Abstract
Uptake of 2,4-dichlorophenoxyacetate (2,4-D) by Ralstonia eutropha JMP134(pJP4) was studied and shown to be an energy-dependent process. The uptake system was inducible with 2,4-D and followed saturation kinetics in a concentration range of up to 60 μM, implying the involvement of a protein in the transport process. We identified an open reading frame on plasmid pJP4, which was designated tfdK, whose translation product TfdK was highly hydrophobic and showed resemblance to transport proteins of the major facilitator superfamily. An interruption of the tfdK gene on plasmid pJP4 decimated 2,4-D uptake rates, which implies a role for TfdK in uptake. A tfdA mutant, which was blocked in the first step of 2,4-D metabolism, still took up 2,4-D. A mathematical model describing TfdK as an active transporter at low micromolar concentrations fitted the observed uptake data best.
Ralstonia eutropha JMP134(pJP4) can utilize 2,4-dichlorophenoxyacetate (2,4-D) as the sole carbon and energy source for growth (5). The tfd genes encoding the 2,4-D pathway are located on plasmid pJP4 (4) and have been studied extensively. The genes tfdA (24), tfdB, and tfdCDEF (21) encode enzymes that are responsible for the conversion of 2,4-D via 2,4-dichlorophenol (2,4-DCP) and 3,5-dichlorocatechol to the central metabolite 3-oxoadipate. In addition, there are two genes with a regulatory function, namely, tfdR and tfdS (15, 18, 30).
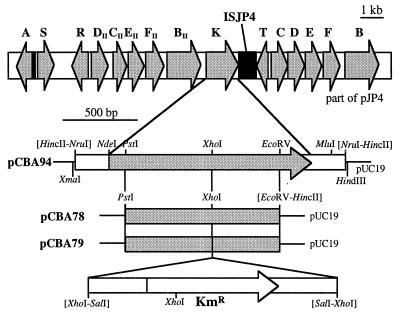
Lately, our laboratory has been interested in ISJP4, an insertion sequence which is located in monocopy on plasmid pJP4 (16). This element seems to have had quite an impact on the genetic organization of the plasmid (Fig. 1). Firstly, it inserted itself into the tfdT gene and thereby inactivated the gene product as a regulator of tfdCDEF expression (15). Secondly, the presence of a small piece of ISJP4 near tfdS may be reminiscent of the element’s involvement in the duplication of tfdR and tfdS, which are actually two identical copies of the same gene (30). Thirdly, the copy of ISJP4 which is inserted in tfdT and its small remnant near tfdS can form a composite transposon and together are able to mobilize intervening DNA sequences (16). It is quite possible that the approximately 11-kb fragment that lies trapped between the two pieces of ISJP4 was actually mobilized onto an early version of pJP4 (16).
FIG. 1.
(Top) Overview of the genetic organization on plasmid pJP4. Arrows, positions and orientations of the various tfd genes; black boxes, ISJP4 DNA. (Bottom) Restriction maps of plasmids used in this study. Plasmid pCBA94 contains a 1.9-kb NruI fragment of pJP4 in pUC19 (29). The sequence of this DNA fragment is presented in Fig. 2. Plasmid pCBA78 contains a 1.1-kb PstI-EcoRV fragment of tfdK in pUC19. A 1.7-kb SalI fragment carrying the kanamycin cassette from plasmid pUT/Km (12) was inserted into the XhoI site of pCBA78, resulting in plasmid pCBA79. The latter was used to construct a tfdK mutant of R. eutropha JMP134(pJP4). pUC-derived plasmids were propagated in cells of E. coli DH5α (23) cultivated at 37°C on Luria broth medium (23) in the presence of 100 μg of ampicillin per ml.
The latter idea made us curious about the information content of this mobile-mobilized DNA fragment. Therefore, we decided to determine its nucleotide sequence and discovered a whole new additional set of tfd genes (Fig. 1), i.e., tfdDIICIIEIIFII, tfdBII, and tfdK. The tfdK gene, its product, and its putative function are described in the present communication. The resemblance of the gene product to various transport proteins led us to investigate 2,4-D uptake by R. eutropha JMP134 and the possible role of tfdK in that process.
The tfdK gene encodes a member of the major facilitator superfamily of transporter proteins.
