ABSTRACT
Bacterial chromosome segregation is facilitated by the ParABS system. The ParB protein binds centromere-like parS sequences and forms nucleoprotein complexes. These nucleoprotein complexes are segregated by the dynamic ATPase ParA. In mycobacteria, ParA also interacts with the polar growth determinant DivIVA (Wag31). This interaction was earlier shown not only to facilitate the segregation of ParB complexes but also to affect cell extension. Here, we identify an additional partner of ParA in Mycobacterium smegmatis, named PapM. Using E. coli-based analysis, we show that PapM likewise interacts with DivIVA and that the tripartite interaction of ParA-PapM-DivIVA is phosphorylation dependent: ParA binding to DivIVA is diminished, while PapM binding is promoted upon phosphorylation of DivIVA. The presence of PapM enhances the dissociation of ParA from the DivIVA complex upon its phosphorylation. Studies of M. smegmatis mutant strains reveal that altered PapM levels influence chromosome segregation and cell length. The elimination of PapM affects ParA dynamics. Furthermore, ParA and, to a lesser extent, PapM modulate the subcellular distribution of DivIVA. Altogether, our studies show that the tripartite interplay between ParA-DivIVA and PapM controls the switch between cell division and cell elongation and, in this way, affects the mycobacterial cell cycle.
IMPORTANCE
The genus of Mycobacterium includes important clinical pathogens (M. tuberculosis). Bacteria of this genus share the unusual features of their cell cycle such as asymmetric polar cell elongation and long generation time. Markedly, control of the mycobacterial cell cycle still remains not fully understood. The main cell growth determinant in mycobacteria is the essential protein DivIVA, which is also involved in cell division. DivIVA activity is controlled by phosphorylation, but the mechanism and significance of this process are unknown. Here, we show how the previously established protein interaction partner of DivIVA in mycobacteria, the segregation protein ParA, affects the DivIVA subcellular distribution. We also demonstrate the role of a newly identified M. smegmatis DivIVA and ParA interaction partner, a protein named PapM, and we establish how their interactions are modulated by phosphorylation. Demonstrating that the tripartite interplay affects the mycobacterial cell cycle contributes to the general understanding of mycobacterial growth regulation.
KEYWORDS: chromosome segregation, mycobacteria, polar growth, ParA
INTRODUCTION
Key processes of the cell cycle must be precisely coordinated. In bacteria, chromosome replication overlaps with chromosome segregation. Both processes are accompanied by cell elongation and must be completed before cell division (1 – 3). Most bacterial species employ a system based on ParA and ParB proteins to actively segregate their chromosomes (3 – 5). ParAB proteins move and re-position chromosomal replication initiation regions (oriC) soon after they are duplicated. ParB binds to oriC proximal parS sites to form nucleoprotein segregation complexes named segrosomes, which are moved toward cell poles due to interactions with the nucleoid-bound ATPase ParA (5, 6). Upon interaction with segrosomes, ParA dimers hydrolyze ATP and are released from the nucleoid, while the ParB complex moves toward a higher concentration of the nucleoid-bound ParA (7, 8). Surprisingly, the ParAB-mediated segregation shows some bacterial species-specific features (4, 9, 10). These differences involve the interactions of the segregation proteins with other proteins or protein complexes, like the cell elongation complex which governs polar cell extension in Actinobacteria, such as mycobacteria.
Mycobacteria are well known as slow-growing, clinically important pathogens (e.g., Mycobacterium tuberculosis), but this genus also contains relatively fast-growing saprophytes (e.g., Mycobacterium smegmatis, whose division time is approximately 150 min) (11, 12). Mycobacteria are rod-shaped bacilli, characterized by a thick, lipid-rich cell wall and uncommon features of the cell cycle (13 – 15). Their slow growth as well as the ability to remain in a dormant, nonreplicating state for years increase their potential of being successful intracellular pathogens (11). Interestingly, the key mechanisms of mycobacterial cell cycle control are still not well understood.
One of the unique mycobacterial features is their asymmetric polar cell extension: the cell pole inherited from the mother cell (the old cell pole) extends faster than the newly established cell pole (the new cell pole) (11, 16 – 19). The asymmetry of the mycobacterial cell is conferred by the polar protein complex that governs cell extension. The key component of this polar complex is the essential, polarity determination protein DivIVA (in mycobacteria also called Wag31 or Antigen 84) (20 – 23). Changes in the DivIVA level modulate polar cell wall synthesis and affect cell shape (20, 21, 23, 24). DivIVA was shown to control the activity of the enzymes involved in the cell wall synthesis (25, 26). At the latest stages of cell division, DivIVA is recruited to the division site where it interacts with divisome protein PBP3 (27, 28). It was also suggested that the other DivIVA interaction partners CwsA and CrgA may contribute to DivIVA relocation (29). The late recruitment of DivIVA at the cell division site suggests its involvement in constituting the new cell poles (11, 19). DivIVA mutational analysis suggested that DivIVA activity involves different interactions at the new pole, the old pole, and the septum (27). However, the details of the DivIVA role in the formation of the new poles are still to be investigated.
Mycobacterial DivIVA was shown to be phosphorylated at a single threonine (Thr73 in M. tuberculosis, Thr74 M. smegmatis) by PknA and PknB kinases in response to environmental conditions (20, 25, 30). DivIVA phosphorylation is further controlled by MtrB, which is the sensor kinase of the two-component system MtrAB (31). DivIVA phosphorylation reportedly increased its polar accumulation. The modification was elevated during the exponential growth phase and was linked to enhanced cell wall synthesis leading to faster cell extension (20, 25, 32). Thus, phosphorylation-dependent DivIVA activity was suggested to link the rate of the mycobacterial cell extension to environmental conditions. However, the exact mechanism of this process remains underexplored.
Markedly, the asymmetry of the mycobacterial cells is also manifested by the asymmetry of their cell division, chromosome positioning, and segregation. Mycobacteria, like other bacteria, use ParA and ParB proteins to segregate the newly replicated oriC regions (33, 34). While in M. tuberculosis, parA and parB genes are essential, in M. smegmatis, elimination of ParA or ParB is possible but leads to severe chromosome segregation defects and alters the cell length (34, 35). In M. smegmatis, ParB binds three parS sites located in the proximity of the oriC region to form a segrosome (35, 36). Upon initiation of the DNA replication, duplicated segrosomes move asymmetrically—the one that moves toward the new pole travels faster and covers a greater distance than the one that moves toward the old cell pole. The positioning and movements of the segrosomes are governed by their interactions with the ParA protein (35). In M. smegmatis, EGFP-ParA is observed to accumulate transiently in proximity to the new cell pole in the new-born cell. During the progress of cell cycle, ParA-EGFP forms a fluorescent patch that extends along the cell, toward the old pole and ParB complex. In the absence of ParA, segrosomes are mislocalized, and cell division is often misplaced, resulting in the frequent formation of anucleate cells and the increased variation of the progeny cell length (35).
The unique features of mycobacterial chromosome segregation further include the interaction between ParA and DivIVA (9, 34, 37). Disruption of ParA-DivIVA binding results in modest segregation defects (37). However, the lack of ParA recruitment to DivIVA impairs segrosome movement and accelerates the cell elongation, implying that ParA affects DivIVA function. Interestingly, a point mutation that disrupts ParA binding to DNA leads to increased ParA accumulation at the cell poles, suggesting a competition between DivIVA and the nucleoid for ParA binding. Our studies have also suggested that polar recruitment of ParA may be induced by unfavorable environmental conditions (37).
Here, we further elucidate the mechanism of mycobacterial chromosome segregation. We identified, via a bacterial two hybrid screen, the novel component of M. smegmatis chromosome segregation machinery, protein MSMEG_5597 named PapM (ParA partner in M. smegmatis) that interacts with both ParA and DivIVA. We investigated how PapM modulates the interaction between ParA and DivIVA and how phosphorylation of the latter affects this interaction. We demonstrate that PapM cooperates with ParA in chromosome segregation and controls cell length. Finally, we explore PapM’s influence on ParA dynamics and the impact of both partners on DivIVA subcellular distribution.
RESULTS
An interaction screen identified the novel ParA interaction partner named PapM
We sought to identify novel components of M. smegmatis chromosome segregation machinery. To this end, we screened the bacterial two-hybrid (BTH) library of M. smegmatis genome in the pKT25 vector using T18-ParA (pUT18CparA) as the bait. In the BTH system, the interaction between a bait and a pray, both fused to adenyl cyclase subunits (T18 and T25), restores adenyl cyclase activity in E. coli BTH101 cells and leads to development of colored colonies on an indicator media (38).
In the library screening among the positive clones, we identified one containing the fragment (encoding 8–206 aa) of the msmeg_5597 gene hereafter named papM (ParA partner in M. smegmatis). The papM gene encoded for a 22.5-kDa protein consisting of 206 amino acids and belonging to a family of TetR regulators which are characterized by the presence of N-terminal HTH motif and ability to dimerize (39). To confirm the interaction of PapM with M. smegmatis ParA, we have cloned the entire papM gene into BTH vectors obtaining pUT18CpapM and pKT25papM. BTH analysis confirmed the interaction between T25-ParA and T18-PapM, as well as between T25-PapM and T18-ParA (Fig. 1A; Fig. S1A). Additionally, we noted the interaction between T18-PapM and T25-PapM (Fig. 1A), which indicates the dimerization of PapM protein consistent with dimerization of TetR family regulators (39).
Fig 1.
