ABSTRACT
The polymorphism at amino acid 17 of quinolone resistance-determining region of GyrA has been stated with a potential role in fluoroquinolone susceptibility in different mycobacterial species. However, no study has provided dependable evidence so far. Here, we verified that gene-edited Mycobacterium abscessus mutants bearing Ser/Gly at this position were more susceptible to fluoroquinolones than their parent strain and the revertant that supports mycobacteria containing Ser/Gly at this position were more susceptible to fluoroquinolones than those containing Ala.
IMPORTANCE
Fluoroquinolones (FQs) play a key role in the treatment regimens against tuberculosis and non-tuberculous mycobacterial infections. However, there are significant differences in the sensitivities of different mycobacteria to FQs. In this study, we proved that this is associated with the polymorphism at amino acid 17 of quinolone resistance-determining region of Gyrase A by gene editing. This is the first study using CRISPR-associated recombination for gene editing in Mycobacterium abscessus to underscore the contribution of the amino acid substitutions in GyrA to FQ susceptibilities in mycobacteria.
KEYWORDS: fluoroquinolone, mycobacteria, intrinsic resistance, gene editing, Mycobacterium abscessus
OBSERVATION
Fluoroquinolones (FQs) are very important and extensively used class of synthetic antibacterial agents that are recommended for drug-resistant Mycobacterium tuberculosis and some non-tuberculous mycobacteria such as macrolide-resistant Mycobacterium abscessus complex (1 – 3). Moxifloxacin (MOX) is known to be an effective FQ and a key component of the new first-line regimen which can shorten the treatment duration of drug-sensitive tuberculosis from 6 months to 4 months (4). The target of the FQs in mycobacteria is type II topoisomerase, which consists of two subunits, GyrA and GyrB, that form the catalytically active A2B2 heterotetrameric structure (5). In mycobacteria, the FQs can interact with the cleaved DNA together with the GyrA and GyrB proteins to stabilize a cleavage complex and inhibit the religation of the cleaved DNA which potentially results in lethal double-strand DNA breaks in the genome (6). Mutations in quinolone resistance-determining region (QRDR) of gyrA and gyrB genes have been proven to lead to FQ resistance (7, 8).
Interestingly, most mycobacteria are intrinsically less susceptible to FQs than other bacteria, such as Escherichia coli, and the levels of susceptibility to FQs differ markedly for different mycobacterial species (9). Previous studies found the amino acid substitution at position 17 in QRDRGyrA (83 in E. coli; 90 in M. tuberculosis; 92 in M. abscessus of GyrA) may be involved in the FQ susceptibility by analyzing the sequences of the QRDR in GyrA and GyrB in different mycobacterial species as well as other bacteria (9, 10). The presence of an Ala at position 17 of QRDRGyrA in most of the mycobacterial species (such as M. tuberculosis and M. abscessus) and a Ser in the three other mycobacterial species (Mycobacterium peregrinum, Mycobacterium fortuitum, and Mycobacterium aurum) or E. coli associated with the minimal inhibitory concentrations (MICs) of quinolones, suggesting this amino acid residue might be a crucial determinant of different susceptibilities to quinolones among mycobacteria (Fig. 1A) (9, 10). However, to date, no direct molecular experimental evidence supports this hypothesis. The success of mycobacteria gene editing with the development of CRISPR (11) allowed us to apply this new tool for exploring the correlation between FQ susceptibility patterns and the amino acid substitutions in mycobacteria.
Fig 1.
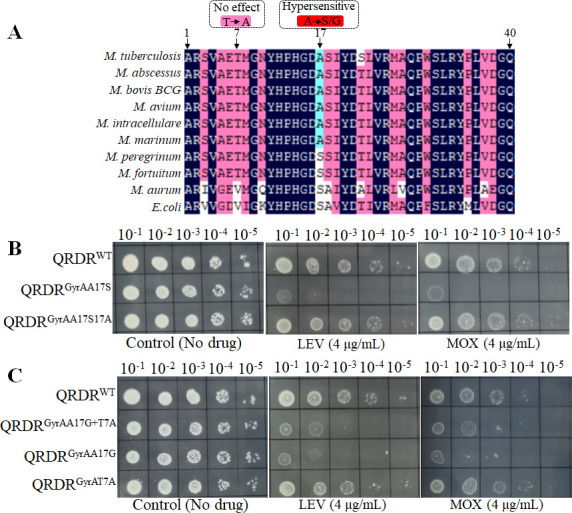
The amino acid 17 in QRDR of GyrA contributes to intrinsic susceptibility of FQs in mycobacteria. (A) Alignment of the peptide sequences of the QRDR of GyrA from mycobacterial species and E. coli. (B and C) Drug susceptibility of different M. abscessus strains to MOX and levofloxacin. Tenfold serial dilutions of M. abscessus strains grown to OD600 of 0.7 were spotted on Middlebrook 7H10 containing indicated concentrations of antibiotics. Plates were incubated for 3 days. Representative data from three independent experiments are shown.
