Abstract
Escherichia coli transiently accumulates large amounts of inorganic polyphosphate (polyP), up to 20 mM in phosphate residues (Pi), in media deficient in both Pi and amino acids. This transient accumulation is preceded by the appearance of nucleotides ppGpp and pppGpp, generated in response to nutritional stresses. Mutants which lack PhoB, the response regulator of the phosphate regulon, do not accumulate polyP even though they develop wild-type levels of (p)ppGpp when subjected to amino acid starvation. When complemented with a phoB-containing plasmid, phoB mutants regain the ability to accumulate polyP. PolyP accumulation requires high levels of (p)ppGpp independent of whether they are generated by RelA (active during the stringent response) or SpoT (expressed during Pi starvation). Hence, accumulation of polyP requires a functional phoB gene and elevated levels of (p)ppGpp. A rapid assay of polyP depends on its adsorption to an anion-exchange disk on which it is hydrolyzed by a yeast exopolyphosphatase.
Inorganic polyphosphate (polyP), linear polymers of hundreds of phosphates linked by high-energy phosphoanhydride bonds, are ubiquitous in nature, having been found in all organisms examined (16, 17, 38). Various biological functions have been demonstrated or considered for polyP depending on the levels present in different species or types of cells or subcellular compartments and on the metabolic status. These functions include actions as a substitute for ATP, as a chelator of divalent metal ions (e.g., Mn2+ and Ca2+), as a reservoir of phosphate, and in physiological adjustments to growth, development, stresses, and deprivations (5, 15, 27).
Escherichia coli responds to stresses, acute and prolonged, in a variety of ways. One of the best understood is the stringent response to the depletion of an amino acid (3, 4), which leads to the activation of relA and consequent massive accumulations of pppGpp and ppGpp. These nucleotides repress many genes, including those for ribosome synthesis, and activate (directly or indirectly) 50 or more genes responsible for coping with stress and starvation (4, 10).
Another example of nutritional stress is depletion of orthophosphate in the medium, which is sensed by a phosphate regulon (Pho) (23, 26, 34–37). Cells which enter a phase of starvation for Pi induce accumulation of ppGpp (32). This induction is relA independent and depends on the spoT gene product. Low phosphate levels are detected by PhoR, which then activates PhoB, the principal phosphate regulator (22, 23, 26). PhoB in turn activates 30 or more genes, including phoA, which encodes alkaline phosphatase (AP) (31, 35, 36). The product of spoT produces ppGpp and also degrades ppGpp to GDP and inorganic pyrophosphate (9, 29).
The phenotype of the E. coli mutant lacking polyP kinase (PPK), the enzyme responsible for the synthesis of polyP, is a failure to survive in stationary phase, attributable to the lack of stationary-phase gene expression (27). In this report, we describe the accumulation of polyP during nutritional stress imposed by amino acid and Pi limitation and the dependence of this accumulation on PhoB and high levels of ppGpp in vivo.
MATERIALS AND METHODS
Reagents.
The sources of the reagents and related supplies used in this study are as follows: ATP, creatine kinase, DNase I, and RNase IIIa were from Boehringer Mannheim; creatine phosphate, MOPS [3-(N-morpholino)propanesulfonic acid], kanamycin, ampicillin, tetracycline, p-nitrophenyl phosphate, amino acids, polyP (type P65), and bovine serum albumin were from Sigma; [γ-32P]ATP and carrier-free 32Pi were from Amersham; polyethyleneimine-cellulose F thin-layer chromatography plates were from Merck; DE81 filter paper was from Whatman International Ltd., Maidstone, England; rPPXI, used in the polyP assay, was prepared and assayed as described previously (39).
Bacterial strains, plasmids, and phages.
All bacterial strains used in this study are derivatives of E. coli K-12 (Table 1); the K10 strains contain a missense mutation in the pit gene, which encodes a low-efficiency Pi transport system (8) and displays an arsenate-resistant phenotype. Bacteriophage P1vir was used for all transduction experiments (24).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| AT833 | K10 Hfr (PO2A) pit-10 fhuA22 fadL701 cre-510 relA1 metB1 mcrB1 thi garB10 ompF627 spoT1 | 14 |
| AT13 | K10 pit10 phoB63 | A. Torriani |
| AT854 | K10 ΔphoU::KanrrecA1 pit-10 | A. Torriani |
| AT857 | K10 recA1 pit-10 phoU35 | A. Torriani |
| AT280 | K10 C2 phoR68 | A. Torriani |
| AT776 | K10 C3 phoR69 | A. Torriani |
| NR21 | W3110 F− IN(rrnD-rrnE)I | 12 |
| AT25 | LEP1 F−phoB23 | 2 |
| AT30 | G206 pit10 phoB62 spoT1 | A. Torriani |
| AT884 | GP33 K-12 phoB::Tn5 | A. Torriani |
| AT401 | W3110 phoR68 creC452 | A. Torriani |
| MG1655 | CF1648 F−arcA1655 fnr-1655 | M. Cashel |
| NR2 | MG1655 phoB::Tn5 | Present work |
| CF1652 | MG1655 ΔrelA251 | M. Cashel |
| CF1693 | MG1655 ΔrelA251 ΔspoT207 | M. Cashel |
| Plasmids | ||
| AT508 | W3110 pHI4 (phoB+) | A. Torriani |
| NR25 | CF1648 pALS13(relA′+) | 33 |
| NR26 | CF1648 pALS14 (relA′) | 33 |
Media and growth conditions.