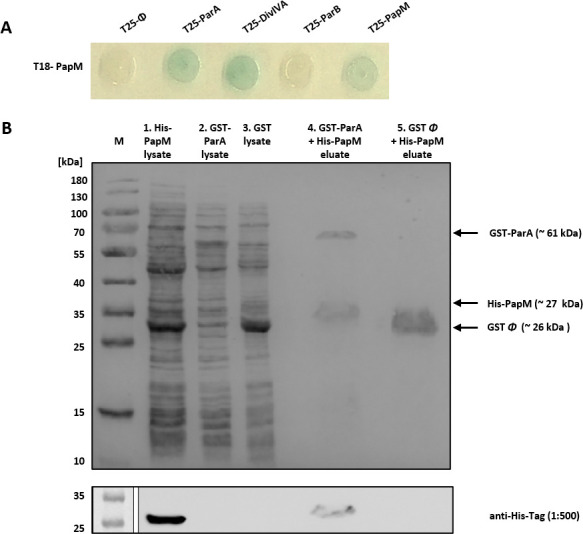
Identification of PapM as a novel M. smegmatis ParA interaction partner. (A) BTH analysis shows T18-PapM interactions. The blue color of the colony indicates interaction. (B) Affinity chromatography confirmation of PapM-ParA interactions. The SDS-PAGE showing the His-PapM binding to GST-ParA but not to the GST column. First, the lysate of the E. coli BL21 producing GST-ParA (lane 2) or GST (lane 3) was loaded on GSH-Sepharose; next, after the washing of unbound proteins, lysate of the E. coli BL21 producing His-PapM (lane 1) was loaded on both columns. The proteins retained at resin were eluted with glutathione (lanes 4 and 5). The bottom panel shows the Western blotting confirmation of His-PapM presence in eluate in the presence of GST-ParA but not in control GST eluate. Anti-His antibody was used for His-PapM detection. Presented result is the representative example of independent four experimental repeats.
To confirm the observed interaction between ParA and PapM, we performed an affinity chromatography pulldown. In this approach, the glutathione-S-transferase (GST)-ParA fusion protein or the GST as the negative control was bound to GSH-Sepharose. Next, the lysate of E. coli BL21 overproducing His-PapM was applied to the column. Analysis of the affinity chromatography eluates using SDS-PAGE and Western blot with anti-His antibodies showed that His-PapM was retained on the resin in presence of GST-ParA but not in presence of GST alone (Fig. 1B). This experiment confirmed the direct interaction between PapM and ParA.
Having confirmed ParA-PapM interaction in vitro, we used BTH to examine if PapM preferably interacted with ParA dimer or monomer and if ParA’s binding to DNA could influence its interaction with PapM. To this end, we tested the interaction of PapM with the following previously described ParA variants: ATP binding deficient monomer ParAK44A; dimerization-impaired ParAG40V; ATP hydrolysis deficient and DNA-associated dimer ParAD68A; DNA interaction deficient dimer ParAR219E; and ParAT3A, which is unable to interact with DivIVA (37). The only T25-ParA variant that interacted with T18-PapM was ParAR219E, suggesting that PapM preferably binds ParA dimer in the absence of DNA (Fig. S1A). This notion was reinforced by the lack of detected interactions with monomer version ParAK44A and ParAG40V (Fig. S1A). Interestingly, an alphafold multimer model of a dimer of PapM and a dimer of ParA (40) predicted a PapM interaction at the DNA binding site on ParA, which could indicate PapM interfering with the ParA-DNA interaction (Fig. S1B).
Summarizing, the BTH library screening identified a novel ParA partner named PapM, while an affinity chromatography pulldown approach confirmed a direct interaction between ParA and PapM.
PapM, ParA, and DivIVA form a tripartite complex, which is affected by DivIVA phosphorylation
In mycobacteria, ParA interacts, apart from ParB, also with DivIVA (34, 37). Therefore, we tested whether PapM could interact with other ParA partners. Indeed, BTH analysis revealed T18-PapM interaction with T25-DivIVA but not with T25-ParB (Fig. 1A). Further testing of the PapM interactions using M. smegmatis DivIVA subdomains fused to the T18 domain of adenylate cyclase indicated that interaction interface may encompass the linker between DivIVA coiled-coil domains and fragment of second coiled coil, the region that contains a single phosphorylation site—Thr 74. Interestingly, the same fragment of DivIVA was earlier shown to be involved in the interactions with ParA (34).
Next, based on the obtained BTH results, we set out to examine PapM interactions with ParA and DivIVA using protein colocalization assays in E. coli. Earlier, we have shown that EGFP-ParA colocalized at the poles of E. coli cells with mCherry-DivIVA when coproduced (9, 37). Here, we tested whether PapM fused to fluorescent protein mTurquoise2 (PapM-mT2) would colocalize with the polar mCherry-DivIVA complex and whether ParA recruitment to polar DivIVA complex would be affected by the presence of PapM-mT2. Additionally, we investigated whether DivIVA phosphorylation would influence ParA or PapM interactions.
In E. coli cells, PapM-mT2 localized at the poles in the presence of polarly localized mCherrry-DivIVA but not in the control strain with polarly localized Ics-mCherry [Ics protein fragment targeted the mCherry to the cell poles (41)] (Fig. 2A and B). In E. coli coproducing PapM-mT2, EGFP-ParA, and mCherry-DivIVA, the three fusion proteins colocalized at the cell poles (Fig. 2D). This result indicates the engagement of PapM in the formation of the tripartite complex. Measurements of the ratio of EGFP-ParA fluorescence at the poles to its fluorescence at the mid-cell indicated that EGFP-ParA recruitment to mCherry-DivIVA was not visibly affected by the presence of PapM-mT2 (Fig. 2I).
Fig 2.
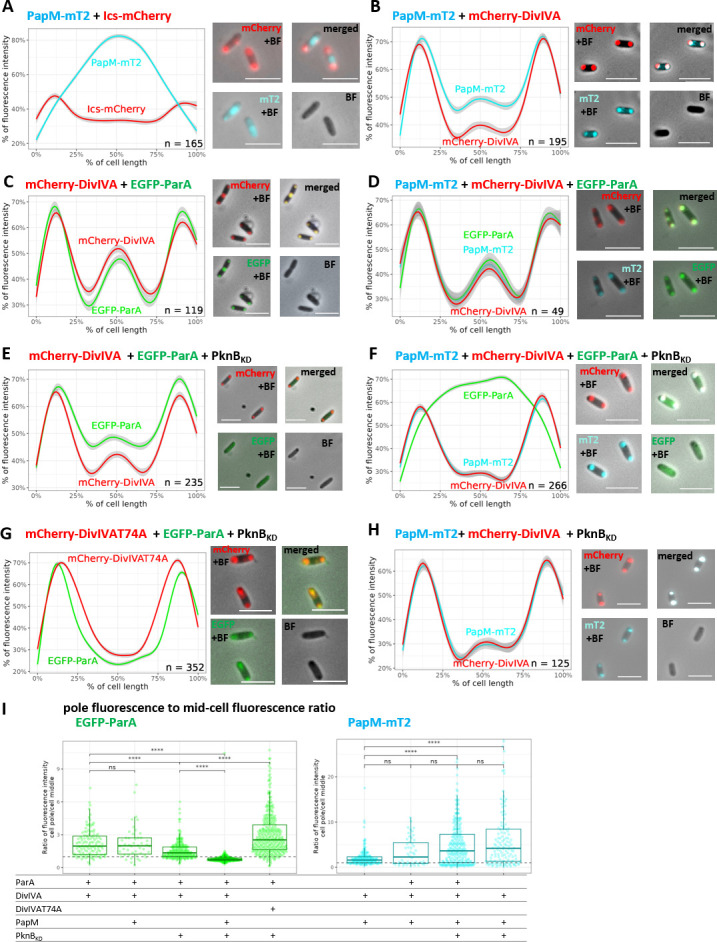
Phosphorylation enhances PapM binding to DivIVA while diminishing ParA recruitment to DivIVA in E. coli. (A) Colocalization of PapM-mTurquoise2 (MT2) with Ics-mCherry. (B) Colocalization of PapM-mT2 with mCherry-DivIVA. (C) Colocalization of mCherry-DivIVA and EGFP-ParA. (D) Colocalization of mCherry-DivIVA, EGFP-ParA and PapM-mT2. (E) Colocalization EGFP-ParA with mCherry-DivIVA in presence of His-PknBKD. (F) Colocalization of mCherry-DivIVA, EGFP-ParA, and PapM-mT2 in the presence of His-PknBKD. (G) Colocalization of phosphoablative mCherry-DivIVAT74A variant and EGFP-ParA in the presence of His-PknBKD. (H) Colocalization of PapM-mT2 with mCherry-DivIVA in the presence of His-PknBKD. Left panels show the profiles of fluorescence measured along the E. coli BL21 cells, and the right panels show the representative images of E. coli cells producing EGFP-ParA (green), mCherry-DivIVA (red), and PapM-mT2 (blue). The number of the cells used for analysis (n) is indicated in the plots; scale bar, 5 µm. (I) The ratio of EGFP-ParA (left panel) and PapM-mT2 (right panel) fluorescence at the pole to the fluorescence at the mid-cell. The ratio higher than 1 (dashed line) means more fluorescence signal at the poles; the ratio lower than 1 illustrates the fluorescence in the mid-cell. The data come from at least two independent biological replicates. The statistical significance between strains determined by Wilcoxon test (two-sided) with Holm method used for multiple comparisons is marked with asterisks: P-values ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), and ≤0.0001 (****).
