TDP43 (TARDBP) | cNLS SLiM (IDR)e | 0.066 | Nuclear import, RBP chaperoning/disaggregation | |
| Motor proteins | Myo4p(yeast) | C terminus (SD) | She3p(yeast) | Pseudocoiled-coil (SD) | 0.058 | mRNA transport |
| She3p-She2p(yeast)f | P/R sites (IDR)-structure (SD) | 1.6 | ||||
| KIF11 | Tail domain (IDR/SD) | IGF2BP1 (ZBP1) | RRM domains (SD) | |||
| KIF3C | C terminus (IDR) | FMRP | NR | |||
| UPF1 | UPF1 | Helicase domain (SD) | SLBP | N-terminal half (IDR) | mRNA stability | |
| RecA domain (SD) and Thr28 (IDR) | ZC3H12A (regnase 1) | Internal segment (IDR) and RNase domain (SD) | ||||
| N terminus | STAU1/2 | Tubulin-binding domain (IDR) | ||||
| CCR4-NOT | CNOT1 | HEAT domain (SD) | ZFP36 (TTP) | SLiM (IDR) | 2 | mRNA stability, translation |
| ZFP36L1 (BRF1) | ||||||
| NR | TNRC6C (GW182)g | W-containing motifs (IDR) | ||||
| CNOT9 | Concave surface (SD) | Bam | CBM SLiM (IDR) | 0.183 | ||
| RC3H1/2 (Roquin 1/2) | mRNA stability | |||||
| CNOT4 | mRNA stability | |||||
| Convex surface (W pockets) (SD) | ZFP36 (TTP) | Isolated W residues (IDR) | 5.5 | mRNA stability | ||
| TNRC6A/C (GW182)g | mRNA stability, translation | |||||
| NOT module | NR (SD) | RC3H1/2 (Roquin 1/2) | C terminus (IDR) | mRNA stability | ||
| HELZ | mRNA stability, translation | |||||
| PUM1/2 | N-terminal domain (IDR) | |||||
| Bicaudal C | KH domains (SD) | |||||
| SHD (SD) | NANOS1–3 | SLiM (IDR) | Translation | |||
| YTHDF2 | P/Q/N-rich region (IDR) | RNA stability | ||||
| CNOT4 | C terminus (IDR) | mRNA stability | ||||
| PABPC | PABPC1 | MLLE domain (SD) | ATXN2 | PAM2 SLiM (IDR) | 0.7 | RBP chaperoning |
| HELZ | NR | |||||
| NR | USP10 | 26 | NR | |||
| Multiple domains | MKRN1 | RQC, translation | ||||
| PABPC1/4 | NR | NFX1 | mRNA stability | |||
| 4EHP | 4EHP | Dorsal surface (SD) | Bicoid (Bcd) | SLiM (IDR) | Translation | |
| NR | PKNOX1 (Prep1) | |||||
| eIF4G | TIF4631/2(yeast) | C terminus | Scd6(yeast) | RGG box (IDR) | ||
| Khd1(yeast) | NR | |||||
| Internal region | Sbp1(yeast) | RGG box (IDR) | ||||
| Internal region and C terminus | Npl3(yeast) |
CBM, CAF40-binding motif; IDR, intrinsicaLLy disordered region; Kd, dissociation constant; NIR, nuclear import receptor; NLS, nuclear localization sequence; NR, not reported; PABPC, cytoplasmic poly(A)-binding protein; PY-NLS, proline-tyrosine-rich NLS; RBP, RNA-binding protein; RQC, ribosome quality control; RRM, RNA recognition motif; SD, structured domain; SLiM, short linear motif; SMN, survival motor neuron protein; snRNP, small nuclear ribonucleoprotein; STAU, Staufen; TNPO1, transportin 1.
Only those direct RBP-effector interactions are listed where a contact region has been mapped in at least one of the interacting proteins. For a fully referenced list of direct and indirect interactions, see Supplementary Table 2. Effectors and RBPs are named by their official gene symbol with text in brackets indicating popular aliases. All interacting proteins are metazoan except for the indicated yeast proteins.
IDR (or its part) or SD (or its part) in brackets indicates peptide organization of the contact region in its non-bound state. Where not reported, this information was obtained through modelling of the protein structure using the structure prediction algorithm AlphaFold173.
The Kd in most cases pertains to an in vitro interaction between SLiM of an RBP and the interacting effector domain or the full-length effector protein.
Part of U1 snRNP.
Both She2p and She3p are RBPs in contact with the transported mRNA62.
RBPs and survival motor neuron protein.
Survival motor neuron protein (SMN) is an essential effector protein the canonical role of which is in the assembly of nuclear RNPs. This protein is notorious for its reduced function causing spinal muscular atrophy (SMA), a fatal autosomal recessive disorder characterized by degeneration of lower motor neurons43,44. Although the reason for the specific manifestation of SMN deficiency in motor neurons remains unclear, several studies have pointed to additional roles of SMN in neuronal mRNA trafficking and control of local translation; these processes rely on and are regulated by direct interactions between SMN and various RBPs43,45,46 (Table 1). Curiously, unlike most other RBP–effector contacts, these interactions rely primarily on recognition of a single residue, a mono- or dimethylated but not non-methylated arginine embedded in an arginine–glycine (RGG/RG)-rich IDR of an RBP, by the conserved Tudor domain of SMN43,47,48 (Fig. 3c and Table 1). Little is known about the exact regulatory role of these contacts. However, intragenic SMA-associated mutations within the Tudor domain of SMN that disrupt RBP–SMN interactions point to their direct involvement in disease pathogenesis45,49,50.
RBPs and nuclear import receptors.
Most RBPs with a nuclear role require an active, carrier-mediated process for their import into the nucleus. This is facilitated by nuclear import receptors (NIRs) that bind RBPs exposing a nuclear localization sequence (NLS) and ferry them across a nuclear pore complex, while securing directionality of transport into the nucleus through competitive interactions with a small GTPase, Ran51 (Fig. 3d). NIRs such as Kapβ1/Impα and transportin 1 (TNPO1) specifically recognize the classical, lysine-rich NLS (cNLS) and proline-tyrosine (PY)-NLS, respectively. Remarkably, beyond their role in nucleocytoplasmic trafficking, NIRs have recently been found to moonlight as RBP chaperones, having the capacity to prevent cytoplasmic condensation of aggregation-prone RBPs, and as ‘disaggregases’, denoting their ability to dissolve preformed, including aberrant, RNP condensates52–55 (Fig 1). How do these unexpected moonlighting activities of NIRs come about? Akin to most other short linear motifs (SLiMs) of RBPs that mediate PPIs with effectors, NLSs are largely disordered, flanked by IDRs, and bind a structured surface of NIRs. However, most NLSs exhibit significantly higher effector-binding affinities than several other characterized SLiMs of RBPs (Table 1). The high-affinity interaction with NLS in turn facilitates secondary, low-affinity contacts that a NIR makes with other IDR domains, especially prion-like and RGG/RG-rich domains of the cargo RBP. These weak and dynamic interactions are thought to be essential in antagonizing homeostatic and pathological RNP condensate formation53–55. We discuss their therapeutic potential below.
Cytoplasmic activities of the nexus
RBPs and motor proteins.
Subcellular localization of mRNA enables precise control over the site of protein synthesis56. This in turn allows a cell to configure different subcellular domains for specialized functions to support biological processes as diverse as budding in yeast, embryonic patterning in the fruitfly and synaptic activity in mammalian neurons57. Molecular motor-based transport of membraneless RNA granules has emerged as the prevailing mechanism of mRNA localization, and much of the mRNA transport was found to occur through a process called ‘hitchhiking’, whereby RNA granules dock onto membrane-bound organelles that in turn couple to motor proteins for transport58,59. However, certain RNA granules were found to directly connect to motor proteins via RBPs for autonomous transport of their RNA cargo, offering a plausible explanation for the observed diversity of mRNA localizations60. For instance, direct RBP–motor protein PPIs were found to mediate mating type-defining asymmetrical localization of ASH1 mRNA in budding yeast61–63, transport of Actb mRNA to cellular leading edge to support migration of a mouse fibroblast20 or dendritic translocation of neurospecific mRNAs for their localized translation at the synapse64 (Fig. 4a, Table 1 and Supplementary Table 2). A deeper understanding of this nexus should help to clarify how hundreds of different mRNAs are distinctly localized in the cell to allow for an exquisite spatial and temporal control of gene expression.
RBPs and the RNA helicase UPF1.
Half-lives of cellular mRNAs are heavily dependent on stability-linked cis-acting RNA elements. A handful of RBPs with specificity for stem–loop structures or extended double-stranded RNA stretches are known to interact with a unique effector, the RNA helicase UPF1, to promote decay of mRNAs that harbour such elements65. UPF1, which is best known as a central component of a major quality control pathway known as nonsense-mediated decay6, thus operates as a point of convergence for functionally diverse RBPs, each of which regulates the stability of its targeted mRNAs (Fig. 4b). To stimulate the RNA helicase activity of UPF1, the recruiting RBPs must bypass the intramolecular autoinhibitory interaction between the CH domain and the helicase domain of UPF1 (refs.66,67). How this is achieved remains unclear, although several reports point to an activating role of direct, IDR-mediated contacts between RBPs and the inhibitory domains of UPF1 (refs.22,65,68,69) (Table 1 and Supplementary Table 2). Upon activation, UPF1 is believed to unwind the RBP-bound RNA structure to facilitate local endonucleolytic cleavage, which, along with a decapping complex and exonucleases, ensures rapid destruction of the targeted transcript70. Additional RBPs are speculated to engage this pathway of mRNA decay65.
RBPs and the CCR4–NOT complex.
