ABSTRACT
During infections, Staphylococcus aureus is exposed to hypochlorous acid (HOCl) and hypothiocyanous acid (HOSCN), which are produced by the neutrophil myeloperoxidase as potent antimicrobial killing agents. In this work, we applied RNAseq transcriptomics, Brx-roGFP2 biosensor measurements, and phenotype analyses to investigate the stress responses and defense mechanisms of S. aureus COL toward HOSCN stress. Based on the RNAseq transcriptome profile, HOSCN exerts strong thiol-specific oxidative, electrophile, and metal stress responses as well as protein damage in S. aureus, which is indicated by the strong induction of the HypR, TetR1, PerR, QsrR, MhqR, CstR, CsoR, CzrA, AgrA, HrcA, and CtsR regulons. Phenotype analyses of various mutants in HOSCN-responsive genes revealed that the HOSCN reductase MerA conferred the highest resistance toward HOSCN stress in S. aureus COL, whereas the QsrR and MhqR electrophile stress regulons do not contribute to protection. Brx-roGFP2 biosensor measurements and bacillithiol (BSH)-specific Western blot analyses revealed a strong oxidative shift of the bacillithiol redox potential (E BSH) and increased S-bacillithiolations in S. aureus, indicating that BSH is oxidized to bacillithiol disulfide (BSSB) under HOSCN stress. While the ΔmerA mutant was delayed in recovery of the reduced E BSH, overproduction of MerA in the ΔhypR mutant enabled faster recovery of E BSH due to efficient HOSCN detoxification. Moreover, both MerA and BSH were shown to contribute to HOSCN resistance in growth assays. In summary, HOSCN provokes a thiol-specific oxidative, electrophile, and metal stress response, an oxidative shift in E BSH and increased S-bacillithiolation in S. aureus.
IMPORTANCE
Staphylococcus aureus colonizes the skin and the airways but can also lead to life-threatening systemic and chronic infections. During colonization and phagocytosis by immune cells, S. aureus encounters the thiol-reactive oxidant HOSCN. The understanding of the adaptation mechanisms of S. aureus toward HOSCN stress is important to identify novel drug targets to combat multi-resistant S. aureus isolates. As a defense mechanism, S. aureus uses the flavin disulfide reductase MerA, which functions as HOSCN reductase and protects against HOSCN stress. Moreover, MerA homologs have conserved functions in HOSCN detoxification in other bacteria, including intestinal and respiratory pathogens. In this work, we studied the comprehensive thiol-reactive mode of action of HOSCN and its effect on the reversible shift of the E BSH to discover new defense mechanisms against the neutrophil oxidant. These findings provide new leads for future drug design to fight the pathogen at the sites of colonization and infections.
KEYWORDS: Staphylococcus aureus, HOSCN, transcriptome, MerA, bacillithiol
INTRODUCTION
Staphylococcus aureus is a commensal bacterium and part of the human skin and airway microbiota (1). At the same time, S. aureus can be a major human pathogen, leading not only to local skin and soft-tissue infections but also to life-threatening systemic and chronic infections, e.g., septic shock, pneumonia, and osteomyelitis, especially in immunocompromised patients (2 – 4). Due to the high use of antibiotics in hospitals, the emergence of multi-resistant S. aureus (MRSA) isolates poses a high risk for treatment failure of S. aureus infections. Thus, the understanding of the adaptation and defense mechanisms of S. aureus during infections is an urgent need to identify new drug targets to combat life-threatening MRSA infections (5, 6).
After infection of the host, the pathogens are phagocytosed by the cells of our innate immune system, such as macrophages and neutrophils. The phagocytosis of the bacteria is associated with an oxidative burst of activated neutrophils, which is aimed to kill the invading bacteria (7 – 10). First, the NADPH oxidase is assembled in the phagosomal membrane, which transfers electrons to oxygen to produce reactive oxygen species (ROS), such as superoxide anions at concentrations of 2 mM/s (8 – 12). Upon fusion of the phagosomal membrane with azurophilic granules, the myeloperoxidase (MPO) is released into the phagosomal lumen. MPO dismutates superoxide to H2O2, which is used by MPO for the oxidation of chloride to produce the highly microbicidal hypochlorous acid (HOCl) (7 – 10). In the presence of the pseudohalide thiocyanate (SCN-), MPO generates the hypothiocyanous acid (HOSCN), which is a similar powerful oxidant and antimicrobial agent as HOCl (7, 13). Although SCN- is the preferred substrate for MPO, the chloride outcompetes SCN- in the neutrophil phagosome, indicating that HOCl is the main oxidant produced by MPO (14). Apart from MPO, two other heme peroxidases, including lactoperoxidase (LPO) and eosinophil peroxidase (EPO) catalyze the reaction of SCN- with H2O2 to produce high levels of 1–3 mM HOSCN in the airways and saliva and 5–50 µM in the blood plasma of the human body (13, 15 – 17). Thus, S. aureus encounters HOSCN, especially at the sites of infections and colonization, including the airway mucosa of the upper respiratory tract where high SCN- levels are secreted (13).
While HOCl has been regarded as the most potent killing agent produced by neutrophils, its reaction with cellular macromolecules is rather non-specific, leading to thiol-oxidation and chlorination of proteins and subsequent protein aggregation as mechanisms of bacterial death (7, 13, 18, 19). In contrast, HOSCN is a more thiol-specific oxidant (20, 21), which reacts with protein thiols and low molecular weight (LMW) thiols, leading to the formation of unstable sulfenyl thiocyanate intermediates (R-S-SCN) and subsequently to protein disulfides, such as S-thiolations as well as intramolecular and intermolecular disulfides (7, 9, 10, 13, 22).
The bacterial responses and defense mechanisms against HOSCN stress have been investigated in important human pathogens, such as Escherichia coli, Streptococcus pneumoniae, Pseudomonas aeruginosa, and S. aureus (10, 13). While P. aeruginosa is highly sensitive toward HOSCN stress, the Gram-positive respiratory pathogens S. pneumoniae and S. aureus were found to be much more HOSCN resistant (23, 24). In buffer-killing assays, more than 50% of S. aureus and S. pneumoniae cells survived the treatment with 800 µM HOSCN after 2 hours, whereas P. aeruginosa was rapidly killed within 30 min (25). The HOSCN resistance of S. pneumoniae is mediated by the major LMW thiol glutathione (GSH), which is imported from the host via the GshT ABC transporter binding protein and maintained in the reduced state by the glutathione reductase Gor (24, 26 – 28). In addition, the flavin disulfide reductase (FDR) Har was identified as the main HOSCN reductase in S. pneumoniae, conferring GSH-independent resistance toward HOSCN (29), and deletion of both the GSH system and Har resulted in hypersensitivity toward HOSCN (29). Moreover, the Har homologous class-II FDR enzymes MerA and RclA were shown to confer strong resistance toward the neutrophil oxidants HOCl and HOSCN in S. aureus and E. coli (25, 30 – 32), supporting that MerA homologs represent the main defense mechanisms enabling survival during neutrophil infections.
Additionally, S. aureus encodes several antioxidant enzymes, such as the catalase KatA and the peroxiredoxin AhpCF, which play compensatory roles in the detoxification of the majority of H2O2 and confer high aerobic peroxide resistance (33, 34). The LMW thiol bacillithiol (BSH) and its associated bacilliredoxin (Brx)/BSH/bacillithiol disulfide reductase (YpdA) pathway function in the defense against ROS and HOCl stress in S. aureus (35 – 38). Under HOCl stress, BSH is involved in widespread S-bacillithiolation of the thiol proteome, leading to thiol-protection and redox-regulation of redox-sensitive enzymes and transcription factors, such as the glyceraldehyde-3-phosphate dehydrogenase GapA of S. aureus and the OhrR repressor in Bacillus subtilis (39 – 42). The Brx/BSH/YpdA pathway is involved in the redox control and reversibility of S-bacillithiolations and is required for the survival of S. aureus during macrophage infections and HOCl stress (35, 36).
Furthermore, S. aureus encodes specific redox-sensing transcriptional regulators, such as PerR, MgrA, SarZ, HypR, QsrR, and MhqR, which respond to different redox-active compounds, e.g., ROS, HOCl, HOSCN, or reactive electrophile species by post-translational thiol-modifications, leading to conformational changes and inactivation or activation of the regulatory proteins (35, 43, 44). These redox regulators control adaptation and defense mechanisms, which are upregulated under specific redox stress conditions to degrade the redox-active species or to restore redox homeostasis (35, 43, 44). Recently, we have shown that the Rrf2-family repressor HypR senses neutrophil oxidants, such as HOCl and HOSCN stress, via Cys33-Cys99′ intersubunit disulfide formation, leading to inactivation of its repressor activity and upregulation of the HOSCN reductase MerA (25, 31). While the functions of MerA homologs toward HOSCN resistance have been explored in different pathogens (13, 23, 25, 29, 30), the genome-wide changes in gene expression and possible functions of other defense mechanisms and HOSCN-sensing redox regulators have not been systematically investigated in S. aureus.
In this study, we used global RNA-seq gene expression profiling to investigate the stress responses, which are upregulated under HOSCN, and to identify further defense mechanisms. HOSCN caused a strong thiol-specific oxidative, electrophile, and metal stress response as well as protein damage as revealed by the induction of the HypR, TetR1, PerR, QsrR, MhqR, CstR, CsoR, CzrA, AgrA, HrcA, and CtsR regulons. Thus, the gene expression profiles overlap strongly between HOCl and HOSCN stress, indicating similar adaptation and defense mechanisms in S. aureus against neutrophil oxidants. Using the Brx-roGFP2 biosensor, we further showed that HOSCN leads to a rapid reversible oxidative shift of the BSH redox potential (E BSH), confirming its thiol-specific mode of action. The recovery of the reduced E BSH was impaired in the absence of MerA, whereas constitutive MerA expression in the ΔhypR mutant resulted in faster E BSH regeneration. Deletion of both MerA and BSH increased the susceptibility toward HOSCN stress during the growth, indicating that MerA and BSH confer independent HOSCN resistance. Thus, our study revealed the thiol-specific mode of action of HOSCN in S. aureus COL and the importance of MerA and the LMW thiol BSH in the defense against HOSCN stress.
