Abstract
Radiomics is an emerging field that aims to extract and analyse a comprehensive set of quantitative features from medical images. This scoping review is focused on MRI‐based radiomic features for the molecular profiling of breast tumours and the implications of this work for predicting patient outcomes. A thorough systematic literature search and outcome extraction were performed to identify relevant studies published in MEDLINE/PubMed (National Centre for Biotechnology Information), EMBASE and Scopus from 2015 onwards. The following information was retrieved from each article: study purpose, study design, extracted radiomic features, machine learning technique(s), sample size/characteristics, statistical result(s) and implications on patient outcomes. Based on the study purpose, four key themes were identified in the included 63 studies: tumour subtype classification (n = 35), pathologically complete response (pCR) prediction (n = 15), lymph node metastasis (LNM) detection (n = 7) and recurrence rate prediction (n = 6). In all four themes, reported accuracies widely varied among the studies, for example, area under receiver characteristics curve (AUC) for detecting LNM ranged from 0.72 to 0.91 and the AUC for predicting pCR ranged from 0.71 to 0.99. In all four themes, combining radiomic features with clinical data improved the predictive models. Preliminary results of this study showed radiomics potential to characterise the whole tumour heterogeneity, with clear implications for individual‐targeted treatment. However, radiomics is still in the pre‐clinical phase, currently with an insufficient number of large multicentre studies and those existing studies are often limited by insufficient methodological transparency and standardised workflow. Consequently, the clinical translation of existing studies is currently limited.
Keywords: biomarkers, breast cancer, MRI, radiomics, texture analysis
This article contributes to the ongoing development of breast cancer detection and classification by discussing the novel field of radiomics, which can aid in personalised, precision medicine for patients. This review presents current research that considers radiomics for breast cancer subtype classification, prediction of pathologically complete response, detection of lymph node metastasis and recurrence prediction, all of which impact treatment and patient outcomes.
Introduction
Breast tumours are characteristically heterogeneous in biological makeup, contributing to treatment failure and poor prognosis for patients. They also have a variety of clinical presentations and subtypes, each responding uniquely to different treatments. 1 Current methods of molecular and pathological testing through biopsy sampling are limited in demonstrating the entire tumour and intratumoral heterogeneity as they only extract part of the tumour's tissue. Biopsies are invasive procedures that cannot be repeated on the same sample for monitoring disease progression. 2 Hence, there is an emerging need to identify newer, non‐invasive imaging techniques, such as radiomics, to provide a better understanding of tumour biology and molecular characteristics.
Due to its wide availability and relatively low cost, mammographic imaging is the method of choice for screening and prevention at a national level; however, studies have shown that its sensitivity and specificity range from fair to good, measuring between 70% and 90%. 2 Its positive predictive value can be as low as 15%, which indicates how many patients with positive results actually have cancer. 2 On the other hand, magnetic resonance imaging (MRI) has excellent soft tissue contrast and high sensitivity that can reach 100%. 3 Unlike mammography, MRI does not rely on ionising radiation to produce images, rather it uses a magnetic field to capture interactions with structures in the body and is unaffected by tissue density. 4 MR breast imaging is multiparametric, meaning a range of sequences are included, such as dynamic contrast‐enhanced (DCE), diffusion‐weighted imaging (DWI) and T1‐ and T2‐weighted (W) images. Each sequence presents information on the functional and anatomical properties of the tumour, including the mobility of water molecules within tissue and angiogenesis. 5 These characteristics can be indicative of tumour growth, spread and malignancy, all of which assist in making prognostic predictions and determining the aggressiveness of treatment required. 6 Conventionally, image‐based features are qualitatively evaluated by considering size, shape and type of enhancement, leaving behind hidden and unused data. 7 Quantitative analysis of image data is required to predict the tumour's molecular profile and treatment response and achieve personalised medicine for breast cancer patients. This can be accomplished through radiomics. 1
Radiomics is an emerging field of research that extracts existing medical imaging features through mathematical analysis and computer‐aided detection. 8 Its application allows for tumour characterisation and molecular profiling, optimises patient outcome through personalised treatment plans and monitors tumour evolution. 9 Radiomics offers advantages through improved sensitivity and specificity of diagnosis, prognosis predictions, disease management and disease evolution. It is particularly valuable in non‐invasive applications such as breast cancer screening and prevention. 2 The radiomics process is complex and variable, beginning with acquiring high‐quality images that do not contain artefacts or motion. Although obtaining images without artefacts is ideal, it is challenging to achieve. Therefore, features could be developed and selected that are more robust to artefacts and motion. A region of interest (ROI) containing the tumour is then segmented by one of three methods outlined in Table 1.
Table 1.
Method of image segmentation. 5
| Segmentation method | Description |
|---|---|
| Manual | Radiologist manually traces the outline of the tumour but it is time‐consuming for large datasets |
| Semi‐automatic | Manual seed planting in the tumours’ centre followed by automatic border drawing by computer algorithms |
| Automatic | Uses highly trained computer algorithms to delineate the tumour and is more accurate for detecting the lesion versus background. Features are more reproducible with automatic methods |
The ROI is analysed to enable quantitative features to be extracted. Current tumour evaluation focuses on qualitative features (although quantitative parameters can also be extracted), including density, microcalcifications and necrosis, which is limited by minimalistic sampling of tumour tissue. 10 On the other hand, radiomic features analyse the pixel gray values and patterns within the medical image to provide information on the tumours’ makeup, its relationship to surrounding tissues and its microenvironment. 10 There are a number of radiomic features, which are categorised into groups, such as those listed in Table 2.
Table 2.
Radiomic features. 11
| Feature category | Description |
|---|---|
| Morphology or shape | Geometric properties of the ROI, for example, volume, maximum surface, tumour compactness and sphericity |
| First‐order statistics or histogram‐based features | Distribution of pixel/voxel values, for example, mean, median, uniformity, kurtosis, skewness and entropy |
| Second‐order statistics or texture | Contrast values between neighbouring pixels and measures intratumoral heterogeneity, for example, gray‐level co‐occurrence matrix (GLCM) and gray‐level run matrix (GLRM) |
| Higher‐order statistics | Requires filters or transforms to be applied to images and assesses patterns and details, for example, Fourier analysis |
| Structural features | Describing the complexity of the structures, for example, fractal dimension |
| Kinetic | Assesses washout rates and heterogeneity of contrast enhancement, for example, time to peak enhancement |
As thousands of features can be extracted from a medical image, they are often limited or clustered to meet the desired aim, which can include disease diagnosis, predicting recurrence or metastatic spread. 12 These quantitative features can be combined with clinicopathological information (e.g. age, histological grade and biomarkers) to form a radiomics signature or nomogram. This signature/nomogram can provide further information on the tumour and improve the model's predictive ability.
Currently, the majority of radiomic studies in breast cancer research focus on the classification of tumours into benign or malignant lesions. Given the increasing need and desire for personalised medicine, there is a growing interest in the use of radiomics for molecular classification of breast tumours based on biological markers. These are the oestrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor 2 (HER2) and Ki67 proliferation marker. Through immunohistochemical (IHC) testing, numeric values and measures of positivity can be made to classify tumours into four subtypes: luminal A, luminal B, HER2‐enriched and triple‐negative breast cancer. Each of these subtypes has different treatment options that they best respond to and differing rates of aggression. For instance, luminal A tumours have a better prognosis and long‐term disease‐free survival compared to luminal B. 13
The past few decades have seen significant improvements in the survival rates of breast cancer patients. However, there are still patients who are ineffectively treated or remain undertreated, creating additional physical and emotional burdens. Given the fact that anticancer medications usually have severe side effects, identifying patients who would not benefit from specific treatments and avoiding needless exposure to toxicity is critical. 14 Radiomics has this potential and can also identify other prognostic and predictive factors (e.g. aggressiveness and metastatic risk). Furthermore, identifying normal versus pathogenic biological processes and pharmacological responses to a specific treatment allows for proper risk stratification of patients to occur. 15 Therefore, the purpose of this scoping review was to investigate the range of existing studies in the field of radiomics for breast cancer classification in magnetic resonance imaging. Focus was placed on four themes of research derived from the literature, which were radiomics for tumour subtype classification, prediction of pathologically complete response (pCR), detection of lymph node metastasis and predicting recurrence.
Methods
Protocol
The protocol used for this review was the Preferred Reporting Items for Systematic Reviews and Meta‐Analysis extension for Scoping Reviews (PRISMA‐ScR) checklist.