A MOPS-buffered minimal medium (25) contained glucose (4 mg/ml) as a carbon source and K2HPO4 as the Pi source; the low-Pi medium contained 0.1 mM Pi, and the high-Pi medium contained 2 mM Pi. A mixture of 20 amino acids was omitted or supplied either at 2 μg of each/ml (for the amino acid-limited medium) or at 20 μg of each/ml (for the amino acid-enriched medium). For studies on growth and polyP accumulation, cells were grown overnight (∼18 h) at 37°C in MOPS minimal medium with all 20 amino acids (20 μg of each/ml), glucose (4 mg/ml), and 2 mM K2HPO4. The cultures were then diluted in fresh, prewarmed (37°C) MOPS minimal medium containing 0.1 mM Pi and supplemented with either 2 or 20 μg of each amino acid per ml, as indicated for specific experiments. Cultures were also inoculated into both low-Pi and high-Pi MOPS minimal medium without the addition of amino acids. 32Pi was added at 20 μCi/ml of medium. The cultures were grown aerobically at 37°C and monitored by turbidity at 540 nm.
Extraction of polyP for assay.
The 32Pi-labeled E. coli culture (0.5 to 1.0 ml) was centrifuged at 14,000 rpm for 5 min at 4°C in an Eppendorf Microfuge. The supernatant was discarded and the pellet was suspended in a 50-μl solution containing 1.5 M formic acid, 1 M urea, 0.5% sodium dodecyl sulfate, and 10 mM EDTA. PolyP, type P65 (about 65 residues long), was used as the carrier at 50 μg/tube. The cell suspension was homogenized by sonication (Sonifier cell disruptor; Heat Systems-Ultrasonics, Inc.) at 0 to 3°C for three 30-s bursts at 80% output.
Enrichment of polyP for assay.
An aliquot of the cell lysate (20 μl) was loaded onto a DE81 disk (1.3-cm diameter) and set on a paper towel for 2 to 5 min to dry. Each disk was washed three times by shaking in 10 ml of THK buffer (10 mM Tris-HCl [pH 8.0], 100 mM KCl, 5 mM KPO4 [pH 8.0]). Partially air-dried disks were then placed in a 0.5-ml Eppendorf tube without the cap and with a tiny hole at the bottom; the tube with disk was then placed inside a 1.5-ml Eppendorf tube. PolyP was eluted with 50 μl of elution buffer (10 mM Tris-HCl [pH 8.0], 500 mM KCl) by soaking the disk in the buffer for 10 min at room temperature and then spinning for 5 s in a Microfuge; elution with 50 μl of buffer was repeated three times. To the combined 200 μl of eluate was added 2 μl of polyP, type P65 (20 μg), and 6 μl of a 15% Norit suspension in water. The suspension was mixed and centrifuged for 5 min at 14,000 rpm in a Microfuge. The supernatant was transferred to a fresh Eppendorf tube and the pellet was washed twice with 400 μl of 10 mM Tris-HCl buffer, pH 8.0. The supernatants were pooled for a total volume of 1.0 ml, which constituted the purified polyP extract. The DE81 adsorption of polyP and the removal of 32P contaminants by Norit each produced a 10-fold or greater enrichment of polyP.
Enzymatic assay of polyP.
To each DE81 disk in 4 wells of a 24-well Multiwell tissue culture plate (Becton Dickinson Labwares, Lincoln Park, N.J.) was added 200 μl of a sample of the enriched [32P]polyP; polyP was readsorbed by shaking the plates for 60 min at room temperature; supernatants were removed and discarded. The disks were rinsed three times with 1 ml of deionized water, and the washings were discarded. PPX buffer (50 mM Tris-HCl [pH 7.4], 100 mM ammonium acetate, 4 mM magnesium chloride, 10 mg of bovine serum albumin per ml) containing purified rPPX (5 μg of protein/ml) was added to each of two assay wells (200 μl/well), and 200 μl of PPX buffer lacking the rPPX enzyme was added to the other two wells. After incubation at 37°C for 60 min, the reaction mixtures were transferred to scintillation vials. The disks were rinsed three times with 0.1 ml of THK buffer, and the washings were transferred with a multiple-channel pipette into the same scintillation vial. DE81 disks were rinsed once with 1 ml of THK buffer, dehydrated with 200 μl of 100% (vol/vol) ethyl alcohol, and subsequently dried under an infrared light for 5 min. Both the pooled reaction mixture and the DE81 disk were counted separately for radioactivity with a scintillation counter. Thus, each sample was measured in duplicate, for radioactivity both released by rPPX (i.e., Pi) and retained by the disk (non-polyP). Background values were supplied by the duplicate samples which were not treated with rPPX.
Calculation of polyP levels in the assay.
The average of the two enzyme-treated samples was corrected by the average background of the two untreated controls. The values calculated based on the release of Pi from the disk matched within 5% those for the radioactivity lost from the disk. Background values relative to the enzyme-treated samples varied from about 95%, when polyP was abundant in the culture, to only about 5%, when polyP was at minimal levels in the culture.
Measurement of 32Pi-labeled nucleotide levels.