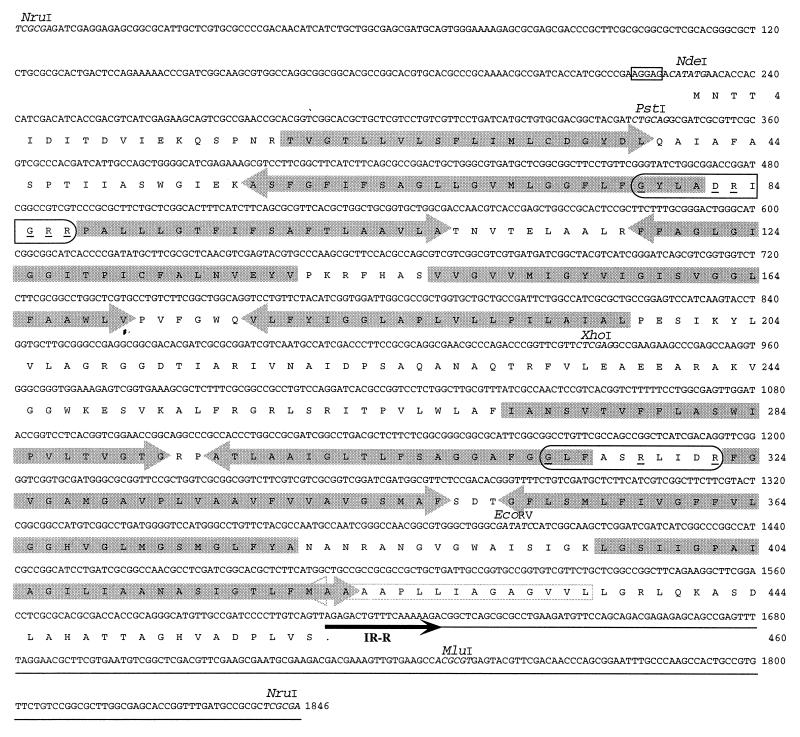
The sequence of the tfdK gene and its flanking regions on plasmid pJP4 is presented in Fig. 2. The gene starts with an ATG codon, ends with TAG, and is 1,380 bp long. Interestingly, the stop codon (TAG) overlaps with the TTA duplicated target site of ISJP4 and its right-hand inverted repeat (GAGACTGTTTCAAAAAGA) (16). The tfdK gene is preceded by an AGGAG ribosome binding site. The polypeptide that it putatively encodes is 460 amino acids (aa) in length, with a predicted molecular mass of 47.6 kDa. More than half (54%) of the amino acids encoded by tfdK are hydrophobic (A, I, L, F, W, and V). Comparison of TfdK to entries in various protein sequence databases revealed homology to members of the so-called major facilitator superfamily (MFS) (17). This family consists of eubacterial, archaeal, and eukaryotic membrane proteins which facilitate the transport of various compounds such as sugars and antibiotics. Typical for members of the MFS is a structure of 12 membrane-spanning α-helices, which were also identified for TfdK (Fig. 2). Furthermore, TfdK contained the GXXX[D/E][R/K]XG[R/K][R/K] motif between the second and third transmembrane domains (GYLADRIGRR; Fig. 2) and a partial repetition of this motif between domains 8 and 9 (GLFASRLIDR), which is another characteristic of MFS members (13).
FIG. 2.
Nucleotide sequence of the 1,846-bp NruI fragment of plasmid pJP4 containing the tfdK gene. DNA sequencing of subclones of this fragment was performed in a Thermosequenase reaction (Amersham, Little Chalfont, United Kingdom) with IRD-41-labeled primers (MWG Biotech, Ebersberg, Germany). DNA fragments were separated and analyzed on a LiCOR Automated Sequencer (model 400L; LiCOR, Lincoln, Nebr.). Relevant restriction sites are indicated in italics. A putative ribosome binding site is boxed. ISJP4 DNA is underlined, and its border is indicated by a black arrow (IR-R). The translation product of tfdK is given below the nucleotide sequence. Two characteristic MFS motifs are circled (13); underlined within these are conserved amino acid residues. Arrows on the amino acid sequence indicate predicted transmembrane α-helices (see text); arrows that point in the same direction as the tfdK reading frame represent helices with an inside-to-outside orientation, oppositely pointed arrows represent outside-to-inside membrane helices. Eleven such helices were identified by the TMpred program of the ISREC Bioinformatics Group (Epalinges, Switzerland). The position of a 12th hydrophobic α-helix is also shown (open arrow), but this helix was not recognized by the program as a membrane-spanning one.
TfdK was most related to proteins for the transport of carboxylated aromatic substances (also see reference 28). The highest degree of resemblance (33% identity and 59% similarity in a 449-aa overlap with one gap) was found with PcaK, a transporter of 4-hydroxybenzoate in Pseudomonas putida (11). Other proteins homologous to TfdK included four from Acinetobacter calcoaceticus, namely, PcaK (31% identity, 56% similarity, 355 aa, and 3 gaps) (14); the benzoate transporter BenK (29% identity, 58% similarity, 473 aa, and 6 gaps) (3); MucK (28% identity, 53% similarity, 467 aa, and 11 gaps) which has been implicated in the transport of cis,cis-muconate (28); and the vanK gene product (30% identity, 55% similarity, 466 aa, and 7 gaps) (GenBank accession no. AF009672). Furthermore, there was similarity with two putative transporters for 3-(3-hydroxyphenyl)propionate, i.e., MhpT (32% identity, 59% similarity, 472 aa, and 7 gaps) from Escherichia coli (GenBank accession no. AE000142) and HppK (27% identity, 53% similarity, 472 aa, and 3 gaps) from Rhodococcus globerulus (GenBank accession no. U89712). It should be noted that transport activity has been demonstrated only for PcaK from P. putida and BenK from A. calcoaceticus (3, 19).