Next, using the same E. coli colocalization approach, we set out to examine if DivIVA interaction with ParA and PapM may be affected by DivIVA phosphorylation. We confirmed DivIVA phosphorylation by coproduction with either His-PknA (His-PknAKD) or His-PknB kinase domain (His-PknBKD) in E. coli (Fig. S2). Microscopy analysis showed that, in the presence of His-PknBKD, recruitment of EGFP-ParA to mCherry-DivIVA was diminished and an increased EGFP-ParA fluorescence signal along the cell was observed (Fig. 2C, E, and I). Markedly, when mCherry-DivIVA was produced in the presence of both His-PknBKD and PapM-mT2, the signal from EGFP-ParA was fully dispersed along the cell, while PapM-mT2 still colocalized with mCherry-DivIVA (Fig. 2F and I). Next, using phosphoablative mCherry-DivIVAT74A variant, we found that EGFP-ParA fully colocalized with this variant in the presence of His-PknBKD (Fig. 2G). This analyses confirmed that colocalization of mCherry-DivIVA and EGFP-ParA is disrupted by DivIVA phosphorylation.
The analysis of the ratio of EGFP-ParA fluorescence at the poles to its fluorescence at the mid-cell confirmed that PapM reduced the polar EGFP-ParA recruitment to mCherry-DivIVA in the presence of PknB kinase domain (Fig. 2E, F, and I). Thus, PapM enhanced the DivIVA phosphorylation-dependent release of ParA from the polar DivIVA complex. We also noted that, in the presence of His-PknBKD (in the absence of ParA), the ratio of PapM-mT2 fluorescence at the pole to its fluorescence at the mid-cell increased (Fig. 2 B, H, and I). This indicated that PapM-mT2 colocalized more efficiently with phosphorylated mCherry-DivIVA than with unphosphorylated.
Additionally, we tested whether PapM-mT2 colocalized with nucleoid-bound EGFP-ParA in E. coli (in the absence of mCherry-DivIVA). The nucleoid staining confirmed that EGFP-ParA colocalized with DNA in the absence and the presence of PapM-mT2 (Fig. S3A through D). PapM-mT2 colocalized with EGFP-ParA and the nucleoid (Fig. S3D). Finally, since PapM was annotated as the protein belonging to TetR regulators family, we also tested PapM interactions with DNA. The electrophoretic mobility shift assay (EMSA) experiment with supercoiled pUC19 plasmid did not indicate PapM-DNA binding (Fig. S4A), which could be due to structural alteration within the putative HTH motif in PapM or the lack of specific binding site in the tested DNA (Fig. S4B and C).
To summarize, the E. coli colocalization approach confirmed the engagement of PapM in the interaction between ParA and DivIVA. This analysis also indicated that ParA recruitment to DivIVA was diminished by DivIVA phosphorylation, while PapM recruitment was boosted. We also found that PapM enhanced the phosphorylation-dependent release of ParA from the polar DivIVA complex.
Modifications of papM levels affect the M. smegmatis growth, chromosome segregation, and cell length in a ParA-dependent manner
Having established that PapM interacts directly with ParA, we set out to investigate the biological function of PapM in M. smegmatis. To this end, we constructed M. smegmatis strains with papM deletion and a papM overexpression in the wild-type and ΔparA background. Since parA deletion was earlier shown to slow the M. smegmatis culture growth, leading to the formation of anucleate cells and altering the cell length (34), we examined the impact of modified papM expression on the above-mentioned processes.
Deletion of papM in the wild-type background did not affect the growth rate; however, papM deletion in the ΔparA background slowed the growth of the mutant strain in comparison to ΔparA parental strain (Fig. 3A). Next, we used the pMVpami papM construct in which papM was placed under inducible but leaky pami promoter [which triggers expression at a significant level even without adding inducer (Fig. S4A)]. This construct introduced to the ΔparAΔpapM background successfully complemented the papM deletion (Fig. S5B). Using the pMVpami papM construct, we further tested the growth of the strains with an increased papM expression in wild-type and ΔparA genetic background. In absence of an inducer, the overexpression of papM in the wild-type background [at approximately three to five times level of the wild type (Fig. S5A)] did not alter the growth rate in comparison to the wild-type control strain (with empty pMVpami) (Fig. 3B). Also, upon addition of the inducer to the papM, overexpressing strain did not affect the growth rate compared to wild-type control strain (Fig. S5C). Interestingly, the increased level of papM expression in the ΔparA background fully restored the growth rate of the parental ΔparA strain to the growth rate of wild-type control strain (Fig. 3B; Fig. S4C). The overexpression of papM in the ΔparAΔparM background also restored the even slower growth rate of the parental double deletion strain confirming full complementation of papM deletion.
Fig 3.
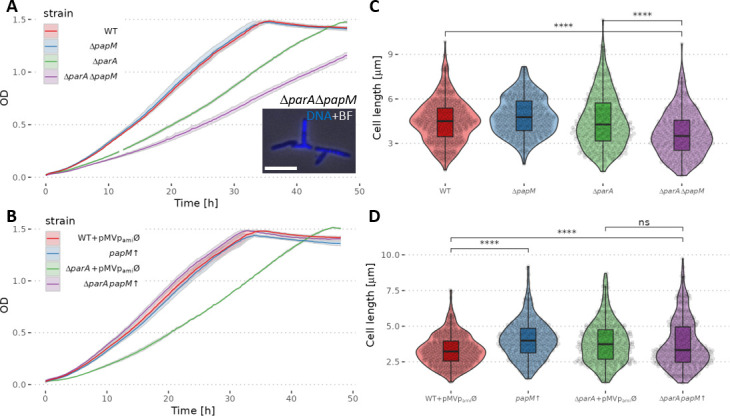
papM deletion or its overexpression in wild-type and ΔparA background affects M. smegmatis culture growth rate and cell length. (A) The growth curves of ΔpapM, ΔparA, and ΔparAΔpapM strain compared to the wild-type (WT) strain; inset, example image of branched ΔpapM, ΔparA M. smegmatis cells. The image shows the overlay of DAPI fluorescence (nucleoid stain, blue) and brightfield. Scale bar, 5 µm. (B) The growth curve of papM overexpressing strain in wild-type (papM↑) and ΔparA background (ΔparA papM↑) as compared to control strains with empty pMVpami plasmid (WT + pMVpamiØ and ΔparA + pMVpamiØ). (C) Cell length distribution in ΔpapM (478 cells), ΔparA (346 cells), and double mutant ΔparAΔpapM (350 cells) strain, compared to wild-type strain (WT) (482 cells). (D) Cell length distribution in papM overexpressing strain in wild-type (433 cells) or ΔparA background (392 cells) as compared to controls with empty pMVpami vector in the wild-type (488 cells) or ΔparA background (351 cells). The data come from at least two independent biological replicates. The statistical significance between strains determined by Student’s t-test (two-sided) with Holm method used for multiple comparisons is marked with asterisks: P-values ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), and ≤0.0001 (****).
Next, we investigated whether papM deletion or overexpression affected chromosome segregation. We found that papM deletion in the wild-type background led to a modest chromosome segregation defect (5.6% anucleate cells in ΔpapM compared to 0.6% in the wild-type strain) (Table 1). Deletion of papM in the ΔparA background had negligible influence on the segregation defect resulting from the lack of ParA (31.9% anucleate cells in the ΔparAΔpapM strain, compared to 30.8% of anucleate cells in the ΔparA strain); however, changes of the cell shape, such as bulging or branching, were detectable (Fig. 3A, inset). A significant disturbance of chromosome segregation was caused by papM overexpression in the wild-type background (13.4% of anucleate cells). Surprisingly, the segregation phenotype resulting from papM overexpression seemed to be not reflected in the growth rate of the mutant strain. Notably, the overexpression of papM in ΔparA partially restored the chromosome segregation defect caused by deletion of parA, lowering the fraction of anucleate cells when compared to the parental strain (21.6% at papM overexpression in ΔparA background compared to 30.8% in parental ΔparA strain). The PapM-dependent reduction of chromosome segregation defect in the ΔparA background was consistent with the observed increased growth rate of the strain overexpressing papM in the ΔparA background.
TABLE 1.
Chromosome segregation defects and cell length in ΔpapM strain or papM overexpression in wild-type (papM↑) or ΔparA background (ΔparA papM↑)
| WT | ΔpapM | ΔparA | ΔparA ΔpapM | WT pMVØ | papM↑ | ΔparA pMVØ | ΔparA, papM↑ | |
|---|---|---|---|---|---|---|---|---|
| Anucleate cells % | 0.6 | 5.6 | 30.8 | 31.9 | 2.2 | 13.4 | 29.2 | 21.6 |
| Cell length mean ± 95% CI, median | 4.52 ± 0.1, 4.5 |
4.85 ± 0.1, 4.77 |
4.51 ± 0.14, 4.28 |
3.66 ± 0.13, 3.51 |
3.32 ± 0.09, 3.23 |
4.05 ± 0.11, 3.99 |
3.86 ± 0.13, 3.73 |
3.74 ± 0.13, 3.32 |
The chromosome segregation defect resulting from parA deletion was earlier noted to be reflected by the altered cell length (34). The increased variation of the cell length was attributed to mispositioned cell division and the formation of short, anucleate cells, while the length of the other daughter cells increased. Markedly, the abolished interaction between ParA and DivIVA also increased the rate of cell elongation (37). Therefore, here, we tested whether papM deletion or its overexpression in wild-type and parA deletion background influenced the cell length.