Cellular levels of ppGpp and pppGpp were measured in 32Pi-labeled cultures grown in MOPS minimal medium with or without amino acid supplementation (2 μg/ml), as described previously (28, 40).
PolyP accumulation and (p)ppGpp synthesis during stringent response.
E. coli MG1655 and a phoB mutant (MG1655 phoB::Tn5) were grown in MOPS medium containing 0.4 mM Pi (10 μCi of [32P]orthophosphate per ml) and 20 μg of amino acids per ml. At an optical density at 540 nm (OD540) near 0.2, serine hydroxamate (SHX) was added (0.5 mg/ml) for induction of amino acid starvation. At time intervals after the addition of SHX, samples were removed and analyzed for polyP content. The cellular levels of (p)ppGpp were also analyzed in these samples.
Other methods.
PPK activity in cell extracts was assayed as described previously (1). AP activity was measured in permeabilized cells (21). One unit of AP activity corresponded to hydrolysis of 1 nmol of p-nitrophenyl phosphate per min per mg of protein. Protein concentrations in cell extracts were determined according to Lowry et al. (20).
RESULTS
Influence of PhoB on accumulation of polyP.
Pho regulon mutants affected in PhoB synthesis were tested for accumulation of polyP. It had been observed that in a MOPS-buffered minimal medium containing low levels of Pi (0.1 mM) and amino acids (2 μg/ml), large amounts of polyP accumulated in a narrow growth range (OD540, about 0.3) in wild-type cells (25a). The amounts of polyP accumulated by different wild-type and mutant strains grown in an amino acid-supplemented, low-Pi (0.1 mM) medium to an OD540 near 0.3 are recorded in Table 2. A lack of PhoB due either to missense mutations (phoB63, phoB23, phoB62) or to insertional inactivation of phoB (phoB::Tn5) resulted in low levels of polyP (0.3 to 1.9 nmol/mg of protein). Inactivation of the protein kinases PhoR and CreC, which activate PhoB by phosphorylation (23, 35), led to even lower levels (0.1 nmol/mg of protein). Mutations in phoU (ΔphoU, phoU35) and in phoR (phoR68, phoR69), which lead to constitutive expression of the Pho regulon (31, 34, 35), resulted in polyP accumulations comparable to those in wild-type cells (Table 2). PolyP accumulation showed variations in different strain backgrounds. The MG1655 and W3110 strains accumulated more polyP than the K10 strains.
TABLE 2.
Effects of phoB on polyP accumulationa
| Strain | Relevant genotype | PolyP (nmol/mg of protein) |
|---|---|---|
| AT833 | K10 wild type | 10.8 |
| AT13 | K10 phoB63 | 1.8 |
| AT854 | K10 ΔphoU | 7.7 |
| AT857 | K10 phoU35 | 15 |
| AT280 | K10 phoR68 | 16.9 |
| AT776 | K10 phoR69 | 17.6 |
| AT884 | GP3 phoB::Tn5 | 1.8 |
| AT25 | LEP1 phoB23 | 1.8 |
| AT30 | G206 phoB62 | 1.9 |
| NR21 | W3110 wild type | 115 |
| AT401 | W3110 phoR creC | 0.1 |
| CF1648 | MG1655 wild type | 48 |
| NR2 | MG1655 phoB::Tn5 | 0.3 |
MOPS-buffered minimal medium, with 0.1 mM Pi and 2 μg of each amino acid per ml, was used. The OD540 values at the time of sampling were near 0.3.
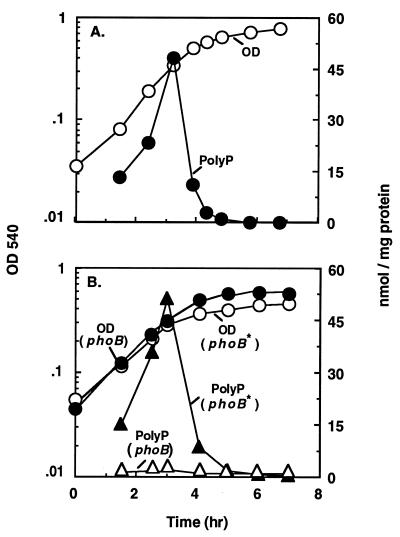
To observe the overall effect of PhoB on growth and polyP accumulation in Pi-limited medium supplemented with inadequate amounts of amino acids, a wild-type strain, MG1655 (pit+ relA+), and an isogenic phoB mutant strain (MG1655 phoB::Tn5) were compared (Fig. 1). The wild-type strain transiently accumulated up to 48 nmol/mg, compared to 0.3 nmol/mg or less in the phoB mutant. When the mutant was complemented with a multicopy phoB+ plasmid, the transient accumulation of polyP was restored to the wild-type level (Fig. 1B).
FIG. 1.
Complementation of a phoB mutant with PhoB restores polyP accumulation. Wild-type E. coli MG1655 (A), a corresponding phoB::Tn5 mutant, and the mutant complemented with a phoB plasmid (phoB*) (B) were grown in MOPS-buffered minimal medium containing 0.1 mM Pi (20 μCi of 32Pi per ml) and 2 μg of each of the 20 amino acids per ml. Cultures (1 ml) were sampled for measurement of growth and polyP content (see Materials and Methods). Growth (OD) and polyP accumulation (PolyP) of the wild type (A) and of the phoB::Tn5 and plasmid-complemented phoB::Tn5 strains (B) are shown.