RESULTS
S. aureus COL is more resistant toward HOSCN when grown in RPMI compared to LB medium
First, we determined the sub-lethal concentration of HOSCN, which impairs the growth of S. aureus COL, when cultivated in Luria-Bertani (LB) and RPMI medium, without killing effects. We used the LPO-SCN- system to generate HOSCN, which was purified and quantified as described (23, 25, 31). S. aureus was grown to the mid-log phase and treated with increasing doses of 94–250 µM purified HOSCN to monitor the decrease of the growth rate (Fig. 1A and B). We noted that HOSCN has stronger inhibitory effects on the bacterial growth when S. aureus was cultivated in LB (Fig. 1A), whereas in RPMI medium even high concentrations of 250 µM HOSCN did not cause notable growth delays (Fig. 1B). When applied to the LB culture, the duration of the growth delay (60–120 min) increased with the HOSCN dose (136–250 µM), but even with 250 µM, the bacteria were able to succeed in growth after 2 hours with the same growth rate as the untreated cells. Thus, HOSCN detoxification must occur fast, allowing complete recovery of growth after the removal of HOSCN. For further transcriptome experiments, we have chosen 176 µM HOSCN as a sublethal concentration in LB medium causing significant growth delays without killing effects in S. aureus cells.
Fig 1.
Effect of HOSCN stress on the growth of S. aureus COL in LB (A) and RPMI medium (B). Growth curves of S. aureus COL in LB medium (A) and RPMI (B) were monitored after exposure to increasing doses of 94–250 µM HOSCN stress at an OD540 and OD500 of 0.5, respectively. S. aureus is more resistant toward HOSCN stress when grown in RPMI medium compared to LB. The results are from three biological replicate experiments. Error bars represent the standard deviation (SD).
RNA-seq transcriptome analysis indicates a strong thiol-specific oxidative, electrophile, and metal stress response and protein damage under HOSCN stress in S. aureus
Next, we used RNA-seq transcriptome analysis to investigate the changes in gene expression in S. aureus COL, which was cultivated in LB in three biological replicates and exposed to 176 µM HOSCN for 30 min at an OD540 of 0.5. To select genes with significant fold changes, we have chosen the M-value cutoff (log2-fold change HOSCN/control) of ≥1 and ≤−1. According to this cutoff, 649 and 702 genes were significantly more than twofold up- and downregulated, respectively, under HOSCN stress in S. aureus COL (Fig. 2; Tables S1 and S2).
Fig 2.
RNA-seq transcriptomics indicates that HOSCN induces a thiol-specific stress response in S. aureus COL. For RNA-seq transcriptome profiling, S. aureus COL was grown in LB medium to an OD540 of 0.5 and treated with sublethal 176 µM HOSCN stress for 30 min. The gene expression profile in response to HOSCN stress is shown as a ratio/intensity scatter plot (M/A-plot), which is based on the differential gene expression analysis using DeSeq2. The M-value represents the log2 fold change, and the A-value is the log2 average intensity (log2 base mean) of each transcript under HOSCN stress versus the untreated control. Light gray symbols denote transcripts with no fold changes (P > 0.05). Colored symbols and dark gray symbols indicate significantly induced or repressed transcripts (M-value ≥ 1.0 or ≤−1.0; P ≤ 0.01). Light gray symbols denote transcripts with no fold changes (P > 0.05). The significantly up- and downregulated regulons were functionally classified into the HOSCN/ROS defense (HypR, TetR, PerR, QsrR, and MhqR), proteostasis (CtsR, HrcA), sulfide detoxification (CstR), Cys biosynthesis (T-box Cys), metal stress responses (CsoR, CzrA, Fur, Zur), glucose catabolism (GapR), and virulence (SigB, AgrA) regulons, which revealed a strong thiol-specific oxidative and electrophile stress response and protein damage under HOSCN stress. These differentially expressed regulons with significant fold changes were color coded as indicated in the legend. The transcriptome analysis was performed from three biological replicates. The complete RNA-seq expression data of all genes after HOSCN stress and their regulon classifications are listed in Tables S1 and S2.
Overall, the HOSCN transcriptome clearly indicates a characteristic thiol-specific oxidative stress signature, as observed also by other thiol-reactive compounds, including HOCl, allicin, and AGXX (31, 45, 46) (Tables S1 to S4). Among the top scorers were the 1,059–1,304-fold upregulated SACOL2588-89 operon (TetR1 regulon) of unknown function and the HypR-controlled hypR-merA operon (77–154-fold), encoding the conserved HOSCN reductase MerA, conferring high resistance against HOSCN, HOCl, and allicin stress in S. aureus (25, 31, 46) (Fig. 2; Tables S1 and S2). In addition, the quinone-sensing QsrR and MhqR regulons respond strongly (7.5–38.7-fold) to HOSCN stress, including the catE-SACOL0409-azoR1, yodC, catE2, frp, and mhqRED operons, which function in the detoxification of quinones and confer resistance toward quinones, antibiotics, and oxidants (47, 48). The oxidative stress-specific PerR regulon, including the genes for antioxidant enzymes (katA, ahpCF, and tpx) and iron storage proteins (dps and ftnA), was strongly upregulated by HOSCN (3.4–40-fold) (49) (Fig. 2; Tables S1 and S2). Collectively, the strong induction of the HypR, TetR1, QsrR, MhqR, and PerR regulons revealed that HOSCN stress provokes a thiol-specific oxidative and electrophile stress response in S. aureus.
Among the HOSCN-responsive regulons were further the CtsR and HrcA regulons (13–270-fold), which control ATP-dependent chaperones (DnaK, GrpE, and GroESL) and proteases (ClpB, ClpC, and ClpP) to repair or degrade damaged proteins due to HOSCN-induced protein thiol-oxidation (50, 51) (Fig. 2; Tables S1 and S2). Additionally, HOSCN upregulates the transcription of the reactive sulfur species (RSS)-sensing CstR regulon, comprising the cstAB-sqr and cstR-tauE operons (3–33-fold), which encode the multidomain sulfurtransferase (CstA), persulfide dioxygenase-sulfurtransferase (CstB), and sulfide:quinone oxidoreductase (Sqr), involved in the detoxification of hydrogen sulfide in S. aureus (52 – 54). Thus, the sulfur compound HOSCN might resemble RSS since protein thiols are oxidized first to the intermediate sulfenyl thiocyanate (55 – 57). The thiol-specific oxidation by HOSCN further leads to the depletion of Cys and LMW thiols, indicated by the strong (13.8–31.5-fold) upregulation of the T-box Cys-controlled cysE-cysS-mrnC-yacO-SACOL0579 operon, which is involved in the synthesis of O-acetyl-cysteine and cysteinyl-tRNA (58). In addition, the cysK gene encoding the cysteine synthase and bshA and bshC involved in BSH biosynthesis were 3.4–4.9-fold induced by HOSCN stress, indicating an enhanced requirement of Cys and BSH due to the thiol-specific oxidative mode of action of HOSCN (Fig. 2; Tables S1 and S2).
HOSCN causes the induction of the metal stress responsive CsoR and CzrA regulons (16–89-fold), controlling metal homeostasis by the upregulation of uptake systems for Cu+ and efflux systems for Zn2+/Co2+, respectively (59 – 61) (Fig. 2; Tables S1 and S2). The metal-sensing repressors CsoR and CzrA share metal-binding Cys residues, which are most likely oxidized upon HOSCN, leading to the loss of their repressor activity and transcriptional upregulation of their regulons. In addition, some members of the Fur and Zur regulons, which respond to iron and Zn2+ starvation, are weakly upregulated by HOSCN stress. Among the energy metabolism, the GapR-controlled gapR-gap-pgk-tpiA-pgm eno operon involved in glycolysis was significantly upregulated (6–21-fold), while operons encoding the pyruvate dehydrogenase and TCA cycle enzymes were not differentially transcribed (Fig. 2; Tables S1 and S2). Additionally, we noticed the upregulation of the Rex-dependent pyruvate-formate lyase-encoding pflAB operon (3–10-fold) and the SrrAB-regulated SACOL0219-hmp operon (3.7–4.5-fold) encoding a flavohemoglobin under HOSCN stress, suggesting oxygen limitation and the switch to anaerobiosis.
Furthermore, the large GraRS cell wall stress regulon and the virulence regulons controlled by the accessory gene regulator A (AgrA) and the general stress and starvation sigma factor B (SigB) were differentially transcribed under HOSCN stress, with the majority of genes downregulated under HOSCN stress. Among the most strongly downregulated transcripts are the atpCDGAHFEBI operon encoding the ATP synthase (0.1–0.7-fold), the sigB-rsbW-rsbV-rsbU operon encoding the regulatory genes of SigB (0.13–0.16-fold), the epiGEFPDCBA operon for epidermin biosynthesis (0.1–0.5-fold), and the crtNMQIO operon for staphyloxanthin biosynthesis (0.06–0.27). In agreement with the HOCl-induced transcriptome changes, the purEKCSQLFMNHD operon for purine biosynthesis was partly downregulated (0.07–0.6-fold) under HOSCN stress, which could be due to limiting ATP levels since the ATP synthase operon was repressed (Fig. 2; Tables S1 and S2). However, while the PyrR and PurR regulons for pyrimidine and purine nucleotide biosynthesis and ribosomal operons involved in translation were strongly downregulated under HOCl stress, translation-associated genes were weakly 1.5–2.75-fold upregulated under HOSCN stress (Fig. 2; Tables S1 to S4). The downregulation of translation under stress and starvation conditions is associated with the synthesis of the alarmones (p)ppGpp, leading to a stringent response by the inhibition of processes required for active growth, while amino acid biosynthesis and stress response pathways are activated to promote bacterial survival (62). This lack of the stringent response by HOSCN stress might be due to the lower toxicity of HOSCN, which only causes reversible thiol-oxidation of proteins and LMW thiols, but cells are able to recover quickly in growth after detoxification of HOSCN (Fig. 1A).
Altogether, the HOSCN-specific transcriptome signature resembles that of other strong thiol-specific oxidants, such as HOCl, allicin, and AGXX stress, as revealed by the thiol-specific oxidative (HypR, PerR), electrophile (QsrR, MhqR), sulfur (CstR, T-box Cys), and metal stress response (CsoR, CzrA, Fur, Zur) as well as impaired proteostasis (HrcA, CtsR).
