Abstract
Objective
Checkpoint immunotherapy unleashes T-cell control of tumours but is suppressed by immunosuppressive myeloid cells. The transmembrane protein MS4A4A is selectively highly expressed in tumour-associated macrophages (TAMs). Here, we aimed to reveal the role of MS4A4A+ TAMs in regulating the immune escape of tumour cells and to develop novel therapeutic strategies targeting TAMs to enhance the efficacy of immune checkpoint inhibitor (ICI) in colorectal cancer.
Design
The inhibitory effect of MS4A4A blockade alone or combined with ICI treatment on tumour growth was assessed using murine subcutaneous tumour or orthotopic transplanted models. The effect of MS4A4A blockade on the tumour immune microenvironment was assessed by flow cytometry and mass cytometry. RNA sequencing and western blot analysis were used to further explore the molecular mechanism by which MS4A4A promoted macrophages M2 polarisation.
Results
MS4A4A is selectively expressed by TAMs in different types of tumours, and was associated with adverse clinical outcome in patients with cancer. In vivo inhibition of MS4A4A and anti-MS4A4A monoclonal antibody treatment both curb tumour growth and improve the effect of ICI therapy. MS4A4A blockade treatment reshaped the tumour immune microenvironment, resulting in reducing the infiltration of M2-TAMs and exhausted T cells, and increasing the infiltration of effector CD8+ T cells. Anti-MS4A4A plus anti-programmed cell death protein 1 (PD-1) therapy remained effective in large, treatment-resistant tumours and could induce complete regression when further combined with radiotherapy. Mechanistically, MS4A4A promoted M2 polarisation of macrophages by activating PI3K/AKT pathway and JAK/STAT6 pathway.
Conclusion
Targeting MS4A4A could enhance the ICI efficacy and represent a new anticancer immunotherapy.
Keywords: colorectal cancer, macrophages, immunotherapy, T lymphocytes
WHAT IS ALREADY KNOWN ON THIS TOPIC
Immunosuppressive myeloid cells, including macrophages, are abundant in most types of tumours and are thought to be closely related to immunotherapy resistance.
MS4A4A is selectively expressed by macrophage-lineage cells and specifically highly expressed in tumour-associated macrophages (TAMs), but its function remains unknown.
WHAT THIS STUDY ADDS
MS4A4A promotes M2 macrophage polarisation and induces CD8+ T-cell dysfunction.
In vivo inhibition of MS4A4A and anti-MS4A4A monoclonal antibody (mAb) treatment both curb tumour growth.
Anti-MS4A4A mAb treatment remodels the immunosuppressive tumour immune microenvironment.
Targeted MS4A4A treatment enhances the efficacy of programmed cell death protein 1 (PD-1) blockade.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our results may open a new window on the role of TAMs in modulating antitumour immunity and provide new directions for the development of more efficient tumour immunotherapy strategies.
Introduction
The advent of immunotherapy has ushered in a new era of therapies for cancer.1 Nonetheless, there is an unmet clinical need to achieve benefits from immunotherapy in patients with colorectal cancer (CRC). Currently, only subgroups with mismatch repair-deficient or microsatellite instability-high (MSI-H) CRC can benefit from immune checkpoint inhibitors (ICIs).2 However, MSI-H CRC accounts for only 15% of all CRC cases. In advanced CRC, the proportion is even lower, accounting for only 4~5%.3 Therefore, there is still a pressing need to extend CRC immunotherapy further to benefit broader patient populations.
The tumour microenvironment (TME) is an important mediator of tumour immunotherapy resistance. Many studies have shown that various immunosuppressive cells in the TME are closely related to immunotherapy resistance.4 5 For example, myeloid cells can suppress T-cell function through direct ligand-receptor binding, release of soluble inhibitory cytokines and sequestration of T-cell essential amino acids.6 Therefore, relieving the immunosuppression in the TME is an important means of improving the effect of immunotherapy.
Macrophages are among the most abundant immune cells observed in CRC, and those infiltrating the TME are usually defined as tumour-associated macrophages (TAMs).7 High TAM density correlates with a poor prognosis in several cancers, but its role in CRC is controversial.8 One study found that tumour front infiltration by CD68+ TAMs was associated with improved survival in a patient with CRC.9 However, CD68+ TAMs density was not a significant prognostic biomarker in another study.10 Such conflicting data are attributed to the high plasticity of macrophages.11 TAMs can polarise toward pro-inflammatory M1 or anti-inflammatory M2 phenotypes. However, TAMs in tumours are often acclimated by tumour cells and polarise toward the M2-phenotype. Moreover, M2-TAMs suppress cytotoxic T cells through a variety of mechanisms. The most important pathway by which M2-TAMs exert immunosuppressive effects is the induction of T-cell exhaustion. When T cells are exhausted, they show loss of the proliferative potential, decreased and modified effector function and increased coexpression of inhibitory receptors (IRs).12 How to inhibit or reverse T-cell exhaustion in the TME is an intensely studied and difficult issue.
Using public databases and bioinformatic analysis, we identified a target gene, a membrane spanning four domains A4A (MS4A4A), closely related to M2 polarisation and T-cell exhaustion. MS4A4A belongs to the membrane-spanning 4A protein family and has been identified as an M2 macrophages marker.13 MS4A4A is selectively expressed in tissue-resident macrophages under homoeostatic and inflammatory conditions and is highly expressed in TAMs.14 Previous studies have shown that high MS4A4A expression is significantly associated with poor prognosis in several cancers.15 16 However, the function of MS4A4A in TAMs and the relationships of TAM-expressed MS4A4A with CRC development remain elusive.
Here, we used anti-MS4A4A monoclonal antibody (mAb) for the first time to investigate the therapeutic potential of MS4A4A blockade in the treatment of CRC. Our work shows that anti-MS4A4A treatment can not only delay tumour progression, reshape the tumour immune microenvironment (TIME), but also enhance the efficacy of anti-programmed cell death protein 1 (PD-1) therapy.