The resemblance with other aromatic acid transporter proteins and the observation that the tfdK gene is located within one large region on plasmid pJP4 with structural and regulatory genes for the utilization of 2,4-D led us to speculate that it codes for a 2,4-D transporter protein. A preliminary indication that R. eutropha has an uptake system for 2,4-D has been reported elsewhere (6). We constructed a tfdK mutant and compared its uptake behavior to that of the wild-type strain. The mutant R. eutropha JMP134(pJP4::cba79) was constructed by insertion of a kanamycin cassette into the tfdK gene on pJP4 as follows. Cells of R. eutropha JMP134 carrying pJP4 were electrotransformed with plasmid pCBA79 (Fig. 2) as described elsewhere (25) with a Gene Pulser (BioRad, Glattbrugg, Switzerland). Plasmid pCBA79 is pUC derived and cannot replicate in R. eutropha. Transformants were selected for growth on nutrient broth (NB) (Biolife, Milan, Italy) plates (10 g/liter) in the presence of 50 μg of kanamycin per ml, which was indicative of recombinational events between plasmids pJP4 and pCBA79. Total DNA of kanamycin-resistant transformants was isolated and analyzed by Southern hybridization with probes for the kanamycin resistance gene, for tfdK, and for pUC (not shown). A plasmid with a double recombination, in which the tfdK gene had become interrupted by the Km cassette, was designated pJP4::cba79.
We determined uptake rates of 14C-labeled 2,4-D by both wild-type and tfdK mutant cells at different 2,4-D concentrations in a rapid filtration assay. The cells were grown at 30°C in 50 ml of Pseudomonas mineral medium (MM) (10) containing 5 mM 2,4-D as the sole carbon and energy source to an optical density at 600 nm of 0.35 to 0.40, after which they were washed three times in an equal volume of ice-cold MM. After the third wash, the cells were resuspended in 1 ml of ice-cold MM and kept on ice until they were used in the uptake assay. The cells were then diluted to a final optical density of 1 in a total reaction volume of 1.8 ml MM supplemented with 160 mg of NB per liter. This mix was incubated at 30°C for 5 min before the addition of 2,4-D. The reaction was started by adding 200 μl of defined mixtures of [ring-UL-14C]-labeled (specific activity, 18.2 mCi/mmol [Sigma Chemical Co., St. Louis, Mo.]) and unlabeled 2,4-D (Aldrich-Chemie, Steinheim, Germany) to a final concentration of 1.0, 4.9, 9.8, 20, 57, 127, 250, 510, 1,010, or 1,885 μM. Directly after the addition of substrate, 200-μl samples were taken at different time intervals and were immediately filtered on a multiple filtration unit (Millipore, Volketswil, Switzerland) through 0.45-μm-pore-size filters (Sartorius AG, Goettingen, Germany) which had been presoaked in MM plus 50 μM 2,4-D. The cells on the filters were then immediately washed with 5 ml of MM plus 50 μM 2,4-D, and the filters were transferred to scintillation tubes, after which 3 ml of Filter-Count (Packard Instrument Co., Downers Grove, Ill.) scintillation cocktail was added. Radioactivity was determined with a BETAmatic I (Kontron Analytical, Zürich, Switzerland) liquid scintillation counter.
Uptake of 2,4-D by wild-type cells of R. eutropha JMP134(pJP4).