Northern blot analyses verified the induction of the thiol-specific stress response by HOSCN stress in S. aureus
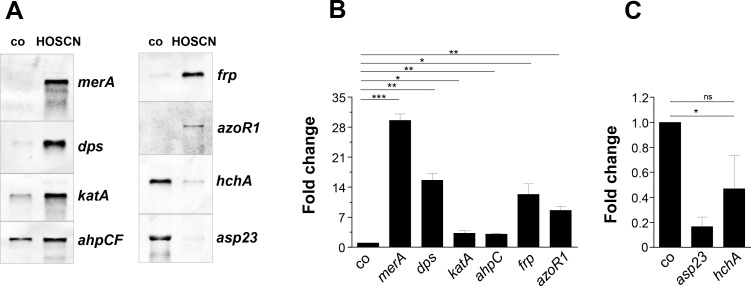
To confirm the RNA-seq expression data, we performed Northern blot analysis in S. aureus under HOSCN stress. We focused on the most strongly upregulated thiol-specific stress HypR, PerR, and QsrR regulons and the downregulated SigB regulon (Fig. 3A through C). Transcription of the HypR-controlled merA gene was most strongly 30-fold induced by HOSCN in Northern blots, followed by the 10–14-fold induction of the QsrR-regulated azoR1 and frp transcripts. The PerR-dependent dps, katA, and ahpCF transcripts were similarly highly expressed under HOSCN stress, but katA and ahpCF had higher basal transcription levels, explaining their lower 3–3.5-fold induction ratios (Fig. 3A and B). The SigB-dependent genes hchA and asp23 were strongly downregulated in the Northern blot analyses (Fig. 3A and C). Thus, the Northern blot data confirmed the strong induction of the thiol-specific stress response and downregulation of the SigB regulon under HOSCN stress as revealed by RNA-seq transcriptome data.
Fig 3.
Northern blot analysis confirmed the strong induction of the thiol-specific HypR, PerR, and QsrR regulons, while the SigB regulon is downregulated under HOSCN stress in S. aureus. (A) Transcription of the genes merA (HypR regulon), dps, katA, ahpC (PerR regulon), frp, azoR1 (QsrR regulon), hchA, and asp23 (SigB regulon) was analyzed in Northern blot experiments using RNA isolated from S. aureus COL WT 30 min after exposure to 176 µM HOSCN at an OD540 of 0.5. (B and C) Quantification of the transcriptional induction of the genes after HOSCN stress in S. aureus was performed from the Northern blot images using ImageJ. HOSCN-induced fold changes were calculated from three biological replicates, and error bars represent the SD. For the calculation of the fold changes after HOSCN stress, the transcript intensities of each gene after HOSCN stress were normalized to the mRNA intensity of the untreated control, which was set to 1. The statistics of the fold changes for each gene under HOSCN stress versus the control were calculated using a Student’s unpaired two-tailed t-test for two samples with unequal variance by the GraphPad Prism software. ns, P >0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001.
The role of the HOSCN reductase MerA, antioxidant enzymes, and the bacillithiol system in the defense of S. aureus against HOSCN stress
Previously, we have shown that the NADPH-dependent flavin disulfide reductase MerA functions as HOSCN reductase and major defense mechanism against neutrophil oxidants in S. aureus USA300 JE2 (25, 31). In the RNA-seq transcriptome of S. aureus COL, the HypR, PerR, QsrR, and MhqR regulons were most strongly upregulated upon HOSCN stress, suggesting possible protective functions upon HOSCN stress (Fig. 2; Tables S1 and S2). Thus, we aimed to elucidate the contribution of the HypR-controlled MerA, the PerR-dependent antioxidant response, the Brx/BSH/YpdA pathway, and the electrophile-sensing QsrR and MhqR regulons in the defense against HOSCN stress in S. aureus COL.
We analyzed the growth phenotypes of various mutants deficient in the thiol-specific oxidative and electrophile stress response, including the ∆merA, ∆hypR, ∆perR, ∆katA, ∆ahpC, ∆katA∆ahpC, ∆dps, ∆brxAB, ∆bshA, ∆ypdA, ∆qsrR, ∆mhqR, ∆frp, and ∆gbaA deletion mutants after exposure to sublethal doses of 176 µM HOSCN stress (Fig. 4; Fig. S1). The growth comparison of the S. aureus COL WT with the ∆merA and ∆hypR mutants revealed an increased susceptibility of the ∆merA mutant, while the ∆hypR mutant displayed increased resistance toward HOSCN due to the constitutive overexpression of MerA in the absence of the HypR repressor (31) (Fig. 4A and B). The mutant phenotypes could be restored to wild-type (WT) level in the hypR+ and merA+ -complemented strains, leading even to better growth with HOSCN upon ectopic expression of merA from the xylose-inducible promoter of plasmid pRB473 (Fig. 4A and B). These results in the S. aureus COL background are consistent with the phenotypes of the S. aureus USA300 JE2 ∆merA and ∆hypR mutants (25). However, systematic functional analyses of other single mutants deficient in the PerR-dependent antioxidant response, the Brx/BSH/YpdA redox system, or the QsrR and MhqR electrophile stress response did not show any significant growth phenotypes after HOSCN exposure in comparison to the WT (Fig. S1). This indicates that the HOSCN reductase MerA is the major HOSCN defense mechanism in S. aureus.
Fig 4.
The HOSCN reductase MerA plays a major role in the protection of S. aureus against HOSCN stress (A, B), and the absence of KatA and AhpC (C) also leads to HOSCN sensitivity. (A-C) The S. aureus COL WT, ∆merA, ∆hypR, and ∆ahpC∆katA mutants and the merA+ and hypR+ -complemented strains were grown in LB until an OD540 of 0.5 and treated with 176 µM HOSCN. Mean values were calculated from three biological replicates, and error bars represent the SD. The statistics of growth differences between the HOSCN-treated mutants or complemented strains versus the HOSCN-treated WT were calculated using a Student’s unpaired two-tailed t-test for two samples with unequal variance by the GraphPad Prism software. ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001.
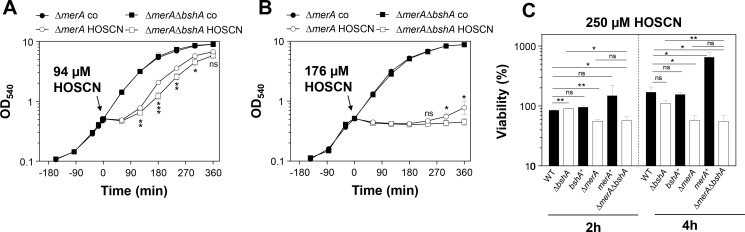
Previous analyses showed that the catalase KatA and the peroxiredoxin AhpCF could compensate for each other in H2O2 detoxification in S. aureus (33, 34). Indeed, the ∆katA∆ahpC double mutant was significantly impaired in growth upon HOSCN stress in relation to the WT, supporting that antioxidant enzymes protect against HOSCN-induced oxidative stress. Previous studies in S. pneumoniae revealed that the absence of both the MerA-homolog Har and the glutathione system leads to hypersensitivity toward HOSCN stress, whereas the ∆har single mutant did not display increased HOSCN sensitivity (29). Our growth analysis showed slightly enhanced sensitivity of the ∆merA∆bshA double mutant versus the ∆merA mutant upon exposure to 94 and 176 µM HOSCN stress, supporting that both MerA and the LMW thiol BSH contribute to HOSCN resistance (Fig. 5A and B).
Fig 5.
In the absence of MerA, BSH significantly contributes to HOSCN resistance in growth assays but not in viability assays in S. aureus. (A and B) For growth curves, the S. aureus COL ∆merA and ∆merA∆bshA mutants were grown in LB until an OD540 of 0.5 and exposed to 94 or 176 µM HOSCN. Mean values were calculated from three biological replicates, and error bars represent the SD. The statistics of growth phenotypes were calculated between the HOSCN-treated ∆merA∆bshA mutant versus the HOSCN-treated ∆merA mutant using a Student’s unpaired two-tailed t-test for two samples with unequal variance by the GraphPad Prism software. (C) For viability assays, the S. aureus WT, ∆bshA, ∆merA, and ∆merA∆bshA mutants and the merA +-complemented strain were treated with 250 µM HOSCN during the log phase and plated for CFUs after 2 and 4 hours of stress exposure. The percentage viability rate was calculated in relation to that of the untreated WT, which was set to 100%. Mean values were calculated from three biological replicates, and error bars represent the SD. The statistics of viability differences were calculated between the HOSCN-treated mutants or complemented strains versus the HOSCN-treated WT as well as between the HOSCN-treated ∆merA∆bshA double mutant versus the ∆merA or ∆bshA mutants using a Student’s unpaired two-tailed t-test for two samples with unequal variance by the GraphPad Prism software. ns, P > 0.05; *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001.
Furthermore, viability assays of ∆merA, ∆bshA, and ∆merA ∆bshA mutants were conducted in the presence of high concentrations of 250 µM HOSCN, which caused a strong growth delay in the WT (Fig. 1A). Consistent with the growth phenotypes, the ∆merA mutant was significantly impaired in viability after 2 and 4 hours, whereas the ∆bshA mutant showed no viability defect compared to the WT (Fig. 5C). The merA +-complemented strain showed a strongly increased viability compared to the WT due to the constitutive expression of MerA from plasmid pRB473. However, the ∆merA∆bshA double mutant was similarly impaired in viability as the ∆merA single mutant (Fig. 5C), indicating that BSH only makes a minor contribution toward HOSCN resistance to improve the growth of S. aureus. The susceptibility of the ∆merA mutant was also not affected in the ∆merA∆katA or ∆merA∆frp double deletion mutants, indicating that the catalase and the NADPH-dependent flavin reductase (Frp) cannot compensate for the absence of MerA in the defense against HOSCN stress (Fig. S2A through D).
Altogether, our phenotype analysis revealed that the HOSCN reductase MerA is the most important HOSCN defense mechanism of S. aureus, while the KatA/AhpCF antioxidant enzymes together provide some growth advantage under HOSCN stress. In addition, the LMW thiol BSH was shown to promote growth under HOSCN stress in the absence of MerA in S. aureus.
