Main
In the recent study by Rosati et al, we described a novel unconventional T cell population enriched in the peripheral blood of patients with Crohn’s disease (CD) and characterised by a semi-invariant T cell receptor (TCR) repertoire.1 However, the specificity of these Crohn’s-associated invariant T (CAIT) cells was not defined. Identifying the specificity of CAIT cells is essential to understand the origin of the antigen triggering their enrichment in CD.
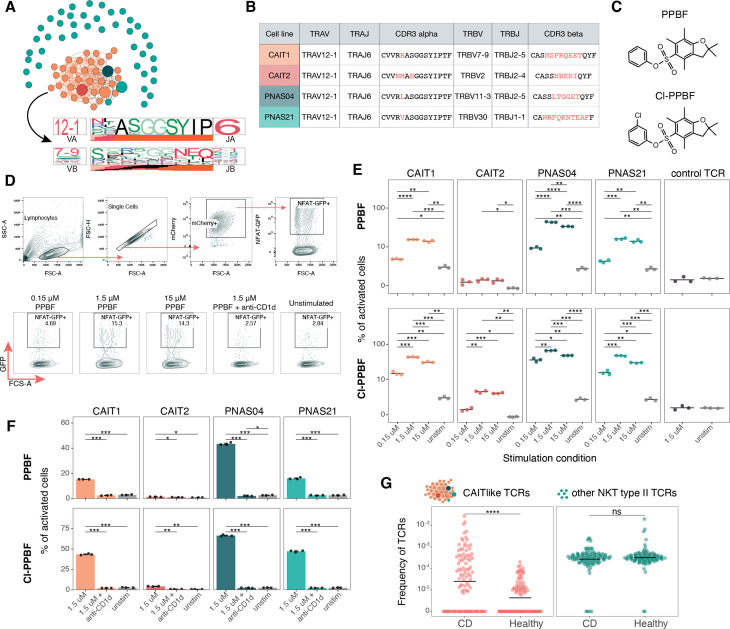
In our previous study, we observed that CAIT cells have TCRs similar to those reported for some natural killer T (NKT) type II cells.2 3 Here, we performed a sequence similarity analysis4 and identified a large cluster composed of CAIT clonotypes and three reported NKT type II clonotypes (figure 1A). While the NKT type II and CAIT clonotypes all had highly similar TCR alpha chains carrying TRAV12-1/TRAJ6 genomic segments, their beta chains were highly diverse (figure 1A, bottom). Dash et al have shown that TCRs with similar sequences frequently have the same specificity.5 6 In the original publications describing these clonotypes, the authors reported that the NKT type II cells recognise small molecules of the pentamethylbenzofuransulfonates (PBFs) family presented by the invariant HLA-like CD1d protein.2 3 Thus, we investigated whether CAIT TCRs shared the specificity of the NKT type II cells.
Figure 1.
Comparison of natural killer T (NKT) type II and Crohn’s-associated invariant T (CAIT) cells. (A) Sequence similarity analysis of NKT type II2 3 and CAIT T cell receptors (TCRs). Each node corresponds to the unique alpha/beta TCR sequence and edges connect highly similar TCRs (tcrdist metric<150). Larger nodes indicate TCRs used for cloning. TCRdist sequence logos for TCRalpha and TCRbeta chains of TCRs from the cluster are shown at the bottom. (B). Genomic segments and amino acid CDR3 sequences of TCRs picked for experimental validation. Red font indicates differences in CDR3 regions. (C). Chemical structures of phenyl-pentamethyldihydrobenzofuransulfonate (PPBF) and chlorophenyil-pentamethyldihydrobenzofuransulfonate (ClPPBF). (D.) Gating strategy and representative flow plots for pentamethylbenzofuransulfonates (PBFs) stimulation experiment. (E). Frequency of activated cells reactive to PPBF (top) and ClPPBF (bottom). Only significant p values from a T-test with Holm method for multiple testing correction are shown (*<0.05, **<0.01, ***<0.001, ****<0.0001). (F). Anti-CD1d antibody prevents activation of all four cell lines with PPBF (top) and ClPPBF (bottom). Only significant p values from a t-test with Holm method for multiple testing correction are shown (*<0.05, **<0.01, ***<0.001, ****<0.0001). (G.) Frequency of CAITlike NKT type II TCRs from Almeida et al (attached to a large cluster on figure 1A) and non-CAIT like NKT type II TCRs in Crohn’s disease and healthy cohorts from Rosati et al .1 Only significant p value (****<0.0001) from a Mann-Whitney U-test is shown.
We transduced TCR-null NFAT-GFP reporter Jurkat cells with constructs encoding two representative CAIT TCRs and two coclustering NKT type II TCRs (figure 1B). We synthetised two of the PBF compounds, phenyl-pentamethyldihydrobenzofuransulfonate (PPBF), the original compound identified as a CD1d-dependent activator of NKT type II cells,2 and a more potent PPBF analogue chlorophenyil-pentamethyldihydrobenzofuransulfonate (ClPPBF)3 (figure 1C). Importantly, Jurkat cells naturally express CD1d and thus can act as antigen-presenting cells for CD1d-dependent antigens. To evaluate TCR activation, we cultured Jurkat cell lines with incremental concentrations of PPBF and ClPPBF (figure 1D).