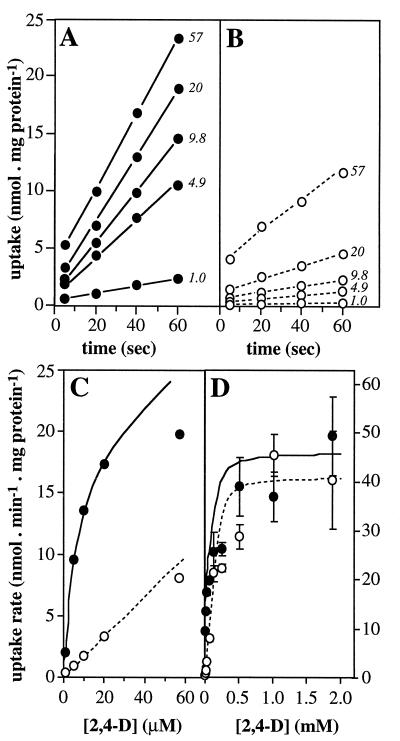
Wild-type JMP134(pJP4) cells that had been grown on 5 mM 2,4-D took up 2,4-D (Fig. 3A). Uptake rates were derived from the slope of the lines through each set of data points. The rates of 2,4-D uptake showed saturation kinetics with 2,4-D concentrations of as much as 60 μM (Fig. 3C), which suggests the involvement of a protein in the transport process. The uptake of 2,4-D was inducible; when cells growing on 10 mM fructose were spiked with 0.2 mM 2,4-D at mid-log phase and were allowed to continue growing for another 45 min before harvesting, we found uptake rates comparable to those of 2,4-D grown cells, whereas fructose-grown cells showed no significant uptake activity in the assay (not shown). Furthermore, the uptake was sensitive to incubation with metabolic inhibitors. When cells were preincubated with 0.5 mM potassium cyanide (Merck, Darmstadt, Germany), which prevents the formation of a proton motive force, uptake (at an initial 2,4-D concentration of 9.8 μM) was reduced to 17% of normal activity. Preincubation with the protonophore carbonylcyanide-m-chlorophenylhydrazone (CCCP) at 20 μM (Fluka, Buchs, Switzerland) or α-dinitrophenol (DNP) (Merck) at 0.5 mM reduced uptake activities to 20 or 4%, respectively. The effect of 1 mM tetraphenylphosphonium (Fluka), which dissipates the membrane potential, was less severe; about half of the activity (44%) remained.
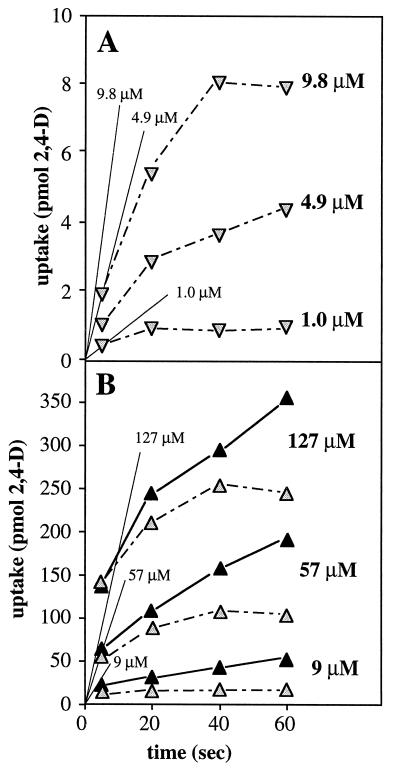
FIG. 3.
Uptake of 2,4-D by 2,4-D-cultivated R. eutropha JMP134 cells harboring pJP4 or pJP4::cba79 at different substrate concentrations. (A and B) Uptake of 2,4-D (in nanomoles · milligram of protein−1) for wild type and the tfdK mutant, respectively. Numbers accompanying a set of data points represent the 2,4-D concentration (micromolar) in that particular uptake assay. Data points were corrected for aspecific binding of 2,4-D to the filters by control experiments in the absence of cells. The slope of the line through a set of data points was taken as the uptake rate. For higher 2,4-D concentrations, lines extrapolated to time zero did not pass through a zero 2,4-D concentration. This phenomenon was observed only with cells grown on 2,4-D; perhaps such cells have an altered cell surface that stimulates adsorption of 2,4-D. The uptake rates were plotted versus corresponding 2,4-D concentrations in the assay as shown in panels C (0 to 60 μM) and D (0 to 2 mM). Bars in panel D represent standard errors of the measurements; error bars at lower concentrations (panel C) fell within the symbols. Protein concentrations of the original cell suspensions were determined by the Bradford method (2) after boiling cells for 10 min in 0.1 N NaOH. Uptake experiments with 2,4-D-grown wild-type or tfdK mutant cells were repeated three times with independently cultured cells; uptake data for one culture only are shown. Solid lines and dotted lines in panels C and D represent modelled uptake behavior of wild-type cells and tfdK mutant cells, respectively (see text). The following parameters were used: 2,4-D permeability constant, 1.8 × 10−4 liters · min−1 · mg of protein−1 (both strains); Vmax,TfdA, 46 (wild-type) or 41 (tfdK mutant) nmol · min−1 · mg of protein−1; Vmax,TfdK of active transport, 20 (wild type) or 0 (tfdK mutant) nmol · min−1 · mg of protein−1; Km,TfdK, 5 μM (wild type only; not applicable for the tfdK mutant).
Uptake of 2,4-D by the tfdK mutant.