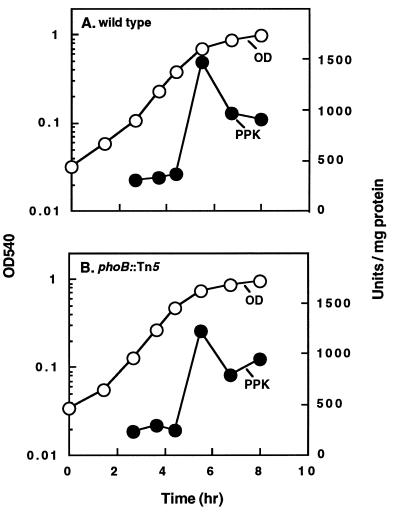
PPK activity and polyP accumulation.
A phoB mutant (MG1655 phoB::Tn5) and the corresponding wild-type strain were compared for PPK activity during growth in a low-Pi (0.1 mM) MOPS medium containing limited amounts (2 μg/ml) of amino acids (Fig. 2). In the wild type, PPK activity was at its lowest level when the accumulation of polyP was at its peak (Fig. 2A); PPK activity increased when the cells entered the stationary phase but were unaffected by the mutation in phoB (Fig. 2B).
FIG. 2.
PPK activity of the wild type and a phoB mutant. A phoB mutant (MG1655 phoB::Tn5) (B) and the corresponding wild-type strain (A) were grown in low-Pi (0.1 mM) MOPS-buffered minimal medium with 2 μg of each of the 20 amino acids per ml. Cultures (1 ml) were sampled periodically for measurement of growth and PPK activity.
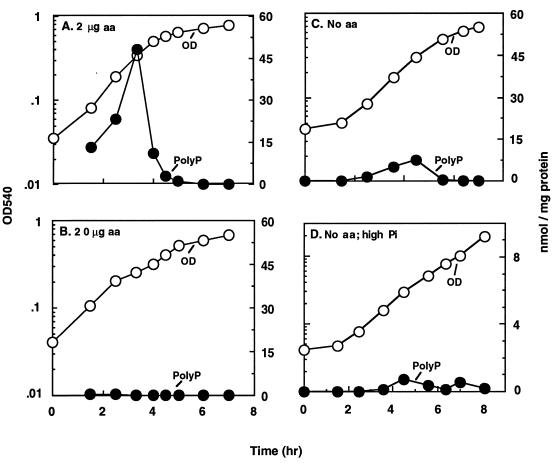
PolyP accumulation depends on an insufficient supply of amino acids in a Pi-limited medium.
When the concentration of amino acids in the Pi-limited medium was increased from 2 to 20 μg/ml, the accumulation of polyP in the wild-type cells was almost completely abolished (Fig. 3B). A corresponding culture grown in the presence of 2 μg of amino acids per ml accumulated over 45 nmol of polyP per mg of protein (Fig. 3A). Cells growing in Pi-limited medium without amino acid supplementation accumulated polyP up to a maximum of 8 nmol/mg of protein (Fig. 3C), whereas the cells growing in high-Pi (2 mM) medium accumulated only about 1 nmol/mg of protein (Fig. 3D). Thus, the supply of a limited amount of amino acids (2 μg/ml) to prototrophic cells may create transient starvation due to nutritional imbalances, leading to (p)ppGpp accumulation by the stringent response. Changes in Pi and/or amino acid concentration in the growth medium did not result in a significant amount of polyP accumulation in phoB mutants (data not shown).
FIG. 3.
PolyP accumulation depends on amino acid (aa) concentration in the growth medium. E. coli MG1655 was grown in minimal medium containing 0.1 mM Pi (20 μCi of 32Pi per ml) and the following concentrations of each of the 20 amino acids: 2 (A), 20 (B), or 0 (C) μg/ml. (D) Cells grown in minimal medium with excess Pi (2 mM). Growth and polyP content of the cells were measured as described in the legend to Fig. 1.
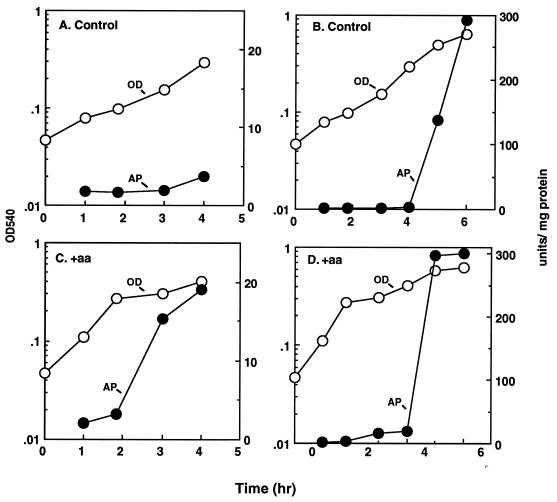
Effects of amino acid limitation on AP induction.