Results
MS4A4A is selectively upregulated in TAMs and associated with a poor prognosis in patients with cancer
Macrophages are one of the most abundant immune cell types infiltrating CRC (online supplemental figure S1A). Accumulated TAMs in CRC are associated with tumour progression and therapeutic efficacy.10 17 Many studies have shown that TAMs are often subjected to tumour cell acclimation and polarisation toward the M2-phenotype,18 19 which in turn induces T-cell exhaustion to mediate resistance to ICI therapy.20 21 In a search for key regulatory molecules related to the M2-type polarisation and T-cell exhaustion, we first used the Gene Expression Omnibus microarray data for macrophages to screen the differentially expressed genes (DEGs) in M2 macrophages. Subsequently, we comprehensively analysed the expression features of IRs, including PDCD1, TIGIT, CTLA4, LAG3, HAVCR2 and CD160, in The Cancer Genome Atlas (TCGA) and the GSE17538 CRC data set and identified a series of DEGs highly correlated with IRs. Finally, we performed the intersection of the above DEGs and identified one gene: MS4A4A (online supplemental figure S1B).
gutjnl-2022-329147supp001.pdf (1,012.9KB, pdf)
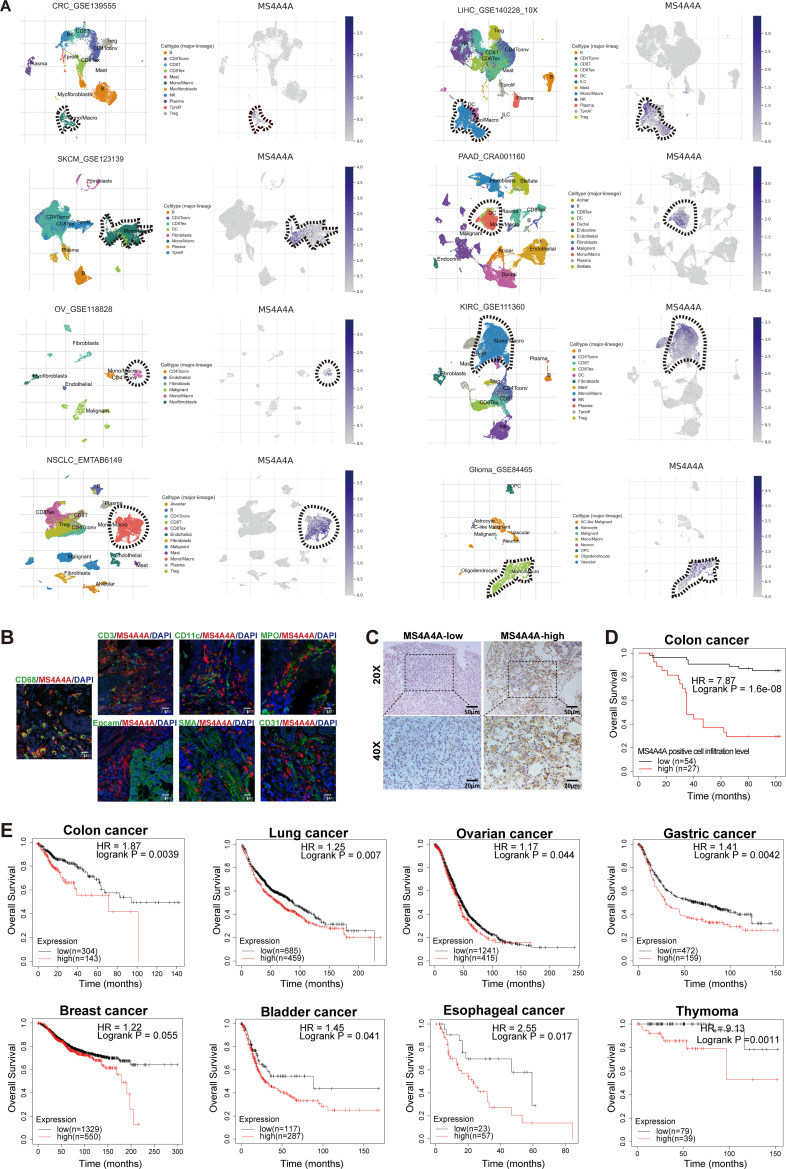
In the GSE39582 CRC data set, we further verified that MS4A4A was significantly positively correlated with the expression of various M2 markers and T-cell exhaustion markers (online supplemental figure S1C). To confirm whether the association of MS4A4A with T-cell exhaustion is general, we analysed the association of MS4A4A expression with T-cell exhaustion signatures in multiple tumours. Strikingly, MS4A4A expression was highly correlated with the T-cell exhaustion signature, indicating that the presence of MS4A4A+ TAMs is indicative of an immunosuppressed and exhausted CD8+ tumour-infiltrating lymphocyte compartment (online supplemental figure S1D). Using the Tumor Immune Single-cell Hub (TISCH) database and immunofluorescence analysis, we found that MS4A4A gene was specifically expressed in TAMs in various tumours, including CRC, and MS4A4A protein colocalised with TAMs in CRC tissues (figure 1A,B and online supplemental figure S1E). A recent study showed that TAMs could be divided into two cell subpopulations, C1QC+ TAMs and SPP1+ TAMs.22 Here, MS4A4A was predominantly expressed in C1QC+ TAMs, a subgroup reported to be associated with T-cell exhaustion,23 and their expression of M2 markers was higher than in SPP1+ TAMs (online supplemental figure S2A–H). Furthermore, using the GEPIA2021 online tool, we found that MS4A4A gene expression was significantly upregulated in M2 macrophages in various tumours (online supplemental figure S2I).
Figure 1.