Interruption of the tfdK gene had a clearly negative effect on the ability to transport 2,4-D in the micromolar range (Fig. 3B). Rates of uptake of 2,4-D-grown tfdK mutant cells were up to 10 times lower than those of the wild type and increased linearly with the 2,4-D concentration in this range (Fig. 3C). This suggests diffusion as a mechanism for 2,4-D transport. To substantiate this, we calculated the permeability coefficient for 2,4-D from the data obtained with the tfdK mutant, assuming diffusion as the sole transport mode. Uptake by simple diffusion (in micromoles · minute−1 · milligram of protein−1) occurs at a rate (Vdiff) which is equal to
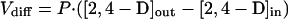 |
1 |
where P is the permeability coefficient (in liters · minute−1 · milligram of protein−1) and [2,4-D]out and [2,4-D]in are the external and intracellular concentrations (micromolar) of 2,4-D, respectively (22). Since intracellular concentrations are kept low in 2,4-D-grown cells because of metabolic activity, we also included a term for the activity of TfdA, the first enzyme of the 2,4-D pathway (8, 9, 24). We measured TfdA activities of 41 nmol · min−1 · mg of protein−1 in extracts of 2,4-D-grown tfdK mutant cells, as determined by the method described by Nickel et al. (20) and 46 nmol · min−1 · mg of protein−1 in extracts of wild-type cells. The activity of the TfdA enzyme (VTfdA) (in micromoles · minute−1 · milligram of protein−1) equals
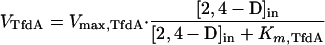 |
2 |
where Vmax,TfdA is the maximum rate of conversion (in this case, 0.041 μmol · min−1 · mg of protein−1, see above) and Km,TfdA is the enzyme’s half-saturation constant (17.5 μM) (9). When diffusion and conversion of 2,4-D are in equilibrium (i.e., Vdiff = VTfdA), equations 1 and 2 can be combined, and the uptake rate (Vupt) (in micromoles · minute−1 · milligram of protein−1), which is equal to Vdiff and VTfdA, can be expressed as a function of the extracellular 2,4-D concentration as follows (1):
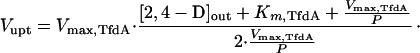 |
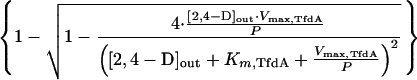 |
3 |
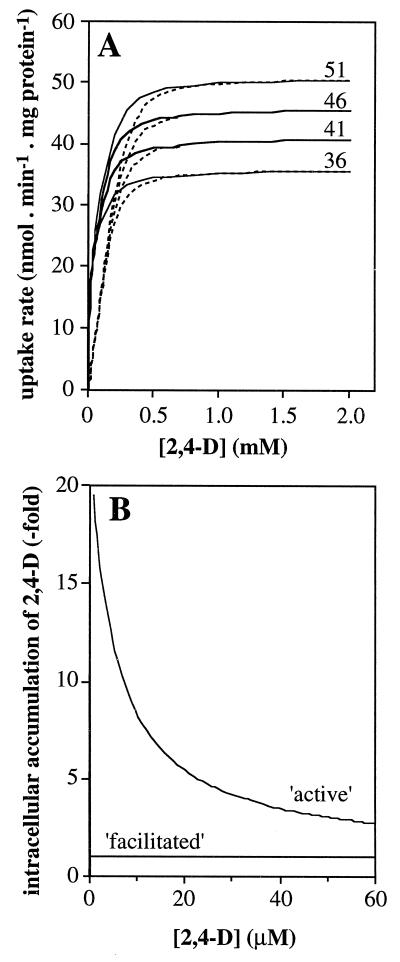
The 2,4-D uptake rates of the tfdK mutant were fitted in equation 3, and we could calculate an apparent permeability coefficient (P) for 2,4-D of 1.8 × 10−4 liters · min−1 · mg of protein−1. By substituting this value in equation 1, one can calculate for any given extracellular 2,4-D concentration the highest possible uptake rate, i.e., Vupt = P · [2,4-D]out (with [2,4-D]in = 0), through diffusion alone. For example, at an extracellular 2,4-D concentration of 1 μM and with approximately 0.025 mg of protein per 0.4 × 109 cells (determined in our assays), and an estimated surface area of 6 × 10−8 cm2 per cell (cells were monad shaped, 2 μm long, and 1 μm in diameter), the maximal number of 2,4-D molecules that can enter a single cell per second is 110. At high extracellular 2,4-D concentrations, the diffusion model predicts saturation of uptake to rates that approximate the Vmax of TfdA (Fig. 4A); this is also what we found experimentally (Fig. 3D). At low 2,4-D concentrations, the rate of transport is indirectly driven by TfdA activity but is limited by the permeability of 2,4-D across the membrane. From these model considerations, we conclude that uptake of 2,4-D by the tfdK mutant most likely occurs through simple diffusion.
FIG. 4.
