ABSTRACT
Turnercyclamycin A and B lipopeptides exhibit Gram-negative bacteria-specific toxicity. This includes CDC urgent threat organisms such as multidrug-resistant Acinetobacter baumannii. Like the last-line agent colistin, turnercyclamycins interact with the lipopolysaccharide (LPS) pathway, but they remain active against clinical isolates that are colistin resistant. Here, we aimed to determine why turnercyclamycins A and B show little cross-resistance with colistin despite some potential mechanistic and structural similarities. The outer membrane was important in the actions of all three agents, since the deletion of lpxC that synthesizes LPS and the addition of exogenous LPS led to resistance to turnercyclamycins and colistin. Even so, it was much more difficult to generate resistance to turnercyclamycin A than turnercyclamycin B. In Escherichia coli, disruption of the mlaA gene, which is involved in outer membrane homeostasis, resulted in resistance to turnercyclamycin B. However, mlaA disruption blocked neither turnercyclamycin A nor colistin activity. This activity was recapitulated in A. baumannii, where transposon mutants were more resistant to turnercyclamycin B. Moreover, the common A. baumannii colistin resistance gene mcr-1 blocked colistin activity in E. coli but did not affect turnercyclamycins. These results demonstrate a unique resistance profile for turnercyclamycins A and B, in addition to suggesting differences in the mechanism of action. Further, turnercyclamycin A was effective in a mouse model of A. baumannii infection, indicating that this compound class may have potential promise in treating drug-resistant infections.
IMPORTANCE
Bacterial resistance to antibiotics is a crisis. Acinetobacter baumannii is among the CDC urgent threat pathogens in part for this reason. Lipopeptides known as turnercyclamycins are produced by symbiotic bacteria that normally live in marine mollusks, where they may be involved in shaping their symbiotic niche. Turnercyclamycins killed Gram-negative pathogens including drug-resistant Acinetobacter, but how do the mechanisms of resistance compare to other lipopeptide drugs? Here, we define resistance from a truncation of MlaA, a protein involved in regulating bacterial membrane phospholipids. Intriguingly, this resistance mechanism only affected one turnercyclamycin variant, which differed only in two atoms in the lipid tail of the compounds. We could not obtain significant resistance to the second turnercyclamycin variant, which was also effective in an infection model. This study reveals an unexpected subtlety in resistance to lipopeptide antibiotics, which may be useful in the design and development of antibiotics to combat drug resistance.
KEYWORDS: lipopeptides, antibiotics, Gram-negative bacteria, Acinetobacter, colistin, drug resistance, turnercyclamycin
INTRODUCTION
Antimicrobial resistance is a global problem with Gram-negative pathogens dominating the list of urgent threats that needs to be addressed (1 – 4). For example, Acinetobacter is a particularly challenging pathogen that is intrinsically hard to combat due to the stability of its outer membrane and the ability to infect multiple sites (5 – 9). Multiple strains of drug-resistant Acinetobacter have emerged, and the underlying mobile resistance elements are easily transferred between strains (10 – 13). Acinetobacter is increasingly resistant to the last line of treatment, colistin (polymyxin E) (14 – 16). The polymyxin mechanism of action involves a portion of lipopolysaccharide (LPS), lipid A, in the outer membrane (OM) of Gram-negative bacteria. Polymyxins bind to lipid A through both lipid-lipid affinity and electrostatic interactions between polycationic polymyxins and the negatively charged phosphate groups of lipid A. The current model is that this interaction disrupts the outer membrane, displacing cations and lysing cells (17 – 20). As such, Gram-negative pathogens usually evade polymyxins by modifying negatively charged points of interaction (17, 18).
However, it is not clear that this simple mechanism fully explains the effectiveness of colistin and other lipopeptides. Recent studies have shown that lipopeptide antibiotics do not share a single mechanism but instead have different targets (17, 18). For example, colistin exhibited antibacterial activity by interacting with LPS in the inner membrane (IM), implying a different mechanism of action than direct interaction with OM LPS (21). Another study suggested that colistin forms free radicals, which would eventually induce oxidative damage to the cell (22). The lipopeptide brevicidine was recently found to interact with OM LPS but also to associate with phosphatidylglycerol and cardiolipin in the IM, resulting in bacterial stress leading to death (23). Other recent studies describe new lipopeptides with distinct mechanisms of action (24, 25). The unclear mechanism of colistin, the diversity of lipopeptide targets, and the prevalent resistance against colistin strengthen the need to discover lipopeptides with high therapeutic indices and distinct mechanisms of action.
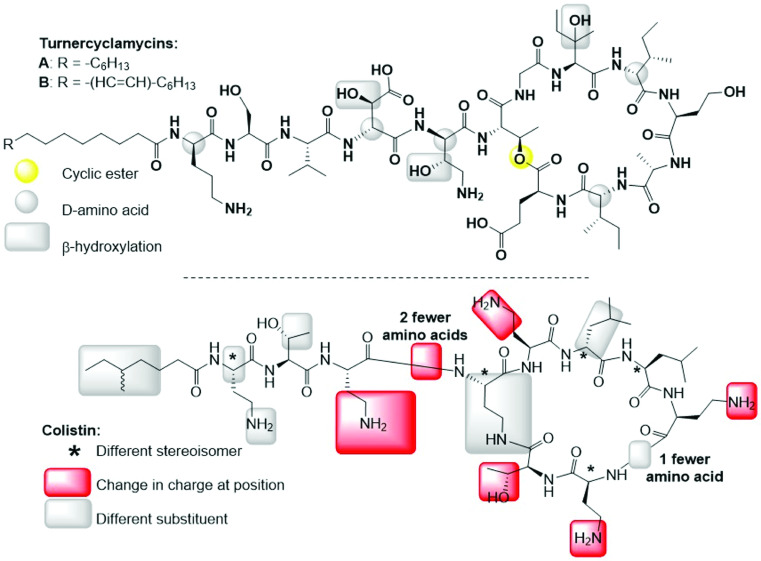
We have sought novel antibiotics by investigating natural biological interactions where antibiosis is likely to be important. One such model is shipworm mollusks, which harbor intracellular symbiotic bacteria rich in antibiotics (26 – 30). A common shipworm symbiont, Teredinibacter turnerae, is essential to the nutrition of the host mollusk, producing cellulases used for wood consumption (31). All T. turnerae strains examined so far also have the genetic capacity to produce lipopeptide antibiotics, the turnercyclamycins (Fig. 1) (30). While turnercyclamycins generally share identical peptide motifs, they differ in the length and unsaturation level of the lipid chain (30).