The impact of MerA on the oxidation of the BSH redox potential under HOSCN stress in S. aureus
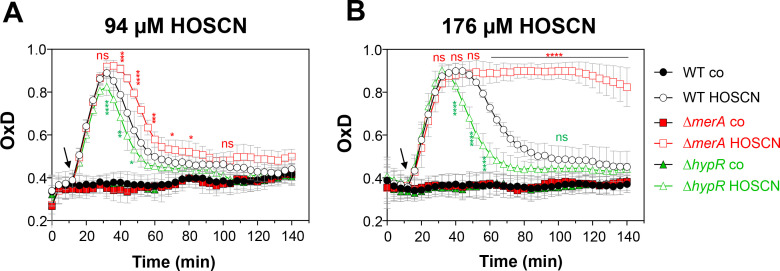
HOSCN acts as a thiol-specific oxidant, leading to reversible oxidation of protein thiols and LMW thiols, such as GSH and BSH (9, 13, 20, 21). Thus, we were interested in investigating whether HOSCN leads to the oxidation of BSH in S. aureus cells. Using the genetically encoded Brx-roGFP2 biosensor, the changes of the BSH redox potential (E BSH) were measured as biosensor oxidation degree (OxD) inside S. aureus cells after HOSCN stress (Fig. 6A and B). The exposure of S. aureus WT to 94 and 176 µM HOSCN stress led to complete oxidation of the Brx-roGFP2 biosensor within 20 min, but the cells recovered very quickly to the fully reduced state of E BSH (Fig. 6A and B). The recovery phase was dose-dependent and required 60 and 100 min after the exposure to 94 and 176 µM HOSCN stress, respectively (Fig. 6A and B). These results revealed that HOSCN causes a strong reversible oxidative shift of the E BSH due to increased BSSB levels, which can be reduced by the BSSB reductase YpdA as shown previously (36).
Fig 6.
HOSCN causes a strong oxidative shift in the BSH redox potential (E BSH) as revealed by the Brx-roGFP2 biosensor. The S. aureus COL WT, ∆merA, and ∆hypR mutants expressing the Brx-roGFP2 biosensor were treated with 94 µM HOSCN (A) or 176 µM HOSCN (B) and the biosensor oxidation degree (OxD) was monitored using the CLARIOSTAR microplate reader as described (63). The Brx-roGFP2 biosensor shows a fast oxidative shift of E BSH upon HOSCN stress in all strains. The ΔmerA mutant was delayed in the recovery of reduced E BSH after treatment with 176 µM HOSCN, while the recovery of the ∆hypR mutant was faster than the WT. Mean values and SD of three to four biological replicates are presented. The statistics of the OxD values were calculated for the HOSCN-treated mutants versus the HOSCN-treated WT at the 30-, 40-, 50-, 60-, 70-, 80-, and 100-min time points using a Student’s unpaired two-tailed t-test for two samples with unequal variance by the GraphPad Prism software. ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001.
To analyze the impact of HOSCN detoxification by MerA on the changes in E BSH upon HOSCN stress, the Brx-roGFP2 biosensor response was monitored in the ΔmerA and ΔhypR mutant backgrounds (Fig. 6A and B). The ΔmerA and ΔhypR mutants showed a similar fast oxidative shift of the E BSH upon HOSCN stress as the WT. However, the ΔmerA mutant was significantly impaired in the regeneration of reduced E BSH after 176 µM HOSCN, whereas the ΔhypR mutant recovered much faster from HOSCN stress than the WT due to constitutive upregulation of MerA (Fig. 6A and B). These results strongly support the important role of MerA in HOSCN detoxification in vivo, and its contribution to the maintenance of cellular redox homeostasis. Since MerA also functions in the defense against HOCl stress (31), we analyzed next the Brx-roGFP2 biosensor response in the ΔmerA and ΔhypR mutants after exposure to sublethal 100 µM HOCl stress. However, the ΔmerA and ΔhypR mutants showed similar Brx-roGFP2 biosensor oxidation and recovery of reduced E BSH after exposure to 100 µM HOCl stress within 80 min as the WT (Fig. S3), indicating that MerA does not contribute alone to HOCl detoxification and the maintenance of reduced E BSH under HOCl stress.
HOSCN causes increased S-bacillithiolations in the proteome of S. aureus
Previously, we showed that HOCl stress leads to strongly increased protein thiol-oxidation, including S-bacillithiolation of redox-sensitive proteins in S. aureus (40). Using BSH-specific non-reducing Western blot analysis, the glyceraldehyde dehydrogenase GapA was the most abundant S-bacillithiolated protein in S. aureus after HOCl stress (40). Thus, we applied non-reducing BSH-specific Western blot analysis to analyze the extent of protein S-bacillithiolation under HOSCN stress in S. aureus. Since MerA was shown to impact the recovery of E BSH after HOSCN stress (Fig. 6A and B), we analyzed the profile of S-bacillithiolations in the WT and ΔmerA mutant after exposure to 176 µM HOSCN stress (Fig. 7A). The HOSCN treatment of the S. aureus WT and the ΔmerA mutant resulted in increased protein S-bacillithiolations, but there was no difference between the WT and the ΔmerA mutants, indicating that the deletion of merA does not affect protein S-bacillithiolations (Fig. 7A). The HOSCN-induced bands of S-bacillithiolations were not detected in the HOSCN-treated ΔbshA mutant and reversible in the WT with dithiothreitol (DTT) in the reducing Western blot analysis (Fig. 7A and B), confirming that HOSCN stress leads to increased protein S-bacillithiolations in the proteome of S. aureus. As noted earlier, the polyclonal BSH rabbit antiserum shows cross-reactivity with abundant cellular proteins (40), detected as background in the untreated WT and the ΔmerA mutant as well as in the HOSCN-treated ΔbshA mutant. However, increased levels of specific bands of S-bacillithiolated proteins were reproducibly detected in the WT and the ΔmerA mutant under HOSCN stress. The specific targets for reversible thiol-oxidation including S-bacillithiolations under HOSCN stress will be elucidated using quantitative redox proteomics approaches in our future research.
Fig 7.
HOSCN stress induces S-bacillithiolation in S. aureus. (A and B) The S. aureus COL WT, ∆merA, and ∆bshA mutants were exposed to 176 µM HOSCN for 30 min in BMM. The NEM-alkylated protein extracts were subjected to BSH-specific non-reducing and reducing Western blot analysis. HOSCN stress causes increased S-bacillithiolation in S. aureus WT and the ∆merA mutant. Two biological replicates are shown for the WT and the ∆merA mutant, denoted with co-1, co-2 and HOSCN-1, HOSCN-2, respectively.
DISCUSSION
During infections and colonization of the upper respiratory tract, S. aureus encounters high concentrations of the pseudohypohalous acid HOSCN, which is generated by different host peroxidases, such as MPO, LPO, and EPO from H2O2 and SCN- in the airway, saliva, and plasma fluids (13, 15). Understanding the adaptation mechanisms of S. aureus toward HOSCN stress is important to tackle life-threatening MRSA infections and to improve human health. In this work, we have used RNA-seq transcriptomics to elucidate the global changes in gene expression of S. aureus upon HOSCN stress, identified new defense mechanisms, and the effect of HOSCN on the changes of the E BSH in S. aureus WT and mutants.
In the transcriptome, HOSCN stress provoked a strong thiol-specific oxidative, electrophile, and metal stress response as well as protein unfolding as revealed by the strong induction of the HypR, TetR1, PerR, QsrR, MhqR, CstR, CsoR, CzrA, AgrA, HrcA, and CtsR regulons. The oxidative and electrophile stress-specific HypR, TetR1, QsrR, and MhqR regulons were most strongly upregulated in the transcriptome under HOSCN, which clearly supports the thiol-reactive mode of action of HOSCN (Fig. 8). These redox regulons were also the top hits in the transcriptome of other thiol-reactive compounds, including HOCl, allicin, and AGXX stress (31, 45, 46). The detailed comparison of the gene expression changes upon HOCl and HOSCN stress (Table S4) further supports the strong overlap between the thiol-stress responses induced by both neutrophil oxidants.
Fig 8.
The thiol-reactive mode of action of the neutrophil oxidants HOSCN in S. aureus. HOSCN is generated during infections inside neutrophils by the myeloperoxidase using H2O2 and SCN-. HOSCN causes a strong thiol-specific oxidative, electrophile, and metal stress response and oxidative protein unfolding in S. aureus as revealed by the induction of the HypR, PerR, QsrR, MhqR, CzrA, CsoR, CtsR, and HrcA regulons in the transcriptome (Fig. 2). The HypR repressor is oxidized to HypR intermolecular disulfides by HOSCN stress, leading to its inactivation and upregulation of the flavin disulfide reductase MerA, which functions in HOSCN detoxification and contributes to the maintenance of the cellular redox homeostasis. Brx-roGFP2 biosensor measurements showed a reversible oxidative shift of the BSH redox potential upon HOSCN stress, which can be influenced by the expression level of MerA. In addition, the levels of S-bacillithiolated proteins increased after HOSCN stress. The changes in the cellular redox homeostasis cause ROS formation, which are detoxified by the catalase KatA and the peroxiredoxin AhpCF. The increased protein thiol-oxidation leads to protein unfolding, which requires the Clp proteases for degradation.
The redox-sensing Rrf2-family HypR repressor controls the flavin disulfide reductase MerA, which confers resistance against HOSCN, HOCl, and allicin stress in S. aureus (25, 31, 46). The HypR repressor senses HOCl and HOSCN stress by intersubunit disulfide formation between Cys33 and Cys99′ of opposing subunits, leading to inactivation of its repressor activity and derepression of transcription of the hypR-merA operon (25, 31) (Fig. 8). Similarly, the QsrR repressor senses quinones and oxidants via thiol-switch mechanisms in vivo, leading to induction of the dioxygenases and quinone reductases, which confer resistance against quinones and oxidants (48, 64). Under diamide stress, QsrR was reversibly oxidized to intersubunit disulfides between Cys4 and Cys29′, while the organic sulfur compound allicin caused S-thioallylation of the Cys residues of QsrR, leading to its inactivation (48). Thus, the strong induction of the HypR and QsrR regulons supports the reversible thiol-oxidation of redox-sensing regulators under HOSCN stress.