Predictions of 2,4-D uptake rates by the diffusion model for the tfdK mutant and by the two models for TfdK-mediated transport in wild-type R. eutropha JMP134(pJP4) cells. (A) Influence of TfdA activity for different values of Vmax,TfdA (51, 46, 41, and 36 nmol · min−1 · mg of protein−1) on uptake rates by the tfdK mutant (dotted lines) and wild-type cells (solid lines) at different assay concentrations. For the wild type, the curves for active transport and facilitated diffusion overlap; therefore, only one is shown. (B) Intracellular accumulation of 2,4-D, i.e., [2,4-D]in/[2,4-D]out, in the tfdA mutant (Vmax,TfdA of 0 nmol · min−1 · mg of protein−1) and with an apparent permeability coefficient for 2,4-D of 1.8 × 10−4 liters · min−1 · mg of protein−1, assuming either active transport (Vmax,TfdK, 20 nmol · min−1 · mg of protein−1; Km,TfdK, 5 μM [equation 6]) or facilitated diffusion (Vmax,TfdK, 160 nmol · min−1 · mg of protein−1; Km,TfdK, 4 μM [equation 5]).
Modelling TfdK transport activity.
Since the only distinction between the wild-type strain and the tfdK mutant was the presence of an intact versus interrupted tfdK gene, and since TfdA activities did not differ significantly between these two strains, the apparently higher rates of uptake by the wild type at lower concentrations must be due to a higher transport rate of 2,4-D across the membrane. Therefore, we propose that the product of the tfdK gene, TfdK, facilitates 2,4-D uptake in wild-type R. eutropha JMP134(pJP4). We tested two hypotheses for the function of TfdK: (i) TfdK is an active transporter protein capable of accumulating 2,4-D against a concentration gradient and (ii) TfdK is a facilitator of downhill, i.e., outside-to-inside, diffusion. Both functions were superimposed on the diffusion model (equation 3) and were fitted by numerical iteration (i.e., determining changes in extra- and intracellular 2,4-D concentrations and in the flux of 2,4-D over time intervals of 0.01 s during 30 s as a function of diffusion and TfdK and TfdA activity) to the observed 2,4-D uptake rates of the wild type.
If TfdK represents an active transport system, it would catalyze 2,4-D transport with a rate equal to
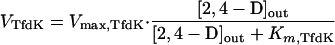 |
4 |
The best-fitting parameters derived for these assumptions (a combination of equations 1, 2, and 4) were an apparent maximal transport rate, Vmax,TfdK, of 20 nmol · min−1 · mg of protein−1 and a half-saturation constant, Km,TfdK, of 5 μM (Fig. 3C). The maximal (apparent) capacity of TfdK transport would thus be 12,600 2,4-D molecules per s per cell, whereas at, e.g., 1 μM [2,4-D]out, TfdK would transport 2,100 molecules. According to this combined model for TfdK active transport plus simple diffusion, 2,4-D uptake of the wild type at higher concentrations should resemble that of the tfdK mutant, since TfdA activity would become the rate-limiting step of transport (Fig. 4A). Indeed, wild-type cells did not differ significantly in uptake behavior from that of mutant cells (Fig. 3D). This seems to be in agreement with our observation that the mutation in tfdK had no effect on the growth of R. eutropha JMP134(pJP4) on 5 mM 2,4-D as the sole carbon source; the growth curves of the wild-type and mutant strains were nearly identical (not shown) with specific growth rates of 0.050 ± 0.003 h−1 and 0.048 ± 0.003 h−1, respectively.
If TfdK catalyzed facilitated diffusion, with equal maximal rates and substrate affinities applying on both sides of the membrane, the mathematics would be as follows:
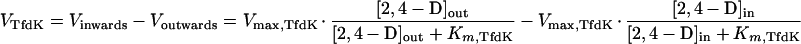 |
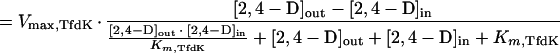 |
5 |
With a Vmax,TfdK of 160 nmol · min−1 · mg of protein−1 (or 2.8 pmol · s−1 · cm−2) and a Km,TfdK of 4 μM, the data for the wild type could be fitted, both at low and high 2,4-D concentrations. In fact, the calculated curve for the uptake rate as a function of extracellular 2,4-D concentration resembled that of the model for active transport closely (and, therefore, it is not shown separately). The facilitated diffusion model predicts that at an extracellular concentration of, e.g., 1 μM, TfdK would be able to transport a maximum of 20,100 2,4-D molecules per s per cell. Similarly to TfdK as an active transporter, uptake rates become rate limited by TfdA at high 2,4-D concentrations (i.e., higher than 1 mM).