Fig 1.
Structural comparison of turnercyclamycins and colistin (polymyxin E), the last-line agent for treating Acinetobacter infections. A key difference is that colistin is thought to act in part because it is polycationic, which might be important in binding to negatively charged lipids. In contrast, turnercyclamycins are net neutral in charge.
Turnercyclamycins showed promise in initial studies where they were active against Gram-negative pathogens without hemolysis or toxicity to kidney cells, which are major liabilities of lipopeptides (30). More importantly, both turnercyclamycins A and B were active against all tested clinical multidrug-resistant Acinetobacter strains (30). Despite their ability to kill colistin-resistant strains, initial studies revealed some effects that were very similar to those of colistin, potentially involving LPS and OM disruption, while other effects appeared to be unique to turnercyclamycins (30). Unlike polymyxins, where positive charge is thought to be important, turnercyclamycins are net neutral, implying either that the polycationic nature of polymyxins is unimportant or that turnercyclamycins may have different mechanisms in comparison to polymyxins. Thus, it was clear that despite some similarities, turnercyclamycins and colistin had differences in mechanisms of action and resistance.
Because antibiotic resistance is a crucial problem in therapy, here, we aimed to determine why turnercyclamycins and colistin did not show cross-resistance in clinical Gram-negative pathogens that we have tested previously. We show that the resistance mechanism is different not just for turnercyclamycins compared to colistin but that all three compounds have different resistance profiles and mechanisms. It was difficult to isolate mutants resistant to turnercyclamycin A, which is effective in vivo, suggesting a potential role for the turnercyclamycin scaffold in therapeutic development.
RESULTS
Turnercyclamycins are active against MCR-1-expressing bacteria
The plasmid-borne mobile colistin resistance gene (mcr-1) is a common mechanism of resistance against the polymyxin class of antibiotics (32, 33). The MCR-1 protein adds phosphoethanolamine to a phosphate group in lipid A. Because colistin is positively charged, this is thought to create an unfavorable charge interaction. In contrast, turnercyclamycins are net neutral (Fig. 1) (30). In our study, an mcr-1-expressing plasmid (Addgene pGDP2-MCR-1) was transformed into two strains of Escherichia coli: DH5α and C600. E. coli was used in place of Acinetobacter because both colistin and turnercyclamycins are very effective at killing E. coli (30), because mcr-1 is reported to work by the same mechanism to provide E. coli with resistance against colistin (34), and because of the ease and relative safety of working with E. coli. The wild-type and mcr-1 strains were treated with various concentrations of colistin, turnercyclamycin A, and turnercyclamycin B.
Colistin killed both E. coli parent and vector-control strains at low concentrations (MIC90 0.5 µg/mL), but when tested against strains expressing mcr-1, it was 16-fold less potent (MIC90 8 µg/mL) (Table 1). Both turnercyclamycins exhibited MIC90s of 2 µg/mL against the wild-type strains, which was not significantly changed in strains expressing mcr-1. From these results, we inferred that turnercyclamycins differ in mechanism compared to colistin, in terms of MCR-1-induced resistance. Additionally, these results paved the way for the possibility of turnercyclamycins being an alternate compound to combat colistin-resistant bacterial infections.
TABLE 1.
Effect of the mcr-1 resistance gene on colistin and turnercyclamycin susceptibility in E. coli DH5α and E. coli C600
| Compounds | MIC90 (µg/mL) | |||||
|---|---|---|---|---|---|---|
| DH5α | pBR322 in DH5α | pGDP2-MCR-1 in DH5α | C600 | pBR322 in C600 | pGDP2-MCR-1 in C600 | |
| Colistin | 0.5 | 0.5 | 8 | 0.5 | 1 | 8 |
| Turnercyclamycin A | 2 | 2 | 2 | 2 | 4 | 4 |
| Turnercyclamycin B | 2 | 2 | 4 | 4 | 4 | 4 |
LPS loss attenuates turnercyclamycin potency
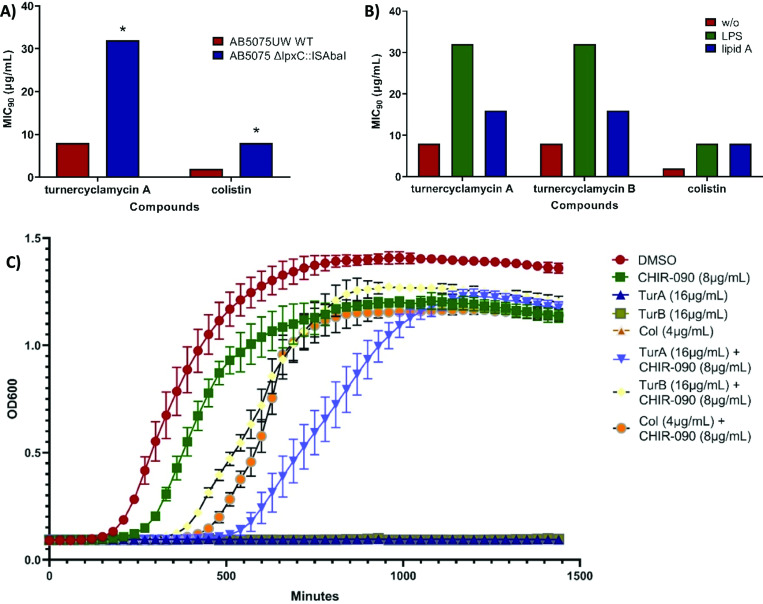
Initial studies suggested that LPS and lipid A might be potential targets of turnercyclamycins, since the susceptibility of Yersinia pestis to both colistin and turnercyclamycins was blocked by LPS modification with 4-amino-4-deoxy-L-arabinose (30). To test if LPS or lipid A plays a role in turnercyclamycin potency, as is the case with colistin, we determined the MIC of turnercyclamycin A against an Acinetobacter baumannii transposon mutant that is devoid of LpxC function (AB5075UW ΔlpxC::ISAbaI). LpxC performs the first committed step in lipid A biosynthesis, and therefore, its disruption produces a bacterium that is devoid of an LPS layer (35). Unlike many bacteria that require LPS/lipooligosaccharide (LOS) for survival, Acinetobacter remains viable without these lipids (36). When treated with turnercyclamycin A or colistin, A. baumannii ΔlpxC exhibited a greater than fourfold loss in susceptibility in comparison to its actions on the wild-type strain (Fig. 2A). This result is consistent with a role for LPS and lipid A in turnercyclamycin’s mechanism, similar to colistin.