MS4A4A was highly expressed in tumour-associated macrophages and associated with a poor prognosis in patients with cancer. (A) Analysis of MS4A4A gene expression in various tumours using the pan-cancer single-cell sequencing data set from the TISCH database (http://tisch.comp-genomics.org/). (B) Representative images of immunofluorescence costaining for MS4A4A (red) with cell-type markers (green) in clinical human CRC tissue specimens. (C) IHC staining with MS4A4A-specific antibodies to detect MS4A4A+ cells infiltration in a human CRC tissue microarray (n=81). (D) Overall survival curves based on MS4A4A+ cells infiltration level using the Kaplan-Meier method. (E) The Kaplan-Meier Plotter online tool (https://kmplot.com/analysis/) was used to analyse the association between MS4A4A mRNA expression levels and overall survival in various tumour (including colon cancer, lung cancer, ovarian cancer, gastric cancer, breast cancer, bladder cancer, oesophageal cancer and thymoma). CRC, colorectal cancer; IHC, immunohistochemistry; mRNA, messenger RNA; TISCH, Tumour Immune Single-cell Hub.
gutjnl-2022-329147supp002.pdf (1.1MB, pdf)
Subsequently, human peripheral blood mononuclear cells, human THP-1 monocytic cells and mouse bone marrow-derived macrophages (BMDMs) were stimulated with different cytokines to induce M1 or M2 polarisation. Our results showed that the messenger RNA (mRNA) and protein levels of MS4A4A were significantly upregulated in M2 macrophages (online supplemental figure S3A–C). We then used CRC cell line culture supernatants to stimulate the above-mentioned macrophages to differentiate into TAMs. Quantitative real-time PCR (qRT-PCR) assays showed that MS4A4A mRNA levels were significantly upregulated in TAMs compared with controls (online supplemental figure S3D–F).
gutjnl-2022-329147supp003.pdf (437KB, pdf)
We evaluated the level of MS4A4A+ cells infiltration in a human CRC tissue microarray by immunohistochemistry (IHC) staining, and then divided patients with CRC into high-density or low-density groups of MS4A4A+ cells based on the mean MS4A4A+ cells density (13.26 cells/high-power field (HPF)) in the whole cohort. Kaplan-Meier analysis showed that high MS4A4A+ cell density was associated with poor overall survival (OS) (figure 1C,D). External data sets also validated that high MS4A4A mRNA level could predict significantly poor OS in various tumours (figure 1E and online supplemental figure S4A–D). These data thus indicate that upregulation of MS4A4A in TAMs is correlated with tumour development.
gutjnl-2022-329147supp004.pdf (505.4KB, pdf)
MS4A4A promotes M2 macrophage polarisation and induces CD8+ T-cell dysfunction
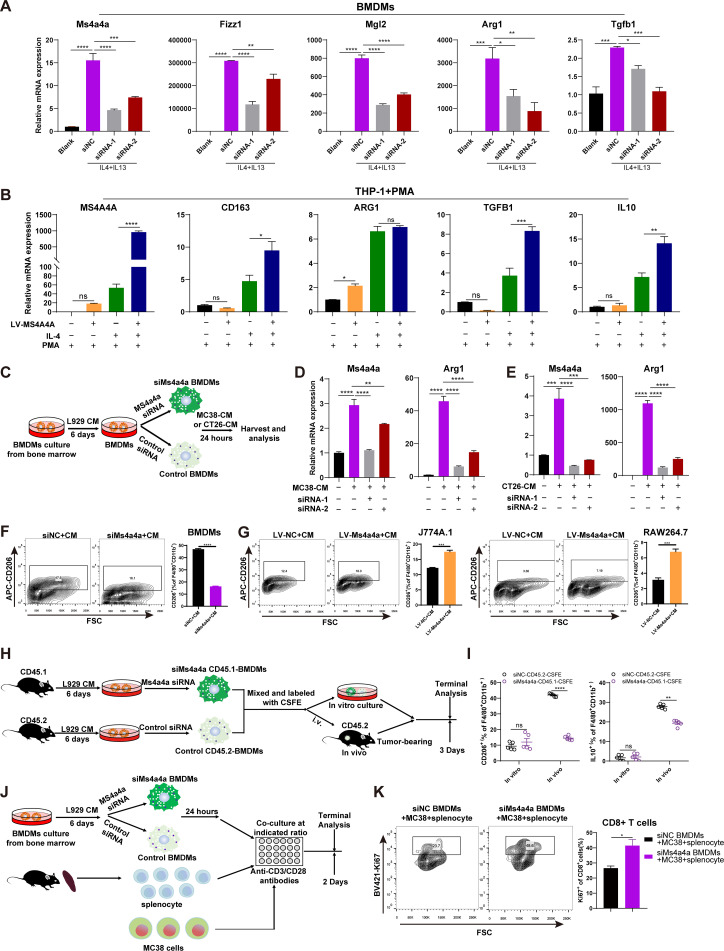
To investigate whether MS4A4A regulates M2 polarisation, we interfered MS4A4A expression in BMDMs using MS4A4A-specific small interfering RNA (siRNA) (siMs4a4a) and polarised them to M2-phenotype by adding interleukin (IL)-4/IL-13. We observed that compared with that in the control group (control siRNA (siNC)), the expression of M2 markers (ie, Fizz1, Mgl2, Arg1 and Tgfb1) in the siMs4a4a group was significantly reduced (figure 2A and online supplemental figure S5A). Next, we used a lentivirus to construct a THP-1 cell line stably overexpressing MS4A4A, induced THP-1 cells to differentiate into M0 macrophages with PMA and then induced M2 polarisation with IL-4. The results showed that most M2 markers were upregulated in the MS4A4A-overexpressing group compared with the control group (figure 2B).
Figure 2.