While both of the models described above for TfdK describe uptake of 2,4-D equally well for 2,4-D-grown wild-type cells with fully induced 2,4-D metabolism, they predict quite different uptake behaviors when metabolism is lacking. In the case of (facilitated) diffusion, 2,4-D would accumulate to intracellular concentrations approximating those outside, whereas an active transport system would accumulate 2,4-D to levels that are in equilibrium with diffusion of 2,4-D out of the cell again, or, in mathematical terms:
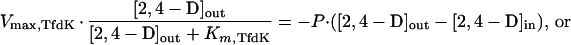 |
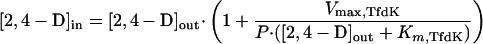 |
6 |
This difference between the two models for TfdK activity is shown in Fig. 4B. To experimentally test which of the two models would be more likely, we performed 2,4-D uptake experiments with a tfdA mutant of R. eutropha JMP134(pJP4) (i.e., R. eutropha pBH501aE [27]; a kind gift of Eva Top, University of Ghent, Belgium). Since this strain cannot grow on 2,4-D, we cultivated it on 10 mM fructose instead and induced the tfd genes with either 2,4-D or 2,4-DCP. The actual inducing agent of the 2,4-D pathway is not 2,4-D or 2,4-DCP but the intermediate 2,4-dichloromuconate (7). Therefore, induction with 2,4-DCP activates the expression of all 2,4-D pathway genes in the wild type and of all genes except tfdA in the tfdA mutant, whereas with 2,4-D all tfd genes become activated in the wild-type and none become activated in the tfdA mutant. We tested this effect on expression of the tfdK gene; by hybridization of total RNA with an antisense tfdK probe, we could indeed demonstrate that the tfdK gene was expressed in wild-type cells after induction with 2,4-D or 2,4-DCP and also in 2,4-DCP-induced tfdA mutant cells, but not in tfdA mutant cells that had been exposed to 2,4-D (not shown). With the 2,4-D-induced tfdA mutant, we studied uptake in the absence of both TfdA and TfdK activities, while the 2,4-DCP-induced tfdA mutant cells allowed us to study uptake in the absence of TfdA activity but with TfdK expressed. 2,4-D uptake rates were compared with those of identically treated wild-type cells.
After growth on fructose plus 2,4-D, tfdA mutant cells showed no significant uptake activity compared to the wild type, which took up 2,4-D at rates that were similar to those of 2,4-D-grown wild-type cells (not shown). Close up, however, tfdA mutant cells appeared to accumulate 2,4-D, but only for a short while (Fig. 5A). The initial uptake rates were as expected for simple diffusion with the calculated apparent permeability coefficient of 2,4-D (Fig. 5A). When assuming a cell volume of 1.5 fl per cell (as for benzoate-grown P. putida cells [26]), more or less stable inside concentrations of 1.6, 7, and 13 μM were reached at extracellular 2,4-D concentrations of 1.0, 4.9, and 9.8 μM, respectively. These findings confirm diffusion to be the mode of transport under these conditions. Wild-type and tfdA mutant cells that were induced with 2,4-DCP took up 2,4-D at almost identical initial rates (Fig. 5B). In contrast to the wild type, however, the rates of uptake by the tfdA mutant slowed down eventually and reached zero (Fig. 5B). This time, 2,4-D was accumulated to intracellular concentrations (35, 178, and 420 μM) that were significantly higher than extracellular concentrations (9, 57, and 127 μM, respectively). At these extracellular concentrations and with TfdK expressed to a level that equals the Vmax,TfdK of that observed for 2,4-D-grown wild-type cells, the active transport model predicted intracellular concentrations of 80, 159, and 234 μM, respectively (calculated by equation 6). These observations favor the model describing TfdK as an active transporter of 2,4-D. The sensitivity of 2,4-D uptake to agents such as CCCP and DNP which dissipate the electrochemical gradient across the membrane support the theory that TfdK mediates an active process. The benzoate transporter PcaK of P. putida uses the energy conserved in the Δψ component of the proton motive force to energize transport (19), and the same is probably also true for BenK (3).
FIG. 5.
2,4-D uptake by 2,4-D-induced tfdA mutant cells (A) and 2,4-DCP-induced (B) wild-type (black symbols) and tfdA mutant (grey symbols) cells of R. eutropha JMP134. Cells were grown on 10 mM fructose to mid-log phase, after which 2,4-D or 2,4-DCP was added to 0.4 or 0.1 mM, respectively, and the cells were allowed to continue growth for another 45 or 70 min, respectively, before harvesting, washing, and use in the uptake assay. Uptake assays were performed as described in the text. The 2,4-D concentration in a particular uptake assay is shown. Uptake values are given in picomoles of 2,4-D (one-time sample of 0.4 × 109 cells) and not in nanomoles per milligram of protein, since the protein content of the tfdA mutant was lower than that of wild-type cells. Thin lines in both panels indicate the predicted initial rates of uptake for tfdA mutant cells at the given assay concentrations either by diffusion (A; equation 1, with [2,4-D]in = 0) or by diffusion plus TfdK-mediated active transport (B; equations 1 plus 4, with [2,4-D]in = 0). Parameters: P = 1.8 × 10−4 liters · min−1 · mg of protein−1; Vmax,TfdK = 20 nmol · min−1 · mg of protein−1; Km,TfdK = 5 μM, with 0.025 mg of protein per filter or total cell surface area of 24 cm2 per filter.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited with GenBank under accession no. U16782.