Fig 2.
LPS is involved in turnercyclamycin activity. (A) MIC90s of turnercyclamycin A and colistin against wild-type (WT) and ΔlpxC A. baumannii 5075UW. * indicates that MIC90s are greater than or equal to the value given. (B) MIC90s of turnercyclamycins and colistin against A. baumannii after the addition of exogenous purified LPS and lipid A, both at 100 µg/mL. (C) Growth curve of A. baumannii measured using OD600, treated with the corresponding compounds. Each curve represents experiments performed using three biological replicates, with error bars indicating three replicate wells per biological replicate. The traces for turnercyclamycins A and B and colistin without other additives are difficult to see because they overlap on the baseline due to a lack of observable growth when treated with antibiotics. TurA and TurB, turnercyclamycins A and B; Col, colistin.
To confirm this finding, we tested whether turnercyclamycins would kill wild-type A. baumannii following LpxC inhibition by a small molecule, CHIR-090 (37). As expected, CHIR-090 alone did not cause the death of A. baumannii since LPS is not required for survival, while turnercyclamycins and colistin were effective as single agents. However, when A. baumannii was treated with both CHIR-090 and either turnercyclamycins or colistin, the bacteria survived (Fig. 2B). Thus, both genetic knockout and chemical inhibition of LpxC yielded bacteria resistant to both families of lipopeptides.
Colistin loses efficacy in the presence of exogenously supplied LPS or lipid A, presumably because of competition with the OM-associated LPS for colistin binding (38, 39). Here, as expected, treatment of A. baumannii with 500 µg/mL of exogenous lipid A or LPS increased the MIC90 of colistin by approximately fourfold (Fig. 2C). In contrast, the MIC90s for turnercyclamycins A and B were increased by fourfold in the presence of intact LPS but only by twofold in the presence of purified lipid A. This result, coupled with the mcr-1 resistance assay, led us to further investigate the resistance mechanisms through the generation of turnercyclamycin-resistant strains.
Disruption of MlaA confers resistance to turnercyclamycin B but not A
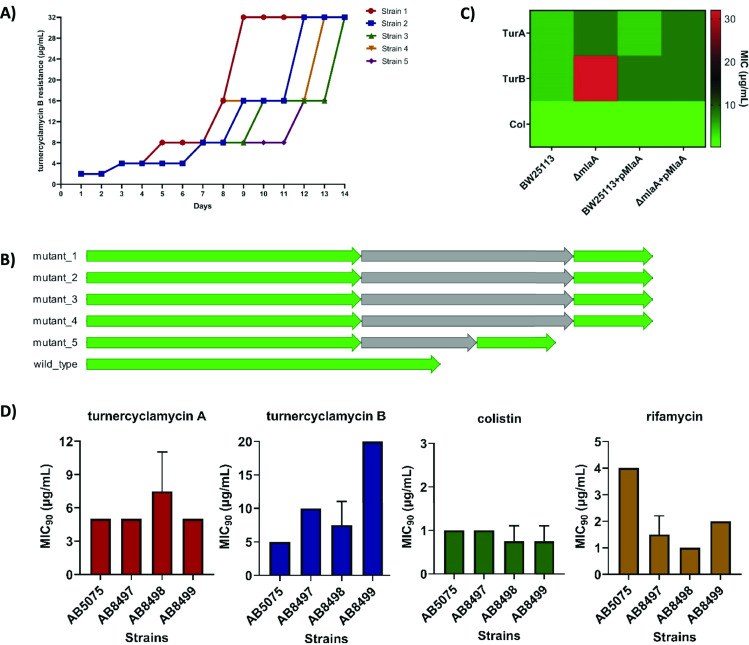
We used E. coli in our efforts to generate turnercyclamycin-resistant strains due to the large database of characterized genes and proteins when compared to Acinetobacter. Furthermore, turnercyclamycins are more potent against E. coli, making them preferred in assays requiring large amounts of compound. We aimed to perform serial inoculation of E. coli cultures into media with gradually increasing levels of turnercyclamycins per passage until an MIC90 of 32× the original MIC90 was achieved. E. coli was passaged daily for up to 63 d. While E. coli readily achieved this 32× target after 8–14 d of exposure to turnercyclamycin B (Fig. 3A), E. coli did not achieve the target level of resistance to turnercyclamycin A after 63 d (only 8× more resistant; Fig. S2). This suggests that turnercyclamycin A does not readily select for resistant mutants. Growth curves generated for the resistant strains revealed that they grow more slowly than the wild type (Fig. S1). Interestingly, even though turnercyclamycins A and B are extremely similar chemically, the turnercyclamycin B-resistant strains were actually more sensitive to turnercyclamycin A than was the wild type (Table 2; Fig. S3). Of note, despite the MIC90 of turnercyclamycin B being 32× that of the original, the MIC50 was still ~2 µg/mL (Fig. S3), indicating residual growth inhibition.
Fig 3.
Disruption of MlaA confers resistance to turnercyclamycin B. (A) Turnercyclamycin B-resistant mutants were generated by daily serial passaging of E. coli DH5α at sub-MIC concentrations of turnercyclamycin, leading to a steady shift in MICs of turnercyclamycin B. This experiment was performed in five biologically independent samples. All mutants conferred early truncation mutations in MlaA. (B) Alignment of the mlaA gene in turnercyclamycin B-resistant mutant strains and WT, viewed using Geneious v10.2.2. All mutants showed early truncation caused by an insertion sequence (gray arrows) in the middle of the mlaA protein coding sequences (green arrows). (C) MICs of Keio E. coli mlaA knockouts treated with turnercyclamycins A and B and colistin, as well as rescue of antibiotic activity by introduction of the pMlaA complementation plasmid. All assays were done in triplicate and three biological replicates. (D) MICs of WT (AB5075) A. baumannii and mlaA knockout strains (AB8497–AB8499) treated with turnercyclamycins A and B, colistin, and rifamycin. All assays were done in triplicate and two biological replicates.
TABLE 2.