Poly(A) tails are required for stability and translational efficiency of mRNAs, but are also subject to enzyme-mediated shortening, or deadenylation, a process that is key for initiating mRNA decay71. The CCR4–NOT deadenylase trims adenosines proximally to the 3′ untranslated region (UTR) with a major effect on mRNA stability. Notably, in addition to its role in ‘baseline’ deadenylation of bulk mRNA, CCR4–NOT is widely thought of as a hub of regulated, mRNA-specific deadenylation, much of which is driven by RBPs, microRNAs (miRNAs) and codon optimality71,72. In the multi-subunit CCR4–NOT complex, the CNOT9 subunit and the CNOT2–CNOT3 heterodimer constitute the major regulatory hotspots through which several RBPs are thought to recruit CCR4–NOT to select mRNAs for deadenylation15,73–77 (Fig. 4c and Table 1). Interestingly, the concave surface of CNOT9 non-specifically binds RNA, albeit in a mutually exclusive manner with RBPs (Fig. 4d). This has led to an intriguing hypothesis that, through competition for CNOT9 binding with mRNA, RBPs might divert the deadenylating activity of CCR4–NOT from bulk mRNA to specific mRNA targets72. Notably, most, if not all, studied PPIs seem to conform to the general trend whereby an IDR of an RBP, often a defined SLiM, interacts with a structured domain of a CCR4–NOT subunit (Fig. 4c and Table 1). A particular case is the convex surface of CNOT9, which recognizes individual tryptophan residues present in the IDR of ZFP36 or the TNRC6 (also known as GW182) adaptor proteins; the latter recruit CCR4–NOT to Argonaute (AGO)–miRNA-targeted mRNAs and have a crucial role in miRNA-mediated gene silencing75,78–81 (Figs. 4c,4f).
RBPs and the cytoplasmic poly(A)-binding protein.
Although neither a classic effector nor a transcript-specific RBP, the abundant cytoplasmic poly(A)-binding protein (PABPC) is central to promoting efficient translation and surveillance of translation and decay rates of nearly every mRNA71. Canonical functions of PABPC encompass controlling the length of the poly(A) tail, physical communication between the two ends of the mRNA, and recruitment of various effectors and regulators of mRNA processing71,82 (Fig. 4d). The latter function of PABPC is mediated via its main protein-binding platform, the MLLE domain, which specifically recognizes a SLiM, known as PAM2, found in IDRs of diverse proteins, including a handful of RBPs82 (Fig. 4d and Table 1). Curiously, in contrast to the expected roles in regulation of mRNA stability and translation82–84, a couple of rather unexpected roles have recently been found for RBP–PABPC interactions (Table 1). In particular, PABPC has been found to moonlight as an RNA-dependent chaperone that prevents spontaneous condensation of the intrinsically disordered RBP ataxin 2 (ATXN2)85. The chaperoning role of the ATXN2–PABPC interaction is especially notable because aberrant condensation of ATXN2 has been associated with two neurodegenerative disorders, spinocerebellar ataxia-2 and amyotrophic lateral sclerosis (ALS)86. Through a separate PAM2-mediated interaction, PABPC helps to position makorin 1 (MKRN1) upstream of premature poly(A) tails to block mRNA translation and facilitate ribosome-associated quality control87. Interestingly, an orthologous Mkrn1–pAbp PPI in the fruitfly is required for derepression of oskar mRNA translation during embryonic patterning84.
Given the precedent of ATXN2 and MKRN1, it is tempting to speculate about other non-canonical functions of RBP–PABPC interactions. These would be important to understand especially when one considers the large number of RBPs that might potentially be in close contact with PABPC via PAM2 motif–MLLE domain interactions. Specifically, our reanalysis of published data indicates that 77 human PABPC1-bound proteins detected by in vivo proximity-dependent biotinylation (BioID) analysis (77/120, 64%)88 are RBPs that harbour either a consensus PAM2 motif or a PAM2-like peptide sequence that is no more divergent from the consensus than the PAM2 motif of MKRN1, which differs from the consensus by three mismatched residues11,87,88 (Fig. 4d and Supplementary Table 3). Much larger numbers of candidate MLLE domain-binding RBPs with or without mismatches to the consensus PAM2 motif are obtained if all annotated human RBPs are considered11 (Fig. 4d and Supplementary Table 3). This is notable given that the MLLE domain is unique to PABPC and just one other, much less abundant human protein, the E3 ubiquitin ligase UBR5 (ref.82). It would thus be of interest to probe potential interactions of these RBPs with PABPC and determine their functions.
RBPs and translation initiation factors.
Rare examples of direct RBP–effector interactions with primary roles in controlling mRNA translation indicate various possible regulatory modes, most of which entail 3′ UTR-bound RBPs and components of the translation initiation machinery (Fig. 4e). For instance, in yeast, a set of RBPs directly associate with eIF4G to block recruitment of the preinitiation complex89, whereas in higher organisms, RBP-dependent recruitment of the translational repressor 4EHP (also known as eIF4E2) prevents eIF4E from binding to the 5′ cap on targeted transcripts (Fig. 4e and Table 1), a mechanism that sustains normal oogenesis in the fruitfly and in mammals19,90,91. Several other translationally linked RBP–effector interactions lack evidence of convergence; however, they largely seem to conform to the general SLiM- or IDR-structure type of interaction (Table 1). We also note that gene-specific translational control by RBPs is often coupled with regulation of mRNA stability and involves indirect, adaptor-mediated interactions with effectors, as is the case for AGO–miRNAs and several other RBPs79,90,92 (Fig. 4f and Supplementary Table 2).
Plasticity and dynamics of connections
An overview of the converging RBP–effector contacts shows that the large majority of them form between a short stretch of an IDR of an RBP, or a defined SLiM residing in it, and a structured part of an effector protein (Table 1). Such interactions not only confer high specificity and span a range of affinities to satisfy the required dynamics of PPIs, but also can be regulated and exhibit significant evolutionary plasticity93. Notably, because SLiMs are short, harbour a small number of essential residues and often reside in IDRs, they can rapidly evolve de novo from random peptide sequences and rewire protein interaction networks93–95. These properties allow novel cases of a particular SLiM to evolve independently in different RBPs for binding to the same structured binding site in a given effector, thus contributing to the convergent evolution of the nexus (Fig 1). A classic example is provided by independently evolved, dissimilar-in-sequence SLiMs of RBPs and competing non-RBPs that bind to the same contact site in the concave surface of CNOT9 (refs.15,74,96) (Fig. 4c).
Dynamics of RBP–effector interactions should be considered together with the events that occur at the level of RBPs binding to RNA, as RNA processing by many RBP-regulated effectors eventually depends on their recruitment to RNA (Fig. 2e). Notably, a recent kinetic analysis suggested that, in cells, RBPs bind to their cognate RNA sites transiently and infrequently, that individual binding events may last only seconds or shorter, and that few if any regulatory RBPs might be bound to a given mRNA at any given time97. If true, then how might adding another layer of transient and possibly infrequent interactions, in this case between RBPs and their effectors, still allow for assembly of functional RNA–RBP–effector modules to secure timely processing of RNA?
The answer might depend on the RNA process in question. For instance, several forms of RBP-regulated RNA processing might require only infrequent formation of relatively short-lived RNA–RBP–effector assemblies to initiate a process that can be propagated by effector-independent means. This may apply to processes such as decapping-triggered mRNA decay, stimulation of alternative splicing and, conceivably, mRNA clamping-dependent translational repression98. By contrast, processes such as TNPO1-mediated RBP extraction and nuclear import, or motor protein-driven mRNP transport may require more stable associations. Indeed, the reported affinities of PY-NLS motifs for TNPO1 are generally one to two orders of magnitude higher than most other measured SLiM/IDR–effector interactions (Table 1). Alternatively, in mRNA transport granules, high concentration of RBPs could fortify their sometimes already superstoichiometric binding to motor proteins via avidity effects62,99 (Fig. 4a).
Regulation of RBP–effector interactions
The formation and break-up of RBP–effector interactions do not rely solely on their strength and the local availability of interacting molecules in the cell but can additionally be regulated by cellular cues. The amenability to regulation can serve a host of biological processes that may require anything from fine-tuning RNA processing, enabling rapid, switch-like responses to intracellular or extracellular stimuli, to permanent rewiring of RNA networks. To accommodate such broad regulatory flexibility, RBP–effector interactions can be modulated either pre-translationally through alternative splicing (Fig. 5a), or post-translationally, by post-translational modifications (PTMs) or competitive interactions (Fig. 5b–d). Below, we discuss the principles of these regulatory strategies and list examples linked to regulation of diverse biological processes.
Fig. 5 |. Physiological regulation at the interface of RBPs and their effectors.