MS4A4A promotes M2 macrophage polarisation and induces CD8+ T-cell dysfunction. (A) BMDMs were transfected with Ms4a4a-specific siRNA or negative control siRNA and polarised into the M2-phenotype using IL-4 (20 ng/mL) and IL-13 (20 ng/mL). The interference efficiency of Ms4a4a and the expression of M2 markers (Fizz1, Mgl2, Arg1 and Tgfb1) were measured by qRT-PCR. (B) An MS4A4A-overexpressing cell line was constructed using the human monocyte cell line THP-1, and then the engineered THP-1 cells were induced to differentiate into M0 macrophages with PMA (50 ng/mL). IL-4 (20 ng/mL) was used to stimulate M0 macrophages to polarise into the M2-phenotype, and the expression levels of MS4A4A and M2 markers (CD163, VEGFA, IL-10, ARG1 and TGFB1) were measured by qRT-PCR. (C–G) Study of the effect of MS4A4A expression on macrophage polarisation in vitro. (C) Bone marrow cells from C57BL/6 mice were extracted in vitro and induced into BMDMs using L929 cells conditioned medium (L929-CM). The BMDMs were then transfected with MS4A4A-specific siRNA or control siRNA on day 6. After 48 hours, the macrophages were cultured in MC38 cells conditioned medium (MC38-CM) or CT26 cells conditioned medium (CT26-CM) for 24 hours. (D–E) The expression levels of Ms4a4a and Arg1 were measured by qRT-PCR (n=3). (F–G) The proportion of M2 macrophages in each group of macrophages was detected by flow cytometry (n=3). (H–I) In vivo confirmation of the effect of MS4A4A on the M2 polarisation function of TAMs. (H) The BMDMs with MS4A4A differential expression (siNC-CD45.2 and siMs4a4a-CD45.1) were labelled with CFSE, and then the two types of cells were mixed at a ratio of 1:1. Some of these cells were cultured in vitro, and others were transferred into tumour-bearing mice. After 3 days, FACS was performed on the above donor cells. (I) FACS analysis of CD206 and IL-10 expression in two types of donor cells (siNC-CD45.2 and siMs4a4a-CD45.1) in vitro and in vivo (n=5). (J–K) Study of the effect of macrophage MS4A4A expression on CD8+ T-cell function in vitro. (J) Experimental design. (K) FACS analysis of Ki67 expression on the indicated CD8+ T cells (n=3). Results are represented as mean±SEM. BMDMs, bone marrow-derived macrophages; CFSE, carboxyfluorescein succinimidyl ester; CM, conditioned medium; FACS, flow cytometry; FSC, forward scatter; IL, interleukin; LV, lentiviral vectors; mRNA, messenger RNA; PMA, phorbol 12-myristate 13-acetate; qRT-PCR, quantitative real-time PCR; siMs4a4a, MS4A4A-specific siRNA; siNC, control siRNA; siRNA, small interfering RNA; TAMs, tumour-associated macrophages.
gutjnl-2022-329147supp005.pdf (625.8KB, pdf)
To better mimic the effect of tumour cells on macrophages in the TME, we prepared mouse CRC cell-derived conditioned medium (CM) and then cultured BMDMs expressing MS4A4A at different levels with CM to induce them to become TAMs (figure 2C). Our results showed that adding CM alone significantly upregulated MS4A4A and ARG1 mRNA expression in macrophages compared with blank control. However, when the expression of MS4A4A was interfered, the regulation of macrophage ARG1 mRNA expression by CM was significantly rescued (figure 2D–E). Furthermore, flow cytometry (FACS) analysis showed that interference with MS4A4A expression decreased CD206 protein expression in TAMs, suggesting that MS4A4A deficiency reduces the M2-like macrophage fraction among TAMs (figure 2F). Additionally, we constructed MS4A4A overexpressing macrophage cell lines for the same experiment and obtained consistent results (figure 2G and online supplemental figure S5B,C).
Next, we used CM to stimulate the conversion of BMDMs with different MS4A4A expression levels into TAMs and collected the TAMs culture supernatant. Our results showed that compared with the control group, the group with MS4A4A expression interference exhibited significantly downregulated expression of the anti-inflammatory cytokines (IL-10, TGF-β1) in the TAM culture supernatant (online supplemental figure S5D). In contrast, MS4A4A overexpression significantly upregulated the levels of both cytokines in TAM culture supernatant (online supplemental figure S5D), suggesting that MS4A4A+ macrophages have greater immunosuppressive capacity. To further confirm the effect of MS4A4A on TAMs functional conversion, we labelled MS4A4A differentially expressed BMDMs (siNC-CD45.2 and siMs4a4a-CD45.1) with carboxyfluorescein succinimidyl ester (CFSE), and then transferred a mixture of both cell types into MC38 tumour-bearing mice. FACS analysis revealed M2 markers expression was significantly downregulated in siMs4a4a-CD45.1-TAMs compared with siNC-CD45.2-TAMs in MC38 tumour (figure 2H,I).
Since a major pathogenic activity of TAMs is to suppress antitumour immune responses,24 we then tested the inhibitory effects of TAMs MS4A4A expression on T-cell proliferative capacity. Compared with control treatment, inhibition of MS4A4A expression significantly enhanced the proliferation ability of cocultured CD8+ T-cells, suggesting that the inhibitory effect of TAMs on T cells was alleviated (figure 2J,K). Together, these results demonstrate that MS4A4A significantly promotes M2-TAM polarisation, leading to a more immunosuppressive TME.