Acknowledgments
We thank Wolfgang Schumacher for advice on and providing of metabolic inhibitors; Moni Bunk, Christian Zipper, Hanspi Kohler, Hauke Harms, and Mario Snozzi for helpful discussions; and Kathrin Nickel for help with the TfdA protocol. We also appreciate the constructive criticism of five anonymous reviewers.
REFERENCES
- 1.Bosma T N P, Middeldorp P J M, Schraa G, Zehnder A J B. Mass transfer limitation of biotransformation: quantifying bioavailability. Environ Sci Technol. 1997;31:248–252. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the determination of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Don R H, Pemberton J M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid degradative plasmid pJP4. J Bacteriol. 1985;161:466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Don R H, Weightman A J, Knackmuss H J, Timmis K N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134 (pJP4) J Bacteriol. 1985;161:85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filer K, Harker A R. Identification of the inducing agent of the 2,4-dichlorophenoxyacetic acid pathway encoded by plasmid pJP4. Appl Environ Microbiol. 1997;63:317–320. doi: 10.1128/aem.63.1.317-320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukumori F, Hausinger R P. Alcaligenes eutrophus JMP134 2,4-dichlorophenoxyacetate monooxygenase is an α-ketoglutarate-dependent dioxygenase. J Bacteriol. 1993;175:2083–2086. doi: 10.1128/jb.175.7.2083-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukumori F, Hausinger R P. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem. 1993;268:24311–24317. [PubMed] [Google Scholar]
- 10.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Briggs Phillips G. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. [Google Scholar]
- 11.Harwood C S, Nichols N N, Kim M K, Ditty J L, Parales R E. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J Bacteriol. 1994;176:6479–6488. doi: 10.1128/jb.176.21.6479-6488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jessen-Marshall A E, Paul N J, Brooker R J. The conserved motif, GXXX(D/E)(R/K)XG[X](R/K)(R/K), in hydrophilic loop 2/3 of the lactose permease. J Biol Chem. 1995;270:16251–16257. doi: 10.1074/jbc.270.27.16251. [DOI] [PubMed] [Google Scholar]
- 14.Kowalchuk G A, Hartnett G B, Benson A, Houghton J E, Ngai K L, Ornston L N. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene. 1994;146:23–30. doi: 10.1016/0378-1119(94)90829-x. [DOI] [PubMed] [Google Scholar]
- 15.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leveau J H J, van der Meer J R. Characterization of insertion sequence ISJP4 on plasmid pJP4 from Ralstonia eutropha JMP134. Gene. 1997;202:103–114. doi: 10.1016/s0378-1119(97)00460-5. [DOI] [PubMed] [Google Scholar]
- 17.Marger M D, Saier M J. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 18.Matrubutham U, Harker A R. Analysis of duplicated gene sequences associated with tfdR and tfdS in Alcaligenes eutrophus JMP134. J Bacteriol. 1994;176:2348–2353. doi: 10.1128/jb.176.8.2348-2353.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols N N, Harwood C S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickel K, Suter M J, Kohler H P. Involvement of two α-ketoglutarate-dependent dioxygenases in enantioselective degradation of (R)- and (S)-mecoprop by Sphingomonas herbicidovorans MH. J Bacteriol. 1997;179:6674–6679. doi: 10.1128/jb.179.21.6674-6679.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry R H, Chilton C H, Kirkpatrick S D. Chemical engineers’ handbook. New York, N.Y: MacGraw-Hill; 1963. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Streber W R, Timmis K N, Zenk M H. Analysis, cloning, and high-level expression of 2,4- dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taghavi S, van der Lelie D, Mergeay M. Electroporation of Alcaligenes eutrophus with (mega)plasmids and genomic DNA fragments. Appl Environ Microbiol. 1994;60:3585–3591. doi: 10.1128/aem.60.10.3585-3591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thayer J R, Wheelis M L. Active transport of benzoate in Pseudomonas putida. J Gen Microbiol. 1982;128:1749–1753. doi: 10.1099/00221287-128-8-1749. [DOI] [PubMed] [Google Scholar]
- 27.Top E M, Maltseva O V, Forney L J. Capture of a catabolic plasmid that encodes only 2,4-dichlorophenoxyacetic acid α-ketoglutaric acid dioxygenase (TfdA) by genetic complementation. Appl Environ Microbiol. 1996;62:2470–2476. doi: 10.1128/aem.62.7.2470-2476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams P A, Shaw L E. mucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J Bacteriol. 1997;179:5935–5942. doi: 10.1128/jb.179.18.5935-5942.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 30.You I S, Ghosal D. Genetic and molecular analysis of a regulatory region of the herbicide 2,4-dichlorophenoxyacetate catabolic plasmid pJP4. Mol Microbiol. 1995;16:321–331. doi: 10.1111/j.1365-2958.1995.tb02304.x. [DOI] [PubMed] [Google Scholar]