Eligibility criteria
Articles were included in this review if they were peer‐reviewed, full text and written in English to facilitate data collection. Only original research studies were included. Given that radiomic methods, MRI sequences and MRI scanners are continually improving, articles published from 2015 onwards were contemporary for this review. Additionally, this review was limited to MRI imaging only as radiomic techniques have been widely applied to this imaging modality and have produced significant results. MRI radiomics have been used to successfully describe phenotype characteristics from DCE and DWI imaging and have accurately predicted prognostic status in breast cancer patients. 3
Included studies utilised radiomic methods or, its synonym, texture analysis, for the purpose of classifying breast tumours into molecular subtypes: predicting pCR, detecting lymph node metastasis or predicting recurrence rates. All studies included patients who had a breast MRI scan performed prior to commencing any treatment (e.g. chemotherapy or surgery) and before a biopsy sample was taken. Hence, this review could investigate the role of radiomics in pre‐treatment predictions, which reduce the burden of unnecessary treatments and toxins for breast cancer patients.
Sources and search strategy
Three databases were explored to find eligible articles, which were Medline, Embase and Scopus. Three databases were deemed sufficient as a saturation point was reached, whereby no original articles were found upon investigating further databases. Additional studies were found by searching for grey literature on Google Scholar and examining reference lists of included studies. Any inconsistencies in the inclusion and classification of articles were resolved through discussion between authors.
The search strategy for Medline and Embase included the key terms and their combinations, as listed in Table 3.
Table 3.
Medline and embase search strategy.
| 1 | Breast cancer .mp | 12 | ?estrogen receptor .mp |
| 2 | Breast neoplasm/ | 13 | 5 or 6 or 7 or 8 or 9 or 10 or 11 or 12 |
| 3 | Breast tumo?r* .mp | 14 | Radiogenomic* .mp |
| 4 | 1 or 2 or 3 | 15 | Radiomic* .mp |
| 5 | Ki67 antigen/ or Ki67.mp | 16 | Texture analysis .mp |
| 6 | Ki 67 .mp | 17 | 14 or 15 or 16 |
| 7 | HER2 .mp | 18 | MRI .mp |
| 8 | Human epidermal growth factor 2 .mp | 19 | Magnetic resonance imaging .mp |
| 9 | Immunohistochemistry/ | 20 | 18 or 19 |
| 10 | Biomarkers, tumour/ or tumo?r* biomarker* .mp | 21 | 4 and 13 and 17 and 20 |
| 11 | ER .mp | ||
Scopus was searched using an advanced search strategy; (TITLE‐ABS‐KEY (radiomic* OR radiogenomic* OR ‘texture analysis’) AND TITLE‐ABS‐KEY (MRI OR ‘magnetic resonance imaging’) AND TITLE‐ABS‐KEY (Ki67 OR HER2 OR ‘? estrogen receptor’ OR ‘progesterone receptor’) AND TITLE‐ABS‐KEY (‘breast cancer’) AND PUBYEAR >2014).
A combination of these keywords was used to conduct searches on Google Scholar and select relevant articles from reference lists.
Data charting
A descriptive summary of reported results was presented in data charting tables (Tables 4a–4d) to reflect both the heterogenous findings and common themes. Included articles were divided into four themes: subtype classification, pCR prediction, lymph node metastasis detection and recurrence prediction, each being charted separately. The data charting table was drafted and tested on five articles before edits were made by all reviewers. Once the final design was agreed upon, the remainder of the data charting process was completed.
Synthesis of results
Critical appraisal of included studies and results is not the aim of scoping reviews. 16 Hence, study characteristics of all included articles were described and presented in the data charting table to identify common themes and highlight numeric findings. A thematic analysis was also performed for all four key themes to discuss the implications of the radiomics technique in clinical practice and to identify areas of future research.
Results
Selection of sources of evidence
The final literature search resulted in 252 articles from database searching and an additional nine from other sources. All articles were exported to Endnote X9, where duplicates were removed, leaving 134 articles for screening. They were first screened based on their title and abstract, where a further 56 articles were excluded. The full texts of the remaining 78 articles were read to determine eligibility based on the specified criteria (Section “Eligibility criteria”). All four reviewers were in consensus about the excluded 15 articles, resulting in 63 articles remaining to be included in the scoping review.
The selection process is outlined in Figure 1.
Figure 1.
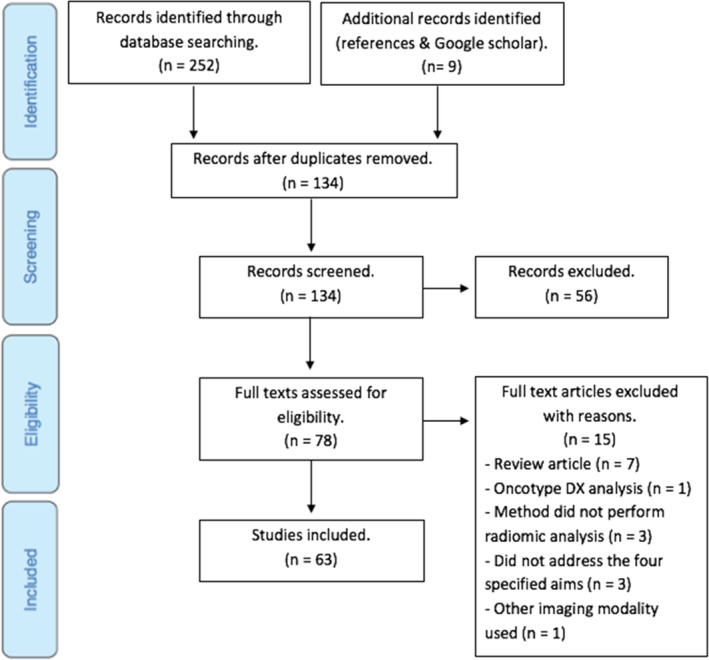
PRISMA flow chart of study selection process.
Characteristics of sources of evidence
Table 4.
(a) Subtype classification (n = 35), (b) pCR prediction (n = 15), (c) lymph node metastasis detection (n = 7), (d) recurrence prediction (n = 6).