MIC90s of turnercyclamycins A and B against evolved turnercyclamycin B-resistant E. coli DH5α
| DH5α strain | MIC90 (µg/mL) | ||
|---|---|---|---|
| Turnercyclamycin A | Turnercyclamycin B | Colistin | |
| WT | 2 | 2 | 0.5 |
| Strain 1 | ≤0.5 | ≥64 | 0.5 |
| Strain 2 | ≥64 | ≥64 | 0.5 |
| Strain 3 | ≤0.5 | ≥64 | 0.5 |
| Strain 4 | 1.0 | ≥64 | 0.5 |
| Strain 5 | 1.0 | ≥64 | 0.5 |
Illumina sequencing of turnercyclamycin B-resistant strains revealed that they all shared a mutation in a single gene encoding the MlaA protein (Fig. 3B). This mutation comprised an early termination of the mlaA gene caused by an insertion sequence, similar to hypothetical proteins found in E. coli, at site 589 (Fig. 3B). MlaA is an OM protein that maintains the asymmetry of the Gram-negative bacterial OM via retrograde trafficking of phospholipids from the OM to the IM (40, 41). To test whether disruption of MlaA was solely responsible for the loss of turnercyclamycin B activity, we tested both turnercyclamycin analogs and colistin against mlaA knockouts of E. coli from the Keio repository (42). Both turnercyclamycin A and colistin retained activity against the knockouts, with MIC90s of 8 and 0.25 µg/mL, respectively (Fig. 3C; Fig. S5). On the other hand, the turnercyclamycin B’s MIC90 was ≥32 µg/mL, with the same incomplete inhibition phenotype that was observed in the spontaneous mutants (Fig. 3C; Fig. S5).
We next transformed both wild-type (WT) and ΔmlaA E. coli with an MlaA expression plasmid and used the resulting strains in tests of antibiotic susceptibility. Expression of MlaA in the ΔmlaA line restored the activity of turnercyclamycin B (MIC90 4 µg/mL), reinforcing the role of MlaA in resistance to turnercyclamycin B. We also obtained knockouts of the E. coli proteins OmpC and OmpF, which form a complex with MlaA (40, 41). These knockouts were not resistant to turnercyclamycins or colistin, suggesting that the resistance profile observed was specifically due to the interruption of MlaA.
Inspired by these results, we attempted similar experiments with a pldA knockout in Acinetobacter baylyi, a non-pathogenic strain that is closely related to A. baumannii. PldA is the phospholipase that is responsible for degrading mislocalized phospholipids and is independent of the Mla system (43). The ΔpldA strain did not significantly demonstrate resistance to turnercyclamycins or colistin and showed similar activity compared to its WT counterpart, A. baylyi ADP1 (Fig. S4). Overall, these results revealed that, so far in all mutants tested, only truncation of MlaA was significantly related to resistance to turnercyclamycin B. Further, it was difficult to generate resistance to turnercyclamycin A.
Finally, we aimed to determine whether mlaA would similarly affect potency in A. baumannii. We obtained three transposon mutants in which the insertion took place at mlaA positions 132, 221, and 507 (AB8497, AB8498, and AB8499), respectively. All three insertions were validated both by PCR and by the increase in sensitivity to rifamycin in comparison to the wild type (Fig. 3D). On average, the mutants were potentially slightly more sensitive to colistin in comparison to the wild type. The MIC90 of turnercyclamycin B against AB8497 was doubled, while it was increased by fourfold against mutant AB8499. In contrast, turnercyclamycin A retained wild-type potency against these strains. Strain AB8498 had low to no change in MIC90s against both turnercyclamycins. These results may indicate that the point of insertion in the mlaA gene is important in achieving resistance, as AB8499, which has an insertion at site 507, was the most resistant strain of the three, which is akin to the turnercyclamycin B-resistant DH5α strains, which have an insertion at site 589. The increased resistance when mlaA is disrupted in E. coli when compared with A. baumannii should be addressed in future studies.
Turnercyclamycin A in vivo efficacy
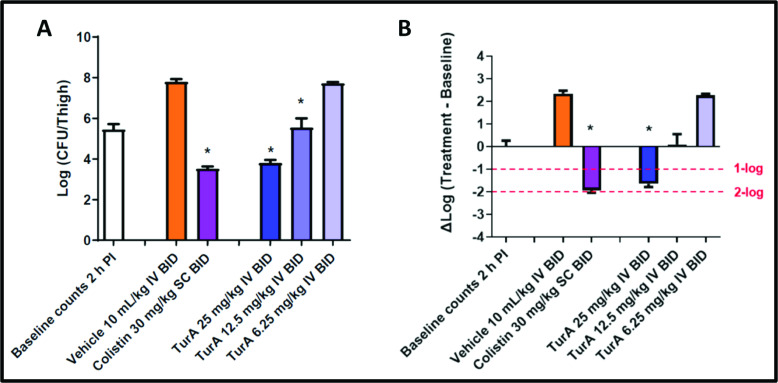
Turnercyclamycin A was produced by fermentation of T. turnerae and rigorously purified and quantified. Animal studies were performed at Eurofins. A preliminary pharmacokinetic (PK) and toxicity profile suggested twice-daily dosing at a maximum of 25 mg/kg. The thigh model of infection was employed using A. baumannii ATCC 17978, comparing turnercyclamycin A (25, 12.5, and 6.25 mg/kg, i.v. at 2 and 14 h after infection) to colistin (30 mg/kg, s.c. at 2 and 14 h after infection). At 26 h after infection, a dose-dependent response was observed, with a statistically significant reduction in bacterial counts for the higher two doses and a response comparable to colistin for the highest dose (Fig. 4). Further work will elaborate the potential efficacy and any potential liabilities of turnercyclamycins and derivatives.
Fig 4.
Turnercyclamycin A shows antimicrobial potency in vivo. Effects of turnercyclamycin A and colistin in the A. baumannii ATCC 17978 thigh infection model with neutropenic BALB/c female mice. (A) Bacterial counts from excised thigh tissue for each treatment group. * indicates a significant difference (P < 0.05) compared to the respective vehicle control determined by one-way ANOVA followed by Dunnett’s test. (B) Change in bacterial counts in thigh tissue at the 26-h sacrifice time point relative to the initial 2-h counts at the time of dosing. * indicates a greater than 1-log10 reduction in counts relative to the baseline at the time of the first dose administration, 2 h after infection, with a significant difference (P < 0.05) based on one-way ANOVA followed by Dunnett’s test.