M. abscessus is a rapidly growing non-tuberculous mycobacterium responsible for a wide variety of human diseases, including chronic pulmonary diseases and several extrapulmonary diseases such as soft tissue, skin, and central nervous system infections (12). These infections are difficult to treat with the standard antibacterial therapy due to their high-level intrinsic resistance to most antibiotics (13). Here, we used M. abscessus as a model organism for studying the correlation between FQ susceptibility patterns and amino acid sequences of QRDRGyrA in mycobacteria. To elucidate the contribution of the amino acid substitution located at position 17 of QRDRGyrA, the GyrA was edited to alter the Ala to Ser at position 17 of QRDRGyrA (QRDRGyrAA17S) in M. abscessus using CRISPR-associated recombineering as described previously for other mycobacteria (11). The primers used in this study are listed in Table S1. Interestingly, QRDRGyrAA17S exhibited a markedly enhanced sensitivity to levofloxacin (LEV) and MOX compared to its parent M. abscessus strain (QRDRWT) as shown in Fig. 1B and Table 1. The MICs of both LEV and MOX to the QRDRGyrAA17S were 1/4 of that of MabWT (Table 1), whereas QRDRGyrAA17S17A, the GyrA of which was edited to alter the Ser back to Ala at position 17 of QRDRGyrA in QRDRGyrAA17S, recovered the FQ resistance level to that of QRDRWT (Fig. 1B and Table 1), thus confirming that Ala17 in QRDRGyrA is critical for the FQs resistance in M. abscessus. Susceptibility of QRDRGyrAA17S to other two types of non-FQ antibiotics remained unchanged (Table 1), indicating that the substitution of this amino acid specifically affects the LEV and MOX susceptibility. Additionally, Aubry et al. found that some M. tuberculosis isolates bearing a combination of T80A and A90G (Thr7Ala and Ala17Gly in QRDRGyrA) substitutions were hypersusceptible to ofloxacin (14). Therefore, we also constructed a M. abscessus mutant (QRDRGyrAA17G+T7A) containing Thr7Ala and Ala17Gly in QRDRGyrA double mutations in GyrA. Similar to QRDRGyrAA17S, QRDRGyrAA17G+T7A also showed the significantly increased sensitivity to LEV and MOX (Fig. 1C and Table 1). To further investigate the contribution of Thr7Ala and Ala17Gly in QRDRGyrA to the susceptibility of FQs, the GyrA was edited to alter the Thr to Ala at position 7 in QRDRGyrA (QRDRGyrAT7A) and Ala to Gly at position 17 in QRDRGyrA (QRDRGyrAA17G) separately. We observed that the Ala17Gly mutation in QRDRGyrA confers hypersensitivity to FQs but not Thr7Ala (Fig. 1C and Table 1). These results imply that Ala17 in QRDRGyrA of mycobacteria plays a key role in susceptibility to FQs and hints that the mycobacterium bearing Ala17Gly or Ala17Ser amino acid substitution in QRDRGyrA may be hypersensitive to FQs.
TABLE 1.
MICs of various drugs for different M. abscessus strains a
| Antibiotics | M. abscessus strains/MICs (μg/mL) | |||||
|---|---|---|---|---|---|---|
| QRDRWT | QRDRGyrAA17S | QRDRGyrAA17S17A | QRDRGyrAA17G+T7A | QRDRGyrAA17G | QRDRGyrAT7A | |
| Levofloxacin | 16 | 4 | 16 | 4 | 4 | 16 |
| Moxifloxacin | 8 | 2 | 8 | 4 | 4 | 8 |
| Rifabutin | 4 | 4 | 4 | 4 | 4 | 4 |
| Amikacin | 4 | 4 | 4 | 4 | 4 | 4 |
Broth microdilution method was used to determine the MICs. The MIC was defined as the lowest drug concentration that prevented visible bacterial growth. The experiment was performed in triplicate and repeated twice.
To the best of our knowledge, this is the first detailed study to underscore the contribution of the amino acid substitutions in GyrA to FQs resistance in mycobacteria using the CRISPR-associated recombintion for gene editing in M. abscessus. Our observations are in strong agreement with a previous study in which it was found by peptide sequences alignment that the amino acid at position 17 of QRDRGyrA was likely involved in the intrinsic resistance of mycobacteria to quinolones (9). In addition, although both Ala17Gly and Ala17Ser of QRDRGyrA could cause hypersensitivity of M. abscesses to FQs, the susceptibility of MOX to QRDRGyrAA17S and QRDRGyrAA17G is different. Following the Clinical and Laboratory Standards Institute (CLSI) guidelines, the breakpoint for MOX resistance in M. abscessus was determined to be 4 µg/mL (15). Consequently, QRDRGyrAA17G is still classified as resistant to MOX. The MICs of FQs to the hypersensitive M. abscessus mutants (QRDRGyrAA17S or QRDRGyrAA17G) are still higher than that to M. tuberculosis, which indicates that besides the contribution of the 17Ala of QRDRGyrA in M. abscessus, other factors leading to higher MICs of FQs to M. abscessus may exist. A recent study analyzed the gyrA and gyrB of FQs-resistant M. abscessus isolates but found no mutation in them, which also suggested that besides gyrA and gyrB, other mechanisms also contribute to FQs resistance in M. abscessus (16).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (2021YFA1300904), partially by the National Natural Science Foundation of China (NSFC 81973372, 21920102003), the Joint Research Health Research Council of New Zealand (HRC 20/1211)-NSFC Collaboration grant (82061128001), and the Chinese Academy of Sciences (154144KYSB20190005, YJKYYQ20210026). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
Jinxing Hu, Email: hujinxing2000@163.com.