| Ref | MRI series | Study aim | Sub (V) 1 | Segmentation method | Radiomic features | Results |
|---|---|---|---|---|---|---|
| (a) | ||||||
| 17 | Multiparametric (1.5T) | Predict Ki67 status in breast cancer | 328(Y) | Manual (3D) | Higher‐order statistics 2 | Multiparametric model performed best with AUC 0.888, accuracy 0.971, sensitivity 0.993 and specificity 0.892 |
| 18 | Multiparametric (3T) | Predict Ki67 status in breast cancer | 144(N) | Semi‐automatic (2D) | Morphological; first‐ and second‐order statistics | Combining DWI and DCE (subtracted fifth post contrast image) produces best results with AUC 0.811 |
| 19 | DWI (3T) | Predict histological grade and Ki67 status using radiomic and deep learning methods | 322(Y) | Manual (2D) | Morphological | Prediction Ki67 expression had AUC 0.818 when using DWI and super resolution DWI images |
| 20 | Multiparametric (1.5T) | Differentiate molecular subtypes and predict Ki67 and HER2 status | 98(Y) | Semi‐automatic (3D) | First‐ and higher‐order statistics | Differentiated HER2+ and TNBC with AUC 0.97. Predicting Ki67 expression had AUC 0.81 |
| 21 | DCE (3T) | Predict Ki67 expression | 159(N) | Automatic (2D) | Morphological; first‐ and second‐order statistics | Classifying low and high Ki67 tumours occurred with statistical significance (P < 0.05) using a range of radiomic features |
| 22 | T2W images (1.5T) | Predict Ki67 status in breast cancer | 318(Y) | Manual (2D) | Morphological; first‐ and second‐order statistics | AUC, sensitivity, specificity and accuracy of the training set was 0.762, 0.72, 0.70 and 0.715 respectively |
| 23 | DWI (2 X 3T) | Develop a model for predicting Ki67 index | 128(Y) | Semi‐automatic (3D) | Morphological; first‐order statistics | Training cohort had AUC 0.75, accuracy 0.71, sensitivity 0.78 and specificity 0.72. Validation cohort had AUC 0.72, accuracy 0.7, sensitivity 0.71 and specificity 0.7 |
| 24 | DCE (1.5T) | Predict Ki67 status in breast cancer | 154(Y) | Manual (3D) | First‐ and second‐order statistics | Model produced AUC 0.785 in training and AUC 0.849 in validation cohort |
| 25 | DCE (3T) | Predict Ki67 status in breast cancer | 159(N) | Semi‐automatic (2D) | Morphological; first‐ and second‐order statistics | Texture features were most significant (P < 0.05). Model performed with AUC 0.773, accuracy 0.757, sensitivity 0.777 and specificity 0.769 |
| 26 | DCE (3T) | Predict Ki67 status in ER+ breast cancer patients based on heterogeneous tumour subregions | 77(N) | Semi‐automatic (3D) | Second‐order statistics | Model prediction based on the tumour subregion performed significantly (P < 0.01) better compared to the entire tumour |
| 27 | DCE (3T) | Predict HER2 and Ki67 status based on peritumoral and intratumoral regions | 351(Y) | Semi‐automatic (2D) | Morphological; first‐ and second‐order statistics 2 | Combined peritumoral and intratumoral model did not produce statistically significant improvements. AUC for predicting HER2 and Ki67 status were 0.713 and 0.749 respectively |
| 28 | DCE (1.5T) | Identify HER2 enriched tumours | 259(N) | Semi‐automatic (3D) | Morphological; first‐order statistics | HER2+ tumours had lower volume, smaller longest axial diameter, longest volumetric diameter, higher minimum signal intensity and lower entropy |
| 29 | Multiparametric (3T) | Predict HER2 status of patients with breast cancer | 306(Y) | Manual (3D) | Morphological; first‐ and second‐order statistics | Multiparametric model had the strongest results. AUC 0.8; sensitivity 88.7%; specificity 76.2% and accuracy 79.5% |
| 30 | DCE (3T) | Predict HER2 status based on tumour subregions | 76(N) | Semi‐automatic (2D) | First‐, second‐ and higher‐order statistics | Classifier performed best when analysing tumour subregions compared to the entire tumour, with AUC 0.929 and 0.847 respectively |
| 31 | DCE (3T) | Differentiate HER2 2+ status in breast tumours | 73(N) | Manual (2D) | First‐ and second‐order statistics | AUC 0.865, sensitivity 88.90%, specificity 73% and accuracy 81.06% |
| 32 | DCE (3T) | Predict HER2 2+ status | 92(N) | Semi‐automatic (2D) | Second‐order statistics | Best performance occurred with post‐contrast enhancement images with an AUC of 0.89 |
| 33 | DWI (3T) | Identify triple negative versus non‐triple negative tumours | 390(Y) | Manual (2D) | Morphological; First‐ and higher‐order statistics | Accuracy of discrimination between TNBC and non‐TNBC was 95.40% in training and 83.4% in validation cohort |
| 34 | DCE (3T) | Identify triple negative breast cancer | 84(N) | Semi‐automatic (3D) | First‐, second‐ and higher‐order statistics 2 | Classifying TNBC versus all others with a combined BPE and radiomics model improved prediction from AUC 0.782 to 0.878 |
| 35 | DCE (3T) | Identify ER positive breast tumours | 51(N) | Manual (2D) | Morphological; first‐order statistics | First‐order statistics features were significant for predicting ER+ tumours, with AUC >0.8 |
| 36 | Multiparametric (1.5T) | Identify characteristics of ER+ tumours | 75(N) | Manual (2D) | First‐ and higher‐order statistics | Higher grade tumours had significantly higher uniformity and lower entropy in T1W images, with the opposite true for T2W images |
| 37 | DWI (3T) | Evaluate breast cancer receptor status and molecular subtypes | 91(N) | Manual (2D) | First‐ and second‐order statistics | Accuracy of luminal B vs luminal A, HER2, TNBC and all others was 91.5%, 100%. 89.3% and 91.1% respectively |
| 38 | Multiparametric (3T) | Assess breast cancer receptor status and molecular subtypes | 91(Y 3 ) | Manual (2D) | Morphological; First‐ and second‐order statistics | Accuracy of luminal B vs luminal A, TNBC and all others was 84.2%, 83.9% and 89% respectively. Accuracy of HER2+ vs. all others was 81.3% |
| 39 | Multiparametric (1.5T) | Differentiate ER status in luminal A and luminal B tumours | 27 4 (N) | Manual (2D) | Second‐order statistics | Prediction of luminal B tumours had AUC 0.878, sensitivity 91.7% and specificity 86.7% |
| 40 | Multiparametric (1.5T & 3T) | Predict breast cancer molecular subtypes | 148(Y 5 ) | Manual (2D) | Second‐order statistics | Predictive model had an accuracy of 74.7% and AUC 0.816. In prospective validation cohort, accuracy was 72.5% and AUC 0.823 |
| 41 | Multiparametric (3T) | Differentiate TNBC from other subtypes | 134(N) | Semi‐automatic (3D) | First‐order statistics | Best performance noted when distinguishing between TNBC and HER2+, with AUC 0.763, sensitivity 86.4% and specificity 72.2% |
| 42 | DWI (3T) | Predict clinical‐pathological subtypes | 112(N) | Manual (3D) | First‐ and second‐order statistics | Accuracy for predicting individual subtypes is 97% luminal A, 100% luminal B (HER2−), 94% luminal B (HER2+), 92% HER2+ and 100% TNBC |
| 43 | DCE (1.5T) | Distinguish between molecular subtypes using a multi‐institutional data set | 91 4 (N) | Semi‐automatic (3D) | Morphological; first‐ and second‐order statistics | Model distinguished between ER+ vs ER‐ with AUC 0.89, PR+ vs PR‐ with AUC 0.69, HER2+ vs. HER2− with AUC 0.65 and TNBC vs all other with AUC 0.67 |
| 44 | DCE (1.5T) | Predict molecular subtype | 96(Y) | Semi‐automatic (3D) | Morphological; first‐ and second‐order statistics; kinetic 2 | Model had overall prediction AUC of 0.869. Discrimination among luminal A, luminal B, HER2 and TNBC subtypes had AUC 0.867, 0.786, 0.888 and 0.923 respectively |
| 45 | DCE (1.5T & 3T) | Identify relationship between radiomic features and molecular subtypes | 275(N) | Semi‐automatic (2D) | Morphological; first‐order statistics; kinetic | Model predicted luminal A (P = 0.0007) and luminal B (P = 0.0063) subtypes with statistical significance |
| 46 | DCE (1.5T & 3T) | Association of imaging phenotypes with molecular subtypes | 922 4 (Y) | Semi‐automatic (3D) | Morphological; first‐order statistics | Statistically significant results for distinguishing luminal A vs. all others (AUC 0.697) and TNBC vs. all others (AUC 0.654) |
| 13 | DCE (3T) | Predict histological outcomes and molecular phenotypes | 49(N) | Semi‐automatic (3D) | Morphological; first‐ and second‐order statistics | Discriminating between subtypes produced AUC >0.8 for all categories. Best results for PR+/PR− with AUC 0.875 |
| 47 | Multiparametric (3T) | Classify IHC subtypes based on machine learning analysis | 134(Y) | Semi‐automatic (3D) | Morphological; second‐order statistics | Accuracy of predicting TNBC vs non‐TNBC ranged from 72.4–91% |
| 48 | DCE (1.5T & 3T) | Distinguish between molecular subtypes using deep learning models | 270(N) | Semi‐automatic (2D) | Second‐order statistics | Deep learning model performed better than training from scratch and transfer learning, with AUC's of 0.65, 0.58 and 0.6 respectively |