While papM deletion in the wild-type background did not affect the average cell length, papM deletion in the ΔparA background resulted in the decreased average cell length (Fig. 3C; Table 1). Elevated levels of PapM increased the cell length in the wild-type background when compared to the wild-type control with an empty pMVpami vector. However, in the ΔparA background, moderate PapM overproduction did not affect cell length significantly (Fig. 3D; Fig. S5D; Table 1).
To summarize, the analysis of the M. smegmatis mutant strains showed that PapM affected chromosome segregation and influenced cell length. PapM involvement in the same cell cycle processes as ParA confirms a functional relationship between PapM and ParA. Notably, the absence of PapM also affected the growth and the cell shape of the strain lacking ParA, which may indicate its direct impact on DivIVA.
Deletion of papM alters ParA dynamics
Given that PapM affected ParA-DivIVA interaction in E. coli, to further investigate the impact of PapM on ParA, we analyzed whether the deletion of papM influences ParA localization in M. smegmatis.
Earlier studies showed dynamic EGFP-ParA localization patterns during the M. smegmatis cell cycle (35). In the new-born cells, EGFP-ParA fluorescence signal was first visible at the new cell pole; next, it was dynamically extended as a cloud-like fluorescence along the cell. At the cell division time, ParA was relocated to the mid-cell, being established at the newly formed pole (35). Here, we performed time-lapse fluorescent microscopy analysis of EGFP-ParA during the cell cycle in a papM deletion background (ΔparAΔpapM complemented with the pMV306pnat egfp-parA) and in the control strain (ΔparA complemented with the pMV306pnat egfp-parA). This was followed by measurements of EGFP-ParA fluorescence intensity along the cell during the cell cycle (from the time of cell separation to the progeny cell separation).
The time-lapse analysis showed that the ParA relocation pattern was affected by deletion of papM. The kymograph analysis of average fluorescence intensity along the cell indicated that, in the control strain, EGFP-ParA relocation to the mid-cell position was detected in about 60%–70% of the cell cycle (Fig. 4A, red ellipse, Fig. S6; Movie 1A). In the ΔpapM strain, accumulation of EGFP-ParA signal at the mid-cell preceding cell division was less clearly detectable (Fig. 4A; Fig. S6; Movie 1B). This may indicate that either the EGFP-ParA signal was more dispersed along the cell or its relocation was less tightly coordinated during the cell cycle.
Fig 4.
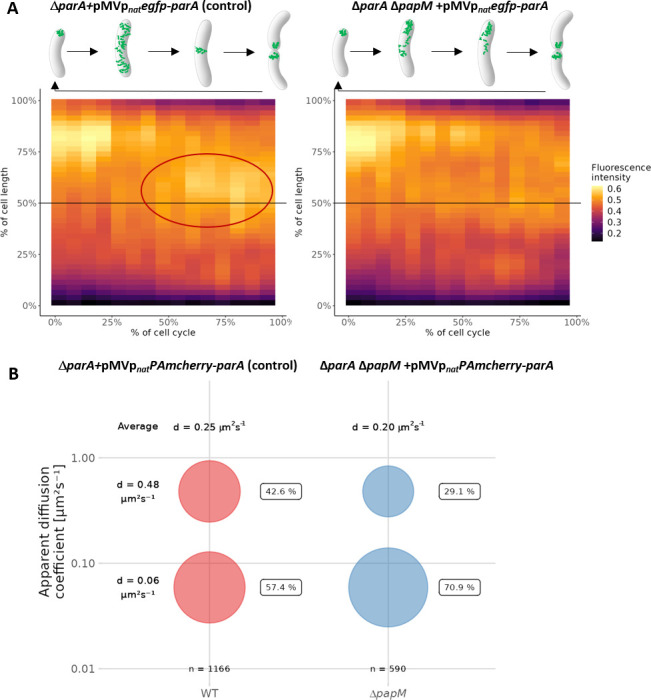
papM deletion alters ParA dynamics. (A) Kymographs showing cumulative EGFP-ParA fluorescence intensity during the cell cycle in the control strain (ΔparA + pMVpnat egfp-parA, 30 cells analyzed) and in the ΔpapM background (ΔparAΔpapM + pMV306pnat egfp-parA, 25 cells analyzed). The time zero is the beginning of the new cell cycle detected as the visible separation of daughter cells accompanied by EGFP fluorescence at the new poles. Mid-cell is marked with a black line, and the ParA assembly at the mid-cell in the control strain is marked with red ellipse. The data come from two independent biological replicates. (B) The bubble plot showing the percentages of mobile and immobile PAmCherry-ParA molecules in control (ΔparA + pMVpnat PAmcherry-parA) and ΔpapM (ΔparAΔpapM + pMV306pnat PAmchery-parA) strain as determined by PALM. The number of tracks analyzed is 1,166 for control (49 cells) and 590 tracks (42 cells) for ΔpapM. The diffusion coefficients are indicated.
Next, to further determine the impact of PapM on ParA dynamics, we used photoactivation localization microscopy (PALM) to track PAmCherry-ParA molecules (in ΔparA strain complemented with the pMV306pnat PAmcherry-parA) and to determine if their mobility was affected by papM deletion. Earlier, it was shown that mutations that affect ParA binding to DNA (but not to DivIVA) had a significant impact on ParA mobility (37).
Here, we found that the elimination of PapM increased the fraction of immobile PAmCherry-ParA molecules (from 57.4% to 70.9% of molecules with diffusion coefficient d = 0.06 µm2 s–1) and decreased the fraction of mobile molecules (from 42.6% to 29.1% of molecules with diffusion coefficient d = 0.48 µm2 s–1), decreasing the average diffusion coefficient from 0.25 µm2 s–1 in wild-type background to 0.20 µm2 s–1 in the absence of PapM (Fig. 4B).
To sum up, these two experiments suggested an impact of PapM on ParA dynamics. Deletion of papM altered the cell cycle-dependent ParA relocation to the newly established cell poles and lowered the mobility of ParA molecules. This altered mobility could possibly result from modified ParA association with DNA which was earlier shown to predominantly affect ParA mobility (37).
Deletion of parA or papM modifies the subcellular distribution of DivIVA, affecting the cell elongation and division
Earlier, we demonstrated that impairing ParA-DivIVA interaction altered the cell elongation rate, which indicated the influence of ParA on DivIVA activity in M. smegmatis (37). Our E. coli colocalization studies presented above indicated that PapM interacted with DivIVA and influenced ParA-DivIVA interaction in the presence of the kinase domain. Additionally, we observed that changes in PapM levels altered the M. smegmatis cell length, which could be due to PapM direct or indirect influence on DivIVA. To investigate PapM’s impact on DivIVA and to compare it to the effect of ParA, we analyzed the polar and mid-cell accumulation of mCherry-DivIVA in ΔparA and ΔpapM strains. Moreover, using the time-lapse microscopy, we analyzed the cell elongation rate in a parA and papM deletion background.
Analysis of mCherry-DivIVA accumulation at the cell poles showed that deletion of parA increased the intensity of the mCherry-DivIVA polar fluorescence signal as compared to fluorescence intensity at the mid-cell (Fig. 5A, B, and D). The increase of mCherry-DivIVA fluorescence was more pronounced at the old pole in comparison to the new pole (Fig. 5A, B, and D). The effect of papM deletion was less significant than that of parA deletion (Fig. 5A, B, and D). Moreover, while papM deletion had no detectable effect on mCherry-DivIVA fluorescence asymmetry, parA deletion increased the ratio of mCherry-DivIVA fluorescence intensity at the old pole to its fluorescence intensity ad the new pole, therefore enhancing the asymmetry of M. smegmatis cells (Fig. 5C).
Fig 5.
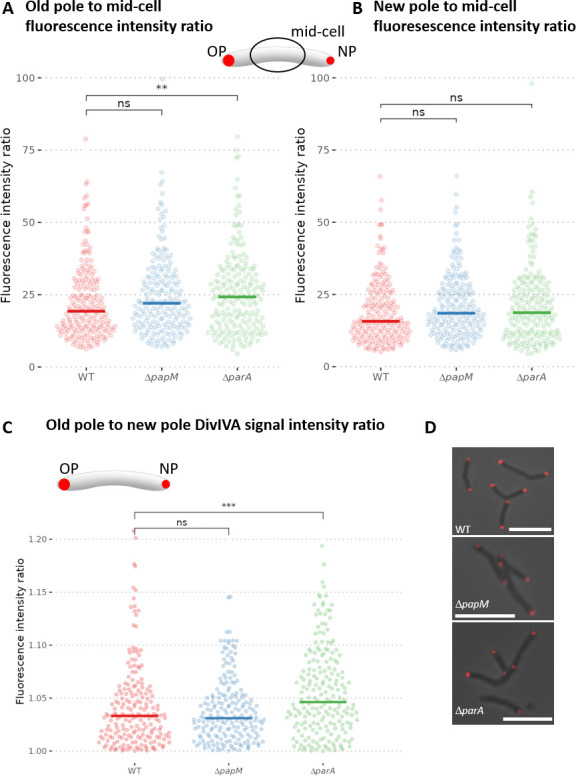
Polar accumulation of mCherry-DivIVA is altered in ΔparA but not ΔpapM M. smegmatis strain as compared to the wild-type control. The analyses were performed in WT control strain (400 cells), ΔpapM (398 cells) and ΔparA (400 cells) strains producing mCherry-DivIVA (apart of the wild-type DivIVA). (A) The ratio of mCherry-DivIVA fluorescence intensity at the old pole to average fluorescence intensity in the mid-cell. (B) The ratio of fluorescence intensity at the new pole to average fluorescence intensity in the mid-cell. (C) The ratio of mCherry-DivIVA fluorescence intensity at the old cell pole to its fluorescence at the new cell pole. (D) Representative images of the cells of each studied M. smegmatis strain. The mCherry-DivIVA fluorescence (red) is merged with the brightfield image. Scale bar, 5 µm. The data come from at least two independent biological replicates. The statistical significance between strains determined by Wilcoxon test (two-sided) with Holm method used for multiple comparisons is marked with asterisks: P-values ≤0.05 (*), ≤0.01 (**), and ≤0.001 (***).