Synthesis of AP in wild-type cells depends solely on the Pi concentration in the growth medium and is under phoB control (34, 35). Evidence that an inadequate supply of both amino acids (2 μg/ml) and Pi (0.1 mM) induced PhoB synthesis was demonstrated by a significant increase in AP activity (∼15 units/mg of protein), which coincided with the accumulation of polyP (Fig. 4C). There was no significant increase in enzyme activity in cells grown in medium without amino acids during this growth period; the cells contained only about 2 to 4 units of enzyme activity (Fig. 4A). The amino acid supplement increased the growth rate initially, but the increase was not sustained after the cells reached an OD540 of about 0.25 (Fig. 4D). Prolonged incubation with or without the amino acid supplement led to full induction of alkaline phosphatase (∼300 units/mg of protein), coincident with complete depletion of Pi from the medium (Fig. 4B and D).
FIG. 4.
Effect of amino acid supplementation on AP induction. E. coli MG1655 was grown in a Pi-limited (0.1 mM) minimal medium without (A and B) or with (C and D) the addition of 2 μg of each of the 20 amino acids per ml. Panels A and C highlight the initial (up to 4-h) growth period and corresponding AP activity of cells represented in panels B and D, respectively. Note the scales on the y-axes.
Buildup of (p)ppGpp and polyP.
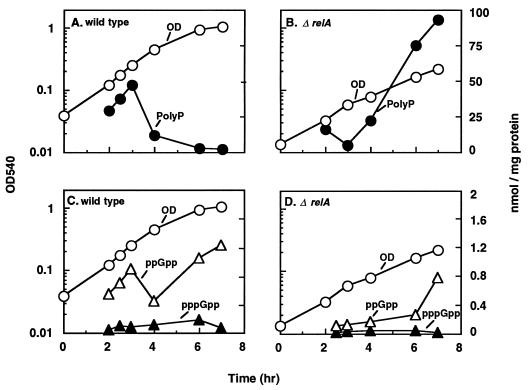
Growth, polyP accumulation, and pppGpp and ppGpp levels were measured in the wild-type strain MG1655 growing in Pi-limited medium containing an inadequate supply of amino acids (Fig. 5). The maximum amount of polyP (ca. 48 nmol/mg of protein) accumulated at an OD540 near 0.3, when growth was decelerating (Fig. 5A). The downshift in the growth rate was due to a short-lived starvation imposed by the limited supply of amino acids in the growth medium. This transient starvation may correspond to the period when the cells derepress amino acid biosynthetic systems to compensate for the limited amino acid supply. The peak of polyP accumulation coincided with a corresponding increase in the level of ppGpp to about 0.9 nmol/mg of protein (Fig. 5C). The concentration of ppGpp increased even more when the cells entered stationary phase: this increase in the level of ppGpp may have been due to SpoT-mediated accumulation, since the cells were starved for Pi at this stage. The changes in the levels of pppGpp during the entire growth period were minimal (Fig. 5C). Under these growth conditions, an isogenic strain without relA (ΔrelA) accumulated large amounts of polyP (ca. 95 nmol) (Fig. 5B). The pattern of polyP accumulation, however, was completely different. This was in response to a delayed accumulation of ppGpp which was not due to the stringent response. A growth rate downshift caused by depletion of both amino acids and Pi allowed the SpoT-mediated induction of ppGpp accumulation (Fig. 5D).
FIG. 5.
Effects of ΔrelA on growth, polyP accumulation, and (p)ppGpp levels. The wild type and a corresponding relA mutant (MG1655 ΔrelA251) were grown in Pi-limited minimal medium containing 2 μg of each of the 20 amino acids per ml and analyzed for polyP accumulation (A and B), and ppGpp and pppGpp levels (C and D).
After the transient accumulation of polyP in wild-type cells, growth presumably resumed at the expense of the accumulated polyP, since the medium was devoid of Pi (<1 μg/ml) at this point of growth (26a). The optical density of this culture after 7 h of growth (Fig. 5A) was the same as that of a similar culture grown for 7 h in low-Pi medium without amino acids (Fig. 3C). Since the wild-type cells reached stationary phase after the complete exhaustion of Pi from the medium, there could be no polyP accumulation at this stage despite the presence of ppGpp (Fig. 5A and 5C). In the case of the relA null mutant, the cells attained a density (OD540) of only about 0.23, compared to over 1.0 for the wild type, and hence continued to synthesize polyP even after 7 h of growth (Fig. 5B). The Pi in the medium in this case was essentially diverted toward polyP synthesis rather than growth for up to 7 h; an overnight (23-h) incubation resulted in depletion of polyP from the cells and an increase in cell density (OD540) to 1.3 (data not shown).
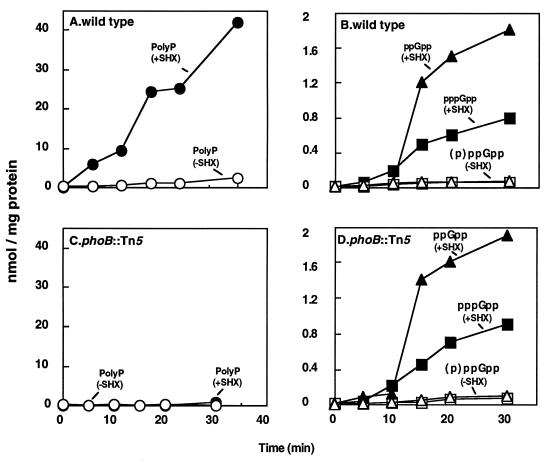
When amino acid starvation was created by incubating cells with SHX, the phoB mutant cells (MG1655 phoB::Tn5) accumulated ppGpp and pppGpp in amounts similar to those observed in wild-type cells (Fig. 6D and B). The wild-type cells had accumulated 45 nmol of polyP at 35 min after the addition of SHX (Fig. 6A); however, phoB cells failed to accumulate significant amounts of polyP (<0.1 nmol/mg of protein) at any time (Fig. 6C). A relA spoT-deleted null mutant did not accumulate either ppGpp or polyP in the presence of SHX and 0.4 mM Pi (18).