| 49 | DCE (1.5T) | Classify IHC subtypes | 60(N) | Semi‐automatic (2D) | Morphological; first‐ and second‐order statistics | Model classified luminal A, HER2+ and TNBC with statistical significance (P < 0.05) |
| 50 | Multiparametric (1.5T & 3T) | Classify molecular subtypes | 107(N) | Manual (2D) | Morphological; first‐ and second‐order statistics | Accuracy of classification was best on the 3T scanner with 86.4% on contrast enhanced T1W and 88.6% on DWI |
| (b) | ||||||
| 51 | DCE (3T) | Predict pCR following NAC in HER2+ tumours | 311(Y) | Manual (3D) | Second‐order statistics 2 | Sensitivity 86.5%; specificity 80%; accuracy 84.9% |
| 28 | DCE (1.5T) | Predict pCR following NAC | 259(N) | Semi‐automatic (3D) | Morphological; second‐order statistics | Model predicted pCR in TNBC with AUC of 0.974 |
| 6 | DCE (1.5) | Predict DFS following NAC in HER2+ tumours | 127(Y) | Manual (2D) | Morphological; second‐order statistics 2 | AUC 0.974 in combined radiomic‐clinicopathological model |
| 52 | DCE (1.5T & 3T) | Pre‐treatment prediction of response to NAC | 158(N) | Semi‐automatic (4D) | Morphological; first‐ & second‐order statistics | Two individual features could predict pCR in pre‐NAC images with AUC of 0.82 and 0.73, which are maximum voxel value and surface area to volume ratio respectively |
| 53 | DCE (3T) | Explore relationship between tumour shrinkage and initial tumour enhancement with pCR in HER2+ breast cancer | 51(N) | Semi‐automatic (2D) | Morphological | Tumour shrinkage pattern was associated with pCR with an AUC 0.778 |
| 54 | DWI (1.5T) | Predict non‐response to NAC | 69(N) | Manual (2D) | Morphological; second‐order statistics; kinetic | Texture (second‐order statistics) and kinetic features could best predict non‐response to NAC, with accuracy, sensitivity and specificity all measuring 74% |
| 55 | DCE (1.5T & 3T) | Predict pCR following NAC by using peritumoral and intratumoral regions | 117(Y) | Manual (2D) | First‐ and second‐order statistics; kinetic | For all subtypes, combined peritumoral and intratumoral model had best results with AUC 0.78 |
| 56 | DCE (1.5T & 3T) | Predict pCR to neoadjuvant therapies | 288(Y) | Manual (3D) | Morphological; first‐ and second‐order statistics | Statistically significant (P < 0.002) results found for predicting pCR in TNBC and HER2+ patients, with AUC 0.707 |
| 57 | DCE (1.5T & 3T) | Characterise HER2+ tumours and estimate response to neoadjuvant therapy | 209 4 (Y) | Manual (2D) | First‐, second‐ and higher‐order statistics | Combined peritumoral and intratumoral model could identify HER2+ tumours and predict response to treatment with AUC 0.89 and 0.80 respectively |
| 58 | Multiparametric (3T) | Predict pCR to NAC | 91(Y) | Manual (3D) | Morphological; first‐ and second‐order statistics | Combined ADC and DCE model had best predictive power with AUC 0.931, accuracy 0.825, specificity 0.766 and sensitivity 1.0 |
| 59 | Multiparametric | Predict pCR to pre‐treatment NAC | 414 4 (Y 3 ) | Manual (2D) | Morphological; first‐ and second‐order statistics | Multiparametric model had highest AUC of 0.79, compared to other models that all had AUC <0.7. Predicting TNBC had AUC 0.96 |
| 60 | DCE (1.5T & 3T) | Predict breast cancers insensitive to NAC | 125(Y) | Manual (2D) | First‐ and second‐order statistics 2 | Combined radiomic‐clinicopathological model produced the best results of AUC 0.986 |
| 61 | Multiparametric (1.5T & 3T) | Predict breast cancer regression patterns following NAC | 144(Y) | Manual (2D) | Morphological; first‐order statistics 2 | Combined radiomic‐clinicopathological model produced the best results of AUC 0.902 |
| 62 | DCE (3T) | Predict efficacy of NAC based on machine learning techniques | 158(Y) | Manual (2D) | First‐ and second‐order statistics 2 | Combined radiomic‐clinicopathological model produced best results with AUC 0.888, sensitivity 86.96% and specificity 79.31% |
| 63 | T2W (3T) | Predict RCB following NAC | 88(N) | Manual (2D) | First‐ and second‐order statistics | Predicting pCR across all lesions had an accuracy of 85.2% |
| (c) | ||||||
| 64 | DCE (3T) | Determine the level of tumour infiltrating lymphocytes | 172(Y) | Semi‐automatic (2D) | First‐ and second‐order statistics 2 | Combining the radiomic and clinical model improved detection of infiltrating lymphocytes from AUC 0.742 to 0.800 |
| 65 | DCE (3T) | Predict axillary sentinel lymph node metastasis | 62(Y) | Manual (3D) | Morphological; first‐, second‐and higher‐order statistics | AUC 0.82; accuracy 0.76; sensitivity 0.75; specificity 0.76 |
| 66 | DCE (3T) | Predict axillary lymph node metastasis | 329(Y) | Manual (3D) | Morphological; first‐ and second‐order statistics 2 | Combined radiomic‐clinicopathological model performed best. AUC 0.894; sensitivity 81.25%, specificity 82.08%; accuracy 81.78% |
| 67 | Multiparametric (1.5T) | Predict sentinel lymph node metastasis | 146(Y) | Manual (3D) | Second‐order statistics | Multiparametric model produced the best results. AUC 0.863; sensitivity 0.663; specificity 0.816 |
| 68 | DCE (1.5T) | Predict lymph node metastasis based on existing dataset | 91 4 (N) | Semi‐automatic (2D) | Morphological; second‐order statistics; kinetic 2 | Genomic features performed best compared to the radiomics model with AUC 0.916 and 0.775 respectively |
| 69 | Multiparametric (3T) | Predict axillary lymph node metastasis | 120(N) | Manual (2D) | Morphological; first‐, second‐and higher‐order statistics; kinetic | Addition of kinetic features improved model performance to AUC 0.91 and accuracy 86.37 |
| 70 | DCE (3T) | Predict axillary lymph node metastasis | 102(N) | Semi‐automatic (2D) | Morphological; second‐order statistics | Combining morphological and texture features improved results. Accuracy 89.54%; sensitivity 94.5%; specificity 80.06% |
| (d) | ||||||
| 28 | DCE (3T) | Predict RCB in patients treated with NAC | 259(N) | Semi‐automatic (2D) | Morphological; first‐order statistics | In TNBC, RCB was associated with mean, minimum, maximum and standard deviation of signal intensity |
| 71 | DCE | Identify imaging phenotypes of heterogeneity and its prognostic ability to predict 10 year recurrence | 95(Y 3 ) | Semi‐automatic (3D) | Morphological; first‐ and second‐order statistics 2 | Nomogram consisting of radiomic features and heterogeneity phenotype improved prediction from c‐statistic 0.55 to 0.73 |
| 11 | DCE (1.5T) | Predict recurrence prognosis in ER+ tumours | 78(N) | Manual (3D) | First‐order statistics; Kinetic | Kurtosis in delayed enhancement phase significantly indicates high recurrence (P = 0.0116). Volume ratio of slow persistent was associated with the low‐risk group (P = 0.041) |
| 72 | DCE (1.5) | Estimate disease free survival and recurrence rates | 294(Y) | Manual (2D) | Morphological; second‐ & higher‐order statistics 2 | Combined radiomic‐clinicopathological model predicted recurrence with c‐index of 0.76 |
| 73 | DCE (1.5T) | Predict recurrence free survival | 162 4 (N) | Semi‐automatic (3D) | Morphological | High METV was predictive of recurrence and statistically significant in the HR+/HER2− (P = 0.012) and HER2+ (P = 0.036) subtypes |
| 74 | DCE (3T) | Predict cancer angiogenesis for tumour biology and patient outcomes | 27(N) | Semi‐automatic (3D) | Morphological; first‐ and second‐order statistics; kinetic 2 | Cancers with increased micro‐vessel density had higher peak signal ratio enhancement (AUC 0.79). Second‐order statistics features could determine angiogenesis with AUC >0.83 |
Note: Predicting Ki67 (red), HER2 (yellow), TNBC (blue), ER and PR (white) and Subtype (green). Assumption is made that when P < 0.05 the null hypothesis is rejected and therefore the results are statistically significant.
Abbreviations: ADC, apparent diffusion coefficient maps; AUC, area under the curve; BPE, background parenchymal enhancement; DCE, dynamic contrast enhanced; DFS, disease‐free survival; DWI, diffusion weighted imaging; ER, oestrogen receptor; HER2, human epidermal growth factor 2; METV, most enhancing tumour volume; pCR, pathologically complete response; PR, progesterone receptor; RCB, residual cancer burden; T, Tesla; TNBC, triple negative breast cancer.
Number of subjects (whether conducted a validation on external set).
Clinical features used.
External validation cohort.
Multicentre study.
Prospective validation cohort.
Results of individual sources of evidence
Of the 63 articles included in this scoping review, 35 addressed subtype classification, 15 for prediction of pathologically complete response, seven for lymph node metastasis detection and six for recurrence prediction. All included studies were retrospective in nature, with only one including a prospective validation cohort. 40 Findings for each category will be discussed.