DISCUSSION
Complications caused by Gram-negative pathogens are problematic worldwide (1 – 4). These include Acinetobacter, which can be difficult to treat, especially in the presence of multidrug resistance (5 – 9). Our initial interest in turnercyclamycins arose because they were specifically effective against Gram-negative pathogens and, more importantly, a panel of resistant A. baumannii strains, even those resistant to the last-line agent colistin (30). Here, we show that turnercyclamycin A is effective in an in vivo model of A. baumannii infection. Because of turnercyclamycin’s clinical potential, it is important to determine why turnercyclamycins had a different resistance profile than colistin. Further, turnercyclamycin A led to less acquired resistance than either colistin or turnercyclamycin B. At a fundamental level, there are several similarities between colistin and turnercyclamycin’s mechanisms of action and resistance, but those were found to lead to cross-resistance under very limited circumstances.
Colistin and turnercyclamycins both required lpxC for activity, and that, along with further results, indicated that LPS was crucial to resistance and potentially to the mechanism of action. Multiple types of modifications to LPS are implicated in polymyxin (colistin) resistance. For example, in Y. pestis, aminoarabinosylation of one LPS phosphate group leads to resistance to polymyxins (and to turnercyclamycins) (30). In A. baumannii, the addition of phosphoethanolamine to a different LPS group by MCR-1 leads to polymyxin resistance but not to turnercyclamycin resistance. We hypothesize that the utility of turnercyclamycins against multidrug-resistant A. baumannii may at least in part result from a more prevalent colistin resistance based upon mcr-1 and not aminoarabinosylation. Further, while loss of LPS itself, as in lpxC mutants, could presumably lead to resistance, in A. baumannii and many other Gram-negative strains, LPS is important for virulence (44, 45). It remains unclear whether LPS itself is directly involved in the molecular mechanism of action or whether a binding event with LPS is required.
In contrast to polymyxins for which resistant strains are readily selected (46, 47), it was more difficult to obtain resistant mutants to turnercyclamycins, especially to turnercyclamycin A. Turnercyclamycin B-resistant strains shared mutations in mlaA, which is involved in the retrograde trafficking of mislocalized phospholipids from the OM to the IM (40, 41). MlaA is not known to directly interact with an antibiotic or to be involved in antibiotic resistance (48 – 51). In a previous study of A. baumannii colistin resistance, the co-deletion of LPS/LOS biosynthesis and mlaA showed greatly increased resistance in comparison to LPS/LOS loss alone (52). However, the effects of mlaA on colistin in A. baumannii are subtle and dose dependent. While the deletion of LPS/LOS confers resistance to colistin in A. baumannii, the resulting growth defects make A. baumannii a less effective pathogen. Recent results show that mutations in other lipid homeostasis genes, especially mlaA, are compensatory, enabling regular growth of LPS/LOS mutants (53). Here, we show that turnercyclamycin B itself also selects for mlaA mutations in E. coli and that turnercyclamycin B loses efficacy in some mlaA mutations in A. baumannii.
It is striking that turnercyclamycin A was not affected by mlaA truncation, when there is only a minor structural difference in the fatty acid tail that distinguishes it from B. This might result either from a direct interaction of MlaA with the lipid portion of the drug or from a differential interaction of turnercyclamycins A and B with phospholipids that are transported by MlaA. In current work in the lipopeptide field, it is common to modify the lipid portion of the molecules and then assume that the underlying mechanisms remain unchanged. Our finding suggests caution, as very simple changes in the turnercyclamycin lipid chains led to a profound change in susceptibility to resistance and to the resistance mechanism itself.
MATERIALS AND METHODS
Bacterial strains
The following strains were used: A. baumannii ATCC 17978 and ATCC 19606 (American Type Culture Collection, Rockville, MD, USA); DH5α competent cells (Thermo Fisher); E. coli C600 (gift of Matthew Mulvey, University of Utah); and E. coli BW25113, ΔompC, ΔompF, and ΔmlaA (National BioResource Project-E. coli, National Institutes of Genetics, Japan). A. baylyi ADP1, ADP1 ΔpldA, A. baumannii 5075 UW, A. baumannii Tn26-mlaA insertion strains 8497, 8498, and 8499, and A. baumannii ΔlpxC::ISAbaI were previously generated as described (gift of Colin Manoil, University of Washington) (54). Strains were cryopreserved as single-use cultures at −80°C.
Antimicrobial broth dilution assay
Cryopreserved strains were streaked on Luria-Bertani (LB) agar (E. coli) and Mueller-Hinton (MH) agar (Acinetobacter). Plates were incubated at 37°C for 8–12 h. Single colonies were transferred into LB or MH broth and incubated for 6–8 h at 30°C and 150 rpm. Cultures were adjusted to match the 0.5 McFarland standard (1 × 108 cells/mL), diluted 200-fold, and used as the inoculum for assays. Test organisms (200 µL) were added to each well of a 96-well plate, and compounds were added using a twofold dilution scheme starting at 32 µg/mL, with eight dilutions per compound. For ΔmlaA A. baumannii strains, 10 µg/mL tetracycline was added, and the compound treatment was a twofold dilution scheme starting at 20 µg/mL, with eight dilutions per compound. After 18–20 h of incubation at 37°C and 150 rpm, 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT; 10 µL; 5 mg/mL) was added to each well, followed by incubation for 2 h. MTT formazan was precipitated and then incubated in DMSO (100 µL) for 1 h. A 570 was measured using a Biotek-Synergy 2 Microplate Reader (Biotek).
LpxC inhibition by CHIR-090 kinetic assay
Single colonies of A. baumannii (ATCC 17978) were grown in MH broth and then adjusted to match the 0.5 McFarland standard (1 × 108 cells/mL), which was diluted 200-fold for use as the inoculum. Turnercyclamycins A and B (16 µg/mL) and colistin (8 µg/mL) were used singly and in combination with CHIR-090 (8 µg/mL). Each assay was performed in triplicate in a 96-well plate. The plate was then incubated statically at 37°C in a Biotek-Synergy 2 Microplate Reader (Biotek). The OD600 of each well was then measured every 30 min for 24 h.
Exogenous addition of purified LPS and lipid A assay
A. baumannii (ATCC 17978) colonies were grown in MH broth, adjusted to match the 0.5 McFarland standard (1 × 108 cells/mL), and diluted 200-fold for use as the inoculum. Prior to the addition of test compounds, either purified LPS (Sigma-Aldrich L2018) or lipid A (Sigma-Aldrich L5399) (100 µg/mL) was added to wells. Antimicrobial broth dilution assay then proceeded as described previously with turnercyclamycins being treated at a twofold dilution scheme starting at 32 µg/mL, with eight dilutions each, and colistin at a twofold dilution scheme starting at 8 µg/mL, with eight dilutions as a control.