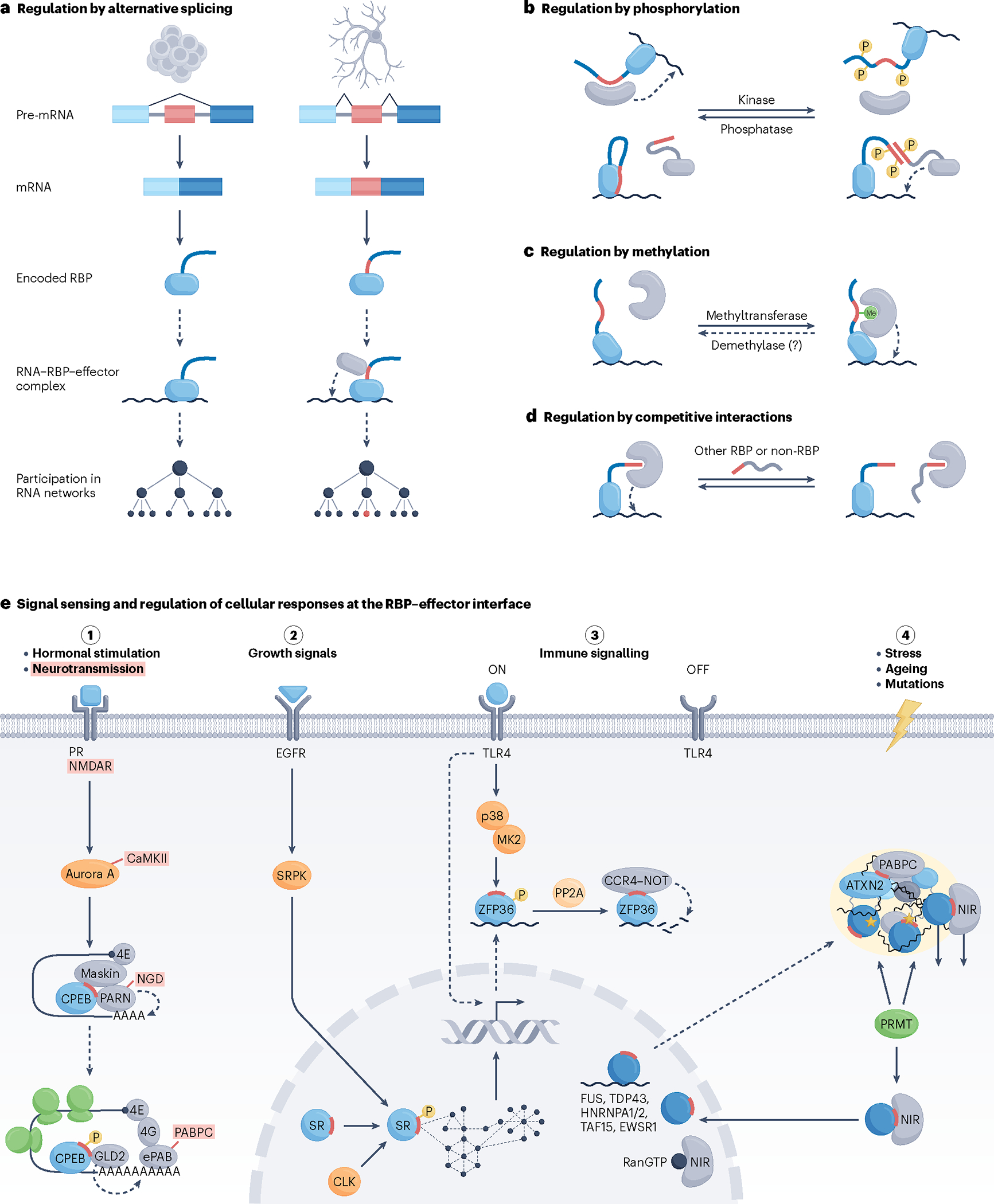
a–d, Modes of pre-translational (panel a) and post-translational (panels b–d) regulation of RNA-binding protein (RBP)–effector interactions involved in the control of biological processes. a, Tissue-specific alternative splicing, illustrated here as skipping or inclusion of the small linear motif (SLiM)-encoding alternative exon (red) in stem cells (left) or neurons (right), respectively, can facilitate rewiring of RNA networks. b, Phosphorylation, which is implemented by kinases and removed by phosphatases, most often disrupts RBP–effector interactions and occurs in RBP regions flanking the SLiM segment (red) that contacts the effector surface (top). Interactions of SR proteins with components of the spliceosome present a particular case in which phosphorylation of the RS domains (red) of both interacting partners stimulates contact establishment (bottom). c, Monomethylation or dimethylation of arginine residues of RBPs can stimulate RBP–effector interactions, typically through recognition of a methylated arginine by the Tudor domain195, as illustrated, or can weaken the affinity of RBPs for effectors, as in the case of the RBP–transportin 1 (TNPO1) interactions (not shown). It is unclear whether demethylation of RBPs also occurs in vivo (denoted by a question mark), although arginine demethylating activity has been ascribed to a handful of enzymes196. d, Competition between an RBP and other RBPs or non-RBPs for a common binding site on an effector can exert a direct regulatory effect on RNA processing. Competition of RanGTP with RNA-free RBPs for binding to nuclear import receptors can be considered as having an indirect effect on RNA processing (Fig. 3d). Curved dashed arrows depict effector activity on RNA in cis. e, The interface of RBPs and their effectors serves as a sensor of intracellular and extracellular signals as well as a regulator of cellular responses to signalling. Illustrated are signal transduction pathways (pathways 1–4) that trigger responses through distinct modes of post-transcriptional RNA processing. Hormonal stimulation of oocytes triggers their maturation, in part, via phosphorylation-dependent reconfiguration of cytoplasmic polyadenylation element binding protein (CPEB)–effector interactions. This turns CPEB from a repressor to an activator of polyadenylation-induced translation, a process that is crucial for germ-cell development. A highly similar pathway leading to CPEB activation is triggered upon synaptic stimulation of neurons and plays a key part in synaptic plasticity (terms in red, where indicated, are specific to the neuronal pathway)197. Green shapes indicate translocating ribosomes (pathway 1). Activities of SR proteins are modulated by external and internal signals via phosphorylation by SR protein kinases (SRPKs) and CDC-like kinases (CLKs), respectively, with the capacity to trigger a systemic response through changes in numerous alternative splicing events (pathway 2). Regulation of RBP–effector interactions via phosphorylation has a central role in securing a timely response to immune signalling, as well as its resolution. Upon stimulation with lipopolysaccharide (LPS), phosphorylation prevents association of ZFP36 with CCR4–NOT to help stabilize the induced and ZFP36-bound pro-inflammatory mRNAs. This response is rapidly reversed once the signalling subsides via protein phosphatase 2A (PP2A)-mediated dephosphorylation of ZFP36 and the ensuing recruitment of CCR4–NOT, which deadenylates the ZFP36-bound transcripts, which are then rapidly degraded (pathway 3). A series of largely nuclear RBPs (dark blue circles) with prion-like domains operate as splicing factors but partially also shuttle to the cytoplasm where they take on additional roles. Upon cellular stress, these and other aggregation-prone RBPs, such as ataxin 2 (ATXN2), potentially with their bound RNA, relocate to ribonucleoprotein (RNP) condensates/stress granules where they are kept functionally inert. Effectors that moonlight as RBP chaperones, including nuclear import receptors (NIRs) and cytoplasmic poly(A)-binding protein (PABPC), assist by preventing irreversible aggregation of RBPs in part through their recognition via nuclear localization sequence (NLS) and PAM2 motifs (red), respectively. Condensation properties as well as NIR interactions and nuclear import of some RBPs are additionally regulated by methylation by protein arginine methyltransferases (PRMTs). Excessive stress, RBP mutations (yellow asterisks) and ageing can prolong the time that RBPs spend in a condensed state, increasing the risk of RBP aggregation and neuronal degeneration. NIRs can act as disaggregases, with an intrinsic capacity to dissolve certain types of aberrant RNP condensates (pathway 4). CaMKII, calcium/calmodulin-dependent protein kinase II; EGFR, epidermal growth factor receptor; ePAB, embryonic poly(A) binding protein; MK2, MAPK-activated protein kinase 2; NMDAR, N-methyl-d-aspartate receptor; PR, progesterone receptor; TLR4, Toll-like receptor 4.
Rewiring of networks through pre-translational control
Alternative splicing, which affects the expression of nearly 95% of human multi-exon genes100, has emerged as a central mechanism for functional diversification of eukaryotic proteomes101. When alternative splicing affects an RBP or an effector of RNA processing, an opportunity arises for the establishment of new or alteration of existing RBP–effector interactions. The observation that IDRs and their embedded SLiMs, but not structured domains of proteins, commonly reside in non-constitutive exons suggests that alternative splicing of IDR-rich RBPs, rather than their SLiM-binding effectors, more commonly impinges on RBP–effector interactions95,102. Pre-translational modulation of RBP–effector interactions via alternative splicing offers additional regulatory opportunities, including stable rewiring of RNA networks (Fig. 5a). Notably, rewiring of PPIs often occurs in a cell- or tissue-specific manner and can crucially contribute to development and tissue identity103.
Several alternative RBP isoforms have been identified that harbour identical RNA-binding domains and differ only in short stretches of IDR, yet exhibit distinct effector roles and biological functions. For instance, two alternative isoforms of the Drosophila How RBP, one restricted to the nuclei of precursor cells and the other found in both the nuclei and the cytoplasm of mature cells, regulate a switch in tendon cell differentiation by exerting opposing effects on mRNA stability104. Interestingly, a similar switch mechanism enforced through differential subcellular localization but applied to the control of splicing and translation is used by alternative isoforms of the orthologous Quaking (QKI) RBP in regulating myelination in the mammalian nervous system104–107. Differential localization also underlies the function of ZAP (also known as ZC3HAV1), whose long isoform targets viral RNA for degradation, with the short isoform assisting by inhibiting programmed ribosomal frameshifting and resolving the antiviral response108,109. By contrast, with no overt difference in localization, one isoform of Musashi 2 (Msi2) RBP sustains a translation-repressive activity to promote anchorage-independent cell growth, whereas the other uniquely responds to differentiation-inducing phosphorylation110. Nonetheless, despite the marked biological impact of rewiring RBP–effector interactions via alternative splicing, our understanding of how such rewiring might lead to system-wide changes that underlie observed phenotypes remains minimal.
Responding to signal transduction via post-translational modulation
Reversible regulation of RBP–effector contacts is typically achieved by PTMs, of which phosphorylation has arguably garnered the most attention, particularly in the context of signal-regulated RNA processing events. The fast turnover of phosphorylation in fact renders this PTM particularly well suited to mediating rapid responses of several types of RNA processing to various signals111,112 (Fig. 5a). For instance, phosphorylation by SR protein kinases (SRPKs) and CDC-like kinases (CLKs) affects pre-mRNA splicing by regulating interactions between SR proteins and components of the spliceosome113. In this manner, SRPKs operate as major transducers of growth signals; upon activation by growth factors, SRPKs relocate to the nucleus and cause widespread changes in alternative splicing114 (Fig. 5e). By contrast, CLKs reside in the nucleus and act, for instance, in restoring the phosphorylation state of SR proteins during recovery from cellular stress115.