In vivo inhibition of macrophage MS4A4A delays CRC progression
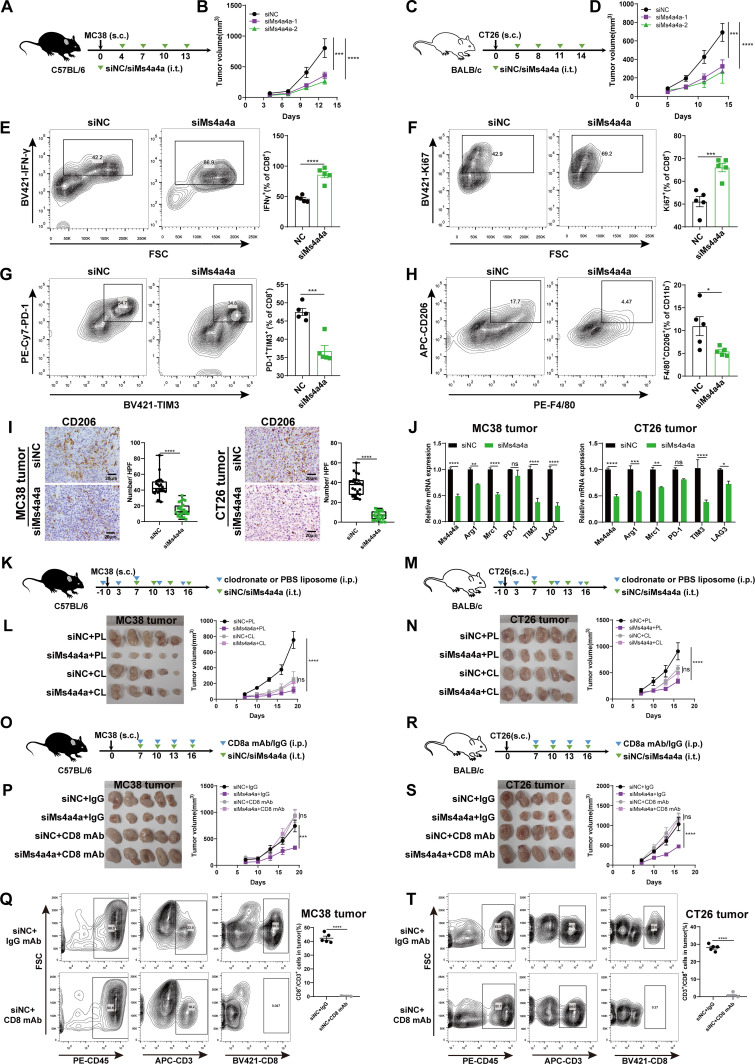
To explore whether MS4A4A regulates macrophage polarisation to affect tumours growth in vivo, we used in vivo-optimised siRNA targeting the MS4A4A gene (siMs4a4a) or control siRNA (siNC) to treat the immunocompetent mice inoculated with MC38 or CT26 cells (figure 3A,C and online supplemental figure S6A,B). The results showed that inhibition of MS4A4A via in vivo-optimised RNA interference (RNAi) significantly delayed tumour growth (figure 3B,D). By analysing immune cell composition in tumour, peripheral blood and spleen, we observed enhanced antitumour immunity in siMs4a4a-treated mice, as evidenced by a marked decrease in macrophages, along with increased CD8+ T-cells (online supplemental figure S7A–E). Further, we found that the activity of tumour-infiltrating CD8+ T-cells (IFN-γ and Ki67) was significantly increased in the siMs4a4a group (figure 3E,F and online supplemental figure S7F,G), while the expression levels of T-cell exhaustion markers (PD-1 and TIM3) and the level of M2-TAM infiltration were significantly reduced (figure 3G–J and online supplemental figure S7H,I).
Figure 3.
In vivo inhibition of macrophage MS4A4A delays CRC progression. (A–J) Tumour growth in mice injected subcutaneously (s.c.) with the MC38 or CT26 cell line and treated with siRNA (siMs4a4a) in vivo (n=5/group). (A) Schematic showing the treatment plan. (B) Tumour growth in MC38 tumour-bearing C57BL/6 mice. (C) Schematic showing the treatment plan. (D) Tumour growth in CT26 tumour-bearing BALB/c mice. (E–H) FACS analysis of specific molecule expression on tumour-infiltrating CD8+ T cells and TAMs from CT26 tumour-bearing BALB/c mice. (I) IHC staining with CD206-specific antibodies to detect CD206+ macrophage infiltration in subcutaneously transplanted MC38 or CT26 tumours. The number of CD206-positive cells per high-power field (HPF) was counted in subcutaneous tumour sections from each group of mice. Five random HPFs were selected for analysis on each slide. (J) Relative expression of the indicated genes determined by qRT-PCR. (K–L) C57BL/6 mice were implanted with MC38 cells and received siNC plus PBS liposome (PL), siMs4a4a plus PL, siNC plus clodronate liposome (CL) or siMs4a4a plus CL treatment (n=5/group). (K) Schematic showing the treatment plan. (L) Left: Representative images of tumours in mice from different treatment groups. Right: Tumour growth. (M–N) BALB/c mice were implanted with CT26 cells and received siNC plus PL, siMs4a4a plus PL, siNC plus CL or siMs4a4a plus CL treatment (n=5/group). (M) Schematic showing the treatment plan. (N) Left: Representative images of tumours in mice from different treatment groups. Right: Tumour growth. (O–Q) C57BL/6 mice were implanted with MC38 cells and received siNC plus IgG antibody, siMs4a4a plus IgG antibody, siNC plus anti-CD8 antibody or siMs4a4a plus anti-CD8 antibody treatment (n=5/group). (O) Schematic showing the treatment plan. (P) Left: Representative images of tumours in mice from different treatment groups. Right: Tumour growth. (Q) FACS analysis of the depletion efficiency for CD8+ T-cells in MC38 tumours. (R–T) BALB/c mice were implanted with CT26 cells and received siNC plus IgG antibody, siMs4a4a plus IgG antibody, siNC plus anti-CD8 antibody or siMs4a4a plus anti-CD8 antibody treatment (n=5/group). (R) Schematic showing the treatment plan. (S) Left: Representative images of tumours in mice from different treatment groups. Right: Tumour growth. (T) FACS analysis of the depletion efficiency for CD8+ T-cells in CT26 tumours. CRC, colorectal cancer; FACS, flow cytometry; FSC, forward scatter; IHC, immunohistochemistry; i.p., intraperitoneal injection; i.t., intratumoral injection; mAb, monoclonal antibody; PBS, phosphate buffer solution; qRT-PCR, quantitative real-time PCR; siMs4a4a, MS4A4A-specific siRNA; siNC, control siRNA; siRNA, small interfering RNA; TAMs, tumour-associated macrophages;
gutjnl-2022-329147supp006.pdf (199.8KB, pdf)
gutjnl-2022-329147supp007.pdf (1.8MB, pdf)
Next, to verify whether the inhibitory effect of MS4A4A inhibition on CRC growth was macrophage-dependent, we depleted macrophages in mice using clodronate liposome. The results showed that the effect of MS4A4A inhibition on the growth of CRC was obviously restored after macrophages were depleted (figure 3K–N and online supplemental figure S8A,B). To further confirm the tumour-promoting role of MS4A4A in TAM, we adoptively transferred GFP-labelled MS4A4A-overexpressing BMDMs (OE-MS4A4A-GFP) and control BMDMs (OE-NC-GFP) into mice pre-challenged with MC38 cells. Our results showed that adoptive transfer of OE-MS4A4A-GFP BMDMs significantly promoted MC38 tumour growth in host mice compared with the control group. However, the tumour-promoting effect of MS4A4A was significantly eliminated by macrophage depletion (online supplemental figure S9A–E). Furthermore, to explore whether the inhibitory effect of MS4A4A inhibition on CRC growth is mediated by CD8+ T-cells, we injected the mouse model with anti-CD8 antibody. We found that the inhibition of tumour growth in siMs4a4a-treated mice was blocked by depleting CD8+ T-cells with anti-CD8 antibody (figure 3O–T). To evaluate whether siRNA injection in vivo can directly inhibit tumour cells proliferation, we treated tumour cells with Ms4a4a-specific siRNA in vitro and found that siRNA treatment did not affect tumour cells proliferation (online supplemental figure S10A–C). The above experiments confirmed that in vivo inhibition of macrophage MS4A4A delays CRC progression and this tumour-suppressive effect was dependent on the presence of macrophages and CD8+ T-cells.