Antibiotic susceptibility testing of MCR-1-expressing E. coli
Chemically competent E. coli DH5α and C600 were transformed with empty vector pBR322 (NEB N3033S) and pGDP2-MCR-1 (Addgene 118404), streaked on LB agar under antibiotic selection (kanamycin, 50 µg/mL), and incubated overnight at 37°C. The resulting strains were used in antimicrobial broth dilution assays.
Evolution of turnercyclamycin-resistant E. coli by sublethal dose passaging
Overnight broth cultures of E. coli DH5α were diluted 1:500 in LB broth (200 µL) containing 0.5×, 1×, and 2× MIC of each turnercyclamycin and incubated at 30°C and 150 rpm for 24 h. The highest concentration that showed growth based upon turbidity was adjusted to the 0.5 McFarland suspension and diluted 200-fold to be used as the inoculum for the next passage. The concentration of the drug that was twofold above the previous inoculum was assigned as the new 1× MIC. This was repeated until an MIC of 32 µg/mL was achieved. Resistance was confirmed and maintained by growing it in LB containing the compound with the new MIC. Experiments were performed in four independent cultures. The purity of the resistant strains was confirmed by wet mount microscopy, 16S rDNA sequencing, and metagenomics analysis via Autometa v2.2.0 (55). Upon purity analysis, one strain was found to be contaminated with Staphylococcus (Strain 2), as determined by Autometa binning.
Next-generation sequencing and mutation analysis
Illumina library preparation and sequencing were performed at the Huntsman Cancer Institute-High-Throughput Genomics (HCI-HTG) shared resource at the University of Utah. Sequencing library preparation used an NEBNext Ultra II DNA Library Prep Kit with a 450-bp mean insert size. Sequencing used an Illumina NovaSeq 6000 sequencer with 2× 150-bp runs. Raw reads were trimmed by Trimmomatic and assembled with metaSPAdes (56). Genes were predicted using Prodigal (57), and coding sequences were compared between wild-type and mutant strains using NCBI blastn tools. The threshold for identifying the same gene was set at 90% identity in the nucleotide sequence. Any gene in the mutant strain genome that had an identity of less than 100% or an alignment coverage of less than 100% when compared to the gene from the wild-type strain was considered a mutated gene. Gene alignment used CLC Genomics Workbench 11 and Geneious v10.2.2.
Plasmid complementation for ΔmlaA E. coli
E. coli BW25113 and ΔmlaA were transformed with a medium copy number pTwist Amp plasmid expressing MlaA (Twist Bioscience, pTwistAmp_MlaA; Fig. S6) and used in antimicrobial broth dilution assays described above, except that kanamycin (0.1 mg/mL) and ampicillin (0.1 mg/mL) were added to the mixtures.
Quantification and statistical analysis
MIC data and growth curves were graphed using GraphPad Prism 9.5.1. Error bars on MIC graphs and growth curves were indicated as mean ± SD of three replicate wells and are done in either two or three biological replicates.
Animal studies
Eurofins used mice from BioLASCO Taiwan. ALB/c female mice (18 ± 2 g) and BALB/c female mice (7 wk old) were used for PK/toxicity and efficacy studies, respectively. The study was conducted at Eurofins (Taiwan), an AAALAC-accredited facility. The protocol was reviewed by the IACUC at Pharmacology Discovery Services Taiwan, Ltd. The University of Utah IACUC number associated with this study is 22-01003.
Pharmacokinetics
Protocols were performed at Eurofins (Taiwan). Turnercyclamycin A in plasma was measured by liquid chromatography-tandem mass spectrometry, and graphs illustrating the mean plasma concentrations (mean ± SD) over time were generated. PK parameters were obtained using non-compartmental analysis of the plasma data with WinNonlin.
Neutropenic thigh infection model
Five neutropenic mice were used in each treatment group. The mice were anesthetized with 3%–5% isoflurane inhalation and then injected intramuscularly in the left thigh with a 0.1-mL suspension of A. baumannii ATCC 17978 [1.9 × 105 colony-forming units (CFU) per mouse]. At 2 and 14 h after infection, turnercyclamycin A was administered intravenously slowly (~30 s, IV bolus) at doses of 6.25, 12.5, and 25 mg/kg. Subcutaneous administration of colistin at a dose of 30 mg/kg was also performed at the same time points. The dosing volumes were 10 mL/kg. An infected control group, which did not receive any treatment, was sacrificed at 2 h after infection to establish the baseline bacterial counts. The remaining groups were sacrificed at 26 h. Bacterial counts in the thigh, standardized to the weight of each thigh, were determined using a serial dilution plating method and calculated using the formula below:
Statistical significance (P < 0.05) was evaluated using one-way ANOVA followed by Dunnett’s test through GraphPad Prism software version 5.0. A significant decrease in bacterial counts, with a P value < 0.05, compared to the vehicle control or baseline groups, was considered statistically significant.
ACKNOWLEDGMENTS
We thank Colin Manoil and Jeannie Bailey for supplying the following strains: A. baumannii 5075UW, ΔlpxC::ISAbaI, AB8497, AB8498, and AB8499 and A. baylyi ADP1 and ADP1 ΔpldA. We would like to acknowledge their help and guidance throughout this project. We would also like to thank Matthew Mulvey for providing E. coli C600.
This work was funded by NIH R01AI162943.
Conceptualization: A.L.L., B.W.M., M.G.H., and E.W.S. Methodology: A.L.L., B.W.M., Z.L., and E.W.S. Software: Z.L. Validation: A.L.L. and E.W.S. Investigation: A.L.L., B.W.M., and Z.L. Resources: E.W.S. Data curation: A.L.L., B.W.M., and Z.L. Writing—original draft: A.L.L., B.W.M., and E.W.S. Writing—review and editing: A.L.L., B.W.M., Z.L., M.A.F., L.R.B., M.G.H., and E.W.S. Visualization: A.L.L., B.W.M., and Z.L. Supervision: M.A.F., L.R.B., M.G.H., and E.W.S. Project administration: E.W.S. Funding acquisition: E.W.S.
Contributor Information
Eric W. Schmidt, Email: ews1@utah.edu.
Ayush Kumar, University of Manitoba, Winnipeg, Manitoba, Canada .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02306-23.