Jianxiong Liu, Email: ljxer64@qq.com.
Tianyu Zhang, Email: zhang_tianyu@gibh.ac.cn.
Gyanu Lamichhane, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02809-23.
Oligonucleotides used in this study.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Jia Y, Zhao L. 2021. The antibacterial activity of fluoroquinolone derivatives: an update (2018-2021). Eur J Med Chem 224:113741. doi: 10.1016/j.ejmech.2021.113741 [DOI] [PubMed] [Google Scholar]
- 2. Mirzayev F, Viney K, Linh NN, Gonzalez-Angulo L, Gegia M, Jaramillo E, Zignol M, Kasaeva T. 2021. World health organization recommendations on the treatment of drug-resistant tuberculosis, 2020 update. Eur Respir J 57:2003300. doi: 10.1183/13993003.03300-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. 2020. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J 56:2000535. doi: 10.1183/13993003.00535-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dorman SE, Nahid P, Kurbatova EV, Phillips PPJ, Bryant K, Dooley KE, Engle M, Goldberg SV, Phan HTT, Hakim J, Johnson JL, Lourens M, Martinson NA, Muzanyi G, Narunsky K, Nerette S, Nguyen NV, Pham TH, Pierre S, Purfield AE, Samaneka W, Savic RM, Sanne I, Scott NA, Shenje J, Sizemore E, Vernon A, Waja Z, Weiner M, Swindells S, Chaisson RE, AIDS Clinical Trials Group, Tuberculosis Trials Consortium . 2021. Four-month rifapentine regimens with or without moxifloxacin for tuberculosis. N Engl J Med 384:1705–1718. doi: 10.1056/NEJMoa2033400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aldred KJ, Blower TR, Kerns RJ, Berger JM, Osheroff N. 2016. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: enhancing drug activity against wild-type and resistant gyrase. Proc Natl Acad Sci U S A 113:E839–46. doi: 10.1073/pnas.1525055113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collin F, Karkare S, Maxwell A. 2011. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl Microbiol Biotechnol 92:479–497. doi: 10.1007/s00253-011-3557-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, Kreiswirth B, Cole ST, Jacobs WR, Telenti A. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother 38:773–780. doi: 10.1128/AAC.38.4.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67:819–831. doi: 10.1093/jac/dkr566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guillemin I, Jarlier V, Cambau E. 1998. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob Agents Chemother 42:2084–2088. doi: 10.1128/AAC.42.8.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guillemin I, Cambau E, Jarlier V. 1995. Sequences of conserved region in the A subunit of DNA gyrase from nine species of the genus Mycobacterium: phylogenetic analysis and implication for intrinsic susceptibility to quinolones. Antimicrob Agents Chemother 39:2145–2149. doi: 10.1128/AAC.39.9.2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan M-Y, Yan H-Q, Ren G-X, Zhao J-P, Guo X-P, Sun Y-C. 2017. CRISPR-Cas12A-assisted recombineering in bacteria. Appl Environ Microbiol 83:e00947-17. doi: 10.1128/AEM.00947-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee M-R, Sheng W-H, Hung C-C, Yu C-J, Lee L-N, Hsueh P-R. 2015. Mycobacterium abscessus complex infections in humans. Emerg Infect Dis 21:1638–1646. doi: 10.3201/2109.141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578 [DOI] [PubMed] [Google Scholar]
- 14. Aubry A, Veziris N, Cambau E, Truffot-Pernot C, Jarlier V, Fisher LM. 2006. Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob Agents Chemother 50:104–112. doi: 10.1128/AAC.50.1.104-112.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown-Elliott BA, Woods GL. 2019. Antimycobacterial susceptibility testing of nontuberculous mycobacteria. J Clin Microbiol 57:e00834-19. doi: 10.1128/JCM.00834-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S-Y, Jhun BW, Moon SM, Shin SH, Jeon K, Kwon OJ, Yoo IY, Huh HJ, Ki C-S, Lee NY, Shin SJ, Daley CL, Suh GY, Koh W-J. 2018. Mutations in gyrA and gyrB in moxifloxacin-resistant Mycobacterium avium complex and Mycobacterium abscessus complex clinical isolates. Antimicrob Agents Chemother 62:e00527-18. doi: 10.1128/AAC.00527-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used in this study.
An accounting of the reviewer comments and feedback.