The differences in DivIVA signal accumulation could be related to the modified cell elongation rate. Therefore, we used time-lapse microscopy to analyze the cell cycle parameters. We measured the cell cycle length of the strains producing mCherry-DivIVA (apart from the wild-type protein) as the time between the appearance of the mCherry-DivIVA signal in the center of the mother cell and the appearance of the mCherry-DivIVA fluorescence in the middle of the daughter cell. We further measured the cell elongation rate of the strains producing mCherry-DivIVA. Additionally, visualization of peptidoglycan with NADA allowed us to detect cell division and measure the cell elongation rate.
The analysis of the cell cycle parameters clearly indicated that parA deletion increased the cell elongation rate and the acceleration was more significant for the old pole inheriting cell (Fig. 6A; Fig. S7). The cell elongation rate of a ΔparA strain was also more varied in comparison to the wild-type control strain, but the average length of the cell cycle was not affected (Fig. 6A and B; Fig. S7). The increased cell elongation rate was consistent with the occurrence of the elongated cells that were observed in the ΔparA strain. Elimination of papM somewhat increased variation of the cell extension rates but did not visibly modify the average length of the cell cycle (Fig. 6A and B). Analysis of ΔparA, ΔpapM, and wild type stained with fluorescent D-amino acid (NBD-amino-d-alanine, NADA) indicated a higher cell extension rate than extension rate of the respective strains producing mCherry-DivIVA (Fig. S7) which may be due to disturbances caused by the production of fusion mCherry-DivIVA apart from wild-type DivIVA. However, the influence of parA deletion and papM deletion on growth rate was the same as in the strains producing mCherry-DiviVA (Fig. S7).
Fig 6.
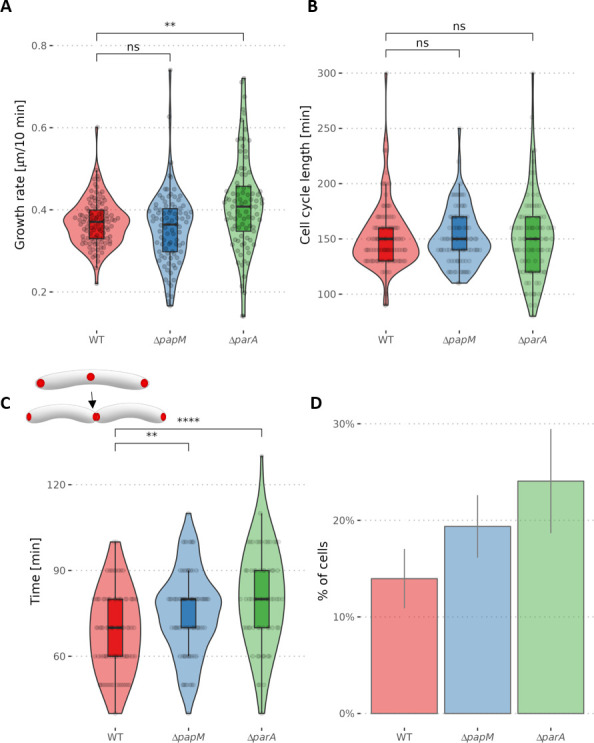
The growth rate and mCherry-DivIVA redistribution during the cell cycle are affected by parA and papM deletion. (A) The cell growth rate measured as the cell length increment (the difference of the cell length of the newly born mother cell and the daughter cell at the time preceding cell division - the mid-cell mCherry-DivIVA signal appearance) divided by cell cycle length shown in B (103, 90, and 89 cells analyzed for WT, ΔpapM, and ΔparA, respectively). (B) The cell cycle length - the time elapsed between the appearance of the mid-cell mCherry-DivIVA signal in the mother cell and the appearance of mid-cell mCherry-DivIVA signal in its daughter cell (the number of cells analyzed as in A). (C) The time between the detection of the mid-cell mCherry-DivIVA signal and detection of the separated cell poles (84, 78, and 65 cells analyzed for WT ΔpapM and ΔparA, respectively). The analyses were performed in the WT control strain and ΔpapM and ΔparA strains producing mCherry-DivIVA (apart of the wild-type DivIVA). (D) The percentage of the cells with the visible mCherry-DivIVA signal in the snapshot images (487, 573, and 241 cells analyzed for WT, ΔpapM, and ΔparA, respectively). The data come from two independent biological replicates. The statistical significance between strains determined by Student’s t-test (two-sided) with Holm method used for multiple comparisons is marked with asterisks: P-values ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), and ≤0.0001 (****).
Next, we focused on the mid-cell mCherry-DivIVA signal which indicated the cell division and DivIVA relocation to the newly formed septum. Time-lapse microscopy allowed us to measure the timespan between the mid-cell mCherry-DivIVA appearance and the visible separation of the two fluorescence foci indicating the establishment of the two daughter cell poles. In the wild-type strain, the mCherry-DivIVA signal was visible for a shorter time than in the ΔparA and ΔpapM strains (69 min on average in wild-type control strain, in comparison to 80 min in the ΔparA strain and 75 min in ΔpapM strain) (Fig. 6C; Movie 2A through C). This analysis was reinforced by timepoint analysis of the mCherry-DivIVA producing strains: in both parA and parM deletion strains, the fraction of cells with the mid-cell mCherry-DivIVA fluorescence signal increased (from 14% in wild type to 24% in ΔparA and 19% in ΔpapM, respectively) (Fig. 6D). This confirms that the timespan required to establish the new pole is increased in both ΔparA and ΔpapM strains.
To summarize, analysis of mCherry-DivIVA fluorescence implied that the absence of ParA increased the DivIVA accumulation at the old pole and increased cell extension rate. Elimination of ParA also affected DivIVA relocation to the cell division site and establishment of the new poles. Deletion of PapM had a marginal impact on DivIVA accumulation at the poles but influenced DivIVA redistribution to the mid-cell. This reinforces the PapM impacts on DivIVA localization and is consistent with the fact that modified PapM levels exhibit an effect on cell length.
DISCUSSION
We have identified a novel ParA and DivIVA partner—named PapM—that contributes to the cell cycle regulation in M. smegmatis. Notably, while in other bacteria, ParA was shown to be engaged in interactions with polar or subpolar proteins, i.e., TipN and PopZ in C. crescentus, HubP in V. cholerae or Scy in S. coelicolor (4, 42 – 45), according to our best knowledge, our findings are the first report of the third partner that interferes with these interactions.
The PapM homologues were found only in several Mycobacterium species, mostly those closely related to M. smegmatis, while the protein has no close homologues in either M. tuberculosis or in many other mycobacteria. This suggest that papM gene could have been eliminated in pathogenic, intracellular mycobacteria whose genomes undergo reduction and which are not exposed to dramatic changes of environmental conditions. That also raises the question if PapM role could be related to fast growth and adjustment to changes of environmental conditions. However, it should be noted that the elimination of PapM had no detrimental effect on the M. smegmatis growth rate under optimal conditions. Interestingly, although PapM was annotated as a protein from the TetR regulator family, it did not show DNA interaction in an EMSA assay. That could be explained either by high specificity toward unknown DNA sequence or by weak DNA binding. Intriguingly, PapM HTH motif analysis and structure prediction indicated structural changes within HTH motif which could interfere with DNA binding. This could also indicate that PapM evolved to play different role than transcriptional regulator.
We show that PapM modulates the interplay between ParA-DivIVA. Our E. coli colocalization analysis indicated that PapM promotes the release of ParA from DivIVA in presence of the PknB kinase domain, which phosphorylates DivIVA. The impact of PapM on ParA dynamics was confirmed by EGFP-ParA localization and the single molecule tracking in M. smegmatis cells. The decreased mobility of ParA molecules in absence of PapM may be due to enhanced binding of ParA to the nucleoid or the DivIVA complex. Earlier, it was shown that the ParAD68A variant that is not able to hydrolyze ATP had significantly reduced mobility due to enhanced nucleoid binding, while the variant ParAR219E had increased mobility due to abolished interaction with nucleoid (37). The detailed analysis of PapM interaction with ParA variants suggested the preferred interaction with ParA dimer that is not able to bind DNA (ParAR219E). The impact of PapM on ParA interactions with DNA and protein dynamics may account for the observed segregation phenotype of PapM overproducing strain. Elevated levels of PapM could interfere with ParA-DNA and/or ParA-DivIVA interaction affecting ParA dynamics and impairing segrosomes separation.