Supplemental Figures and Tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Larsson DGJ, Flach C-F. 2022. Antibiotic resistance in the environment. Nat Rev Microbiol 20:257–269. doi: 10.1038/s41579-021-00649-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chokshi A, Sifri Z, Cennimo D, Horng H. 2019. Global contributors to antibiotic resistance. J Glob Infect Dis 11:36–42. doi: 10.4103/jgid.jgid_110_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Urban-Chmiel R, Marek A, Stępień-Pyśniak D, Wieczorek K, Dec M, Nowaczek A, Osek J. 2022. Antibiotic resistance in bacteria-a review. Antibiotics (Basel) 11:1079. doi: 10.3390/antibiotics11081079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gajdács M, Albericio F. 2019. Antibiotic resistance: from the bench to patients. Antibiotics 8:129. doi: 10.3390/antibiotics8030129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, Lee SH. 2017. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. doi: 10.3389/fcimb.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nowak P, Paluchowska P. 2016. Acinetobacter baumannii: biology and drug resistance - role of carbapenemases. Folia Histochem Cytobiol 54:61–74. doi: 10.5603/FHC.a2016.0009 [DOI] [PubMed] [Google Scholar]
- 7. McConnell MJ, Actis L, Pachón J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- 8. CDC . 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 9. Kadri SS. 2020. Key takeaways from the U.S. CDC’s 2019 antibiotic resistance threats report for frontline providers. Crit Care Med 48:939–945. doi: 10.1097/CCM.0000000000004371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jain R, Danziger LH. 2004. Multidrug-resistant Acinetobacter infections: an emerging challenge to clinicians. Ann Pharmacother 38:1449–1459. doi: 10.1345/aph.1D592 [DOI] [PubMed] [Google Scholar]
- 11. Wang H, Guo P, Sun H, Wang H, Yang Q, Chen M, Xu Y, Zhu Y. 2007. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob Agents Chemother 51:4022–4028. doi: 10.1128/AAC.01259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeon JH, Jang K-M, Lee JH, Kang L-W, Lee SH. 2023. Transmission of antibiotic resistance genes through mobile genetic elements in Acinetobacter baumannii and gene-transfer prevention. Sci Total Environ 857:159497. doi: 10.1016/j.scitotenv.2022.159497 [DOI] [PubMed] [Google Scholar]
- 14. Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- 15. Michalopoulos A, Falagas ME. 2008. Colistin and polymyxin B in critical care. Crit Care Clin 24:377–391, doi: 10.1016/j.ccc.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 16. Nang SC, Azad MAK, Velkov T, Zhou QT, Li J. 2021. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol Rev 73:679–728. doi: 10.1124/pharmrev.120.000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andrade FF, Silva D, Rodrigues A, Pina-Vaz C. 2020. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 8:1716. doi: 10.3390/microorganisms8111716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morrison DC, Jacobs DM. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813–818. doi: 10.1016/0019-2791(76)90181-6 [DOI] [PubMed] [Google Scholar]
- 20. Yin N, Marshall RL, Matheson S, Savage PB. 2003. Synthesis of lipid A derivatives and their interactions with polymyxin B and polymyxin B nonapeptide. J Am Chem Soc 125:2426–2435. doi: 10.1021/ja0284456 [DOI] [PubMed] [Google Scholar]
- 21. Sabnis A, Hagart KLH, Klöckner A, Becce M, Evans LE, Furniss RCD, Mavridou DAI, Murphy R, Stevens MM, Davies JC, Larrouy-Maumus GJ, Clarke TB, Edwards AM. 2021. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 10:e65836. doi: 10.7554/eLife.65836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gogry FA, Siddiqui MT, Sultan I, Haq QMR. 2021. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front Med (Lausanne) 8:677720. doi: 10.3389/fmed.2021.677720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao X, Zhong X, Yang S, Deng K, Liu L, Song X, Zou Y, Li L, Zhou X, Jia R, Lin J, Tang H, Ye G, Yang J, Zhao S, Lang Y, Wan H, Yin Z, Kuipers OP. 2023. Elucidating the mechanism of action of the Gram-negative-pathogen-selective cyclic antimicrobial lipopeptide brevicidine. Antimicrob Agents Chemother 67:e0001023. doi: 10.1128/aac.00010-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cochrane SA, Findlay B, Bakhtiary A, Acedo JZ, Rodriguez-Lopez EM, Mercier P, Vederas JC. 2016. Antimicrobial lipopeptide lridecaptin A1 selectively binds to Gram-negative lipid II. Proc Natl Acad Sci U S A 113:11561–11566. doi: 10.1073/pnas.1608623113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szałaj N, Lu L, Benediktsdottir A, Zamaratski E, Cao S, Olanders G, Hedgecock C, Karlén A, Erdélyi M, Hughes D, Mowbray SL, Brandt P. 2018. Boronic ester-linked macrocyclic lipopeptides as serine protease inhibitors targeting Escherichia coli type I signal peptidase. Eur J Med Chem 157:1346–1360. doi: 10.1016/j.ejmech.2018.08.086 [DOI] [PubMed] [Google Scholar]
- 26. Altamia MA, Lin Z, Trindade-Silva AE, Uy ID, Shipway JR, Wilke DV, Concepcion GP, Distel DL, Schmidt EW, Haygood MG. 2020. Secondary metabolism in the gill microbiota of shipworms (Teredinidae) as revealed by comparison of metagenomes and nearly complete symbiont genomes. mSystems 5:e00261-20. doi: 10.1128/mSystems.00261-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elshahawi SI, Trindade-Silva AE, Hanora A, Han AW, Flores MS, Vizzoni V, Schrago CG, Soares CA, Concepcion GP, Distel DL, Schmidt EW, Haygood MG. 2013. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc Natl Acad Sci U S A 110:E295–E304. doi: 10.1073/pnas.1213892110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lacerna NM, Miller BW, Lim AL, Tun JO, Robes JMD, Cleofas MJB, Lin Z, Salvador-Reyes LA, Haygood MG, Schmidt EW, Concepcion GP. 2019. Mindapyrroles A–C, pyoluteorin analogues from a shipworm-associated bacterium. J Nat Prod 82:1024–1028. doi: 10.1021/acs.jnatprod.8b00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lacerna NM, Ramones CMV, Robes JMD, Picart MRD, Tun JO, Miller BW, Haygood MG, Schmidt EW, Salvador-Reyes LA, Concepcion GP. 2020. Inhibition of biofilm formation by modified oxylipins from the shipworm symbiont Teredinibacter turnerae. Mar Drugs 18:656. doi: 10.3390/md18120656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller BW, Lim AL, Lin Z, Bailey J, Aoyagi KL, Fisher MA, Barrows LR, Manoil C, Schmidt EW, Haygood MG. 2021. Shipworm symbiosis ecology-guided discovery of an antibiotic that kills colistin-resistant Acinetobacter. Cell Chem Biol 28:1628–1637. doi: 10.1016/j.chembiol.2021.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J. 2002. Teredinibacter turnerae gen nov. sp. nov.,a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int J Syst Evol Microbiol 52:2261–2269. doi: 10.1099/00207713-52-6-2261 [DOI] [PubMed] [Google Scholar]