Subtype classification
A total of 35 articles were found that investigate the use of radiomics for subtype classification of breast tumours, with 12 articles using 1.5T scanners, 21 with 3T and five with both 1.5 and 3T MRI scanners. Of the articles that used both MRI scanner strengths, only one compared their performance, noting that accuracy was slightly improved when using a 3T scanner. Classification accuracy was 95% and 97.7% for 1.5T and 3T scanners respectively. 50 This finding supports research suggesting that, due to the higher field strength of 3T scanners, spatial and temporal resolution increases, consequently improving image quality and detection of smaller lesions. 4
Additionally, a multiparametric MRI protocol was used in 11 studies, whereby DCE, DWI and T2W images were included for radiomic analysis and feature extraction. The predictive performance of subtype classification improved when multiparametric MRI models were used compared to single‐series models. Zhou et al. 29 performed radiomic analysis on three models comprised of T2W fat‐saturated images, T1W‐weighted contrast‐enhanced images and a combination of both images to form a multiparametric model. Prediction of HER2 status in breast cancer patients for each model was AUC of 0.74, 0.71 and 0.86, respectively, noting a significant improvement in performance for the multiparametric model. 29 This study also used 3D segmentation and extracted a large range of radiomic features from both the T1 and T2W MR images, hence, could provide a greater range of features to enable more comprehensive information of the tumour's biology to be incorporated into the predictive model.
Furthermore, five studies formed radiomic signatures that comprised both radiomic and clinical features. Wang et al. 34 sought to distinguish triple‐negative breast cancer (TNBC) from all other subtypes and compared the performance of radiomic features alone versus the inclusion of background parenchymal enhancement (BPE), which is known to represent hormonal activity. In each experiment, there was a significant increase in predictive accuracy when using both radiomic and clinical features (Fig. 2), highlighting the importance of BPE heterogeneity on DCE sequences for diagnosing TNBC. 34
Figure 2.
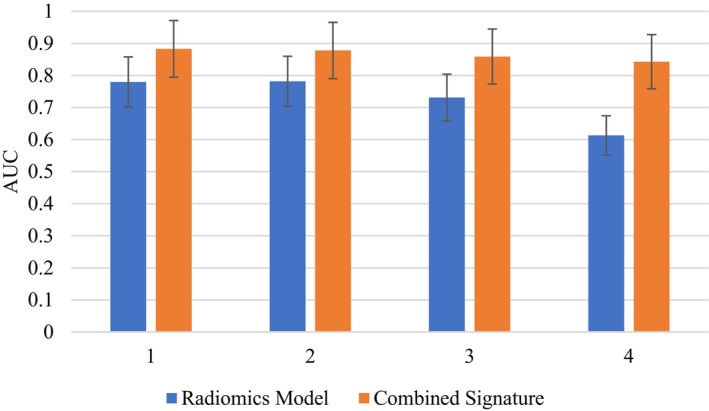
Comparison of predictive performance (AUC) for models using radiomic features alone versus a combined radiomic and clinical features signature, with standard deviation error bar. Experiment 1: TNBC versus ER+. Experiment 2: TNBC versus all other subtypes. Experiment 3: TNBC versus PR+. Experiment 4: TNBC versus luminal B. 34
First‐order statistics features, including kurtosis, entropy and skewness, are investigated in radiomic studies as they are highly predictive of tumour characteristics. Ma et al. 25 sought to determine Ki67 expression in breast tumours and found that high Ki67 lesions have higher kurtosis and lower skewness, which measure the steepness and symmetry of data distribution. Higher kurtosis indicates more heterogeneity, whereas lower skewness suggests the opposite as the dataset appears more homogenous. The homogenous appearance from lower skewness occurs because there are many nearby voxels that have similar uptake, indicating high proliferation. This matches with what are clinically known about high Ki67 tumours, which are highly proliferating and more aggressive compared to low Ki67 tumours. 25 Additionally, lower Ki67 tumours had smaller measures of effective diameter, entropy, kurtosis and homogeneity, all indicating less proliferation. 25 This highlights the ability of a machine learning model to make prognostic predictions relating to tumour aggressiveness and proliferation.
Radiomic analysis of intratumoral subregions was investigated by Fan et al., 26 whereby they sought to determine Ki67 status. As breast tumours are heterogeneous in biological makeup, it is suggested that intratumoral regions can better demonstrate this heterogeneity compared to analysing the entire tumour as one entity. 26 This can reflect biological processes of the tumour to facilitate prognostic predictions. The performance of second‐order statistics features in tumour subregions versus the entire tumour was compared, which predicted Ki67 status with an AUC of 0.807 and 0.748 respectively. 26 This indicates that features from tumour subregions are more representative of and related to the tumour's phenotype and characteristics.
Of note, only three studies explored subtype classification by using a multicentre approach, meaning that the study was heterogenous in the included cohort, imaging protocol and MRI scanners used. Many previous studies note that radiomics research is limited by small patient cohorts, minimal number of radiomic features used and no validation cohorts. 46 To fill these research gaps, Saha et al. 46 performed a study to determine the molecular subtype of breast cancer patients. They included 922 patients who underwent breast MRI scans at a variety of clinics with different imaging protocols. The cohort was divided in half to produce equal‐sized testing and validation groups and used a total of 522 radiomic features for lesion analysis. Moderate‐to‐good results were produced with AUCs of <0.7, resulting in the conclusion that radiomic features alone are insufficient for predicting molecular subtypes. 46 This heterogenous cohort reduces the chance of overfitting and is representative of a broader range of patients, however, it also introduces noise in analysis. 46 Stronger associations between radiomic features and subtype classification of breast tumours could be found with a more uniform cohort and methodology. 46 This finding contradicts conclusions drawn in many other studies, which suggest that heterogenous cohorts and imaging protocols are vital in improving reproducibility in clinical settings. 28
Pathologically complete response
Prediction of pCR included 15 articles, with three articles using 1.5T MRI scanners, four with 3T, six with both 3 and 1.5T and one unknown. Of the studies that compared the performance of both scanners, no significant findings were noted. 52 , 55 , 56 , 57 , 60 , 61 The majority of studies used DCE imaging protocols and produced strong results ranging from AUC of 0.707 to 0.986. 56 , 60 Two studies used T2W images alone for radiomic analysis and could predict pCR following NAC with an AUC of 0.902 61 and accuracy of 85.2%. 63
Of note, five articles utilised radiomics signatures with both clinicopathological and radiomic features, all producing significant results of AUC >0.83. 6 , 51 , 60 , 61 , 62 Interestingly, all articles included validation cohorts to verify initial findings and produced AUCs of >0.79, which is promising for producing reliable results and validating the conclusion that a combined model performs better than radiomics features alone.
Furthermore, Braman et al. 57 was a largely heterogenous study, using multiple MRI scanners from different institutions and variable treatment regimens for patients. Performance among training and validation cohorts remained consistent, with an AUC of 0.89 and 0.80 respectively. 57 Liu et al. 59 was another multicentre study, utilising three external validation cohorts. The radiomics model performed strongly in the training set but noted poor results in the validation cohorts. This study measured pCR following NAC in multiple tumour subtypes but had an unequal distribution of patients in each subtype. The produced radiomics signature was ineffective in predicting pCR for all subtypes, resulting in non‐significant results (i.e. an AUC of <0.6, with a P‐value >0.05 in comparison with the chance level) for some experiments. 59 Hence, it can be concluded that radiomic models cannot be generalised among all patients, rather should be subtype specific. 59
Lymph node metastasis
Seven articles investigating radiomic analysis for the detection of lymph node metastasis were identified. Two studies utilised a multiparametric model to predict lymph node metastasis, meaning the radiomics nomogram was produced based on a combination of MR images, including DCE, DWI and T2W. 67 , 69 In both of these studies, the multiparametric model produced the best results, with AUCs of 0.863 56 and 0.91. 69 This is because, when compared to single‐series models, multiparametric protocols can more accurately represent tumour morphology and pathogenic processes. 67
Additionally, Tan et al. 66 utilised a combined clinicopathological and radiomics model that noted a slight improvement in predictive performance compared to a radiomics‐only model, with AUCs of 0.894 and 0.88 respectively. However, Guo et al. 68 found that a genomic model performed significantly better than the radiomics model, with AUCs of 0.916 and 0.775 respectively.
Another trend noted is that studies utilising a range of radiomic features to develop the radiomic signature produced results of greater statistical significance. For instance, Chai et al. 69 utilised a multiparametric model with morphological, first‐order statistics, second‐order statistics, transform and kinetic features to build the radiomic signature, producing results with an AUC of 0.91. The addition of kinetic features to this model allowed for contrast uptake rates and heterogeneity in contrast enhancement to be analysed. 69 Cui et al. 70 also noted this trend, whereby improved prediction of axillary lymph node metastasis resulted by combining morphological and texture features to form the radiomics signature.