Similarly, the redox-sensing RclR and NemR regulons were upregulated in E. coli and P. aeruginosa upon HOSCN and HOCl stress (32, 65 – 67), whereas the redox-sensitive NmlR regulon and the CTM electron complex (CcdA1, EtrxA1, and MsrAB2) for the repair of oxidized methionine residues (68, 69) were most strongly induced by HOCl in the transcriptome of S. pneumoniae (70). The NemR repressor of E. coli was shown to sense HOCl via thiol-switch mechanisms to control the expression of the glyoxalase GloA and the NEM reductase NemA, which confer resistance toward electrophiles and contribute to survival under HOCl stress (71 – 73). RclR of E. coli controls the rclABC operon, which encodes the MerA homologous flavin disulfide reductase RclA, which functions as highly efficient HOSCN reductase and protects E. coli against neutrophil oxidants, such as HOCl and HOSCN (30, 32, 74). In P. aeruginosa, the RclR homolog controls the peroxiredoxin RclX, which confers resistance toward HOSCN stress (65). However, P. aeruginosa was more sensitive toward HOSCN stress compared to other lung pathogens, such as S. aureus or S. pneumoniae (23). This higher HOSCN sensitivity of P. aeruginosa was proposed to be related to the absence of RclA/MerA/Har homologs, which are conserved in many bacteria and involved in HOSCN detoxification in E. coli, S. aureus, and S. pneumoniae (25, 29, 30, 75). Whether the RclR-dependent peroxiredoxin RclX of P. aeruginosa functions similarly efficiently in HOSCN detoxification remains to be investigated. In addition, many other thiol-dependent redox regulators, such as the metal-coordinating repressors CsoR, CzrA, Zur, and Fur, respond to both HOCl and HOSCN stress in S. aureus (31). Similarly, the SczA-regulated Zn2+ efflux pump CzcD and the CopY-controlled copper-transporter CopA of S. pneumoniae were strongly upregulated under HOCl stress (70, 76, 77), and most likely respond also to HOSCN stress. Overall, there are parallels between the HOCl and HOSCN-induced redox stress regulons in different bacteria, governing the oxidative and electrophile stress responses in E. coli, P. aeruginosa, and S. aureus (67).
Moreover, HOSCN and HOCl stress causes protein thiol-oxidation and oxidative protein unfolding, leading to the strong upregulation of the CtsR and HrcA-regulons in S. aureus (31). The heat-shock-induced CtsR and HrcA repressors control chaperones and chaperonins (DnaK, GrpE, and GroESL) and the Clp proteases, which catalyze the ATP-dependent protein folding and degradation of unfolded proteins (50, 51) (Fig. 8). Similarly, chaperones and proteases controlled by the heat shock-specific sigma factor RpoH were upregulated in the transcriptome of P. aeruginosa after exposure to HOCl and HOSCN stress (65, 66), indicating related protein damage due to protein thiol-oxidation by both neutrophil oxidants. However, while protein aggregation is caused by HOCl stress as a bacterial killing mechanism (7, 18, 19), we did not observe the increased formation of protein aggregates in S. aureus after the exposure to 176 and 250 µM HOSCN stress (Fig. S4), which is consistent with the results obtained in P. aeruginosa WT cells after treatment with sub-lethal HOSCN stress (66).
We further showed that HOSCN exposure affects cellular redox homeostasis, leading to a strong oxidative shift in the E BSH of S. aureus as revealed by the genetically encoded Brx-roGFP2 biosensor. This indicates that HOSCN oxidizes the LMW thiol BSH to BSSB, supporting the thiol-reactive mode of action of HOSCN (Fig. 8). The recovery of the reduced state of E BSH after HOSCN exposure was dose-dependent, but fully reversible with 94 and 176 µM HOSCN. The biosensor oxidation profile was different between HOCl and HOSCN stress since S. aureus was unable to recover the reduced state upon higher doses of HOCl (63). Higher HOCl doses have been shown to cause overoxidation of BSH to form irreversible BSH sulfinic and sulfonic acids and BSH sulfonamide as revealed by thiol metabolomics (78). In contrast, HOSCN caused much lower levels of BSH sulfonamide than HOCl in S. aureus cells (78). Thus, the biosensor oxidation profiles after HOCl and HOSCN stress confirm the different thiol-reactive mode of action of both neutrophil oxidants. While HOSCN leads to reversible biosensor oxidation, higher HOCl concentrations induce irreversible oxidation products (78), which prevent the regeneration of the reduced state of the E BSH of S. aureus (63). The different growth behavior of S. aureus upon HOSCN and HOCl stress further supports their reversible and irreversible thiol-oxidation capabilities (Fig. 1A; Fig. S5). While higher doses of HOCl do not lead to the recovery of growth, S. aureus is able to resume the growth after detoxification of high doses of HOSCN (Fig. 1A; Fig. S5) (25). The depletion of the LMW thiol BSH by HOSCN and HOCl stress results in strong induction of Cys and BSH biosynthesis genes in the transcriptome of S. aureus, including the T-box Cys-controlled cysE-cysS-mrnC-yacO-SACOL0579 operon, cysK, bshA, and bshC. The increased transcription of genes for the biosynthesis of Cys and methionine was also observed upon exposure to the neutrophil oxidants in P. aeruginosa, supporting thiol depletion (65, 66). Furthermore, BSH-specific Western blot analyses confirmed the increased level of protein S-bacillithiolations under both HOCl and HOSCN stress in S. aureus (40). In future studies, we aim to conduct a more detailed redox proteomics analysis to determine the percentage of increased reversible thiol-oxidation upon HOSCN stress.
Based on the transcriptome, we aimed to discover novel HOSCN defense mechanisms of S. aureus. Thus, we analyzed the growth phenotypes of various single mutants involved in the oxidative and electrophile stress response, including the Brx/BSH/YpdA redox system, the peroxide-specific PerR regulon (KatA, AhpC, and Dps), and the QsrR, MhqR, and GbaA electrophile stress regulons. However, we did not detect any growth phenotype of the single mutants after exposure to HOSCN stress. Only the HypR-controlled HOSCN reductase MerA was important for growth under HOSCN stress in S. aureus COL, confirming our previous results in the S. aureus USA300 JE2 background (25). The S. aureus COL ΔmerA mutant was highly sensitive under HOSCN stress, whereas the ΔhypR mutant and the merA +-complemented strain showed constitutive resistance toward HOSCN due to the overexpression of MerA. Thus, the HOSCN reductase MerA was revealed as a major defense mechanism in S. aureus COL. In this study, we further showed that MerA contributed to the regeneration of the reduced state of the E BSH after recovery from HOSCN stress. The regeneration of the reduced state of the E BSH was impaired in the absence of MerA, while constitutive MerA expression in the ΔhypR mutant resulted in faster recovery of reduced E BSH, clearly indicating that MerA is involved in HOSCN detoxification in vivo. Apart from the ΔmerA mutant, only the ΔkatAΔahpC double mutant was sensitive toward HOSCN stress. However, the absence of KatA and AhpC affects aerobic growth due to the hypersensitivity of the ΔkatAΔahpC mutant toward H2O2 stress (33, 34). Since overexpression of KatA and AhpC in the ΔperR mutant did not provide a growth advantage under HOSCN stress, the role of the antioxidant enzymes KatA and AhpC in protection against HOSCN-induced ROS formation remains unclear.
Previously, the roles of the LMW thiol GSH and the MerA homolog Har of S. pneumoniae have been investigated in the defense against HOSCN stress (24, 29). S. pneumoniae does not encode the enzymes for GSH biosynthesis and instead imports GSH from the host using the ABC transporter-binding protein GshT, which is important for growth and oxidative stress resistance (26 – 28). To keep host-derived GSH in a reduced state, S. pneumoniae encodes the GSSG reductase Gor (28). HOSCN leads to the oxidation of GSH to GSSG and the increased formation of S-glutathionylated proteins in S. pneumoniae (24). While the Δhar mutant did not show increased HOSCN susceptibility (29), deletion of either GshT or Gor resulted in impaired growth after HOSCN stress (24). However, the ΔharΔgshT and ΔharΔgor double mutants were hypersensitive toward HOSCN compared to the Δgor and ΔgshT single mutants (29). This indicates that the GSH/Gor system and Har play compensatory roles in the defense against HOSCN stress in S. pneumoniae (29).
In our study, deletion of the Brx/BSH/YpdA system alone did not cause any growth defect under HOSCN, whereas the absence of both MerA and BSH led to slightly reduced growth rates under HOSCN stress compared to the ΔmerA single mutant. However, there was no viability difference between ΔmerA single and ΔmerAΔbshA double mutants, indicating that BSH only contributes to improved growth under HOSCN stress in the absence of MerA. These results indicate that MerA is more important than the Brx/BSH/YpdA system in protection against HOSCN in S. aureus. In contrast, GSH import seems to be more essential for the defense against HOSCN in S. pneumoniae, whereas the contribution of Har to HOSCN detoxification is only relevant in the absence of GSH import or recycling (24, 29). Furthermore, while RclA and MerA both respond strongly to HOSCN and HOCl stress (25, 30 – 32), the homolog Har was not upregulated under HOCl and HOSCN stress in S. pneumoniae (29, 70). Thus, the lack of phenotype of the Δhar single mutant toward HOSCN stress might be related to the fact that Har is not inducible by neutrophil oxidants. Future analyses should investigate in more detail the transcriptome changes upon HOCl and HOSCN stress and the redox-sensing mechanisms involved in the defense against neutrophil oxidants in other pathogenic or colonizing bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The bacterial strains, plasmids, and primers used in this study are described in Tables S5 and S6. For genetic manipulation, E. coli strains were grown in LB medium. While previous phenotype analyses were performed with the S. aureus USA300 JE2 wild type, ΔhypR, and ΔmerA mutants, we have used S. aureus COL in this work, due to the availability of a large mutant collection constructed in the COL background. S. aureus USA300 JE2 is a community-acquired highly virulent MRSA isolate, cured of two antibiotic-resistance plasmids (79). S. aureus COL is an archaic hospital-acquired MRSA isolate of lower virulence compared to USA300 JE2 (80). The S. aureus COL strains were cultivated in LB, Belitsky minimal medium (BMM), or RPMI supplemented with 0.75 µM FeCl2 and 2 mM glutamine as described (81). For the growth of the S. aureus-complemented strains harboring the pRB473-plasmids, the supplementation of the medium with 1% xylose was required for the expression of the protein. For stress experiments, S. aureus strains were treated with sublethal HOSCN concentrations at an optical density at 540 nm (OD540) of 0.5 as described previously (25). Viability assays were performed by plating 100 µL of serial dilutions of S. aureus cultures after treatment with the lethal dose of 250 µM HOSCN on LB agar plates for counting of colony forming units (CFUs). If appropriate, antibiotics were used at the following concentrations: 100 µg/mL ampicillin, 10 µg/mL erythromycin, and 10 µg/mL chloramphenicol. HOSCN was generated using the LPO-SCN- system, where H2O2 and SCN- were converted by the LPO to HOSCN according to previous protocols (23, 25, 31). The compounds sodium hypochlorite (NaOCl), diamide, DTT, and N-ethyl maleimide (NEM) were purchased from Sigma Aldrich. Statistical analysis of the growth curves, viability assays, Northern blot transcription, and Brx-roGFP2 biosensor measurements was performed using Student’s unpaired two-tailed t-test for two samples with unequal variance by the GraphPad Prism software. The specific samples compared in the Student’s unpaired two-tailed t-test are indicated in the figure legends and the P-values are shown for each comparison.