The importance in regulated RBP–effector interactions has also been observed in the control of polyadenylation-induced translation, which has essential roles in diverse biological processes, such as germ-cell development, cell cycle progression and synaptic function116. In these settings, a sequence-specific RBP cytoplasmic polyadenylation element binding protein (CPEB) initially keeps dormant a set of mRNAs with relatively short poly(A) tails. In oocytes stimulated to mature by progesterone, a signalling cascade is initiated that activates the Aurora A kinase, which in turn phosphorylates CPEB and converts it from a repressor into an activator of translation117 (Fig. 5e). This switch in CPEB activity results from molecular rearrangements that affect interactions of CPEB with several effectors involved in the control of poly(A) tail length and mRNA translation118. Remarkably, a highly similar signalling cascade leading to CPEB activation and stimulation of local translation is triggered upon synaptic stimulation of neurons119 (Fig. 5e). In fact, loss-of-function genetic experiments in mice have demonstrated that CPEB-dependent translational control by cytoplasmic polyadenylation has an important role in synaptic plasticity, a process that is believed to form the underlying basis of learning and memory120.
Akin to pre-mRNA splicing and protein synthesis, signalling-mediated regulation of contacts between RBPs and their effectors also has a central role in the control of mRNA stability, the biological significance of which is well illustrated by the regulation of innate immune responses by the AU-rich element (ARE) binding to RBPs such as ZFP36121. In cells that receive an inflammatory stimulus — for instance, lipopolysaccharide (LPS)-activated macrophages — a signalling cascade is triggered that revamps the transcriptional landscape to instate a pathogen-induced gene expression programme typified by induction of short-lived, ARE-containing mRNAs that encode pro-inflammatory cytokines as well as ZFP36 mRNA122. The same stimulus activates a kinase cascade that phosphorylates the ZFP36 protein, preventing it from recruiting the CCR4–NOT deadenylase and allowing for accumulation of ZFP36-bound pro-inflammatory mRNAs and their encoded proteins123 (Figs. 4c,5e). When signalling ceases,PP2A dephosphorylates ZFP36, prompting it to recruit CCR4–NOT to the bound mRNAs, which then undergo rapid deadenylation followed by degradation75,123. Thus, regulation of the interactions between ZFP36 and CCR4–NOT is instrumental in controlling the magnitude and duration of the inflammatory response.
Swift post-transcriptional responses to neurotransmitter release or inflammatory stimuli exemplify biological processes that require quick-acting changes in RBP–effector connectivity; such modulation can be rapidly enacted by phosphorylation. However, in other physiological settings, a longer-lasting action may be required that is less sensitive to speed of onset or termination. Although controlled phosphorylation might again serve such needs, other, generally less-studied PTMs of RBPs can have just as potent regulatory roles at the nexus of RBPs and their effectors111. For instance, in yeast, arginine methylation was found to fortify the interactions between Scd6 or Sbp1 and eIF4G to facilitate translational repression of targeted mRNAs in response to glucose deprivation124,125. Arginine methylation also strengthened interactions between KHSRP and SMN to secure correct localization and mRNA stability in neurites of differentiating mammalian neurons126, but weakened the contact between FUS and TNPO1 to modulate the dynamics of nuclear import in HeLa cells53,127 (Fig. 5e). These findings suggest that methylation, which displays an inherently slow turnover128, can be adept at regulating a wide variety of post-transcriptional events at the RBP–effector interface129. On that note, very little is known about the spatiotemporal control of RBP methylation, including the activities of protein arginine methyltransferases (PRMTs), which catalyse the deposition of this PTM, and it remains unclear whether RBPs are ever demethylated in vivo.
System-wide studies have implicated acetylation as another common PTM of RBPs, and recent evidence suggests that acetylation, akin to phosphorylation and methylation, may also have a regulatory role at the RBP–effector juncture111,130. For instance, similar to arginine methylation, lysine acetylation was found to disrupt the interaction of FUS with TNPO1, reducing nuclear import and stimulating the localization of FUS in cytoplasmic stress granule-like inclusion bodies131. Lysine acetylation was also reported to weaken the interaction of EWSR1 with the U1 snRNP component SNRPC to regulate alternative splicing in response to DNA damage132. Notably, the identification of specific enzymes that catalyse acetylation and deacetylation of these RBPs indicates that the above PTM events might indeed be regulatory131,132.
At the level of RBP–RNA interactions, competition for the same RNA target site between RBPs with different, potentially opposing effects on a given post-transcriptional process, for example, splicing or mRNA stability, leads to distinct regulatory outcomes40,133–137. Similar competition can occur at the level of RBP–effector interactions (Fig. 5d). To date, few if any studies have documented such competition between different RBPs; however, experimental evidence has been gathered for competition between RBPs with non-RBPs for a common binding site on an effector. Examples supported by functionally relevant outcomes include the aforementioned competition between Staufen (STAU) RBPs and UPF2 for binding to UPF1 (ref.138), competition between TOB, PAN3 and eRF3 for binding to PABPC1 (ref.139), or the recently proposed competition between RBPs and RNF219 for binding to CCR4–NOT96. Given the regulatory impact of competitive effector binding on RNA processing, as well as the abundance of converging RBP–effector interactions (Supplementary Table 2), future studies are warranted to shed light on the prevalence and significance of such interactions in various biological processes.
Genetic disorders and potential therapies
Given their central roles in regulating all aspects of gene expression at the RNA level, it is not surprising that compromised function of RBPs underlies the origin of many diseases. According to a recently updated tally, a staggering 30% of all annotated human RBPs (1,054 of 3,470 RBPs) are mutated in Mendelian or somatic genetic diseases, with neurological disorders, metabolic diseases and cancer among the most common associated disease categories10. Similar to our minimal understanding of RBP–effector roles compared with RBP–RNA interactions, we still know very little about the pathomechanisms of RBPs in disease.
The wealth of data garnered through disease association studies suggests that disease-linked mutations occur more frequently in IDRs and RBP domains other than RNA-binding domains10. Furthermore, proteome-wide analyses reveal an enrichment of disease-related mutations in SLiMs within IDRs, especially at functionally important residues of SLiMs140. Given the central role that SLiMs have in establishing RBP–effector interactions (Table 1), it seems plausible that interference with these interactions in disease might contribute to the pathobiology of mutations in RBPs. This consideration becomes particularly worthwhile in light of the observation that the ELM database, the largest repository of experimentally validated SLiMs141, currently lists only around 4,000 out of more than 100,000 SLiMs that a eukaryotic proteome is predicted to contain93. Limited general knowledge of SLiMs and RBP action thus restricts our understanding of the involvement of the RBP–effector nexus in disease to only a handful of documented examples. However, several recent studies demonstrate the therapeutic potential of targeting RBP–effector interactions genetically or pharmacologically, highlighting the regulatory and biological potency of the nexus from a novel perspective.
Mutations that disrupt the nexus
Arguably the best-characterized disease-linked mutations that interfere with RBP–effector interactions cause pathological aggregation of misfolded RBPs, a signature trait of neurodegenerative disorders10. For instance, mutations in FUS can cause familial ALS as well as rare cases of frontotemporal lobar degeneration (FTLD) (Table 2). Remarkably, approximately half of these mutations occur within the PY-NLS and disrupt the high-affinity interaction of FUS with TNPO1 (ref.142) (Table 1). This disruption compromises nuclear import and leads to aberrant cytoplasmic accumulation of the otherwise largely nuclear FUS, which eventually becomes deposited in pathological protein aggregates, as seen in brains of affected individuals143. Notably, the severity of the defect in nuclear import of FUS faithfully tracks with disease onset and progression, supporting a causal relationship between the mutations in PY-NLS and disease pathogenesis143–145.
Table 2 |.
RBP–effector interactions in human disease
| RBP–effector interaction | Genetic disorder or developmental abnormality | Causative or associated mutation | Therapeutic approach |
|---|---|---|---|
|
| |||
| Disease-linked mutations that affect RBP–effector interactions | |||
|
| |||
| FUS–TNPO1 | ALS, rare cases of FTLD143 | Mutations in PY-NLS or | NR |
| HNRNPA1–TNPO1 | ALS175 | cNLS of RBPs | |
| HNRNPH2–TNPO1 | Neurodevelopmental delay, autism148 | ||
| TDP43–Kapβ1/Impα | ALS176,177 | ||
|
| |||
| MATR3–TREX complex | ALS, vocal cord and pharyngeal weakness with distal myopathy149,178 | Mutations in IDRs of MATR3 | NR |
|
| |||
| RBP–SMN | SMA48,179 | Mutations in the Tudor domain of SMN | NR |
|
| |||
| Therapeutically targeted RBP–effector interactions | |||
|
| |||
| SRSF6–U1 snRNP | Familial dysautonomia (Riley–Day syndrome) | Intronic mutation in ELP1 | Small-molecule CLK1 agonist-enhanced SRSF6 phosphorylation to rescue abnormal skipping of the IKBKAP exon 20 (ref.156) |
|
| |||
| SRSFs–U1/U2 snRNPs | Duchenne muscular dystrophy, cystic fibrosis, anhidrotic ectodermal dysplasia with immunodeficiency | Exonic mutation in DMD, intronic mutations in CFTR and IKBKG | Pharmacological CLK inhibition to suppress SRSF phosphorylation and pathogenic exon inclusion158,180,181 |
|
| |||
| SRSFs–U1 snRNP | Leukaemia, prostate cancer, colon cancer, breast cancer, lung cancer, neovascular eye disease | Cancer-associated mutations in tumour suppressor genes and oncogenes | Inhibition of SRPK activity to normalize isoform levels of genes linked to cancer progression, apoptosis and angiogenesis159–162 |
|
| |||
| FUS–TNPO1 EWSR1–TNPO1 TAF15–TNPO1 HNRNPA1–TNPO1 HNRNPA2–TNPO1 TDP43–Kapβ1/Impα |
ALS, FTLD, Alzheimer disease, multisystem proteinopathy | Aggregation-promoting mutations in RBPs or disrupted arginine methylation | Delivery of NIRs to afflicted neurons or modulation of arginine methyltransferase activity52–55,127,164 |
|
| |||
| ATXN2–PABPC | Spinocerebellar ataxia 2, ALS | Expansion of the polyglutamine domain in ATXN2 | Designer PABPC-like chaperones to counter pathogenic RNP condensate formation85 |
|
| |||
| ZFP36–CCR4–NOT | Rheumatoid arthritis, psoriasis, multiple sclerosis, juvenile idiopathic arthritis, inflammatory diseases | Mutations in and/or reduced expression of ZFP36, aberrant signalling | PP2A agonists to activate ZFP36 or forced expression of ZFP36 in peripheral tissues or immune cells165,166,182,183 |
|
| |||
| RBM38–eIF4E | Cancers | NR | SLiM-mimicking synthetic peptide Pep8 increases expression of p53 by blocking the inhibitory RBM38-eIF4E interaction169 |
ALS, amyotrophic Lateral sclerosis; ATXN2, ataxin 2; cNLS, lysine-rich NLS; FTLD, frontotemporal lobar degeneration; MATR3, matrin 3; NIR, nuclear import receptor; NLS, nuclear localization sequence; NR, not reported; PABPC, cytoplasmic poly(A)-binding protein; PP2A, protein phosphatase 2A; PY-NLS, proline-tyrosine-rich NLS; RBP, RNA-binding protein; SLiM, short linear motif; SMA, spinal muscular atrophy; SMN, survival motor neuron protein; snRNP, small nuclear ribonucleoprotein; TNPO1, transportin 1; TREX, transport and export.