FIG. 6.
Effects of phoB on polyP accumulation and (p)ppGpp levels. MG1655 wild-type and MG1655 phoB::Tn5 mutant strains were grown in minimal medium containing 0.4 mM Pi and 20 μg of amino acids per ml. At an OD540 near 0.2, SHX was added to induce amino acid starvation. At the times indicated after the addition of SHX, samples were collected and analyzed for polyP (A and C) and (p)ppGpp (B and D).
Influence of spoT on polyP accumulation.
Inasmuch as the residual ppGpp synthetic activity of relA null mutants can be eliminated by spoT null mutations (40), the double mutant was tested for both polyP accumulation and ppGpp synthesis. Cultures were grown by placing 16-h-old colonies from Luria-Bertani plates directly in minimal medium as opposed to the use of overnight liquid cultures, because the strain tends to accumulate suppressor mutations when grown for long periods. Though the growth rate was very low, the cell density was considerable after 6 h of growth (OD540, 0.25); however, there was neither significant polyP accumulation nor detectable levels of (p)ppGpp (data not shown). These and the previous results indicate the necessity for ppGpp in polyP accumulation during growth in a Pi-limited medium.
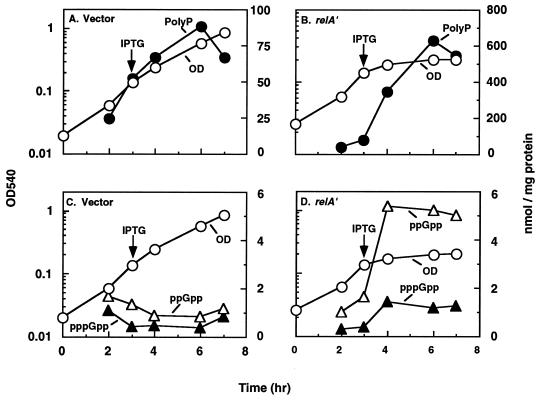
Influence of relA on polyP accumulation.
Direct evidence for the role of relA, and thus for ppGpp, in polyP accumulation was obtained by increasing the expression of a truncated relA gene (relA′), which encodes a smaller but catalytically active peptide with ppGpp synthetic activity (33). Gratuitous induction of ppGpp synthesis by the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible relA′ plasmid resulted in a prompt cessation of growth, highly increased levels of ppGpp and pppGpp (Fig. 7D), and the accumulation of over 600 nmol of polyP per mg (Fig. 7B). The induced level of ppGpp, which exceeded 5 nmol/mg of protein, persisted over the entire period of growth (Fig. 7D). Large amounts of polyP remained in this culture even after overnight incubation (data not shown). In wild-type cells which contained an IPTG-inducible but inactive truncated relA gene, ppGpp levels ranged between 0.8 and 1.7 nmol/mg (Fig. 7C), and far lower levels of polyP accumulated (Fig. 7A).
FIG. 7.
Effect of overexpression of relA on polyP accumulation and (p)ppGpp levels. E. coli MG1655 containing either pALS14 (IPTG-inducible control plasmid with inactive relA′) (A and C) or pALS13 (IPTG-inducible active relA′+) (B and D) was grown in a Pi-limited minimal medium containing 2 μg of each of the 20 amino acids per ml, as described in the legend to Fig. 5. At the times indicated, IPTG (1 mM) was added to the growing culture, which was examined for further growth (OD540), polyP accumulation (A and B), and ppGpp and pppGpp levels (C and D).
DISCUSSION
Accumulation of polyP by E. coli in the stationary phase is often assumed to provide a reservoir of energy convertible to ATP. However, this seems implausible because the metabolic turnover of ATP in E. coli consumes only a fraction of a second (6). Even highly elevated levels of polyP, which, when expressed in phosphoanhydride bonds might be five times or more the level of ATP, could supply ATP for only a second or two. Therefore, other functions for polyP need to be considered, among them a regulatory role. PolyP, even at relatively low levels, has been shown to be essential for adaptation to various stresses and for survival in stationary phase (7, 27).
A major impediment to understanding the physiological roles of polyP has been the inadequacy of good quantitative methods. Previous methods were inexact and nonspecific and required extensive and laborious enrichments of polyP, which limited the number of analyzed samples to just a few a week. The sensitive, rapid, and definitive assay described in this report relies on enrichment of [32P]polyP in E. coli extracts by adsorption on DE81 filter disks, lack of adsorption to Norit, and finally, specific hydrolysis of the polyP adsorbed to the disk by a potent yeast exopolyphosphatase.