gutjnl-2022-329147supp008.pdf (1.2MB, pdf)
gutjnl-2022-329147supp009.pdf (1.3MB, pdf)
gutjnl-2022-329147supp010.pdf (901.4KB, pdf)
MS4A4A promotes M2 macrophage polarisation by activating the PI3K/AKT and JAK/STAT6 pathway
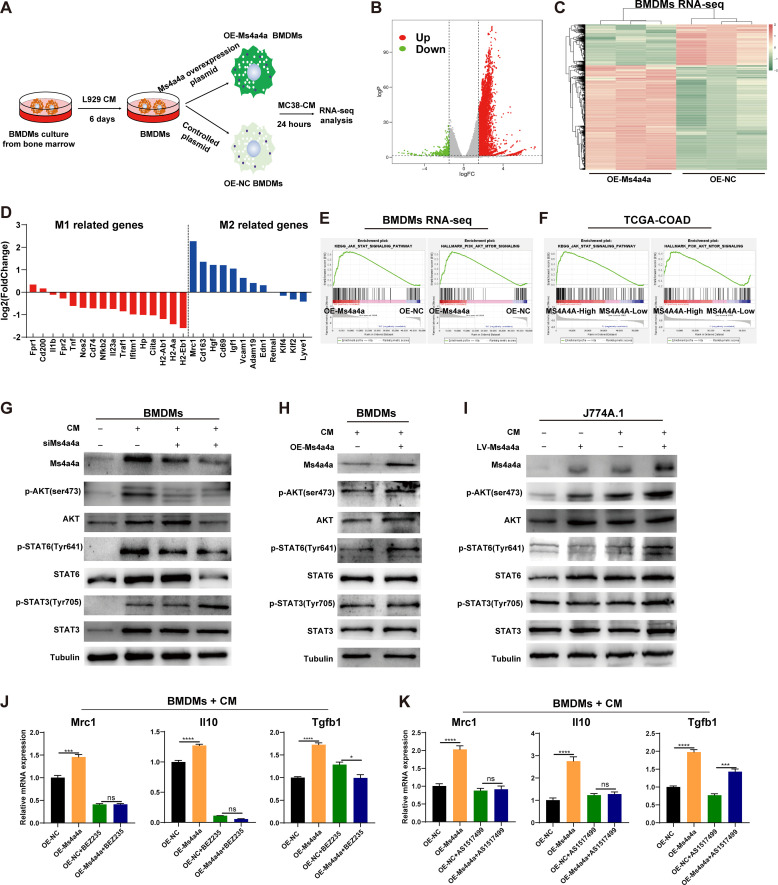
Next, to investigate the molecular mechanism by which MS4A4A promotes M2 polarisation, we transfected an Ms4a4a-overexpression plasmid or control plasmid into BMDMs and stimulated them with MC38 conditioned medium (MC38-CM) (figure 4A). By performing RNA sequencing (RNA-seq) analysis of these TAMs, we found that nearly all M2 macrophage-related genes were upregulated in the MS4A4A-overexpressing group, while most M1 macrophage-related genes were downregulated (figure 4B–D).
Figure 4.
MS4A4A promotes M2 macrophage polarisation by activating the PI3K/AKT and JAK/STAT6 signalling pathway. (A) Bone marrow cells from C57BL/6 mice were induced to differentiate into BMDMs using L929-CM and transfected with an MS4A4A-overexpression plasmid or control plasmid on day 6. After transfection for 48 hours, the cells were stimulated with MC38-CM for 24 hours to induce TAMs and collected for RNA-seq analysis. (B) Volcano plot of differentially and non-differentially expressed genes revealed by RNA-seq analyses comparing Ms4a4a-overexpressing TAMs and control TAMs. (C) Heatmap showing the differential expression of genes in Ms4a4a-overexpressing TAMs versus control TAMs. (D) Gene expression analyses of M1 and M2 macrophage-related genes in Ms4a4a-overexpressing TAMs relative to that in control TAMs. (E) GSEA of RNA-seq data revealed that the JAK/STAT signalling pathway and PI3K/AKT/mTOR signalling pathway were significantly enriched in the Ms4a4a-overexpressing TAM group. (F) GSEA confirmed that the JAK/STAT and PI3K/AKT/mTOR signalling pathway were significantly enriched in patients with CRC with high MS4A4A expression in the CRC data from TCGA. (G–I) Macrophages with different MS4A4A expression levels were stimulated using MC38-CM, followed by western blotting to detect the expression of Ms4a4a, p-AKT (Ser473), AKT, p-STAT6 (Tyr641), STAT6, p-STAT3 (Tyr705) and STAT3. (J) MS4A4A-overexpressing BMDMs and control BMDMs were pretreated with or without the PI3K inhibitor BEZ235 (200 nM) and then stimulated with MC38-CM for 12 hours. qRT-PCR was performed to detect the expression of M2 markers (Mrc1, Il10 and Tgfb1). (K) MS4A4A-overexpressing BMDMs and control BMDMs were pretreated with or without the STAT6 inhibitor AS1517499 (250 nM) and then stimulated with MC38-CM for 12 hours. qRT-PCR was performed to detect the expression of M2 markers (Mrc1, Il10 and Tgfb1). BMDMs, bone marrow-derived macrophages; CM, conditioned medium; COAD, colon adenocarcinoma; CRC, colorectal cancer; GSEA, gene set enrichment analysis; NC, negative control; OE, overexpression; qRT-PCR, quantitative real-time PCR; RNA-seq, RNA sequencing; TAMs, tumour-associated macrophages; TCGA, The Cancer Genome Atlas.