Several other RBPs associated with neurodegenerative disorders, including HNRNPA1, HNRNPH2 and TDP43, can set off pathogenic cascades remarkably similar to that of FUS, with mutations in NLS leading to defective nuclear import, aberrant cytoplasmic accumulation, eventual aggregation and neurological disease146 (Table 2). Aside from their functional similarities, a key property common to these RBPs is their proneness to aggregation, which stems largely from their prion-like domains. Thus, the time, made longer by stress and ageing, that the aberrantly cytoplasmic RBPs spend in a condensed state is believed to be a key parameter in irreversible RBP aggregation and the resulting neuronal degeneration54,147 (Fig. 5e). Interestingly, whereas mutations of NLS in HNRNPA1 or TDP43 can lead to neurodegeneration akin to that caused by mutated FUS, a similar mutation in HNRNPH2 was found to compromise neurodevelopment and manifest in autism148.
Circumstantial evidence also exists for disruption of other types of RBP–effector interaction in human disease. For instance, several mutations in the IDRs of the nuclear matrix RBP matrin 3 (MATR3), which are associated with neuromuscular disorders, were shown to affect interactions of MATR3 with components of the transcription and export (TREX) effector complex and with defects in the nuclear export of mRNAs149. However, the directness of these interactions and their contribution to disease remain unclear, especially given that these mutations also lead to potentially pathogenic associations of MATR3 with other proteins150. Curiously, disease-linked mutations that compromise RBP–effector interactions might also occur in the effector rather than its bound RBPs. Approximately 5% of patients with SMA carry intragenic mutations in the SMN1 gene; a few of these mutations, such as Glu134Lys, locate to the Tudor domain of SMN and weaken its affinity for a series of RBPs43,48 (Fig. 3c and Table 1). Such effector mutations would be expected to broadly compromise RNA metabolism and cellular function; however, their manifestation in a specific phenotype points to the cellular context as an important determinant of disease phenotype151–153. It remains to be seen whether disease-linked alterations identified in other effectors might cause pathology by compromising interactions with RBPs10,154,155. Improved understanding of the RBP–effector nexus is sure to shed light on the underpinnings of various human disorders.
Drugging the nexus
Some recent discoveries have electrified the field by demonstrating a striking potential of manipulating RBP–effector interactions in treatment of some of the most devastating human diseases, including neuromuscular disorders and cancer. Below, we summarize opportunities for therapeutic intervention at the nexus that arise from modulating the strength of existing interactions or through establishment of novel contacts between RBPs and their effectors.
One of the most promising therapeutic modalities harnesses the regulatory role of phosphorylation and the relative ease with which one can pharmacologically manipulate kinase and phosphatase activities. Notably, phosphorylation of SR proteins, which regulates the contacts of RBPs with the spliceosome and thus their activities as splicing factors (Fig. 3b), has been successfully modulated by small molecules to affect therapeutically relevant splicing decisions (Fig. 5e). For instance, the RECTAS compound, an agonist of CLK1, has shown promise for treatment of familial dysautonomia (FD), a fatal recessive neurodegenerative disease caused by a mutation in intron 20 of the ELP1 gene, which leads to abnormal skipping of the ELP1 exon 20 (refs.156,157). RECTAS was found to directly interact with and activate CLK1, leading to enhanced phosphorylation of the SR protein SRSF6 and restorative inclusion of exon 20 in multiple FD disease models156. By contrast, CLK inhibitors have proved effective in reducing phosphorylation of SRSFs and promoting therapeutic skipping of mutation-affected exons. One such example is TG003 inhibitor-promoted skipping of a mutated exon in the dystrophin (DMD) gene, which increases production of the DMD protein in cells of patients with Duchenne muscular dystrophy158 (Table 2). Moreover, synthetic SRPK inhibitors, which likewise suppress phosphorylation of SRSFs and their interactions with the spliceosome, have shown potent tumour-suppressive activities in diverse cancer types that present with pathological exon inclusions in genes such as VEGF, MYB, BRD4 and MED24 (refs.159–162). Together, these studies exemplify how chemical control of RBP–effector interactions might provide a new mechanism-oriented therapeutic opportunity.
Neurodegenerative disorders have long been considered incurable and with few treatment options. Disorders such as ALS, FTLD and Alzheimer disease are characterized by disrupted homeostasis of aggregation-prone RBPs, which is thought to underlie the formation of abnormal inclusion bodies146. The pathogenesis associated with this process may result from toxic properties of the inclusion bodies themselves and/or loss of function due to RBP mislocalization163. The recent discovery that NIRs, including TNPO1 and Kapβ1/Impα, act as chaperones and disaggregases beyond their canonical roles in the nuclear import of RBPs has generated considerable excitement about the prospect of reversing RBP aggregation to mitigate neuronal degeneration52,55,164 (Fig. 5e). These studies provide a rationale for therapeutic strategies that aim to increase the level of NIRs via gene therapy approaches or, conceivably, pharmacological strengthening of the interactions between mislocalized RGG/RG-rich RBPs and TNPO1 (refs.53,127) (Table 2). Although implementation of these strategies or the ATXN2-chaperoning PABPC in human therapy may prove challenging85, the unique potential to combat some of the most devastating genetic diseases warrants further investigation.
Manipulating RBP–effector interactions has also shown promise in treating autoimmune and inflammatory disorders. For instance, targeting the anti-inflammatory RBP ZFP36 with agonists of PP2A, which results in dephosphorylation of ZFP36 and recruitment of the CCR4–NOT deadenylase, can confer significant protection against inflammatory arthritis and bone erosion in mice165 (Fig. 5e). Consistently, adenoviral delivery of ZFP36 protected against bone loss and led to reduced inflammatory cell infiltration in rats166, whereas mice with genetically stabilized ZFP36 exhibited protection against induced forms of arthritis and other immune disorders167 (Table 2).
Aside from modulating the nexus-regulated RNA splicing, stability and RNP condensation discussed above, a case has also been made for the control of mRNA translation. The activity of the cancer-associated RBP RBM38, which suppresses p53 translation by directly interacting with eIF4E on Trp53 mRNA168, could be inhibited by a synthetic, RBM38 SLiM-mimicking peptide, called Pep8, that interferes with the RBM38–eIF4E interaction, derepresses p53 translation and attenuates tumour sphere formation and growth of xenograft tumours169. Incidentally, several small molecules and designer peptides that mimic SLiMs have shown promise as drugs that act by inhibiting various types of PPI, and some have already entered clinical trials93.
Taken together, the above examples illustrate the breadth of therapeutic opportunities arising from manipulation of RBP–effector interactions, which is all the more significant in light of our still rudimentary knowledge of this nexus. Whereas pharmacological targeting approaches seem readily translatable to human therapy, implementation of strategies entailing gene therapy will necessitate further feasibility studies. We note that with expanding understanding of the nexus, additional targeting modalities may emerge; antisense oligonucleotides, several of which are currently being used in the clinic170, might, for instance, find therapeutic utility in the pre-translational manipulation of SLiMs (Fig. 5a).
Conclusions and perspectives
The regulation of RNA networks has largely been understood at the level of RNA recognition by RBPs and more recently at the level of RNP condensate formation. Progress in these and other areas of RNA research has revealed much about the organizational principles of RNA networks, the complexity of which is managed by converging molecular interactions that are apparent at every level of network organization. Despite these insights, relatively little is known about the mechanistic bases of RBP activities or the ways in which effectors, the chief executors of RNA processing, contribute to the overall structure and operation of RNA networks. Interestingly, an integrated view of the binary interactions considered in this Review alone suggests that the regulatory level at which RBPs communicate with their effectors might consist of a network that is perhaps more interconnected than anticipated (Fig. 6). Technological breakthroughs along with accumulating knowledge about the functions and structures of RBPs and their effectors are certain to enable a more expansive view and better understanding of this nexus.
Fig. 6 |. A network view of the nexus.