Recurrence prediction
Radiomics for prediction of cancer recurrence consisted of six eligible studies, with three utilising a combined clinicopathological and radiomics model. The addition of clinical features, including heterogeneity phenotypes, micro‐vessel density and histological grade, saw improvements in radiomic signature performance. For example, Chitalia et al. 71 noted significant improvements in recurrence prediction, with AUC increasing from 0.55 to 0.73. Similar trends were noted in the other two studies, which achieved maximum performances of AUC 0.76 and 0.79 respectively. 72 , 74 All three studies concluded that radiomic signatures based on both radiomic and clinicopathological features improve the overall accuracy of recurrence predictions. The addition of kinetic features to studies provided valuable information on recurrence patterns by measuring the relationship between contrast enhancement and delays in uptake. 11 , 74 Enhancement characteristics provide information on the aggressiveness of the tumours and can be associated with recurrence risk. 11 For example, Tokuda et al. 11 could distinguish between patients with low and high risk of recurrence by analysing the kinetic feature of volume ratio, which relates to the rate of contrast uptake. Slower, persistent volume ratio was associated with a lower risk of recurrence (P = 0.041), highlighting the value of kinetic features for risk stratification of patients. 11 Additionally, one study was multicentre and produced statistically significant (P < 0.05) results for predicting recurrence in the HER+/HER2− and HER2+ subtypes only. 73 Morphological features were the only type of radiomic features used in this study, suggesting that a wider range of features could improve the applicability of the radiomics model across other subtypes. 73
Discussion
The study has highlighted the potential for radiomic techniques to facilitate the diagnosis and classification of breast cancer, as well as predict patient outcomes. Through mathematical analysis of MRI images, an individual tumour's anatomy and pathological processes can be further understood. This allows for improved accuracy of tumour classification and can indicate the most clinically relevant treatment options for personalised medicine. However, it was also recognised that radiomic studies differ significantly and can be flawed in their methodology, consequently inhibiting the clinical application of this technique. Given the emerging nature of radiomics and variations in methodological approaches, this finding was consistent with other reviews in this field. Therefore, future radiomics research should aim to develop a standardised methodology to improve understanding of the basic concepts of radiomics among the medical imaging community. Additionally, there should be a more detailed assessment of the quality of these radiomic studies.
Another potential for future research is that unused quantitative data remain within existing breast images, which can be extracted through radiomic analysis to better understand tumour characteristics. DCE images are highly sensitive to enhancement patterns and angiogenesis as a result of contrast enhancement, which is valuable in describing tumour aggressiveness and malignancy. 5 The value of contrast required for DCE images is an ongoing debate, and the extent to which contrast is necessary should be further investigated. DWI and T2W images are non‐contrast protocols, with DWI assessing water diffusion to reflect pathological processes associated with the tumours microenvironment and membrane integrity. 5 T2W images can assess intracellular and extracellular activity to detect diseased tissue, which appears with increased free water and hyperintensity. 22 T2W images can also assess the morphological characteristics of breast tumours, such as architecture, cystic makeup and heterogeneity, but are currently unable to determine enhancement characteristics. Therefore, multiparametric MRI sequences are advantageous for demonstrating a wider range of tumour characteristics, which can improve accuracy of subtype classification, prediction of treatment response and recurrence or metastatic spread. Furthermore, radiomic features have distinct characteristics and appearances on the different MRI sequences. For instance, Ko et al. 36 investigated the radiomic and clinical features of ER+ tumours. Higher‐grade, more aggressive tumours had greater uniformity and lower entropy (textural features measuring heterogeneity) on contrast‐enhanced (CE) T1W images, whereas the opposite tendency was noted on T2W images. These findings are contradictory as higher uniformity on CE T1W images indicates more homogeneity, whereas decreased uniformity on T2W images suggests heterogeneity. 36 This contradictory finding highlights an important characteristic of aggressive tumours, which is that they enhance homogenously due to increased vascularisation. Multiparametric MRI protocols therefore are critical for improving the accuracy of radiomic models for predicting patient outcomes and the biological and pathological processes of breast tumours.
Intratumour heterogeneity refers to the presence of variations within a tumour and is increasingly recognised as an important factor in understanding tumour behaviour and predicting treatment response. Radiomics can provide valuable insights into intratumour homogeneity, and to fully understand its complexity, the tumour can be divided into subregions to examine the variations in radiomic features within different regions of the tumour. This approach allows for a more detailed assessment of the spatial patterns and distribution of imaging features within the tumour. Voxel‐wise analysis takes the spatial resolution to an even finer level. Examining individual image voxels and their corresponding radiomic features.
Acquisition parameters (e.g. echo time and repetition time) as well as reconstruction algorithms and slice thickness affect MR images produced during an imaging sequence. 75 These factors, in turn, determine image noise and texture, which affects the radiomic features extracted and their quantitative value. 75 Consequently, images captured on different scanners at a variety of institutions will reflect different radiomic properties and produce unstable features. 75 Hence, it is difficult to obtain uniform results that have consistent imaging parameters, which is a barrier to the application of radiomic techniques to clinical settings. Scanner strength is another parameter affecting MR images, with few radiomic studies comparing the radiomic features and predictive performance of 1.5T and 3T scanners. Most studies concluded that no statistically significant findings could be noted, however, Sun et al. 50 found slightly higher accuracy for predicting molecular subtype with 3T scanners compared to 1.5T (accuracy of 86.4% and 82.8% respectively).
ROI segmentation is a vital step in the radiomics method as data are extracted from these defined regions for feature analysis. 75 It can be limited by poorly defined tumour borders and lack of consensus and standardised methodology. Researchers have the option to perform manual, semi‐automatic or automatic segmentation with 2D or 3D reconstructions. Automatic 3D segmentation is the preferred method of segmentation as it is faster, can provide a holistic understanding of the tumour's relationship with surrounding tissues and reduces the chance of bias. On the other hand, semi‐automatic or manual 2D segmentation is prone to errors, inter‐ or intrareader variability and is time‐consuming. 2 Yet, studies using the suboptimal manual 2D segmentation approach are still producing statistically significant results as can be seen with Wang et al., 33 who distinguished TNBC from non‐TNBC with an accuracy of 95.4%. Currently, radiomic features are extracted from the intratumoral (i.e. within the tumour's border) space. However, there is growing interest in the peritumoral space, which is the region immediately surrounding the tumour border that is defined in ROI segmentation. Analysis of radiomic features in the peritumoral space can provide insight into tumour characteristics and biology, such as enhancement patterns, angiogenesis and necrosis. Studies demonstrate improved pCR prediction when using both peritumoral and intratumoral features, resulting in an AUC of 0.78. 5
It is evident that a radiomics signature/nomogram consisting of both radiomic and clinicopathological features improves prediction of anatomical and functional tumour features. Radiomic or clinical features alone lack detail and provide noisy data. 18 However, as each individual feature provides diagnostic and therapeutic information, a more complete depiction of the tumour can be gathered when features are combined, resulting in statistically significant findings and improved positive predictive values. 18 , 60 Therefore, it is advantageous to use a combined radiomic and clinicopathological model with a range of features as predictions on subtype classification and patient outcomes significantly improve, allowing for the possibility of personalised medicine.
Current radiomic studies appear limited by small patient cohorts, with included studies ranging from 27 to 922 patients, with an average of 169. This raises issues including overfitting, unreliable results and poor generalisation to the breast cancer population, limiting the significance of findings from radiomic studies. Therefore, future studies should seek to include larger patient cohorts with a more equal distribution of patients, such that study findings can be more reliable, accurate and applicable to real‐life breast cancer patients.
This review is limited by only investigating radiomic studies using MRI as the imaging modality. The scope of radiomic studies explored was reduced by not considering ultrasound, mammography or computed tomography (CT) scans. However, this limitation is countered by the fact that this study performed an in‐depth discussion on the vast potential MRI images that must predict patient outcomes and perform lesion classification. MRI is well explored in relation to radiomic techniques, with results of existing studies demonstrating highly accurate predictions when using the DCE, DWI and T2W MR images for radiomic analysis. Additionally, the limited range of included studies from 2015 onwards may have resulted in missing key preliminary studies in the field of radiomics.
Conclusion
Personalised treatment, risk stratification and prognostic predictions are highly valued and desired in this current era of personalised medicine, and radiomics provides the potential to avoid unnecessary toxicities, reduce patient burden and improve overall outcomes. Therefore, radiomics has the potential to non‐invasively assess tumour pathophysiology and anatomy to allow for early and accurate diagnosis of breast cancer. Radiomics is still in the pre‐clinical phase, with a lack of prospective studies and standardised methodology, preventing its application in clinical settings. With future studies addressing these methodological pitfalls, radiomics has the potential to revolutionise the management of breast cancer patients and improve both survival rates and quality of life through personalised medicine.