All four transgenic cell lines (CAIT1, CAIT2, PNAS04, PNAS21) reacted to both compounds in a dose-dependent manner (figure 1E). Consistent with the original study, all tested TCRs reacted more strongly to ClPPBF. The CAIT2 cell line, with the lowest level of activation, (figure 1E) carried the most dissimilar TCRalpha sequence compared with the other cell lines. CAIT2 has mismatches at CDR3 positions 4 and 7 (figure 1B), suggesting the importance of these positions for TCR avidity. A control cell line with known TCR specificity was not activated by the compounds, indicating that compound recognition is TCR-dependent (figure 1E). The reactivity of all cell lines dropped to unstimulated levels in the presence of the CD1d-blocking antibody, demonstrating that the TCR interaction is CD1d-restricted (figure 1F).
To further investigate the possible role of NKT type II cells in CD, we searched all NKT type II sequences reported in Almeida et al in our previously published TCR data from patients with CD and healthy controls.1 3 Only TCRs from NKT type II cells with the characteristic CAIT TCRalpha chain motif showed significant enrichment in patients with CD, while other TCRs were found in comparable amounts in patients and controls (figure 1G). Thus, only a subgroup of NKT type II cells with specific TCR features is enriched in CD.
While we show the specificity of CAIT cells for PBF small molecules in the context of CD1d, many questions remain. Many other small molecules similar to PBFs exist, including drug derivatives and microbial metabolites.3 7 8 It is thus reasonable to hypothesise that different small molecules may be triggering CAIT cells in vivo. Importantly, NKT cells can differentiate into opposing phenotypes, from proinflammatory to regulatory,8–10 necessitating further characterisation on the functional profile of the NKT type II CAIT cell subset and its behaviour in patients with CD.
Detailed methodologies are described in online supplemental file 1.
gutjnl-2022-328684supp001.pdf (1.9MB, pdf)
Footnotes
Twitter: @elisarosix
AAM and MVP contributed equally.
Contributors: Conceptualisation: ER, MVP, AAM; analysis: ER, MVP, AAM; Visualisation: AAM; Resources: PGT, AF, FD, SP, UL; Supervision and coordination: AF, PGT, ER. Writing original draft: ER, MVP, AAM. Funding acquisition: PGT, ER, AF. All authors provided discussion, participated in revising the manuscript, and agreed to the final version.
Funding: This work was funded by American Lebanese Syrian Associated Charities (ALSAC) at St. Jude, 5U01AI150747 (PGT), 5R01AI136514 (PGT). This work received infrastructure support from the EU’s Horizon 2020 SYSCID program under the grant agreement No 733100 and the DFG Excellence Cluster Precision Medicine in Chronic Inflammation (EXC2167-390884018)
Competing interests: ER is currently employed in the biopharmaceutical company Evotec SE. PGT has consulted and/or received honoraria and travel support from Illumina, Johnson and Johnson, and 10X Genomics. PGT serves on the Scientific Advisory Board of Immunoscape and CytoAgents .The other authors declare no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Rosati E, Rios Martini G, Pogorelyy MV, et al. A novel unconventional T cell population enriched in Crohn's disease. Gut 2022;71:2194–204. 10.1136/gutjnl-2021-325373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Rhijn I, Young DC, Im JS, et al. Cd1D-Restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci U S A 2004;101:13578–83. 10.1073/pnas.0402838101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almeida CF, Smith DGM, Cheng T-Y, et al. Benzofuran sulfonates and small self-lipid antigens activate type II NKT cells via CD1d. Proc Natl Acad Sci U S A 2021;118:e2104420118. 10.1073/pnas.2104420118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schattgen SA, Guion K, Crawford JC, et al. Integrating T cell receptor sequences and transcriptional profiles by clonotype neighbor graph analysis (CoNGA). Nat Biotechnol 2022;40:54–63. 10.1038/s41587-021-00989-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dash P, Fiore-Gartland AJ, Hertz T, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 2017;547:89–93. 10.1038/nature22383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glanville J, Huang H, Nau A, et al. Identifying specificity groups in the T cell receptor repertoire. Nature 2017;547:94–8. 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zajonc DM, Maricic I, Wu D, et al. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med 2005;202:1517–26. 10.1084/jem.20051625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burrello C, Pellegrino G, Giuffrè MR, et al. Mucosa-Associated microbiota drives pathogenic functions in IBD-derived intestinal iNKT cells. Life Sci. Alliance 2019;2:e201800229. 10.26508/lsa.201800229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao C-M, Zimmer MI, Wang C-R. The functions of type I and type II natural killer T cells in inflammatory bowel diseases. Inflamm Bowel Dis 2013;19:1330–8. 10.1097/MIB.0b013e318280b1e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Middendorp S, Nieuwenhuis EES. Nkt cells in mucosal immunity. Mucosal Immunol 2009;2:393–402. 10.1038/mi.2009.99 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-328684supp001.pdf (1.9MB, pdf)