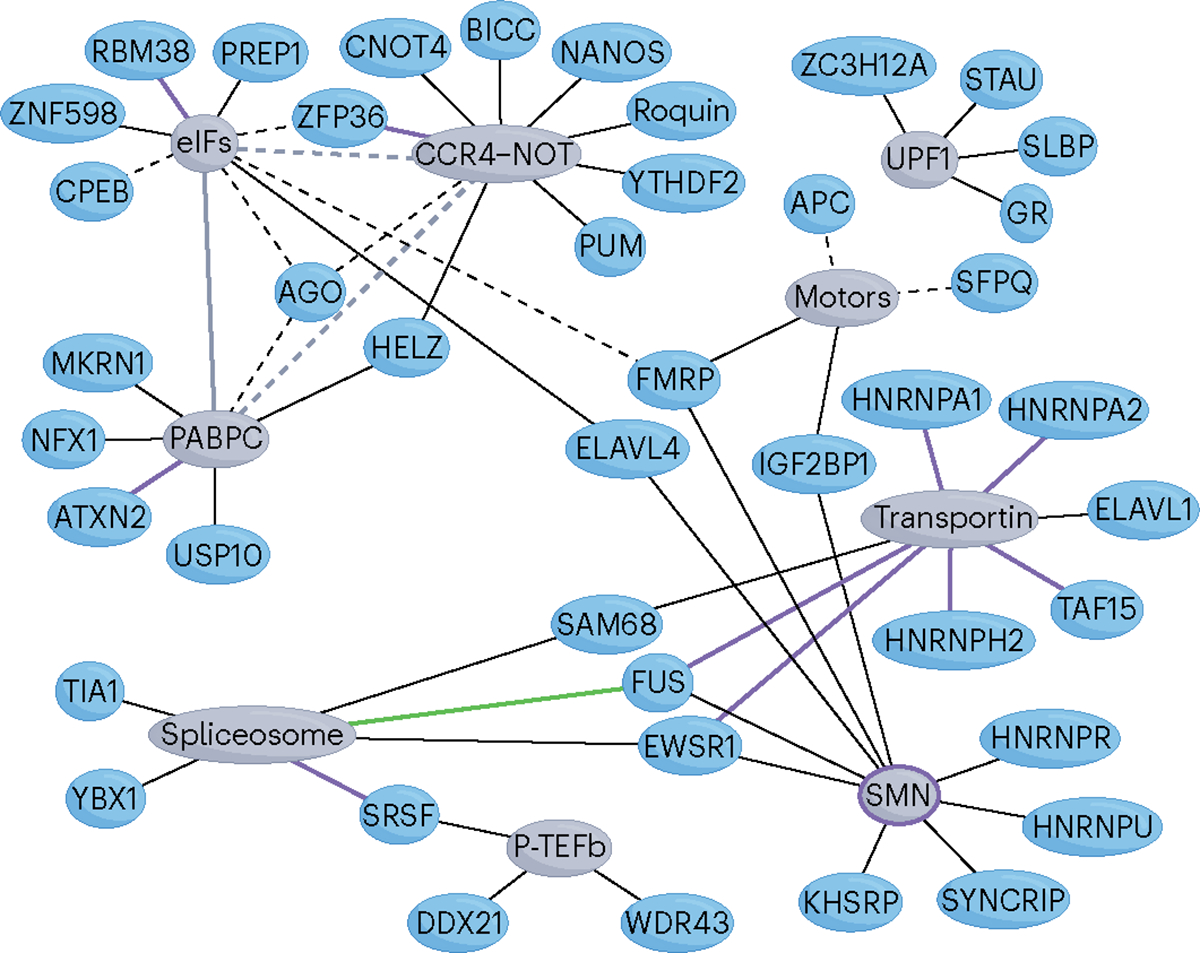
Shown is a compilation of converging RNA-binding protein (RBP)–effector interactions that have been characterized at both the molecular and functional levels. Note that several RBPs (blue shapes) connect (black lines) to more than one effector (grey shapes) and that there also exist RBP-independent interactions between different effectors (grey lines). Solid and dashed lines denote direct and indirect (adaptor-mediated) protein–protein interactions (PPIs), respectively. The green line that connects FUS and the spliceosome represents an RBP–RNA interaction. Purple lines represent disease-associated or therapeutically targeted interactions (Table 2). The purple circle around survival motor neuron protein (SMN) indicates spinal muscular atrophy (SMA)-linked mutations of SMN that weaken its affinity for multiple RBPs. Only mammalian RBPs are shown. Motor proteins (motors; KIF11, KIF3C, KIF5A, KIF3AB) and translation initiation factors (eIFs; eIF4E, eIF4E2 (4EHP), eIF4G) have been grouped together and are shown as a single effector. Non-converging PPIs discussed in the text, TDP43–Kapβ2/a1 and MATR3–TREX, are omitted. CPEB, cytoplasmic polyadenylation element binding protein; MKRN1, makorin 1; PABPC, cytoplasmic poly(A)-binding protein; STAU, Staufen.
Several pertinent questions remain. For instance, oftentimes, a mode of post-transcriptional control by an RBP becomes apparent from meta-analysis of transcriptome-wide data sets, but many, sometimes most, individual transcripts are not regulated as expected, despite being bound. Could this be explained by transcript-dependent efficiency of effector recruitment (or blockage of effector recruitment) by the RBP, alternative RBP isoforms or PTM of effectors? Is regulation of RNA binding by RBPs, as seen in budding yeast, embryonic patterning in the fruitfly or polarization of mammalian neurons, coordinated with the regulation of RBP–effector interactions57? Could functions of RBPs be accurately predicted based on knowledge of their interactions with effectors — or presence of SLiMs alone — in addition to RNA-binding information? Does the condensation propensity of many RBPs affect their ability to recruit effectors? How is the availability of effectors affected when condensate formation becomes pathological? Could we avert neurodegeneration by manipulating RBP–NIR interactions?
Unlike the large arsenal of high-throughput methods tailored to studies of RNA binding by RBPs, investigations of RBP–effector interactions have traditionally been limited to reductionist approaches. Although invaluable to the current understanding of communication between RBPs and effectors, such studies often yield limited and possibly inaccurate information about the dynamic nature, complexity and effects of these interactions in vivo. To tackle this problem, additional approaches that more directly capture the in vivo context of RBP–effector interactions are required. For instance, cryo-electron microscopy (cryo-EM) studies of purified RBP–effector complexes could provide views of concurrently interacting molecules and their conformations to help address the many unanswered questions about the workings of RBP–adaptor–effector conduits. These efforts could be aided by single-molecule biophysical experiments as well as kinetic assays that entail ultra-fast crosslinking strategies to investigate the dynamics of the interactions97. It may also be possible to devise pull-down protocols that combine, for example, enzyme-mediated proximity tagging with CLIP or RIP to study RBP–effector interactions in vivo transcriptome-wide. Such studies could centre on either the effector, adaptor or RBP of interest and explore protein complexes that simultaneously bind to RNA in a position-specific manner.
With the large majority of the annotated RBPs still unstudied, novel adaptor and effector proteins continuing to be discovered171,172 and with fewer than 5% of the expected number of SLiMs being functionally annotated93,141, it is clear that we have only just begun to delve into the converging RBP–effector interface. Expanding this knowledge will be important to better understand the mechanisms of RNA processing as well as the aetiology of human disease.
Supplementary Material
Acknowledgements
The authors thank E. Jankowsky, L. Maquat, A. Ephrussi and members of the authors’ laboratories for helpful discussions. The authors’ work pertaining to the reviewed topic is supported by the Intramural Research Program, Center for Cancer Research (to E.V.) and grant GM144693 (to J.M.) from the US NIH, and grant no. 2054195 (to S.C. and J.M.) from the US National Science Foundation. The authors apologize for the literature omitted owing to space limitations.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41576-022-00550-0.
Peer review information Nature Reviews Genetics thanks Matthias Hentze and Sarath Janga for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Licatalosi DD & Darnell RB RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11, 75–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen M & Manley JL Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scotti MM & Swanson MS RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowsky E & Harris ME Specificity and nonspecificity in RNA–protein interactions. Nat. Rev. Mol. Cell Biol. 16, 533–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramskold D, Wang ET, Burge CB & Sandberg R An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5, e1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurosaki T, Popp MW & Maquat LE Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper TA, Wan L & Dreyfuss G RNA and disease. Cell 136, 777–793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shendure J et al. DNA sequencing at 40: past, present and future. Nature 550, 345–353 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Ramanathan M, Porter DF & Khavari PA Methods to study RNA–protein interactions. Nat. Methods 16, 225–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebauer F, Schwarzl T, Valcarcel J & Hentze MW RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Hentze MW, Castello A, Schwarzl T & Preiss T A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Van Nostrand EL et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forch P, Puig O, Martinez C, Seraphin B & Valcarcel J The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21, 6882–6892 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forch P et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6, 1089–1098 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Sgromo A et al. A CAF40-binding motif facilitates recruitment of the CCR4–NOT complex to mRNAs targeted by Drosophila Roquin. Nat. Commun. 8, 14307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandler H, Kreth J, Timmers HT & Stoecklin G Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 39, 4373–4386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian MR et al. Structural basis for the recruitment of the human CCR4–NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 20, 735–739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leppek K et al. Roquin promotes constitutive mRNA decay via a conserved class of stem–loop recognition motifs. Cell 153, 869–881 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Cho PF et al. A new paradigm for translational control: inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121, 411–423 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Song T et al. Specific interaction of KIF11 with ZBP1 regulates the transport of beta-actin mRNA and cell motility. J. Cell Sci. 128, 1001–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu JY & Maniatis T Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 (1993). This study was the first to functionally characterize RBP–effector interactions and their impact on RNA processing by showing that PPIs between SR proteins and the spliceosome have a regulatory role in alternative splicing.