Furthermore, the above RNA-seq data and the TCGA CRC data set were used for gene enrichment analysis. We found that the JAK/STAT and PI3K/AKT/mTOR pathways were significantly enriched in MS4A4A-overexpressing macrophages (OE-Ms4a4a) and patients with CRC with high MS4A4A expression (figure 4E,F). Previous studies have shown that the PI3K/Akt/mTOR, JAK/STAT6 and JAK/STAT3 pathways play important roles in regulating macrophage M2 polarisation.25 26 To investigate whether these pathways mediate the regulation of M2 polarisation by MS4A4A, we stimulated macrophages differentially expressing MS4A4A with MC38-CM to mimic the polarising effect of tumour cells on macrophages. Our results showed that MS4A4A promoted AKT and STAT6 phosphorylation in macrophages, but did not affect STAT3 phosphorylation (figure 4G–I). Furthermore, using a PI3K inhibitor (BEZ235) or STAT6 inhibitor (AS1517499) to treat MS4A4A-overexpressing macrophages, we found that both inhibitors significantly restrained the effect of MS4A4A in upregulating M2-type marker expression in macrophages (figure 4J,K). To investigate which PI3K isoforms mediate the effect of MS4A4A on M2 polarisation of TAMs, we treated macrophages with inhibitors of different PI3K isoforms. The results showed that PI3Kγ isoform inhibitor could significantly restore MS4A4A-mediated M2 polarisation promotion (online supplemental figure S11A,B).
gutjnl-2022-329147supp011.pdf (416.2KB, pdf)
Anti-MS4A4A mAb treatment delays CRC progression and remodels the immunosuppressive TIME
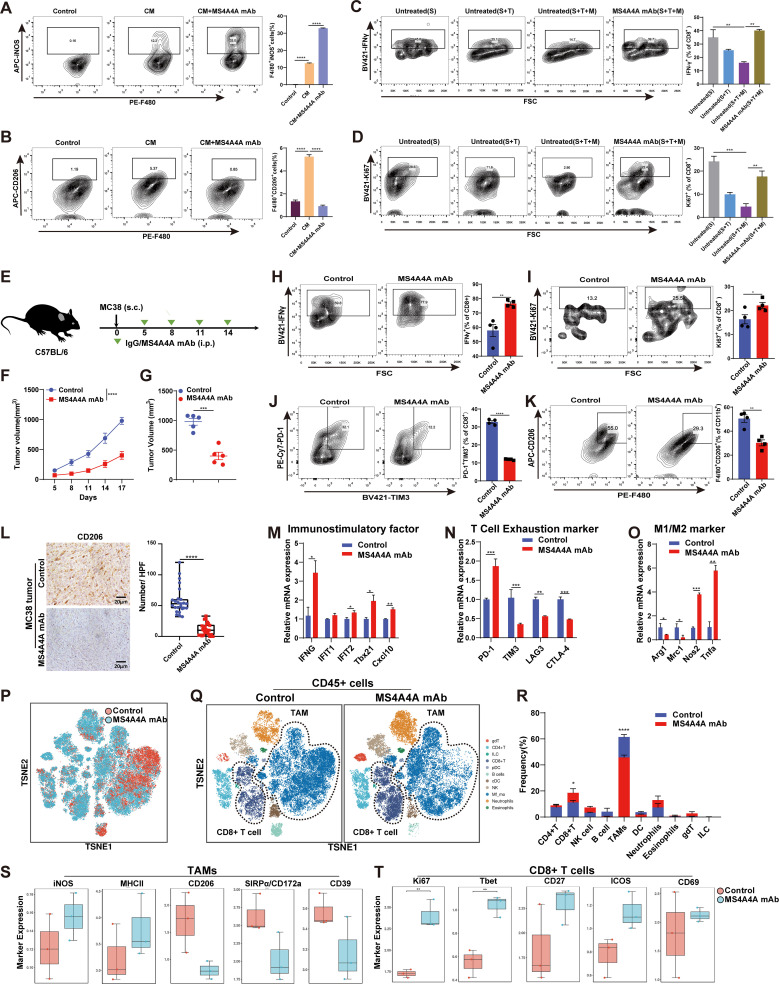
To evaluate the potential of an MS4A4A+ macrophage-targeting strategy to enhance antitumour responses, an anti-MS4A4A mAb recently developed by our group was used, which was able to recognise both human MS4A4A and mouse Ms4a4a (online supplemental figure S12A,B). First, to verify the effect of the anti-MS4A4A mAb on the TAM polarisation in vitro, we treated BMDMs with MC38-CM and the anti-MS4A4A mAb. qRT-PCR and FACS analysis showed that anti-MS4A4A significantly reverted MC38-CM-induced M2 polarisation and promoted M1 polarisation, suggesting that anti-MS4A4A treatment blocked TAMs M2 polarisation (figure 5A,B and online supplemental figure S12C). Furthermore, in vitro coculture experiments showed a marked inhibition of CD8+ T-cell function in coculture with tumour cells and macrophages. However, the addition of anti-MS4A4A mAb could significantly restore the function of CD8+ T-cells (figure 5C,D).
Figure 5.