RNA isolation, library preparation, and next-generation cDNA sequencing
For the HOSCN transcriptome analysis, S. aureus COL was grown in LB medium to the exponential phase of an OD540 of 0.5 and exposed to sublethal 176 µM HOSCN, which was generated freshly using the LPO-SCN- system. The HOCl transcriptome data were generated from S. aureus USA300, which was grown in RPMI to an OD500 of 0.5 and exposed to 1.5 mM HOCl for 30 min. S. aureus cells were harvested before (as control) and 30 min after exposure to HOSCN or HOCl stress. Subsequently, the cells were disrupted in 3 mM ethylenediaminetetraacetic acid (EDTA)/200 mM NaCl lysis buffer using a Precellys24 ribolyzer. RNA isolation was performed using the acid phenol extraction protocol as described (82). The RNA quality was checked by Trinean Xpose (Gentbrugge, Belgium) and the Agilent RNA Nano 6000 kit using an Agilent 2100 Bioanalyzer (Agilent Technologies, Böblingen, Germany). Ribo-Zero rRNA Removal Kit (Bacteria) from Illumina (San Diego, CA, USA) was used to remove the rRNA. TruSeq Stranded mRNA Library Prep Kit from Illumina was applied to prepare the cDNA libraries. The cDNAs were sequenced paired end on an Illumina NextSeq 500 system (San Diego, CA, USA) using 75 bp read length. The transcriptome sequencing raw data files are available in the ArrayExpress database under accession number E-MTAB-13314 for the HOSCN stress data set and as E-MTAB-13313 for the HOCl stress data set used for comparison.
Bioinformatics data analysis, read mapping, data visualization, and analysis of differential gene expression
The paired-end cDNA reads were mapped to the S. aureus COL genome sequence (accession number NC_002951) using bowtie2 v2.2.7 (83) with default settings for paired-end read mapping. All mapped sequence data were converted from SAM to BAM format with SAMtools v1.3 (84) and imported to the software ReadXplorer v.2.2 (85).
Differential gene expression analysis of triplicates including normalization was performed using Bioconductor package DESeq2 (86) included in the ReadXplorer v2.2 software (85). The signal intensity value (A-value) was calculated by the log2 mean of normalized read counts and the signal intensity ratio (M-value) by the log2 fold change. The evaluation of the differential RNA-seq data was performed using an adjusted P-value cutoff of P ≤ 0.01 and a signal intensity ratio (M-value) cutoff of ≥1 or ≤−1. Genes with an M-value outside this range and P ≤ 0.05 were considered as differentially up- or downregulated under HOSCN stress.
Construction of the S. aureus COL Δfrp, Δdps, ΔmerAΔbshA, ΔmerAΔfrp, and ΔmerAΔkatA mutants and the Brx-roGFP2 biosensor strains in the ΔmerA and ΔhypR mutant backgrounds
The S. aureus COL Δfrp (SACOL2534) and Δdps (SACOL2131) single and the ΔmerAΔbshA,ΔmerAΔkatA and ΔmerAΔfrp double deletion mutants were constructed using the temperature-sensitive shuttle vector pMAD as described (31). For the Δdps and Δfrp single mutants, the 500 bp up- and downstream regions of the coding regions were amplified using gene-specific primers (Table S6). Subsequently, the PCR products were fused by overlap extension PCR and ligated into the BglII and SalI sites of plasmid pMAD. The obtained pMAD constructs were electroporated into the intermediate S. aureus RN4220 strain for methylation and further transduced into S. aureus COL using phage 81 (87). The clean marker-less Δfrp and Δdps deletion mutants were selected after plasmid excision as described (31). The ΔmerAΔbshA,ΔmerAΔkatA and ΔmerAΔfrp double mutants were obtained by transducing phage 81, which carried the plasmids pMAD-ΔbshA, pMAD-ΔkatA, or pMAD-Δfrp into the S. aureus COL ΔmerA mutant. Selection of the double deletion mutants was performed as previously described (31). The deletions of internal gene regions were confirmed by PCR and DNA sequencing (31).
The S. aureus COL ΔhypR-pRB473-brx-roGFP2 andΔmerA pRB473-brx-roGFP2 biosensor strains were constructed by transduction of the plasmid pRB473-brx-roGFP2 into the S. aureus COL ΔhypR and ΔmerA mutants using the phage 81 (87). The pRB473-brx-roGFP2 biosensor was confirmed in the ΔmerA and ΔhypR mutants by PCR and fluorescence microscopy.
Northern blot analysis
Northern blot transcriptional analysis was performed using RNA isolated from S. aureus COL before and 30 min after exposure to 176 µM HOSCN as described (82). Hybridizations were conducted using digoxigenin-labeled antisense RNA probes for frp and hchA that were synthesized in vitro using T7 RNA polymerase as in previous studies (31). The RNA probes specific for the genes merA, katA, ahpC, dps, azoR1, and asp23 were generated previously (31, 46, 88).
Western blot analysis to analyze S-bacillithiolations
To analyze protein S-bacillithiolations after HOSCN stress, non-reducing BSH-specific Western blot analysis of S. aureus COL WT strain and the ∆merA and ∆bshA mutants after exposure to HOSCN was conducted as previously described (31). The S. aureus cells were cultivated in LB medium until an OD540 of 2, transferred to BMM, and treated with 176 µM HOSCN for 30 min. Cells were collected in the presence of 50 mM NEM, washed in Tris-EDTA (TE) buffer (pH 8.0), and disrupted using the Precellys24 ribolyzer. Protein amounts of 25 µg were separated under non-reducing conditions using 15% SDS-PAGE and the Western blot analysis was performed as described previously (39). For control purposes, the Western blot analysis was performed with the same samples under reducing conditions (with DTT) to reduce oxidized proteins with reversible S-bacillithiolations. Polyclonal rabbit anti-BSH antiserum was used at a dilution of 1:500 for Western blot analyses.
Brx-roGFP2 biosensor measurement to analyze the changes in the BSH redox potential (E BSH) in S. aureus COL
The S. aureus COL WT, ΔhypR, and ΔmerA mutant strains expressing the Brx-roGFP2 biosensor were grown in LB medium as overnight culture, transferred to BMM, and adjusted to an OD500 of 3. Subsequently, the bacterial cells were transferred to the microplates and the biosensor OxD was monitored after the injection of HOSCN as described previously (63, 89). To obtain the fully reduced and oxidized control samples, the S. aureus COL Brx-roGFP2 expression strains were exposed to 10 mM DTT and 10 mM diamide, respectively. The Brx-roGFP2 biosensor fluorescence emission was measured at 510 nm after excitation at 405 and 488 nm using the CLARIOstar microplate reader (BMG Labtech). The time-dependent changes of the biosensor OxD were measured continuously for each sample and normalized to the fully reduced and oxidized controls as described (63, 89).
Isolation of protein aggregates after HOSCN stress
Intracellular protein aggregates were isolated from S. aureus cells, which were grown in LB medium and exposed to 176 and 250 µM HOSCN stress for 30 min at an OD540 of 0.5, as described in previous protocols (45, 66, 90). In brief, the cells were resuspended in 40 µL buffer A (10 mM potassium phosphate pH 6.5, 1 mM EDTA, 20% sucrose, 5 µg/mL lysostaphin) and 360 µL buffer B (10 mM potassium phosphate pH 6.5, 1 mM EDTA), followed by disruption using the ribolyzer. Subsequently, the insoluble protein aggregates were collected and analyzed using SDS-PAGE as described (45).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft, Germany (AN746/4-1 and AN746/4-2) within the SPP1710 on “Thiol-based Redox switches,” by the SFB973 (project C08) and TR84 (project B06) to H.A.
No competing financial interests exist.
Contributor Information
Haike Antelmann, Email: haike.antelmann@fu-berlin.de.
Artem S. Rogovskyy, Texas A&M University, College Station, Texas, USA
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article and its supplemental materials. The transcriptome sequencing raw data files are available in the ArrayExpress database under accession number E-MTAB-13314 for the HOSCN stress data set and as E-MTAB-13313 for the HOCl stress data set used for comparison.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03252-23.
Growth curves for phenotype analyses, Brx-roGFP2 biosensor experiments, and aggregation assays.
Transcriptome datasets of S. aureus after exposure to HOSCN and HOCl stress.