- 22.Park E, Gleghorn ML & Maquat LE Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl Acad. Sci. USA 110, 405–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberti S & Hyman AA Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Sabari BR, Dall’Agnese A & Young RA Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon AS, Peeples WB & Rosen MK A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roden C & Gladfelter AS RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mengistu H, Huizinga J, Mouret JB & Clune J The evolutionary origins of hierarchy. PLoS Comput. Biol. 12, e1004829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilinski D et al. Recurrent rewiring and emergence of RNA regulatory networks. Proc. Natl Acad. Sci. USA 114, E2816–E2825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore MJ & Proudfoot NJ Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Bose M, Lampe M, Mahamid J & Ephrussi A Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell 185, 1308–1324.e23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keskin O, Gursoy A, Ma B & Nussinov R Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 108, 1225–1244 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Xiao R et al. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adelman K & Lis JT Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Core L & Adelman K Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi X et al. RNA targets ribogenesis factor WDR43 to chromatin for transcription and pluripotency control. Mol. Cell 75, 102–116.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Calo E et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji X et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153, 855–868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tahirov TH et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 465, 747–751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsen TW & Graveley BR Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ule J & Blencowe BJ Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell 76, 329–345 (2019). [DOI] [PubMed] [Google Scholar]
- 41. Xiao SH & Manley JL Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344 (1997). This paper provides early evidence that RBP–effector interactions can be regulated by PTMs; it demonstrates that phosphorylation of RS domains strengthens the interactions between SR proteins and U1 snRNP with relevance to pre-mRNA splicing.
- 42. Jutzi D et al. Aberrant interaction of FUS with the U1 snRNA provides a molecular mechanism of FUS induced amyotrophic lateral sclerosis. Nat. Commun. 11, 6341 (2020). This study demonstrates that the splicing activity of FUS relies on an interaction with spliceosomal RNA rather than a protein subunit, providing a rare case of protein–RNA interactions that connect an RBP and an effector.
- 43.Singh RN, Howell MD, Ottesen EW & Singh NN Diverse role of survival motor neuron protein. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 299–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li DK, Tisdale S, Lotti F & Pellizzoni L SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 32, 22–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubers L et al. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 20, 553–579 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Fallini C et al. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev. Neurobiol. 74, 319–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thandapani P, O’Connor TR, Bailey TL & Richard S Defining the RGG/RG motif. Mol. Cell 50, 613–623 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Tripsianes K et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18, 1414–1420 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Wijaya YOS et al. Phenotypes of SMA patients retaining SMN1 with intragenic mutation. Brain Dev. 43, 745–758 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Kotani T et al. A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J. Neurol. 254, 624–630 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Marfori M et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Qamar S et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofweber M et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Guo L et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoshizawa T et al. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e22 (2018). Together with Qamar et al. (2018), Hofwebe. et al. (2018) and Guo et al. (2018), the authors demonstrate that NIRs can act as RBP chaperones and disaggregases beyond their canonical roles in the nuclear import of RBPs, highlighting the relevance of misregulated RBP–NIR interactions to neurodegeneration.
- 56.Holt CE & Bullock SL Subcellular mRNA localization in animal cells and why it matters. Science 326, 1212–1216 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin KC & Ephrussi A mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao YC et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell 179, 147–164.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pushpalatha KV & Besse F Local translation in axons: when membraneless RNP granules meet membrane-bound organelles. Front. Mol. Biosci. 6, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lecuyer E et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Besse F & Ephrussi A Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9, 971–980 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Edelmann FT et al. Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat. Struct. Mol. Biol. 24, 152–161 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Muller M et al. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 9, e1000611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davidovic L et al. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 16, 3047–3058 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Kim YK & Maquat LE UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chakrabarti S et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41, 693–703 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Clerici M et al. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 28, 2293–2306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim YK, Furic L, Desgroseillers L & Maquat LE Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120, 195–208 (2005). This study identifies the first RBP that directly interacts with the RNA helicase UPF1 to promote decay of targeted mRNAs.
- 69.Mino T et al. Translation-dependent unwinding of stem-loops by UPF1 licenses Regnase-1 to degrade inflammatory mRNAs. Nucleic Acids Res. 47, 8838–8859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanaan J et al. UPF1-like helicase grip on nucleic acids dictates processivity. Nat. Commun. 9, 3752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Passmore LA & Coller J Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 23, 93–106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Raisch T et al. Reconstitution of recombinant human CCR4–NOT reveals molecular insights into regulated deadenylation. Nat. Commun. 10, 3173 (2019). This study proposes that RBPs might, through competition with non-specifically bound mRNAs, divert the deadenylating activity of CCR4–NOT from bulk mRNA to specific mRNA targets.
- 73.Keskeny C et al. A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4–NOT complex. Genes Dev. 33, 236–252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sgromo A et al. Drosophila bag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets. RNA 24, 381–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bulbrook D et al. Tryptophan-mediated interactions between tristetraprolin and the CNOT9 subunit are required for CCR4–NOT deadenylase complex recruitment. J. Mol. Biol. 430, 722–736 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Bhandari D, Raisch T, Weichenrieder O, Jonas S & Izaurralde E Structural basis for the Nanos-mediated recruitment of the CCR4–NOT complex and translational repression. Genes Dev. 28, 888–901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Enwerem III et al. Human Pumilio proteins directly bind the CCR4–NOT deadenylase complex to regulate the transcriptome. RNA 27, 445–464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y et al. A DDX6–CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell 54, 737–750 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Duchaine TF & Fabian MR Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb. Perspect. Biol. 11, a032771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jonas S & Izaurralde E Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433 (2015). [DOI] [PubMed] [Google Scholar]
- 81. Mathys H et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell 54, 751–765 (2014). Together with Chen et al. (2014), this report reveals the location of W-binding pockets in CNOT9, one of the hotspots for interactions of RBPs and TNRC6 adaptor proteins with the CCR4–NOT complex; these studies also present the structure of DDX6 bound to CCR4–NOT, pointing to physical links between miRNA- or RBP-dependent target recognition and regulation of mRNA stability and/or translation.
- 82.Xie J, Kozlov G & Gehring K The “tale” of poly(A) binding protein: the MLLE domain and PAM2-containing proteins. Biochim. Biophys. Acta 1839, 1062–1068 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Katzenellenbogen RA, Vliet-Gregg P, Xu M & Galloway DA NFX1–123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J. Virol. 83, 6446–6456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dold A et al. Makorin 1 controls embryonic patterning by alleviating Bruno1-mediated repression of oskar translation. PLoS Genet. 16, e1008581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Boeynaems S et al. Poly(A)-binding protein is an ataxin-2 chaperone that emulsifies biomolecular condensates. Preprint at bioRxiv 10.1101/2021.08.23.457426 (2021). This report documents the moonlighting activity of PABPC as a chaperone that prevents spontaneous condensation of the intrinsically disordered RBP ATXN2.
- 86.Elden AC et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hildebrandt A et al. The RNA-binding ubiquitin ligase MKRN1 functions in ribosome-associated quality control of poly(A) translation. Genome Biol. 20, 216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Youn JY et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Rajyaguru P, She M & Parker R Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45, 244–254 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Christie M & Igreja C eIF4E-homologous protein (4EHP): a multifarious cap-binding protein. FEBS J. 10.1111/febs.16275 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Villaescusa JC et al. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS ONE 4, e5213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kamenska A, Simpson C & Standart N eIF4E-binding proteins: new factors, new locations, new roles. Biochem. Soc. Trans. 42, 1238–1245 (2014). [DOI] [PubMed] [Google Scholar]
- 93. Van Roey K et al. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 114, 6733–6778 (2014). This comprehensive review considers SLiMs as a distinct class of protein interaction module and provides an in-depth analysis of their properties and functions in the control of cell physiology.
- 94.Davey NE, Cyert MS & Moses AM Short linear motifs – ex nihilo evolution of protein regulation. Cell Commun. Signal. 13, 43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buljan M et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Poetz F et al. RNF219 attenuates global mRNA decay through inhibition of CCR4–NOT complex-mediated deadenylation. Nat. Commun. 12, 7175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma D et al. The kinetic landscape of an RNA-binding protein in cells. Nature 591, 152–156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gotze M et al. Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA 23, 1552–1568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baumann S et al. A reconstituted mammalian APC-kinesin complex selectively transports defined packages of axonal mRNAs. Sci. Adv. 6, eaaz1588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pan Q, Shai O, Lee LJ, Frey BJ & Blencowe BJ Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008). [DOI] [PubMed] [Google Scholar]
- 101.Wright CJ, Smith CWJ & Jiggins CD Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 23, 697–710 (2022). [DOI] [PubMed] [Google Scholar]
- 102. Ellis JD et al. Tissue-specific alternative splicing remodels protein–protein interaction networks. Mol. Cell 46, 884–892 (2012). Together with Buljan et al. (2012), the authors argue that tissue-regulated exons are enriched in PPI-forming IDRs and provide experimental evidence that alternative splicing of these exons often diversifies protein interaction capabilities.
- 103.Baralle FE & Giudice J Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nabel-Rosen H, Volohonsky G, Reuveny A, Zaidel-Bar R & Volk T Two isoforms of the Drosophila RNA binding protein, How, act in opposing directions to regulate tendon cell differentiation. Dev. Cell 2, 183–193 (2002). [DOI] [PubMed] [Google Scholar]
- 105.Yamagishi R, Tsusaka T, Mitsunaga H, Maehata T & Hoshino S The STAR protein QKI-7 recruits PAPD4 to regulate post-transcriptional polyadenylation of target mRNAs. Nucleic Acids Res. 44, 2475–2490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Volk T, Israeli D, Nir R & Toledano-Katchalski H Tissue development and RNA control: “HOW” is it coordinated? Trends Genet. 24, 94–101 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Fagg WS et al. Autogenous cross-regulation of Quaking mRNA processing and translation balances Quaking functions in splicing and translation. Genes. Dev. 31, 1894–1909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schwerk J et al. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat. Immunol. 20, 1610–1620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zimmer MM et al. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat. Commun. 12, 7193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.MacNicol MC et al. Evasion of regulatory phosphorylation by an alternatively spliced isoform of Musashi2. Sci. Rep. 7, 11503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.England WE et al. An atlas of posttranslational modifications on RNA binding proteins. Nucleic Acids Res. 50, 4329–4339 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gelens L & Saurin AT Exploring the function of dynamic phosphorylation-dephosphorylation cycles. Dev. Cell 44, 659–663 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Zhou Z & Fu XD Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Z et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 47, 422–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ninomiya K, Kataoka N & Hagiwara M Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 195, 27–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Richter JD CPEB: a life in translation. Trends Biochem. Sci. 32, 279–285 (2007). [DOI] [PubMed] [Google Scholar]
- 117.Sarkissian M, Mendez R & Richter JD Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev. 18, 48–61 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim JH & Richter JD Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24, 173–183 (2006). [DOI] [PubMed] [Google Scholar]
- 119. Udagawa T et al. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell 47, 253–266 (2012). Together with Sarkissian et al. (2004) and Kim et al. (2006), the authors describe phosphorylation-dependent reconfiguration of CPEB–effector interactions that turn CPEB from a repressor to an activator of polyadenylation-induced translation, a process that is crucial for development of germ cells and for synaptic plasticity in the nervous system.