Markedly, PapM also interacts with DivIVA. E. coli colocalization analysis suggested that PapM recruitment to DivIVA is enhanced in presence of ParA. Interestingly, the analyses of interactions using DivIVA subdomains suggested that the DivIVA fragment that encompasses phosphorylation site Thr74 potentially may be involved in PapM binding. This implies the influence of DivIVA phosphorylation on the interactions with PapM. Indeed, PapM colocalization with DivIVA was enhanced in the presence of the PknB kinase domain. The impact of PapM on DivIVA was also demonstrated by changes in the cell length in strains with modified PapM levels. Overexpression of papM in a wild-type background increased the cell length, while papM deletion in a parA deletion background shortened average cell length. Markedly, the elimination of both ParA and PapM had deteriorating effect on growth and led to appearance of misshaped, branched cells, suggesting DivIVA mislocalization. Intriguingly, increased PapM levels in a parA deletion background also partially suppressed segregation phenotype resulting from the absence of ParA. The direct influence of PapM on DivIVA and its function in coordination of the cell elongation with the cell division are likely to account for the observed phenotypes. The other possible explanation is the impact of PapM on other proteins that contribute to chromosome segregation or cell cycle coordination.
The important finding of this study is the demonstration that ParA-DivIVA interaction is phosphorylation dependent. Taking into account that DivIVA phosphorylation is correlated with fast cell growth (20, 25, 32), the enhanced binding of ParA to non-phosphorylated DivIVA shown here is consistent with our earlier observations. We noted earlier that polar recruitment of ParA may be induced by unfavorable environmental conditions which inhibit cell growth (37). Moreover, we have shown that the absence of ParA increases the polar accumulation of DivIVA and increases the time required to establish the new poles during cell division. This could be resulting from a slower DivIVA accumulation and/or maturation and, speculatively, phosphorylation at the septum. The lack of ParA also enhances the asymmetry of mycobacterial cell poles leading to greater DivIVA accumulation (speculatively increased phosphorylation) at the old cell pole. These observations are consistent with the observed increased cell elongation rate and also the increased variation of the cell extension rate in the absence of ParA. The strain lacking PapM also exhibited increased variation of the cell extension rate. Such a variation is likely to result from increased heterogeneity, i.e., the difference of the growth rate between the old pole and new pole inheriting cell. Since phosphorylated DivIVA is reportedly more active in cell wall synthesis (25), we are inclined to suggest that ParA, similarly to PapM, may affect DivIVA phosphorylation or, alternatively, interaction with its other protein partners.
Based on the presented results, we propose the speculative model of tripartite interactions (Fig. 7). According to our model, ParA preferably binds DivIVA at the nascent septum. The maturation of the DivIVA complex and its phosphorylation release ParA from the new pole to promote chromosome segregation. On the other hand, ParA has an impact on DivIVA complex stability: in the absence of ParA, “old pole” DivIVA complexes are more stable, and formation of the new pole complexes requires more time. PapM enhances phosphorylation-dependent ParA release from polar DivIVA complex. PapM also has a direct effect on DivIVA maturation but to a lesser extent than ParA. In absence of PapM, formation of DivIVA complex is affected. Altered PapM levels in absence of ParA affect DivIVA function and the cell cycle, possibly interfering with switch between cell elongation and cell division. The impact on this cell cycle checkpoint is likely to explain the PapM interference with ParA-dependent chromosome segregation. Thus, PapM influences both partners activity, affecting ParA dynamics and DivIVA function. The impact of PapM on DivIVA is likely to be both direct and indirect, by modification of ParA-DivIVA interaction. The tripartite complex interplay seems to be tightly interlinked and coordinates the switch between cell extension and cell division.
Fig 7.
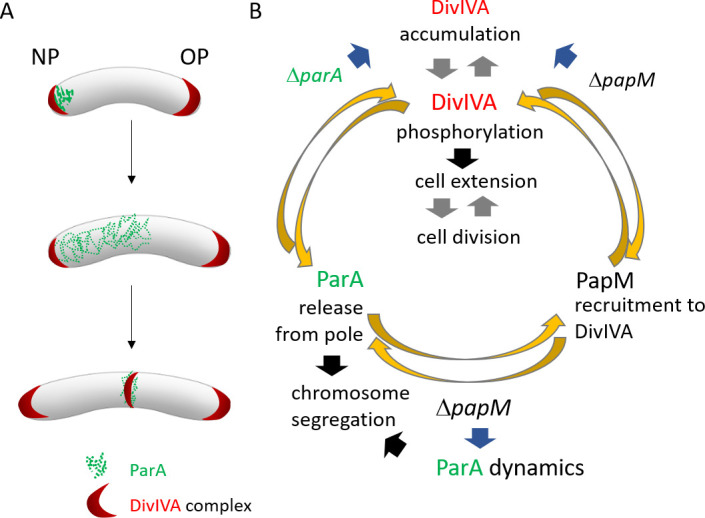
The model of ParA-PapM-DivIVA interplay. (A) The scheme of M. smegmatis cell cycle showing ParA and DivIVA dynamics. (B) Model of ParM-ParA-DivIVA interactions. Black arrows are established relations, gray arrows indicate postulated relations, blue arrows indicate the relations suggested by the mutant M. smegmatis strain phenotypic analyses presented here, and yellow arrows illustrate conclusions based on in E. coli colocalization experiment.
MATERIALS AND METHODS
Cloning and construct preparation
DNA manipulations were performed using standard protocols (46). Reagents and enzymes were supplied by New England Biolabs (NEB), Merck, and Thermo Scientific. Oligonucleotides were synthesized by Merck and Genomed, and sequencing was performed by Microsynth. Escherichia coli strains were grown in lysogeny broth medium at 37°C [DH5α, BL21(DE3)] or 30°C (BTH101). Culture conditions, antibiotic concentrations, and transformation protocols followed standard procedures (46).
Bacterial two-hybrid system
BTH interaction studies were performed as previously described (38). Details of the construction of the M. smegmatis library in the BTH system and the screening of the library as well as the construction of BTH vectors are described in the Supplementary Material. The plasmids used in this study are listed in Table S1.
Affinity chromatography
Affinity chromatography was used to test the interaction between PapM and ParA. The GST-ParA cell lysate [40 mL in phosphate-buffered saline (PBS)] was obtained from an 800-mL culture of BL21(DE3) pGEX-6P-2-parAMs . The clarified lysate was incubated (overnight, at 4°C, with gentle agitation) with 1-mL bed volume of GSH-Sepharose (GE Healthcare). Next, the resin was loaded on the chromatography column, and unbound proteins were removed by washing with cold PBS buffer (at least three resin volumes). In parallel, a control experiment was performed in which the clarified control lysate of BL21(DE3) pGEX-6P-2Φ [prepared according to the same procedure as for BL21(DE3) pGEX-6P-2-parAMs ] was loaded on the column. Next, the His-PapM lysate obtained from the 800-mL culture of BL21(DE3) pET28apapM (40 mL of lysate in PBS) was added to the both columns and incubated overnight at 4°C, with gentle agitation. The unbound proteins were removed by washing with at least three-bed volumes of PBS buffer, and then, proteins bound to resin were eluted with PBS buffer containing 20 mM reduced glutathione. All collected fractions (3 mL) were analyzed using SDS-PAGE and Western blotting with an anti-His-tag antibody (Thermo Fisher Scientific).
E. coli colocalization studies
For assays of EGFP-ParA, mCherry-DivIVA, and PapM-mT2 colocalization in E. coli BL21(DE3) cells, pACYCDuet-1 vector derivatives containing papM-mT2 and/or egfp-parA genes and pETDuet-1 vector derivatives containing mcherry-divIVA or ics-mcherry (as the control) and optionally his-pknB fragment encoding kinase domain (11–274 aa) were used. E. coli BL21(DE3) strains containing either of the above-mentioned constructs were cultured to the log phase in the presence of ampicillin and chloramphenicol. The expression of the cloned genes was induced by the addition of 0.1 mM IPTG for 1 h. After induction, 10 µL of culture was mounted on the microscopic slides, covered with 5 µL of phosphate-buffered saline (PBS)-glycerol (1:1) solution. Cells were examined by a Leica DM6 B fluorescence microscope equipped with a 100× objective with a DFC7000 GT camera. Images were analyzed using LAS X 3.6.0.20104, Fiji, and “R” software (R Foundation for Statistical Computing, Vienna, Austria; https://www.r-project.org/). E. coli cells in phase contrast images were automatically detected using a custom ImageJ script, and fluorescence signal was collected along the long axis of each cell. R software was used to normalize and visualize the fluorescence profiles. The fluorescence intensity was measured at the cell poles (5%–20% and 80%–95% of the cell length) and in the middle of the cell (35%%–65% of the cell length for PapM; 25%–35 and 65%%–75% for ParA).
For analysis of PapM-mT2, EGFP-ParA, and nucleoid colocalization, cultures of BL21(DE3) containing pACYCDuet-1 vector derivatives containing papM-mT2 and/or egfp-parA genes were incubated to the log phase in the presence of chloramphenicol. Then, the expression of papM-mT2 and/or egfp-parA genes was induced by the addition of IPTG to a final concentration of 0.1 mM and further culturing for 1 h. At the same time, to inhibit cell division, nalidixic acid was added up to a final concentration of 25 µg /mL (Chai et al., 2014) (47). Next, the cultures were incubated with DAPI (2 µg/mL) for 20 min, centrifuged (5,000 rpm/5 min) and resuspended in PBS buffer. For SYTO 62 staining, 200 µL of the culture was incubated for 15 min with SYTO 62 (0.25 μM) and then mounted on the microscopic slides, dried, and covered with 5 µL of PBS-glycerol (1:1) solution and coverglass. Specimens were examined by a Leica DM6 B fluorescence microscope equipped with a 100× objective. Pictures were analyzed using a custom ImageJ script as described above and the R software, including the ggplot2 package (48).