Conflicts of Interest
The authors declares no conflict of interest.
Acknowledgement
Open access publishing facilitated by The University of Sydney, as part of the Wiley ‐ The University of Sydney agreement via the Council of Australian University Librarians.
[Correction added on 10 August 2023, after first online publication: The total number of articles[n] has been changed from 37 to 35 throughout the article in this version ].
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.
References
- 1. Pesapane F, Rotili A, Agazzi GM, et al. Recent radiomics advancements in breast cancer: lessons and pitfalls for the next future. Curr Oncol 2021; 28: 2351–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Conti A, Duggento A, Indovina I, Guerrisi M, Toschi N. Radiomics in breast cancer classification and prediction. Semin Cancer Biol 2021; 72: 238–250. [DOI] [PubMed] [Google Scholar]
- 3. Maforo N, Li H, Lan L, Edwards A, Giger ML. Prognostic radiomics of breast cancer on DCE and DWI MR images. Int J Med Phys Res Pract 2016; 43: 3378. 10.1118/1.4955798. [DOI] [Google Scholar]
- 4. Wood R, Bassett K, Spry VC, Tong L. 1.5 Tesla magnetic resonance imaging scanners compared with 3.0 Tesla magnetic resonance imaging scanners: systematic review of clinical effectiveness. Can Agency Drugs Technol Health 2012; 2: 1–5. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3442613/. [PMC free article] [PubMed] [Google Scholar]
- 5. Ye D, Wang H, Yu T. The application of radiomics in breast MRI: a review. Technol Cancer Res Treat 2020; 19: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Q, Xiao Q, Li J, Duan S, Wang H, Gu Y. MRI‐based radiomic signature as a prognostic biomarker for HER2‐positive invasive breast cancer treated with NAC. Cancer Manage Res 2020; 12: 10603–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koçac B, Durmaz ES, Ates E, Kiliçkesmez Ö. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol 2019; 25: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Timmeren JE, Cester D, Tanadini‐Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“how to” guide and critical reflection. Insights Imaging 2020; 11: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crivelli P, Ledda RE, Parascandolo N, Fara A, Soro D, Conti M. A new challenge for radiologists: radiomics in breast cancer. Biomed Res Int 2018; 2018: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tagliafico AS, Piana M, Schenone D, Lai R, Massone AM, Houssami N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020; 49: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tokuda Y, Yanagawa M, Minamitani K, Naoi Y, Noguchi S, Tomiyama N. Radiogenomics of magnetic resonance imaging and a new multi‐gene classifier for predicting recurrence prognosis in estrogen receptor‐positive breast cancer. Medicine 2020; 99: e19664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee S, Park H, Ko ES. Radiomics in breast imaging from techniques to clinical applications: a review. Korean J Radiol 2020; 21: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monti S, Aiello M, Incoronato M, et al. DCE‐MRI pharmacokinetic‐based phenotyping of invasive ductal carcinoma: a radiomic study for prediction of histology outcomes. Contrast Media Mol Imaging 2018; 2018: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Espinosa E, Zamora P, Feliu J, Barón MG. Classification of anticancer drugs—a new system based on therapeutic targets. Cancer Treat Rev 2003; 29: 515–523. [DOI] [PubMed] [Google Scholar]
- 15. Adolfsson J, Steineck G. Prognostic and treatment‐predictive factors‐is there a difference. Prostate Cancer Prostatic Dis 2000; 3: 265–268. [DOI] [PubMed] [Google Scholar]
- 16. Colquhoun HL, Levac D, O'Brien KK, et al. Scoping reviews: time for clarity in definition, methods and reporting. J Clin Epidemiol 2014; 67: 1291–1294. [DOI] [PubMed] [Google Scholar]
- 17. Liu W, Cheng Y, Liu Z, et al. Preoperative prediction of Ki67 status in breast cancer with multiparametric MRI using transfer learning. Acad Radiol 2020; 28: 44–53. [DOI] [PubMed] [Google Scholar]
- 18. Fan M, Yuan W, Zhao W, et al. Joint prediction of breast cancer histological grade and Ki‐67 expression level based on DCE‐MRI and DWI radiomics. Journal of Biomedical and Health Informatics 2019; 24: 1632–1642. [DOI] [PubMed] [Google Scholar]
- 19. Fan M, Liu Z, Xu M, et al. Generative adversarial network‐based super‐resolution of diffusion‐weighted imaging: application to tumour radiomics in breast cancer. NMR Biomed 2020; 33: e4345. [DOI] [PubMed] [Google Scholar]
- 20. Demircioglu A, Grueneisen J, Ingenweth M, et al. A rapid volume of interest‐based approach of radiomics analysis of breast MRI for tumour decoding and phenotyping of breast cancer. PloS One 2020; 15: e0234871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Juan M, Yu J, Peng G, Jun L, Feng S, Fang L. Correlation between DCE‐MRI radiomics features and Ki‐67 expression in invasive breast cancer. Oncol Lett 2017; 16: 5084–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang C, Cheng Z, Huang Y, et al. An MRI‐based radiomics classifier for preoperative prediction of Ki‐67 status in breast cancer. Acad Radiol 2018; 25: 1111–1117. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y, Zhu Y, Zhang K, et al. Invasive ductal breast cancer: preoperative predict Ki‐67 index based on radiomics of ADC maps. Radiol Med 2019; 125: 109–116. [DOI] [PubMed] [Google Scholar]
- 24. Kayadibi Y, Koçak B, Ucar N, Akan YN, Akbas P, Bektas S. Radioproteomics in breast cancer: prediction of Ki‐67 expression with MRI‐based radiomic models. Acad Radiol 2021; 29 Suppl 1: S116–S125. [DOI] [PubMed] [Google Scholar]
- 25. Ma W, Ji Y, Qi L, Guo X, Jian X, Liu P. Breast cancer Ki67 expression prediction by DCE‐MRI radiomics features. Clin Radiol 2018; 73: 1–5. [DOI] [PubMed] [Google Scholar]
- 26. Fan M, Cheng H, Zhang P, et al. DCE‐MRI texture analysis with tumour subregion partitioning for predicting Ki‐67 status of estrogen receptor‐positive breast cancers. J Magn Reson Imaging 2017; 48: 237–247. [DOI] [PubMed] [Google Scholar]
- 27. Li C, Song L, Yin J. Intratumoral and peritumoral radiomics based on functional parametric maps from breast DCE‐MRI for prediction of HER‐2 and Ki‐67 status. J Magn Reson Imaging 2021; 54: 703–714. [DOI] [PubMed] [Google Scholar]
- 28. Choudhery S, Gomez‐Cardona D, Favazza CP, et al. MRI radiomics for assessment of molecular subtype, pathological complete response, and residual cancer burden in breast cancer patients treated with neoadjuvant chemotherapy. Acad Radiol 2020; 4: S145–S154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou J, Tan H, Li W, et al. Radiomics signatures based on multiparametric MRI for the preoperative prediction of the HER2 status of patients with breast cancer. Acad Radiol 2020; 28: 1352–1360. [DOI] [PubMed] [Google Scholar]
- 30. Lu H, Yin J. Texture analysis of breast DCE‐MRI based on intratumoral subregions for predicting HER2 2+ status. Front Oncol 2020; 10: 1–12. 10.3389/fonc.2020.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang Z, Song L, Lu H, Yin J. The potential use of DCE‐MRI texture analysis to predict HER2 2+ status. Front Oncol 2019; 9: 1–7. 10.3389/fonc.2019.002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song L, Lu H, Yin J. Preliminary study on discriminating HER2 2+ amplification status of breast cancers based on texture features semi‐automatically derived from pre‐, pose‐contrast, and subtraction images of DCE‐MRI. PloS One 2020; 15: e0234800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Q, Mao N, Liu M, et al. Radiomic analysis on magnetic resonance diffusion weighted image in distinguishing triple‐negative breast cancer from other subtypes: a feasibility study. Clin Imaging 2021; 72: 136–141. [DOI] [PubMed] [Google Scholar]