Bacterial strains, phages, plasmids, and oligonucleotide primers.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Foster TJ. 2004. The Staphylococcus aureus "superbug" J Clin Invest 114:1693–1696. doi: 10.1172/JCI23825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 3. Boucher HW, Corey GR. 2008. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 46 Suppl 5:S344–9. doi: 10.1086/533590 [DOI] [PubMed] [Google Scholar]
- 4. Archer GL. 1998. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis 26:1179–1181. doi: 10.1086/520289 [DOI] [PubMed] [Google Scholar]
- 5. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vestergaard M, Frees D, Ingmer H. 2019. Antibiotic resistance and the MRSA problem. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0057-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ulfig A, Leichert LI. 2021. The effects of neutrophil-generated hypochlorous acid and other hypohalous acids on host and pathogens. Cell Mol Life Sci 78:385–414. doi: 10.1007/s00018-020-03591-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winterbourn CC, Kettle AJ. 2013. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal 18:642–660. doi: 10.1089/ars.2012.4827 [DOI] [PubMed] [Google Scholar]
- 9. Winterbourn CC, Kettle AJ, Hampton MB. 2016. Reactive oxygen species and neutrophil function. Annu Rev Biochem 85:765–792. doi: 10.1146/annurev-biochem-060815-014442 [DOI] [PubMed] [Google Scholar]
- 10. Hampton MB, Dickerhof N. 2023. Inside the phagosome: a bacterial perspective. Immunol Rev 314:197–209. doi: 10.1111/imr.13182 [DOI] [PubMed] [Google Scholar]
- 11. Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. 2006. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J Biol Chem 281:39860–39869. doi: 10.1074/jbc.M605898200 [DOI] [PubMed] [Google Scholar]
- 12. Kettle AJ, Ashby LV, Winterbourn CC, Dickerhof N. 2023. Superoxide: the enigmatic chemical chameleon in neutrophil biology. Immunol Rev 314:181–196. doi: 10.1111/imr.13183 [DOI] [PubMed] [Google Scholar]
- 13. Meredith JD, Gray MJ. 2023. Hypothiocyanite and host-microbe interactions. Mol Microbiol 119:302–311. doi: 10.1111/mmi.15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Dalen JC, Whitehouse WM, Winterbourn CC, Kettle JA. 1997. Thiocyanate and chloride as competing substrates for myeloperoxidase. Biochem J 327:487–492. doi: 10.1042/bj3270487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arnhold J, Malle E. 2022. Halogenation activity of mammalian heme peroxidases. Antioxidants (Basel) 11:890. doi: 10.3390/antiox11050890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wijkstrom-Frei C, El-Chemaly S, Ali-Rachedi R, Gerson C, Cobas MA, Forteza R, Salathe M, Conner GE. 2003. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol 29:206–212. doi: 10.1165/rcmb.2002-0152OC [DOI] [PubMed] [Google Scholar]
- 17. Lorentzen D, Durairaj L, Pezzulo AA, Nakano Y, Launspach J, Stoltz DA, Zamba G, McCray PB, Zabner J, Welsh MJ, Nauseef WM, Bánfi B. 2011. Concentration of the antibacterial precursor thiocyanate in cystic fibrosis airway secretions. Free Radic Biol Med 50:1144–1150. doi: 10.1016/j.freeradbiomed.2011.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gray MJ, Wholey WY, Jakob U. 2013. Bacterial responses to reactive chlorine species. Annu Rev Microbiol 67:141–160. doi: 10.1146/annurev-micro-102912-142520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winter J, Ilbert M, Graf PCF, Ozcelik D, Jakob U. 2008. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135:691–701. doi: 10.1016/j.cell.2008.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pattison DI, Davies MJ. 2006. Reactions of myeloperoxidase-derived oxidants with biological substrates: gaining chemical insight into human inflammatory diseases. Curr Med Chem 13:3271–3290. doi: 10.2174/092986706778773095 [DOI] [PubMed] [Google Scholar]
- 21. Skaff O, Pattison DI, Davies MJ. 2009. Hypothiocyanous acid reactivity with low-molecular-mass and protein thiols: absolute rate constants and assessment of biological relevance. Biochem J 422:111–117. doi: 10.1042/BJ20090276 [DOI] [PubMed] [Google Scholar]
- 22. Hawkins CL, Pattison DI, Davies MJ. 2003. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25:259–274. doi: 10.1007/s00726-003-0016-x [DOI] [PubMed] [Google Scholar]
- 23. Shearer HL, Kaldor CD, Hua H, Kettle AJ, Parker HA, Hampton MB. 2022. Resistance of Streptococcus pneumoniae to hypothiocyanous acid generated by host peroxidases. Infect Immun 90:e0053021. doi: 10.1128/IAI.00530-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shearer HL, Paton JC, Hampton MB, Dickerhof N. 2022. Glutathione utilization protects Streptococcus pneumoniae against lactoperoxidase-derived hypothiocyanous acid. Free Radic Biol Med 179:24–33. doi: 10.1016/j.freeradbiomed.2021.12.261 [DOI] [PubMed] [Google Scholar]
- 25. Shearer HL, Loi VV, Weiland P, Bange G, Altegoer F, Hampton MB, Antelmann H, Dickerhof N. 2023. Mera functions as a hypothiocyanous acid reductase and defense mechanism in Staphylococcus aureus. Mol Microbiol 119:456–470. doi: 10.1111/mmi.15035 [DOI] [PubMed] [Google Scholar]
- 26. Vergauwen B, Verstraete K, Senadheera DB, Dansercoer A, Cvitkovitch DG, Guédon E, Savvides SN. 2013. Molecular and structural basis of glutathione import in gram-positive bacteria via GshT and the cystine ABC importer TcyBC of Streptococcus mutans. Mol Microbiol 89:288–303. doi: 10.1111/mmi.12274 [DOI] [PubMed] [Google Scholar]
- 27. Brouwer S, Jespersen MG, Ong CY, De Oliveira DMP, Keller B, Cork AJ, Djoko KY, Davies MR, Walker MJ, Kline KA. 2022. Streptococcus Pyogenes hijacks host glutathione for growth and innate immune evasion. mBio 13:0067622. doi: 10.1128/mbio.00676-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Potter AJ, Trappetti C, Paton JC. 2012. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J Bacteriol 194:6248–6254. doi: 10.1128/JB.01393-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shearer HL, Pace PE, Paton JC, Hampton MB, Dickerhof N. 2022. A newly identified flavoprotein disulfide reductase Har protects Streptococcus pneumoniae against hypothiocyanous acid. J Biol Chem 298:102359. doi: 10.1016/j.jbc.2022.102359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meredith JD, Chapman I, Ulrich K, Sebastian C, Stull F, Gray MJ. 2022. Escherichia coli RclA is a highly active hypothiocyanite reductase. Proc Natl Acad Sci U S A 119:e2119368119. doi: 10.1073/pnas.2119368119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loi VV, Busche T, Tedin K, Bernhardt J, Wollenhaupt J, Huyen NTT, Weise C, Kalinowski J, Wahl MC, Fulde M, Antelmann H. 2018. Redox-sensing under hypochlorite stress and infection conditions by the Rrf2-family repressor HypR in Staphylococcus aureus. Antioxid Redox Signal 29:615–636. doi: 10.1089/ars.2017.7354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parker BW, Schwessinger EA, Jakob U, Gray MJ. 2013. The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J Biol Chem 288:32574–32584. doi: 10.1074/jbc.M113.503516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linzner N, Loi VV, Antelmann H. 2022. The catalase KatA contributes to microaerophilic H2O2 priming to acquire an improved oxidative stress resistance in Staphylococcus aureus. Antioxidants 11:1793. doi: 10.3390/antiox11091793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189:1025–1035. doi: 10.1128/JB.01524-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Linzner N, Loi VV, Fritsch VN, Antelmann H. 2021. Thiol-based redox switches in the major pathogen Staphylococcus aureus. Biol Chem 402:333–361. doi: 10.1515/hsz-2020-0272 [DOI] [PubMed] [Google Scholar]
- 36. Linzner N, Loi VV, Fritsch VN, Tung QN, Stenzel S, Wirtz M, Hell R, Hamilton CJ, Tedin K, Fulde M, Antelmann H. 2019. Staphylococcus aureus uses the bacilliredoxin (BrxAB)/bacillithiol disulfide reductase (YpdA) redox pathway to defend against oxidative stress under infections. Front Microbiol 10:1355. doi: 10.3389/fmicb.2019.01355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Posada AC, Kolar SL, Dusi RG, Francois P, Roberts AA, Hamilton CJ, Liu GY, Cheung A. 2014. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect Immun 82:316–332. doi: 10.1128/IAI.01074-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mikheyeva IV, Thomas JM, Kolar SL, Corvaglia A-R, Gaϊa N, Leo S, Francois P, Liu GY, Rawat M, Cheung AL. 2019. YpdA, a putative bacillithiol disulfide reductase, contributes to cellular redox homeostasis and virulence in Staphylococcus aureus. Mol Microbiol 111:1039–1056. doi: 10.1111/mmi.14207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chi BK, Gronau K, Mäder U, Hessling B, Becher D, Antelmann H. 2011. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10:M111.009506. doi: 10.1074/mcp.M111.009506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Imber M, Huyen NTT, Pietrzyk-Brzezinska AJ, Loi VV, Hillion M, Bernhardt J, Thärichen L, Kolšek K, Saleh M, Hamilton CJ, Adrian L, Gräter F, Wahl MC, Antelmann H. 2018. Protein S-bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase gap in Staphylococcus aureus under hypochlorite stress. Antioxid Redox Signal 28:410–430. doi: 10.1089/ars.2016.6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imber M, Pietrzyk-Brzezinska AJ, Antelmann H. 2019. Redox regulation by reversible protein S-thiolation in gram-positive bacteria. Redox Biol 20:130–145. doi: 10.1016/j.redox.2018.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee JW, Soonsanga S, Helmann JD. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104:8743–8748. doi: 10.1073/pnas.0702081104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Antelmann H. 2023. Redox signalling mechanisms during host-pathogen interactions in the human pathogen Staphylococcus aureus, p in press. In Tang YW, Hindiyeh M, Liu D, Sails A, Spearman P, Zhan Z (ed), Molecular medical Microbiology, 3rd edition. vol. Academic Press. [Google Scholar]
- 44. Chandrangsu P, Loi VV, Antelmann H, Helmann JD. 2018. The role of bacillithiol in gram-positive firmicutes. Antioxid Redox Signal 28:445–462. doi: 10.1089/ars.2017.7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loi VV, Busche T, Preuß T, Kalinowski J, Bernhardt J, Antelmann H. 2018. The AGXX antimicrobial coating causes a thiol-specific oxidative stress response and protein S-bacillithiolation in Staphylococcus aureus. Front Microbiol 9:3037. doi: 10.3389/fmicb.2018.03037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loi VV, Huyen NTT, Busche T, Tung QN, Gruhlke MCH, Kalinowski J, Bernhardt J, Slusarenko AJ, Antelmann H. 2019. Staphylococcus aureus responds to allicin by global S-thioallylation - role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress. Free Radic Biol Med 139:55–69. doi: 10.1016/j.freeradbiomed.2019.05.018 [DOI] [PubMed] [Google Scholar]