Here, we have described how massive accumulations of polyP take place during growth in exponential phase, when the cell responds simultaneously to a limitation of amino acids and phosphate. The stringent response of E. coli involves synthesis of ppGpp and pppGpp by the ribosome-bound RelA (3, 4, 19). These nucleotides serve as signals for the transcriptional repression of rRNA and proteins and for activation of the expression of certain biosynthetic genes. Changes in cellular levels of (p)ppGpp have also been observed in response to deficiencies of Pi (11, 32), carbon (19) and nitrogen (13). Very low levels of extracellular Pi (<4 μM) induce the Pho regulon (36). PhoB, the response regulator, turns on several genes, among them the genes for AP (phoA) and for proteins involved in Pi uptake and assimilation.
In the face of low levels of Pi (0.1 mM) and amino acids (by the addition of 2 μg of each per ml) in the medium, polyP accumulates to high levels in the exponential phase of growth (Fig. 1A). Under these conditions, phoB mutants accumulate little or no polyP (<1% of the wild type) (Fig. 1B and Table 2). Complementation of a phoB mutant with a multicopy phoB plasmid restores the capacity to accumulate polyP (Fig. 1B). Hence, PhoB seems to have a direct role in polyP accumulation.
Both low Pi and an inadequate amount of amino acids in the growth medium were required to elicit accumulation of polyP (Fig. 3A). By contrast, there was no accumulation when the wild-type cells were grown in a low-Pi medium supplemented with large amounts of amino acids (Fig. 3B). Also, cells grown in high-Pi medium did not accumulate a significant amount of polyP (Fig. 3D). The phoB response to low Pi and amino acid limitation was also demonstrated by a partial induction of AP and in the accumulations of pppGpp and ppGpp. Inasmuch as these accumulations of polyP and (p)ppGpp can also take place in relA null mutants (Fig. 5A and D), an alternative source of these buildups must be available. Such a source is spoT: a double mutant lacking both relA and spoT accumulates neither (p)ppGpp nor polyP.
Additional evidence of the role of (p)ppGpp in polyP accumulation is demonstrated by an inducible multicopy plasmid which encodes an active RelA peptide. This plasmid carries a truncated relA gene (relA′) which can still encode an active RelA peptide. The massive production of both polyP and (p)ppGpp (Fig. 7B and D) attributed to this plasmid is affirmed by the far lesser responses to a control plasmid lacking the relA′ gene (Fig. 7A and C).
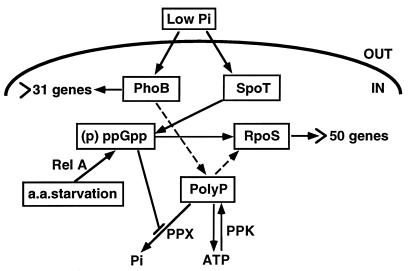
Based on the data presented here and the observations made by Kuroda et al. (18), a tentative model which attempts to explain the mechanism of polyP accumulation in E. coli is presented (Fig. 8). The nucleotide ppGpp, required for polyP accumulation, can be synthesized by either relA or spoT. The response regulator of the Pho regulon, PhoB, is required for polyP accumulation in Pi-limited growth medium. Neither phoB nor relA spoT mutants accumulate polyP in Pi-limited medium. Since a relA mutant does not accumulate (p)ppGpp during growth in a medium exhausted for both amino acids and Pi, we infer that the changes in SpoT activities and/or its synthesis are required during Pi starvation for the accumulation of polyP. This accumulation could occur due to a change in growth rate as a result of a shift from amino acid assimilation to synthesis, as the exogenous amino acids are depleted from the growth medium. Concurrently, a transient Pi starvation is observed as the exogenous Pi is diverted toward polyP synthesis.
FIG. 8.
Hypothetical scheme for polyP accumulation and function. PolyP accumulation depends on PhoB and (p)ppGpp synthesized either by the relA (stringent-response) pathway or by the phosphate starvation-inducible spoT pathway. The inhibition by (p)ppGpp of exopolyphosphatase PPX as one means of polyP accumulation and the dependence of rpoS expression on polyP are mentioned in the text. Dashed arrows indicate either a direct or an indirect effect on the synthesis or accumulation of the gene products.
The activities of the principal enzymes that synthesize and degrade polyP fluctuate only marginally. Therefore, polyP accumulation can be attributed to a singular and profound inhibition by pppGpp and/or ppGpp of the hydrolytic breakdown of polyP by exopolyphosphatase (PPX), thereby blocking the dynamic turnover of polyP (18). The level of polyP in the cell may also be influenced by PPK, the enzyme which generates ATP from polyP. Turnover of polyP (resulting from its cyclic synthesis by PPK and hydrolysis by PPX) was found to be 12 min or less (25a), a result simulated by the accumulation of polyP in a reaction mixture containing purified PPK and PPX responding to the presence of pppGpp and ppGpp (18). Suppression of the ppk feature of heat sensitivity and katE expression by extra copies of rpoS, the gene encoding the RNA polymerase sigma factor that regulates some 50 stationary-phase genes, implicates polyP in the regulation of rpoS expression and function (27). Reduction of in vivo polyP levels by introducing a plasmid bearing a potent yeast polyphosphatase also resulted in decreased expression of RpoS (30). Though phoB mutant cells can accumulate (p)ppGpp in response to amino acid starvation, they fail to accumulate polyP (Fig. 6). It is possible that one or more genes of polyP metabolism are connected to the Pho regulon (via PhoB), involving energy metabolism (37). Clearly, an additional function(s) of phoB in polyP accumulation remains to be discovered. A reduction in the amount of active RNA polymerase can occur in cells growing under nutritional stress; this, in turn, may affect promoter activities (related to polyP synthesis and utilization), depending whether or not the promoter(s) is saturated with polymerase.