- 34. Wang J, Kato F, Oyama‐Manabe N, et al. Identifying triple‐negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast‐enhanced MRI: a pilot radiomics study. PloS One 2015; 10: 1–17. 10.1371/journal.pone.0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H, Hu Y, Li H, Xie Y, Wang X, Wan W. Preliminary study on identification of estrogen receptor‐positive breast cancer subtypes based on dynamic contrast‐enhanced magnetic resonance imaging (DCE‐MRI) texture analysis. Gland Surg 2020; 9: 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ko ES, Kim J, Lim Y, Han B, Cho EY, Nam SJ. Assessment of invasive breast cancer heterogeneity using whole‐tumour magnetic resonance imaging texture analysis. Medicine 2016; 95: e2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leithner D, Bernard‐Davila B, Martinez DF, et al. Radiomic signatures derived from diffusion‐weighted imaging for the assessment of breast cancer receptor status and molecular subtypes. Mol Imaging Biol 2020; 22: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leithner D, Horvat JV, Marino MA, et al. Radiomic signatures with contrast‐enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast Cancer Res 2019; 21: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holli‐Helenius K, Salminen A, Rinta‐Kiikka I, et al. MRI texture analysis in differentiating luminal A and luminal B breast cancer molecular subtypes—a feasibility study. BMC Med Imaging 2017; 17: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waugh SA, Purdie CA, Jordan LB, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol 2016; 26: 322–330. [DOI] [PubMed] [Google Scholar]
- 41. Xie T, Zhao Q, Fu C, et al. Differentiation of triple‐negative breast cancer from other subtypes through whole‐tumour histogram analysis on multiparametric MR imaging. Eur Radiol 2019; 29: 2535–2544. [DOI] [PubMed] [Google Scholar]
- 42. Ni M, Zhou X, Liu J, et al. Prediction of the clinicopathological subtypes of breast cancer using a fisher discriminant analysis model based on radiomic features of diffusion‐weighted MRI. BMC Cancer 2019; 20: 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. Nature Partner Journals 2016; 2: 16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fan M, Li H, Wang S, Zheng B, Zhang J, Li L. Radiomic analysis reveals DCE‐MRI features for prediction of molecular subtypes of breast cancer. PloS One 2017; 12: 1–15. 10.1371/journal.pone.0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging 2015; 42: 902–907. [DOI] [PubMed] [Google Scholar]
- 46. Saha A, Harowicz MR, Grimm LJ, et al. A machine learning approach to radiogenomics of breast cancer: a study of 922 subjects and 529 DCE‐MRI features. Br J Cancer 2018; 119: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie T, Wang Z, Zhao Q, et al. Machine learning‐based analysis of MR multiparametric radiomics for the subtype classification of breast cancer. Front Oncol 2019; 9: 1–10. 10.3389/fonc.2019.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA. Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med 2019; 109: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang S, Fan M, Zhang J, Zheng B, Wang X, Li L. Radiogenomic analysis of breast cancer: dynamic contrast enhanced‐magnetic resonance imaging based features are associated with molecular subtypes. Proc SPIE 2016; 9789: 1–10. 10.1117/12.2217658. [DOI] [Google Scholar]
- 50. Sun X, He B, Luo X, et al. Preliminary study on molecular subtypes of breast cancer based on magnetic resonance imaging texture analysis. J Comput Assist Tomogr 2018; 42: 531–535. [DOI] [PubMed] [Google Scholar]
- 51. Bitencourt AGV, Gibbs P, Saccarelli CR, et al. MRI‐based machine learning radiomics can predict HER2 expression level and pathologic response after neoadjuvant therapy in HER2 overexpressing breast cancer. EBioMedicine 2020; 61: 103042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drukker K, Edwards A, Doyle C, Papioannou J, Kulkarni K, Giger ML. Breast MRI radiomics for the pretreatment prediction of response to neoadjuvant chemotherapy in node‐positive breast cancer patients. J Med Imag 2019; 6: 1–11. 10.1117/1.JMI.6.3.03.034502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heacock L, Lewin A, Ayoola A, et al. Dynamic contrast‐enhanced MRI evaluation of pathologic complete response in human epidermal growth factor receptor 2 (HER2)‐positive breast cancer after HER2‐targeted therapy. Acad Radiol 2019; 27: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Michoux N, Van den Broeck S, Lacoste L, et al. Texture analysis on MR images helps predicting non‐response to NAC in breast cancer. BMC Cancer 2015; 15: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE‐MRI. Breast Cancer Res 2017; 19: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathological response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat 2018; 173: 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Braman N, Prasanna P, Whitney J, et al. Association of peritumoral radiomics with tumour biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)‐positive breast cancer. JAMA Netw Open 2019; 2: 1–18. 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen X, Chen X, Yang J, Li Y, Fan W, Yang Z. Combining dynamic contrast‐enhanced magnetic resonance imaging and apparent diffusion coefficient maps for a radiomics nomogram to predict pathological complete response to neoadjuvant chemotherapy in breast cancer patients. J Comput Assist Tomogr 2020; 44: 275–283. [DOI] [PubMed] [Google Scholar]
- 59. Liu Z, Li Z, Qu J, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicentre study. Clin Cancer Res 2019; 25: 3538–3547. [DOI] [PubMed] [Google Scholar]
- 60. Xiong Q, Zhou X, Liu Z, et al. Multiparametric MRI‐based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy. Clin Transl Oncol 2019; 22: 50–59. [DOI] [PubMed] [Google Scholar]
- 61. Zhuang X, Chen C, Liu Z, et al. Multiparametric MRI‐based radiomics analysis for the prediction of breast tumour regression patterns after neoadjuvant chemotherapy. Transl Oncol 2020; 13: 100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen S, Shu Z, Li Y, et al. Machine learning‐based radiomics nomogram using magnetic resonance images for prediction of neoadjuvant chemotherapy efficacy in breast cancer patients. Front Oncol 2020; 10: 1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Henderson S, Purdie C, Michie C, et al. Interim heterogeneity changes measured using entropy texture features on T2‐weighted MRI at 3.0T are associated with pathological response to neoadjuvant chemotherapy in primary breast cancer. Eur Radiol 2016; 27: 4602–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu N, Zhou J, He X, et al. Radiomics model for evaluating the level of tumour‐infiltrating lymphocytes in breast cancer based on dynamic contrast‐enhanced MRI. Clin Breast Cancer 2020; 21: 440–449. [DOI] [PubMed] [Google Scholar]
- 65. Liu J, Sun D, Chen L, et al. Radiomics analysis of dynamic‐contrast enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front Oncol 2019; 9: 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tan H, Gan F, Wu Y, et al. Preoperative prediction of axillary lymph node metastasis in breast carcinoma using radiomics features based on the fat‐suppressed T2 sequence. Acad Radiol 2020; 27: 1217–1225. [DOI] [PubMed] [Google Scholar]
- 67. Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2‐weighted fat‐suppression and diffusion‐weighted MRI. Eur Radiol 2018; 28: 582–591. [DOI] [PubMed] [Google Scholar]
- 68. Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinoma from the integration of radiomics and genomics data. J Med Imag 2015; 2. 10.1117/1.JMI.2.4.041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chai R, Ma H, Xu M, et al. Differentiating axillary lymph node metastasis in invasive breast cancer patients: a comparison of radiomic signatures from multiparametric breast MR sequences. J Magn Reson Imaging 2019; 50: 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cui X, Wang N, Zhao Y, et al. Preoperative prediction of axillary lymph node metastasis in breast cancer using radiomics features of DCE‐MRI. Sci Rep 2019; 9: 1–10. 10.1038/s41598-019-40831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chitalia RD, Rowland J, McDonald ES, et al. Imaging phenotypes of breast cancer heterogeneity in preoperative breast dynamic contrast enhanced magnetic resonance imaging (DCE‐MRI) scans predict 10‐year recurrence. Clin Cancer Res 2020; 26: 862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Park H, Lim Y, Ko ES, et al. Radiomics signature on magnetic resonance imaging: association with disease‐free survival in patients with invasive breast cancer. Clin Cancer Res 2018; 24: 4705–4714. [DOI] [PubMed] [Google Scholar]
- 73. Drukker K, Li H, Antropova N, Edwards A, Papaioannou J, Giger ML. Most‐enhancing tumour volume by MRI radiomics predicts recurrence‐free survival “early on” in neoadjuvant treatment of breast cancer. Cancer Imaging 2018; 18: 1–9. 10.1186/s40644-018-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xiao J, Rahbar H, Hippe DS, et al. Dynamic contrast‐enhanced breast MRI features correlate with invasive breast cancer angiogenesis. NPJ Breast Cancer 2021; 7: 1–9. 10.1038/s41523-021-00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2018; 2: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