M. smegmatis growth conditions
M. smegmatis strains were cultured either in Middlebrook liquid 7H9 medium (Difco) supplemented with 10% albumin-dextrose-catalase (ADC; BD) and 0.05% Tween 80 or on solid 7H10 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (BD), 0.5% glycerol, and 0.05% Tween 80.
To determine the growth rate of M. smegmatis strain, cultures were inoculated from glycerol stocks and grown to the log phase (OD600, 0.3–0.5). Next, the cultures were diluted in fresh medium with or without the addition of acetamide to concentration 0.1% to an OD600 of 0.05, and the final volume of 300 µL of diluted culture was loaded into the wells of a Bioscreen C-compatible honeycomb plate. The microplate cultures were incubated at 37°C with continuous shaking using Bioscreen C [Automated Growth Curves Analysis System, Growth Curves (Alab)]. The optical density measurements were taken at 20-min intervals for 2–3 days. The results were analyzed in Excel and visualized using R.
For microscopic experiments, M. smegmatis strains from glycerol stocks were used to inoculate starting cultures, which were then grown for 24 h and used for the inoculation of precultures. Next, after being incubated to the log phase (OD600 ~0.5), the precultures were diluted in fresh medium to set up cultures with starting OD600 ~0.05 and cultured to the desired OD.
M. smegmatis modifications
The M. smegmatis strains used in this study are listed in Table S2. The details of the M. smegmatis mutant strains construction are described in the Supplementary Materials. All vectors for M. smegmatis transformations were prepared in E. coli DH5α. To delete papM gene in the M. smegmatis mc2 155 chromosome, targeted gene replacement was performed according to the published protocol (49). The construct based on vectors p2Nil (KanR) and pGOAL17 (sacB, lacZ) (49) was prepared according to the strategy described in Supplementary Materials and integrated into the M. smegmatis chromosome by homologous recombination. For the construction of M. smegmatis complementation and overexpressing strains, derivatives of the mycobacteriophage L5-based integration-proficient vector pMV306pami were used as described in Supplementary Materials. Quantitative reverse transcription PCR (RT-qPCR) was performed to confirm the overexpression of the papM gene in wild-type [MP24 (37) and IM12] and ΔparA genetic background (IM11 and IM13) M. smegmatis strains (as described in detailed in Supplementary Materials).
M. smegmatis microscopy
For snapshot microscopy, M. smegmatis strains were grown to the log phase (OD600 0.4–0.5) in 7H9 medium (as described above). For analysis of the chromosome segregation, cells were incubated with DAPI (2 µg mL–1) for 2 h, centrifuged (5,000 rpm/5 min), and resuspended in PBS buffer. For membrane staining, cells were incubated with FM5-95 (0.5 µg/mL; Thermo Fisher Scientific) for 15 min before the nucleoid staining was completed. Next, bacteria were mounted on the microscopic slides, dried, and covered with 5 µL of PBS-glycerol (1:1) solution and coverglass. Snapshots were taken immediately after sample preparation using a Leica DM6 B microscope with a 100× oil immersion objective.
Time-lapse microscopy analyses were performed in a liquid medium using a CellASIC ONIX platform and compatible B04A plates (Merck), as described previously (50, 51). Early log-phase M. smegmatis cultures (OD600, 0.2–0.4) were grown in liquid 7H9 medium supplemented with 10% ADC and 0.05% Tween 80. All microfluidic experiments were performed under constant pressure (1.5 psi). For NADA-green staining (Torcis Bio-Techne) (52), cells were loaded into the observation chamber and cultured in 7H9 supplemented with ADC and Tween 80 for 1 h. Then, cells were subjected every hour to a 2-min pulse flow of 7H9-ADC-Tween 80 medium supplemented with NADA-green (0.5 mM). Pictures were recorded using differential interference contrast (DIC) and fluorescence channels (490/20 nm excitation filter and 528/38 nm emission filter for GFP and 575/25 nm excitation filter and 632/60 nm emission filter for mCherry) with a CoolSnap HQ2 camera at 10-min intervals using a Delta Vision Elite inverted microscope equipped with a 100× oil immersion objective and an environmental chamber set to 37°C. All images were analyzed using the Fiji and R software packages (R Foundation for Statistical Computing, Austria; http://www.r-project.org), including the ggplot2 package (48). For mCherry-DivIVA polar signal analysis, fluorescence data were normalized by subtracting the minimal value of fluorescence. Then, the 20% lowest values of fluorescence signal from each cell were averaged to obtain background fluorescence value of each cell. Next, maximum fluorescence signal from each cell pole was divided by the fluorescence background to obtain the ratio of pole/background fluorescence. For ParA-EGFP time-lapse analysis, cells were manually detected in the images (25–30 cells), and a fluorescence signal was collected along the long cell axis. R software was then used to normalize fluorescence intensity and draw a cumulative kymograph of the ParA-EGFP fluorescence signal.
Single-molecule tracking PALM was performed using Zeiss Elyra 7 microscope. Photoactivatable mCherry (PAmCherry) was activated with a 405-nm laser, with excitation at 561 nm. For each experiment, 10,000 frames were recorded with images being taken every 30 ms. Molecule tracking and localization analysis were performed using the Trackmate plugin from the Fiji program and SMTracker based upon MATLAB software (MathWorks, Natic, MA, USA). Cell segmentation was performed using Fiji and Oufti software. Mobile and immobile protein molecules were distinguished by calculating the apparent diffusion coefficient using the Gaussian mixture mode in SMTracker. Only tracks with at least four frames were considered for the analysis. Due to cell confinement and motion blurring, D* is an apparent diffusion coefficient (53).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Katarzyna Ginda-Mäkelä for the help with M. smegmatis library preparation and screening and Jolanta Zakrzewska-Czerwińska for comments on the manuscript.
This work was funded by OPUS grant 2017/27/B/NZ1/00823.
Contributor Information
Dagmara Jakimowicz, Email: dagmara.jakimowicz@uwr.edu.pl.
Eric Cascales, Centre national de la recherche scientifique, Aix-Marseille Universite, Marseille, France .
Takehiro Kado, University of Massachusetts Amherst, Amherst, Massachusetts, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.01752-23.
Fig. S1 to Fig. S7.
Supplemental methods.
Legends for Movies S1 and S2.
Constructs used in the study.
Strains used in the study.
Oligonucleotides used in this study.
papM deletion alters EGFP-ParA fluorescence dynamics: control strain.
papM deletion alters EGFP-ParA fluorescence dynamics: papM background.
The mCherry-DivIVA redistribution during the cell cycle is affected by parA and papM deletion: wild type (WT) control strain.
The mCherry-DivIVA redistribution during the cell cycle is affected by parA and papM deletion: parA strain producing mCherry-DivIVA.
The mCherry-DivIVA redistribution during the cell cycle is affected by parA and papM deletion: papM strain producing mCherry-DivIVA.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Reyes-Lamothe R, Sherratt DJ. 2019. The bacterial cell cycle, chromosome inheritance and cell growth. Nat Rev Microbiol 17:467–478. doi: 10.1038/s41579-019-0212-7 [DOI] [PubMed] [Google Scholar]
- 2. Meunier A, Cornet F, Campos M. 2021. Bacterial cell proliferation: from molecules to cells. FEMS Microbiol Rev 45:1–21. doi: 10.1093/femsre/fuaa046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marczynski GT, Petit K, Patel P. 2019. Crosstalk regulation between bacterial chromosome replication and chromosome partitioning. Front Microbiol 10:279. doi: 10.3389/fmicb.2019.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawalek A, Wawrzyniak P, Bartosik AA, Jagura-Burdzy G. 2020. Rules and exceptions: the role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms 8:0. doi: 10.3390/microorganisms8010105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jalal ASB, Le TBK. 2020. Bacterial chromosome segregation by the ParABS system. Open Biol 10:200097. doi: 10.1098/rsob.200097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lutkenhaus J. 2012. The ParA/MinD family puts things in their place. Trends Microbiol 20:411–418. doi: 10.1016/j.tim.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. 2014. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife 3:e02758. doi: 10.7554/eLife.02758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Surovtsev IV, Campos M, Jacobs-Wagner C. 2016. DNA-relay mechanism is sufficient to explain para-dependent intracellular transport and patterning of single and multiple cargos. Proc Natl Acad Sci U S A 113:E7268–E7276. doi: 10.1073/pnas.1616118113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pióro M, Matusiak I, Gawek A, Łebkowski T, Jaroszek P, Bergé M, Böhm K, Armitage J, Viollier PH, Bramkamp M, Jakimowicz D. 2022. Genus-specific interactions of bacterial chromosome segregation machinery are critical for their function. Front Microbiol 13:928139. doi: 10.3389/fmicb.2022.928139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pióro M, Jakimowicz D. 2020. Chromosome segregation proteins as coordinators of cell cycle in response to environmental conditions. Front Microbiol 11:588. doi: 10.3389/fmicb.2020.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baranowski C, Hesper Rego E, Rubin EJ. 2019. The dream of a Mycobacterium. Microbiol Sp 7:1096–1106. doi: 10.1128/9781683670131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown-Elliott BA, Philley JV, Schlossberg D. 2017. Rapidly growing mycobacteria. Microbiol Spectr 5. doi: 10.1128/microbiolspec.TNMI7-0027-2016 [DOI] [PubMed] [Google Scholar]