ACKNOWLEDGMENTS
We are grateful to M. Cashel and A. Torriani-Gorini for technical discussions and for generously providing strains. We thank L. Bertsch for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health.
REFERENCES
- 1.Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli: isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 2.Bracha M, Yagil E. A new type of alkaline phosphatase-negative mutants in Escherichia coli K12. Mol Gen Genet. 1973;122:53–60. doi: 10.1007/BF00337973. [DOI] [PubMed] [Google Scholar]
- 3.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid starved stringent strains. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 4.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 5.Castuma C E, Huang R, Kornberg A, Reusch R N. Inorganic polyphosphates in the acquisition of competence in Escherichia coli. J Biol Chem. 1995;270:12980–12983. doi: 10.1074/jbc.270.22.12980. [DOI] [PubMed] [Google Scholar]
- 6.Chapman A G, Atkinson D E. Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv Microb Physiol. 1977;15:253–306. doi: 10.1016/s0065-2911(08)60318-5. [DOI] [PubMed] [Google Scholar]
- 7.Crooke E, Akiyama M, Rao N N, Kornberg A. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1994;269:6290–6295. [PubMed] [Google Scholar]
- 8.Elvin C M, Hardy C M, Rosenberg H. Molecular studies on the phosphate inorganic transport system of Escherichia coli. In: Torriani-Gorini A, Rothman F G, Silver S, Wright A, Yagil E, editors. Phosphate metabolism and cellular regulation in microorganisms. Washington, D.C: American Society for Microbiology; 1987. pp. 156–158. [Google Scholar]
- 9.Gentry D R, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1371–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 10.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ςS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez F J, Bremer H. Escherichia coli ppGpp synthetase II activity requires spoT. J Biol Chem. 1991;266:5991–5999. [PubMed] [Google Scholar]
- 12.Hill C W, Harnish B W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irr J D. Control of nucleotide metabolism and ribosomal ribonucleic acid synthesis during nitrogen starvation of Escherichia coli. J Bacteriol. 1972;110:554–561. doi: 10.1128/jb.110.2.554-561.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch J P, Hayashi S I, Lin E C C. The control of dissimilation of glycerol and l-α-glycerophosphate in Escherichia coli. J Biol Chem. 1964;239:3106–3108. [PubMed] [Google Scholar]
- 15.Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulaev I S, Vegabov V M. Polyphosphate metabolism in microorganisms. Adv Microbiol. 1983;15:731–738. [Google Scholar]
- 17.Kumble K D, Kornberg A. Inorganic polyphosphate in mammalian cells and tissues. J Biol Chem. 1995;270:5818–5822. doi: 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 19.Lazzarini R A, Cashel M, Gallant J. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains. J Biol Chem. 1971;246:4381–4385. [PubMed] [Google Scholar]
- 20.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:267–275. [PubMed] [Google Scholar]
- 21.Ludke D, Bernstein J, Hamilton C, Torriani A. Identification of the phoM gene product and its regulation in Escherichia coli K-12. J Bacteriol. 1984;159:19–25. doi: 10.1128/jb.159.1.19-25.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K12. J Mol Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 23.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Neidhardt F C, Block P L, Smith D F. Culture media for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Park, S. J., and A. Kornberg. Unpublished results.
- 26.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 26a.Rao, N. N., and A. Kornberg. Unpublished results.
- 27.Rao N N, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarubbi R, Rudd K E, Cashel M. Basal ppGpp level adjustments shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- 29.Sarubbi E, Rudd K E, Xiao H, Ikehara K, Kalman M, Cashel M. Characterization of the spoT gene of Escherichia coli. J Biol Chem. 1989;264:15074–15082. [PubMed] [Google Scholar]
- 30.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao N N, Kornberg A. Inorganic polyphosphate and induction of rpoS expression. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinagawa H, Makino K, Amemura M, Nakata A. Structure and function of the regulatory genes for the phosphate regulon in Escherichia coli. In: Torriani-Gorini A, Rothman F G, Silver S, Wright A, Yagil E, editors. Phosphate metabolism and cellular regulation in microorganisms. Washington, D.C: American Society for Microbiology; 1987. pp. 20–25. [Google Scholar]
- 32.Spira B, Silberstein N, Yagil E. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svitil A L, Cashel M, Zyskind J W. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 34.Torriani-Gorini A. Introduction: the Pho regulon of Escherichia coli. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1994. pp. 1–4. [Google Scholar]
- 35.Wanner B L. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetyl phosphate. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 203–221. [Google Scholar]
- 36.Wanner B L. Multiple controls of the Escherichia coli Pho regulon by the Pi sensor PhoR, the catabolite regulatory sensor CreC, and acetyl phosphate. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1994. pp. 13–21. [Google Scholar]
- 37.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1357–1381. [Google Scholar]
- 38.Wood H G, Clark J E. Biological aspects of inorganic polyphosphates. Annu Rev Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 39.Wurst H, Shiba T, Kornberg A. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J Bacteriol. 1995;177:898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]