- 120.Alarcon JM et al. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn. Mem. 11, 318–327 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carpenter S, Ricci EP, Mercier BC, Moore MJ & Fitzgerald KA Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14, 361–376 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Mahtani KR et al. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell Biol. 21, 6461–6469 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Clement SL, Scheckel C, Stoecklin G & Lykke-Andersen J Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol. Cell Biol. 31, 256–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Poornima G, Shah S, Vignesh V, Parker R & Rajyaguru PI Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 44, 9358–9368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhatter N et al. Arginine methylation augments Sbp1 function in translation repression and decapping. FEBS J. 286, 4693–4708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tadesse H, Deschenes-Furry J, Boisvenue S & Cote J KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum. Mol. Genet. 17, 506–524 (2008). [DOI] [PubMed] [Google Scholar]
- 127.Dormann D et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barth TK & Imhof A Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 35, 618–626 (2010). [DOI] [PubMed] [Google Scholar]
- 129.Biggar KK & Li SS Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16, 5–17 (2015). [DOI] [PubMed] [Google Scholar]
- 130.Castello A et al. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Arenas A et al. Lysine acetylation regulates the RNA binding, subcellular localization and inclusion formation of FUS. Hum. Mol. Genet. 29, 2684–2697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang T et al. Acetylation dependent translocation of EWSR1 regulates CHK2 alternative splicing in response to DNA damage. Oncogene 41, 3694–3704 (2022). [DOI] [PubMed] [Google Scholar]
- 133.Gazzara MR et al. Ancient antagonism between CELF and RBFOX families tunes mRNA splicing outcomes. Genome Res. 27, 1360–1370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zarnack K et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152, 453–466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang Y, Ma M, Xiao X & Wang Z Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 19, 1044–1052 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ivanov P & Anderson P Post-transcriptional regulatory networks in immunity. Immunol. Rev. 253, 253–272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fu XD & Ares M Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gong C, Kim YK, Woeller CF, Tang Y & Maquat LE SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 23, 54–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Funakoshi Y et al. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21, 3135–3148 (2007). Together with Gong et al. (2009), this report provides experimental support for in vivo competition between RBPs and non-RBPs for a common binding site on an effector.
- 140.Uyar B, Weatheritt RJ, Dinkel H, Davey NE & Gibson TJ Proteome-wide analysis of human disease mutations in short linear motifs: neglected players in cancer. Mol. Biosyst. 10, 2626–2642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kumar M et al. ELM — the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 48, D296–D306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ling SC, Polymenidou M & Cleveland DW Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dormann D et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chio A et al. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol. Aging 30, 1272–1275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.DeJesus-Hernandez M et al. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 31, E1377–E1389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim HJ & Taylor JP Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron 96, 285–297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Patel A et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Bain JM et al. Variants in HNRNPH2 on the X chromosome are associated with a neurodevelopmental disorder in females. Am. J. Hum. Genet. 99, 728–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boehringer A et al. ALS associated mutations in Matrin 3 alter protein–protein interactions and impede mRNA nuclear export. Sci. Rep. 7, 14529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Malik AM & Barmada SJ Matrin 3 in neuromuscular disease: physiology and pathophysiology. JCI Insight 6, e143948 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pena V, Liu S, Bujnicki JM, Luhrmann R & Wahl MC Structure of a multipartite protein-protein interaction domain in splicing factor Prp8 and its link to retinitis pigmentosa. Mol. Cell 25, 615–624 (2007). [DOI] [PubMed] [Google Scholar]
- 152.Buskin A et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 9, 4234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Danilova N & Gazda HT Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Model. Mech. 8, 1013–1026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chang MT et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gonatopoulos-Pournatzis T et al. Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Mol. Cell 77, 1176–1192.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 156. Ajiro M et al. Therapeutic manipulation of IKBKAP mis-splicing with a small molecule to cure familial dysautonomia. Nat. Commun. 12, 4507 (2021). This study provides a mechanistic insight into how RBP–effector interactions can be modulated by small synthetic molecules that affect phosphorylation with potential therapeutic effects.
- 157.Yoshida M et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl Acad. Sci. USA 112, 2764–2769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nishida A et al. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat. Commun. 2, 308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tzelepis K et al. SRPK1 maintains acute myeloid leukemia through effects on isoform usage of epigenetic regulators including BRD4. Nat. Commun. 9, 5378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Amin EM et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell 20, 768–780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Batson J et al. Development of potent, selective SRPK1 inhibitors as potential topical therapeutics for neovascular eye disease. ACS Chem. Biol. 12, 825–832 (2017). [DOI] [PubMed] [Google Scholar]
- 162.Hatcher JM et al. SRPKIN-1: a covalent SRPK1/2 inhibitor that potently converts VEGF from pro-angiogenic to anti-angiogenic isoform. Cell Chem. Biol. 25, 460–470.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nussbacher JK, Tabet R, Yeo GW & Lagier-Tourenne C Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guo L, Fare CM & Shorter J Therapeutic dissolution of aberrant phases by nuclear-import receptors. Trends Cell Biol. 29, 308–322 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ross EA et al. Treatment of inflammatory arthritis via targeting of tristetraprolin, a master regulator of pro-inflammatory gene expression. Ann. Rheum. Dis. 76, 612–619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Patil CS et al. Targeting mRNA stability arrests inflammatory bone loss. Mol. Ther. 16, 1657–1664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Patial S et al. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc. Natl Acad. Sci. USA 113, 1865–1870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang J et al. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 25, 1528–1543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lucchesi CA, Zhang J, Ma B, Chen M & Chen X Disruption of the Rbm38-eIF4E complex with a synthetic peptide Pep8 increases p53 expression. Cancer Res. 79, 807–818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yu AM & Tu MJ Deliver the promise: RNAs as a new class of molecular entities for therapy and vaccination. Pharmacol. Ther. 230, 107967 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Morita M et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell Biol. 32, 3585–3593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Naarmann-de Vries IS et al. Methylated HNRNPK acts on RPS19 to regulate ALOX15 synthesis in erythropoiesis. Nucleic Acids Res. 49, 3507–3523 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Braun JE, Huntzinger E & Izaurralde E The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 768, 147–163 (2013). [DOI] [PubMed] [Google Scholar]
- 175.Liu Q et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology 87, 1763–1769 (2016). [DOI] [PubMed] [Google Scholar]
- 176.Winton MJ et al. A90V TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett. 582, 2252–2256 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Doll SG et al. Recognition of the TDP-43 nuclear localization signal by importin alpha1/beta. Cell Rep. 39, 111007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Johnson JO et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17, 664–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Selenko P et al. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8, 27–31 (2001). [DOI] [PubMed] [Google Scholar]
- 180.Shibata S, Ajiro M & Hagiwara M Mechanism-based personalized medicine for cystic fibrosis by suppressing pseudo exon inclusion. Cell Chem. Biol. 27, 1472–1482 e1476 (2020). [DOI] [PubMed] [Google Scholar]
- 181.Boisson B et al. Rescue of recurrent deep intronic mutation underlying cell type-dependent quantitative NEMO deficiency. J. Clin. Invest. 129, 583–597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Carrick DM et al. Genetic variations in ZFP36 and their possible relationship to autoimmune diseases. J. Autoimmun. 26, 182–196 (2006). [DOI] [PubMed] [Google Scholar]
- 183.Patial S & Blackshear PJ Tristetraprolin as a therapeutic target in inflammatory disease. Trends Pharmacol. Sci. 37, 811–821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Keene JD RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 (2007). [DOI] [PubMed] [Google Scholar]
- 185.Terry LJ, Shows EB & Wente SR Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 (2007). [DOI] [PubMed] [Google Scholar]
- 186.Hallegger M et al. TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell 184, 4680–4696.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Snead WT & Gladfelter AS The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell 76, 295–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lucas BA et al. Evidence for convergent evolution of SINE-directed Staufen-mediated mRNA decay. Proc. Natl Acad. Sci. USA 115, 968–973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yue Y et al. Role and convergent evolution of competing RNA secondary structures in mutually exclusive splicing. RNA Biol. 14, 1399–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Calabretta S & Richard S Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 40, 662–672 (2015). [DOI] [PubMed] [Google Scholar]
- 191.Bourgeois CF, Mortreux F & Auboeuf D The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17, 426–438 (2016). [DOI] [PubMed] [Google Scholar]
- 192.Hildebrandt A et al. Interaction profiling of RNA-binding ubiquitin ligases reveals a link between posttranscriptional regulation and the ubiquitin system. Sci. Rep. 7, 16582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rintala-Dempsey AC & Kothe U Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14, 1185–1196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Curtis NJ & Jeffery CJ The expanding world of metabolic enzymes moonlighting as RNA binding proteins. Biochem. Soc. Trans. 49, 1099–1108 (2021). [DOI] [PubMed] [Google Scholar]
- 195.Gayatri S & Bedford MT Readers of histone methylarginine marks. Biochim. Biophys. Acta 1839, 702–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Blanc RS & Richard S Arginine methylation: the coming of age. Mol. Cell 65, 8–24 (2017). [DOI] [PubMed] [Google Scholar]
- 197.Ivshina M, Lasko P & Richter JD Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 30, 393–415 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