- 47. Fritsch VN, Loi VV, Busche T, Sommer A, Tedin K, Nürnberg DJ, Kalinowski J, Bernhardt J, Fulde M, Antelmann H. 2019. The MarR-type repressor MhqR confers quinone and antimicrobial resistance in Staphylococcus aureus. Antioxid Redox Signal 31:1235–1252. doi: 10.1089/ars.2019.7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fritsch VN, Loi VV, Kuropka B, Gruhlke M, Weise C, Antelmann H. 2023. The MarR/DuF24-family QsrR repressor senses quinones and oxidants by thiol switch mechanisms in Staphylococcus aureus. Antioxid Redox Signal 38:877–895. doi: 10.1089/ars.2022.0090 [DOI] [PubMed] [Google Scholar]
- 49. Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69:3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Frees D, Gerth U, Ingmer H. 2014. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int J Med Microbiol 304:142–149. doi: 10.1016/j.ijmm.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 51. Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, gram-positive bacteria. Mol Microbiol 63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x [DOI] [PubMed] [Google Scholar]
- 52. Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. 2015. Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 54:4542–4554. doi: 10.1021/acs.biochem.5b00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Higgins KA, Peng H, Luebke JL, Chang F-MJ, Giedroc DP. 2015. Conformational analysis and chemical reactivity of the multidomain sulfurtransferase, Staphylococcus aureus CstA. Biochemistry 54:2385–2398. doi: 10.1021/acs.biochem.5b00056 [DOI] [PubMed] [Google Scholar]
- 54. Shen J, Peng H, Zhang Y, Trinidad JC, Giedroc DP. 2016. Staphylococcus aureus Sqr encodes a type II sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry 55:6524–6534. doi: 10.1021/acs.biochem.6b00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Luebke JL, Arnold RJ, Giedroc DP. 2013. Selenite and tellurite form mixed seleno- and tellurotrisulfides with CstR from Staphylococcus aureus. Metallomics 5:335–342. doi: 10.1039/c3mt20205d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. 2014. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol 94:1343–1360. doi: 10.1111/mmi.12835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peng H, Shen J, Edmonds KA, Luebke JL, Hickey AK, Palmer LD, Chang F-M, Bruce KA, Kehl-Fie TE, Skaar EP, Giedroc DP, Stallings CL. 2017. Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus. mSphere 2:e00082-17. doi: 10.1128/mSphere.00082-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang J, Ferré-D’Amaré AR. 2015. Structure and mechanism of the T-box riboswitches. Wiley Interdiscip Rev RNA 6:419–433. doi: 10.1002/wrna.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, Skaar EP, Giedroc DP. 2011. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem 286:13522–13531. doi: 10.1074/jbc.M111.220012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grossoehme NE, Giedroc DP. 2009. Energetics of allosteric negative coupling in the zinc sensor Staphylococcus aureus CzrA. J Am Chem Soc 131:17860–17870. doi: 10.1021/ja906131b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee S, Arunkumar AI, Chen X, Giedroc DP. 2006. Structural insights into homo- and heterotropic allosteric coupling in the zinc sensor S. aureus CzrA from covalently fused dimers. J Am Chem Soc 128:1937–1947. doi: 10.1021/ja0546828 [DOI] [PubMed] [Google Scholar]
- 62. Irving SE, Choudhury NR, Corrigan RM. 2021. The stringent response and physiological roles of (Pp)pGpp in bacteria. Nat Rev Microbiol 19:256–271. doi: 10.1038/s41579-020-00470-y [DOI] [PubMed] [Google Scholar]
- 63. Loi VV, Harms M, Müller M, Huyen NTT, Hamilton CJ, Hochgräfe F, Pané-Farré J, Antelmann H. 2017. Real-time imaging of the bacillithiol redox potential in the human pathogen Staphylococcus aureus using a genetically encoded bacilliredoxin-fused redox biosensor. Antioxid Redox Signal 26:835–848. doi: 10.1089/ars.2016.6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ji Q, Zhang L, Jones MB, Sun F, Deng X, Liang H, Cho H, Brugarolas P, Gao YN, Peterson SN, Lan L, Bae T, He C. 2013. Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR. Proc Natl Acad Sci U S A 110:5010–5015. doi: 10.1073/pnas.1219446110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Farrant KV, Spiga L, Davies JC, Williams HD, O’Toole G. 2020. Response of Pseudomonas aeruginosa to the innate immune system-derived oxidants hypochlorous acid and hypothiocyanous acid. J Bacteriol 203:e00300-20. doi: 10.1128/JB.00300-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Groitl B, Dahl JU, Schroeder JW, Jakob U. 2017. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol Microbiol 106:335–350. doi: 10.1111/mmi.13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. da Cruz Nizer WS, Inkovskiy V, Overhage J. 2020. Surviving reactive chlorine stress: responses of gram-negative bacteria to hypochlorous acid. Microorganisms 8:1220. doi: 10.3390/microorganisms8081220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gennaris A, Collet JF. 2013. The 'captain of the men of death', Streptococcus pneumoniae, fights oxidative stress outside the 'city wall' EMBO Mol Med 5:1798–1800. doi: 10.1002/emmm.201303482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saleh M, Bartual SG, Abdullah MR, Jensch I, Asmat TM, Petruschka L, Pribyl T, Gellert M, Lillig CH, Antelmann H, Hermoso JA, Hammerschmidt S. 2013. Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence. EMBO Mol Med 5:1852–1870. doi: 10.1002/emmm.201202435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fritsch VN, Linzner N, Busche T, Said N, Weise C, Kalinowski J, Wahl MC, Antelmann H. 2023. The MerR-family regulator NmlR is involved in the defense against oxidative stress in Streptococcus pneumoniae. Mol Microbiol 119:191–207. doi: 10.1111/mmi.14999 [DOI] [PubMed] [Google Scholar]
- 71. Gray MJ, Li Y, Leichert LI-O, Xu Z, Jakob U. 2015. Does the transcription factor NemR use a regulatory sulfenamide bond to sense bleach? Antioxid Redox Signal 23:747–754. doi: 10.1089/ars.2015.6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gray MJ, Wholey WY, Parker BW, Kim M, Jakob U. 2013. NemR is a bleach-sensing transcription factor. J Biol Chem 288:13789–13798. doi: 10.1074/jbc.M113.454421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lee C, Shin J, Park C. 2013. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol Microbiol 88:395–412. doi: 10.1111/mmi.12192 [DOI] [PubMed] [Google Scholar]
- 74. Königstorfer A, Ashby LV, Bollar GE, Billiot CE, Gray MJ, Jakob U, Hampton MB, Winterbourn CC. 2021. Induction of the reactive chlorine-responsive transcription factor RclR in Escherichia coli following ingestion by neutrophils. Pathog Dis 79:ftaa079. doi: 10.1093/femspd/ftaa079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Derke RM, Barron AJ, Billiot CE, Chaple IF, Lapi SE, Broderick NA, Gray MJ. 2020. The Cu(II) reductase RclA protects Escherichia coli against the combination of hypochlorous acid and intracellular copper. mBio 11:mBio doi: 10.1128/mBio.01905-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. 2011. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol 81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x [DOI] [PubMed] [Google Scholar]
- 77. Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJE, Kuipers OP. 2007. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiol 65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x [DOI] [PubMed] [Google Scholar]
- 78. Dickerhof N, Paton L, Kettle AJ. 2020. Oxidation of bacillithiol by myeloperoxidase-derived oxidants. Free Radic Biol Med 158:74–83. doi: 10.1016/j.freeradbiomed.2020.06.009 [DOI] [PubMed] [Google Scholar]
- 79. Nuxoll AS, Halouska SM, Sadykov MR, Hanke ML, Bayles KW, Kielian T, Powers R, Fey PD. 2012. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog 8:e1003033. doi: 10.1371/journal.ppat.1003033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shafer WM, Iandolo JJ. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect Immun 25:902–911. doi: 10.1128/iai.25.3.902-911.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dörries K, Lalk M. 2013. Metabolic footprint analysis uncovers strain specific overflow metabolism and D-Isoleucine production of Staphylococcus aureus COL and HG001. PLoS One 8:e81500. doi: 10.1371/journal.pone.0081500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wetzstein M, Völker U, Dedio J, Löbau S, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. 1992. Cloning, sequencing, and molecular analysis of the dnak locus from Bacillus subtilis. J Bacteriol 174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hilker R, Stadermann KB, Schwengers O, Anisiforov E, Jaenicke S, Weisshaar B, Zimmermann T, Goesmann A. 2016. Readxplorer 2-detailed read mapping analysis and visualization from one single source. Bioinformatics 32:3702–3708. doi: 10.1093/bioinformatics/btw541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. ROSENBLUM ED, TYRONE S. 1964. Serology density and morphology of staphylococcal phages. J Bacteriol 88:1737–1742. doi: 10.1128/jb.88.6.1737-1742.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fritsch VN, Loi VV, Busche T, Tung QN, Lill R, Horvatek P, Wolz C, Kalinowski J, Antelmann H. 2020. The alarmone (p)ppGpp confers tolerance to oxidative stress during the stationary phase by maintenance of redox and iron homeostasis in Staphylococcus aureus. Free Radic Biol Med 161:351–364. doi: 10.1016/j.freeradbiomed.2020.10.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Van Loi V, Antelmann H. 2020. Method for measurement of bacillithiol redox potential changes using the Brx-rogfp2 redox biosensor in Staphylococcus aureus. MethodsX 7:100900. doi: 10.1016/j.mex.2020.100900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tomoyasu T, Arsène F, Ogura T, Bukau B. 2001. The C terminus of sigma(32) is not essential for degradation by FtsH. J Bacteriol 183:5911–5917. doi: 10.1128/JB.183.20.5911-5917.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curves for phenotype analyses, Brx-roGFP2 biosensor experiments, and aggregation assays.
Transcriptome datasets of S. aureus after exposure to HOSCN and HOCl stress.
Bacterial strains, phages, plasmids, and oligonucleotide primers.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplemental materials. The transcriptome sequencing raw data files are available in the ArrayExpress database under accession number E-MTAB-13314 for the HOSCN stress data set and as E-MTAB-13313 for the HOCl stress data set used for comparison.