- 32. Gao R, Hu Y, Li Z, Sun J, Wang Q, Lin J, Ye H, Liu F, Srinivas S, Li D, Zhu B, Liu Y-H, Tian G-B, Feng Y, Zhang G. 2016. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog 12:e1005957. doi: 10.1371/journal.ppat.1005957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Y. 2018. Transferability of MCR-1/2 polymyxin resistance: complex dissemination and genetic mechanism. ACS Infect Dis 4:291–300. doi: 10.1021/acsinfecdis.7b00201 [DOI] [PubMed] [Google Scholar]
- 34. Delgado-Blas JF, Ovejero CM, Abadia-Patiño L, Gonzalez-Zorn B. 2016. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother 60:6356–6358. doi: 10.1128/AAC.01319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Whittington DA, Rusche KM, Shin H, Fierke CA, Christianson DW. 2003. Crystal structure of LpxC, a zinc-dependent deacetylase essential for endotoxin biosynthesis. Proc Natl Acad Sci U S A 100:8146–8150. doi: 10.1073/pnas.1432990100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson BW, Nieckarz M, Pinedo V, McLean AB, Cava F, Trent MS. 2021. Acinetobacter baumannii can survive with an outer membrane lacking lipooligosaccharide due to structural support from elongasome peptidoglycan synthesis. mBio 12:e0309921. doi: 10.1128/mBio.03099-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barb AW, McClerren AL, Snehelatha K, Reynolds CM, Zhou P, Raetz CRH. 2007. Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry 46:3793–3802. doi: 10.1021/bi6025165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Z, Koirala B, Hernandez Y, Zimmerman M, Park S, Perlin DS, Brady SF. 2022. A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature 601:606–611. doi: 10.1038/s41586-021-04264-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warren HS, Kania SA, Siber GR. 1985. Binding and neutralization of bacterial lipopolysaccharide by colistin nonapeptide. Antimicrob Agents Chemother 28:107–112. doi: 10.1128/AAC.28.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, van den Berg B. 2017. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2:1616–1623. doi: 10.1038/s41564-017-0046-x [DOI] [PubMed] [Google Scholar]
- 41. Yeow J, Tan KW, Holdbrook DA, Chong Z-S, Marzinek JK, Bond PJ, Chng S-S. 2018. The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J Biol Chem 293:11325–11340. doi: 10.1074/jbc.RA118.002441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grant KA, Belandia IU, Dekker N, Richardson PT, Park SF. 1997. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect Immun 65:1172–1180. doi: 10.1128/iai.65.4.1172-1180.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsuura M. 2013. Structural modifications of bacterial lipopolysaccharide that facilitate Gram-negative bacteria evasion of host innate immunity. Front Immunol 4:109. doi: 10.3389/fimmu.2013.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huszczynski SM, Lam JS, Khursigara CM. 2020. The role of Pseudomonas aeruginosa lipopolysaccharide in bacterial pathogenesis and physiology. Pathogens 9:6. doi: 10.3390/pathogens9010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leshaba TMS, Mbelle NM, Osei Sekyere J. 2022. Current and emerging polymyxin resistance diagnostics: a systematic review of established and novel detection methods. J Appl Microbiol 132:8–30. doi: 10.1111/jam.15184 [DOI] [PubMed] [Google Scholar]
- 47. Carrasco LD de M, Dabul ANG, Boralli CMDS, Righetto GM, Carvalho ISE, Dornelas JV, Martins da Mata CPS, de Araújo CA, Leite EMM, Lincopan N, Camargo ILB da C. 2021. Polymyxin resistance among XDR ST1 carbapenem-resistant Acinetobacter baumannii clone expanding in a teaching hospital. Front Microbiol 12:622704. doi: 10.3389/fmicb.2021.622704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Munguia J, LaRock DL, Tsunemoto H, Olson J, Cornax I, Pogliano J, Nizet V. 2017. The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J Mol Med 95:1127–1136. doi: 10.1007/s00109-017-1579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Royer G, Ortiz de la Rosa J-M, Vuillemin X, Lacombe B, Chau F, Clermont O, Mercier-Darty M, Decousser J-W, Ricard J-D, Nordmann P, Denamur E, Poirel L. 2022. Reduced chlorhexidine susceptibility is associated with tetracycline resistance tet genes in clinical isolates of Escherichia coli. Antimicrob Agents Chemother 66:e0197221. doi: 10.1128/AAC.01972-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernier SP, Son S, Surette MG. 2018. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J Bacteriol 200:e00156-18. doi: 10.1128/JB.00156-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kathayat D, Closs G, Helmy YA, Lokesh D, Ranjit S, Rajashekara G. 2021. Peptides affecting the outer membrane lipid asymmetry system (MlaA-OmpC/F) reduce avian pathogenic Escherichia coli (APEC) colonization in chickens. Appl Environ Microbiol 87:e0056721. doi: 10.1128/AEM.00567-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boinett CJ, Cain AK, Hawkey J, Do Hoang NT, Khanh NNT, Thanh DP, Dordel J, Campbell JI, Lan NPH, Mayho M, Langridge GC, Hadfield J, Chau NVV, Thwaites GE, Parkhill J, Thomson NR, Holt KE, Baker S. 2019. Clinical and laboratory-induced colistin-resistance mechanisms in Acinetobacter baumannii. Microb Genom 5:e000246. doi: 10.1099/mgen.0.000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Powers MJ, Trent MS. 2018. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc Natl Acad Sci U S A 115:E8518–E8527. doi: 10.1073/pnas.1806714115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. 2015. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol 197:2027–2035. doi: 10.1128/JB.00131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller IJ, Rees ER, Ross J, Miller I, Baxa J, Lopera J, Kerby RL, Rey FE, Kwan JC. 2019. Autometa: automated extraction of microbial genomes from individual shotgun metagenomes. Nucleic Acids Res. 47:e57–e57. doi: 10.1093/nar/gkz148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures and Tables.