- 13. Hett EC, Rubin EJ. 2008. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 72:126–156, doi: 10.1128/MMBR.00028-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uhía I, Williams KJ, Shahrezaei V, Robertson BD. 2015. Mycobacterial growth. Cold Spring Harb Perspect Med 5:1–14. doi: 10.1101/cshperspect.a021097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Logsdon MM, Aldridge BB. 2018. Stable regulation of cell cycle events in mycobacteria: Insights from inherently heterogeneous bacterial populations. Front Microbiol 9:514. doi: 10.3389/fmicb.2018.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. doi: 10.1126/science.1216166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Logsdon MM, Ho PY, Papavinasasundaram K, Richardson K, Cokol M, Sassetti CM, Amir A, Aldridge BB. 2017. A parallel adder coordinates mycobacterial cell-cycle progression and cell-size homeostasis in the context of asymmetric growth and organization. Curr Biol 27:3367–3374. doi: 10.1016/j.cub.2017.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. 2013. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun 4:2470. doi: 10.1038/ncomms3470 [DOI] [PubMed] [Google Scholar]
- 19. Hannebelle MTM, Ven JXY, Toniolo C, Eskandarian HA, Vuaridel-Thurre G, McKinney JD, Fantner GE. 2020. A biphasic growth model for cell pole elongation in mycobacteria. Nat Commun 11:452. doi: 10.1038/s41467-019-14088-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang C-M, Nyayapathy S, Lee J-Y, Suh J-W, Husson RN. 2008. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology (Reading) 154:725–735. doi: 10.1099/mic.0.2007/014076-0 [DOI] [PubMed] [Google Scholar]
- 21. Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. 2014. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci U S A 111:E3243–E3251. doi: 10.1073/pnas.1402158111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choukate K, Chaudhuri B. 2020. Structural basis of self-assembly in the lipid-binding domain of mycobacterial polar growth factor Wag31. IUCrJ 7:767–776. doi: 10.1107/S2052252520006053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen L, Scherr N, Gatfield J, Walburger A, Pieters J, Thompson CJ. 2007. Antigen 84, an effector of pleiomorphism in Mycobactenum smegmatis. J Bacteriol 189:7896–7910. doi: 10.1128/JB.00726-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Melzer ES, Sein CE, Chambers JJ, Siegrist MS. 2018. DivIVA concentrates mycobacterial cell envelope assembly for initiation and stabliization of polar growth. Cytoskeleton (Hoboken) 75:498–507. doi: 10.1002/cm.21490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jani C, Eoh H, Lee JJ, Hamasha K, Sahana MB, Han J-S, Nyayapathy S, Lee J-Y, Suh J-W, Lee SH, Rehse SJ, Crick DC, Kang C-M. 2010. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol 10:327. doi: 10.1186/1471-2180-10-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. García-Heredia A, Kado T, Sein CE, Puffal J, Osman SH, Judd J, Gray TA, Morita YS, Siegrist MS. 2021. Membrane-partitioned cell wall synthesis in mycobacteria. Elife 10:1–16. doi: 10.7554/eLife.60263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Habibi Arejan N, Ensinck D, Diacovich L, Patel PB, Quintanilla SY, Emami Saleh A, Gramajo H, Boutte CC. 2022. Polar protein Wag31 both activates and inhibits cell wall metabolism at the poles and septum. Front Microbiol 13:1085918. doi: 10.3389/fmicb.2022.1085918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukherjee P, Sureka K, Datta P, Hossain T, Barik S, Das KP, Kundu M, Basu J. 2009. Novel role of Wag31 in protection of mycobacteria under oxidative stress. Mol Microbiol 73:103–119. doi: 10.1111/j.1365-2958.2009.06750.x [DOI] [PubMed] [Google Scholar]
- 29. Plocinski P, Arora N, Sarva K, Blaszczyk E, Qin H, Das N, Plocinska R, Ziolkiewicz M, Dziadek J, Kiran M, Gorla P, Cross TA, Madiraju M, Rajagopalan M. 2012. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J Bacteriol 194:6398–6409. doi: 10.1128/JB.01005-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang C-M, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev 19:1692–1704. doi: 10.1101/gad.1311105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plocinska R, Martinez L, Gorla P, Pandeeti E, Sarva K, Blaszczyk E, Dziadek J, Madiraju MV, Rajagopalan M. 2014. Mycobacterium tuberculosis MtrB sensor kinase interactions with FtsI and Wag31 proteins reveal a role for MtrB distinct from that regulating MtrA activities. J Bacteriol 196:4120–4129. doi: 10.1128/JB.01795-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Botella H, Yang G, Ouerfelli O, Ehrt S, Nathan CF, Vaubourgeix J. 2017. Distinct spatiotemporal dynamics of peptidoglycan synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. mBio 8:e01183-17. doi: 10.1128/mBio.01183-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jakimowicz D, Brzostek A, Rumijowska-Galewicz A, Żydek P, Dołzbłasz A, Smulczyk-Krawczyszyn A, Zimniak T, Wojtasz Ł, Zawilak-Pawlik A, Kois A, Dziadek J, Zakrzewska-Czerwińska J. 2007. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis. Microbiology (Reading) 153:4050–4060. doi: 10.1099/mic.0.2007/011619-0 [DOI] [PubMed] [Google Scholar]
- 34. Ginda K, Bezulska M, Ziółkiewicz M, Dziadek J, Zakrzewska-Czerwińska J, Jakimowicz D. 2013. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol Microbiol 87:998–1012. doi: 10.1111/mmi.12146 [DOI] [PubMed] [Google Scholar]
- 35. Ginda K, Santi I, Bousbaine D, Zakrzewska-Czerwińska J, Jakimowicz D, McKinney J. 2017. The studies of ParA and ParB dynamics reveal asymmetry of chromosome segregation in mycobacteria. Mol Microbiol 105:453–468. doi: 10.1111/mmi.13712 [DOI] [PubMed] [Google Scholar]
- 36. Santi I, McKinney JD. 2015. Chromosome organization and replisome dynamics in Mycobacterium smegmatis. mBio 6:e01999-14. doi: 10.1128/mBio.01999-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pióro M, Małecki T, Portas M, Magierowska I, Trojanowski D, Sherratt D, Zakrzewska-Czerwińska J, Ginda K, Jakimowicz D. 2019. Competition between DivIVA and the nucleoid for ParA binding promotes segrosome separation and modulates mycobacterial cell elongation. Mol Microbiol 111:204–220. doi: 10.1111/mmi.14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu C-H, Yen C-Y, Chen B-W, Lin M-G, Wang L-H, Tang K-Z, Hsiao C-D, Sun Y-J. 2019. Crystal structures of HpSoj–DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation. Nucleic Acids Res 47:2113–2129. doi: 10.1093/nar/gky1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WEE, Shapiro L. 2010. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12:791–798. doi: 10.1038/ncb2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955. doi: 10.1016/j.cell.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134:956–968. doi: 10.1016/j.cell.2008.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamaichi Y, Bruckner R, Ringgaard S, Möll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. 2012. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26:2348–2360. doi: 10.1101/gad.199869.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ditkowski B, Holmes N, Rydzak J, Donczew M, Bezulska M, Ginda K, Kedzierski P, Zakrzewska-Czerwińska J, Kelemen GH, Jakimowicz D. 2013. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in streptomyces coelicolor. Open Biol 3:130006. doi: 10.1098/rsob.130006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Russell DW, Sambrook J. 2001. Molecular cloning: a laboratory manualcold spring harbour. Available from: www.molecularcloning.com
- 47. Chai Q, Singh B, Peisker K, Metzendorf N, Ge X, Dasgupta S, Sanyal S. 2014. Organization of Ribosomes and Nucleoids in Escherichia coli cells during growth and in quiescence. J Biol Chem 289:11342–11352. doi: 10.1074/jbc.M114.557348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wickham H. 2009. Ggplot2: elegant graphics for data analysis (use R!).ggplot2: elegant graphics for data analysis (use R!). Springer, New York, NY. [Google Scholar]
- 49. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology (Reading) 146 ( Pt 8):1969–1975. doi: 10.1099/00221287-146-8-1969 [DOI] [PubMed] [Google Scholar]
- 50. Trojanowski D, Ginda K, Pióro M, Hołówka J, Skut P, Jakimowicz D, Zakrzewska-Czerwińska J. 2015. Choreography of the Mycobacterium replication machinery during the cell cycle. mBio 6:e02125-14. doi: 10.1128/mBio.02125-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hołówka J, Trojanowski D, Ginda K, Wojtaś B, Gielniewski B, Jakimowicz D, Zakrzewska-Czerwińska J. 2017. HupB is a bacterial nucleoid-associated protein with an indispensable eukaryotic-like tail. mBio 8:e01272-17. doi: 10.1128/mBio.01272-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. 2015. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ . Nat Protoc 10:33–52. doi: 10.1038/nprot.2014.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stracy M, Uphoff S, Garza de Leon F, Kapanidis AN. 2014. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Lett 588:3585–3594. doi: 10.1016/j.febslet.2014.05.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to Fig. S7.
Supplemental methods.
Legends for Movies S1 and S2.
Constructs used in the study.
Strains used in the study.
Oligonucleotides used in this study.
papM deletion alters EGFP-ParA fluorescence dynamics: control strain.
papM deletion alters EGFP-ParA fluorescence dynamics: papM background.
The mCherry-DivIVA redistribution during the cell cycle is affected by parA and papM deletion: wild type (WT) control strain.
The mCherry-DivIVA redistribution during the cell cycle is affected by parA and papM deletion: parA strain producing mCherry-DivIVA.
The mCherry-DivIVA redistribution during the cell cycle is affected by parA and papM deletion: papM strain producing mCherry-DivIVA.
An accounting of the reviewer comments and feedback.