Anti-MS4A4A mAb treatment delays CRC progression. (A–B) Mouse bone marrow-derived macrophages were treated with MC38-CM or MC38-CM+anti-MS4A4A mAb (10 µg/mL). FACS analyses of iNOS and CD206 expression by the macrophages (n=3). (C–D) Mouse bone marrow-derived macrophages (M) were cocultured with mouse splenocytes (S) and MC38 tumour cells (T) at a 1:1:1 ratio. The cells were treated with 10 µg/mL anti-MS4A4A mAb for 2 days. FACS analyses of Ki67 and IFN-γ expression by the CD8+ T-cells (n=3). (E–O) Tumour growth in mice injected subcutaneously (s.c.) with murine MC38 colon cancer cells treated with anti-MS4A4A mAb (n=5/group) (E) Schematic showing the treatment plan. (F–G) Tumour growth. (H–K) FACS analysis of specific molecule expression on tumour-infiltrating CD8+ T-cells and TAMs from MC38 tumour-bearing mice. (L) IHC staining with CD206-specific antibodies to detect CD206+ macrophage infiltration in subcutaneously transplanted MC38 tumours. The number of CD206-positive cells per high-power field (HPF) was counted in subcutaneous tumour sections from each group of mice. Five random HPFs were selected for analysis on each slide. (M–O) Relative expression of the indicated genes determined by qRT-PCR. Data depict the mean±SEM (n=3) and are representative of three independent experiments. (P–Q) Murine CT26 colon cancer cells were transplanted subcutaneously (s.c.) into BALB/c mice and treated with an isotype control or anti-MS4A4A mAb (n=3/group). t-SNE analysis of CyTOF data for immune cells from the isotype-treated and anti-MS4A4A mAb-treated CT26 tumours. (R) Histogram showing the quantification of tumour-infiltrating immune cells. (S) CyTOF analysis to compare the differences in the expression of specific molecules in TAMs within CT26 tumours between the control antibody-treated and MS4A4A mAb-treated groups. (T) CyTOF analysis to compare the differences in the expression of specific molecules in CD8+ T-cells within CT26 tumours between the control antibody-treated and MS4A4A mAb-treated groups. CM, conditioned medium; CRC, colorectal cancer; CyTOF, mass cytometry by time of flight; FACS, flow cytometry; FSC, forward scatter; IHC, immunohistochemistry; i.p., intraperitoneal injection; mAb, monoclonal antibody; qRT-PCR, quantitative real-time PCR; TAMs, tumour-associated macrophages; t-SNE, t-distributed stochastic neighbour embedding.
gutjnl-2022-329147supp012.pdf (568KB, pdf)
To test the effect of anti-MS4A4A mAb on CRC growth in vivo, we used anti-MS4A4A mAb to treat tumour-bearing mice. The results showed that anti-MS4A4A significantly inhibits CRC growth in mice (figure 5E–G). FACS analysis showed that the level of infiltrating M2-TAMs (F4/80+CD206+) and expression of CD8+ T-cell exhaustion markers (PD-1 and TIM3) were significantly decreased in the anti-MS4A4A group, while CD8+ T-cell proliferation (Ki67) and effector function (IFN-γ) were significantly enhanced (figure 5H-K). Similarly, IHC staining and qRT-PCR data also suggested a decrease in M2-TAMs and exhausted T cells in the anti-MS4A4A group (figure 5L–O). Subsequently, we established the AOM/DSS-induced CRC mouse model for further validation, and consistent results were also obtained (online supplemental figure S13A–F).
gutjnl-2022-329147supp013.pdf (2.1MB, pdf)
Using mass cytometry by time of flight (CyTOF), we analysed tumour-infiltrating immune cells from CT26 model treated with IgG or anti-MS4A4A mAb. The total cell population was divided into 11 subclusters, and the macrophage population was decreased significantly, while the CD8+ T-cell population was increased in the anti-MS4A4A group (figure 5P–R). Further analysis showed that the increased macrophage population predominantly presented an M2-phenotype (online supplemental figure S14A,B). Consistent with our previous results, CyTOF results showed that M1 markers (iNOS and MHCII) of TAMs, proliferation markers (Ki67) and activation markers (Tbet, CD27, ICOS and CD69) of CD8+ T-cells were upregulated in the anti-MS4A4A group. In contrast, the expression of M2 markers (CD206) and immunosuppressive molecules (CD39, SIRPa) were downregulated in TAMs (online supplemental figure 5S,T). To investigate whether the tumour suppressive effect in the MS4A4A mAb group was related to the antibody-dependent cell-mediated cytotoxicity (ADCC) effect, we injected the mouse model with ADCC-blocking antibody (CD16/CD32 mAb). Our results showed that the addition of CD16/CD32 mAb partially attenuated the reduction in TAM infiltration induced by MS4A4A mAb, but did not affect the MS4A4A mAb-mediated tumour suppression (online supplemental figure S15A–D). These results indicate that MS4A4A blockade in vivo delays tumour progression and remodels the immunosuppressive TIME.
gutjnl-2022-329147supp014.pdf (1.8MB, pdf)
gutjnl-2022-329147supp015.pdf (1.2MB, pdf)
Targeted MS4A4A treatment enhances the efficacy of PD-1 blockade
Although anti-PD-1/programmed death-ligand 1 (PD-L1)-based therapies target CD8+ T-cells, these cells are in fact closely regulated by TAMs in the TME, making targeting TAMs another potential immunotherapeutic approach.24 In this study, we demonstrated that MS4A4A blockade treatment delayed tumour growth and reshapes TIME. Additionally, CyTOF analysis showed that the numbers of PD-1+CD8+ T-cells and PD-L1+ macrophages were significantly increased in the tumour after anti-MS4A4A treatment (online supplemental figure S14C–F). Studies have shown that the levels of tumour-infiltrating CD8+ T-cells and PD-L1 expression in the TME are biomarkers for anti-PD-1/PD-L1–based therapies.27–29 Therefore, we speculated that MS4A4A blockade may have a synergistic effect with anti-PD-1 therapy.
Here, we constructed three distinct tumour models with a range of immunogenicities and sensitivities to anti-PD-1/PD-L1-based therapies. The CT26 and MC38 models are immunogenic colon carcinoma models that are poorly and partially responsive to anti-PD-1/PD-L1 therapy, respectively.30 31 The B16F10 melanoma is a poorly immunogenic tumour model that responds poorly to anti-PD-1/PD-L1 therapy.32 First, we used Ms4a4a-specific siRNA combined with the anti-PD-1 mAb to treat MC38 tumour-bearing mice and found that the anti-PD-1 or siMs4a4a treatment alone could inhibit tumour growth. However, the combined treatment of both resulted in further suppression of tumour growth and improved the survival of mice (online supplemental figure S16A–E). Similar results were obtained in the CT26 model (online supplemental figure S16F–J). FACS analysis showed that the combination therapy could significantly reduce the coexpression of T-cell exhaustion markers in the tumour and enhance CD8+ T-cell effector function and proliferation (online supplemental figure S16K–N).
gutjnl-2022-329147supp016.pdf (1.1MB, pdf)
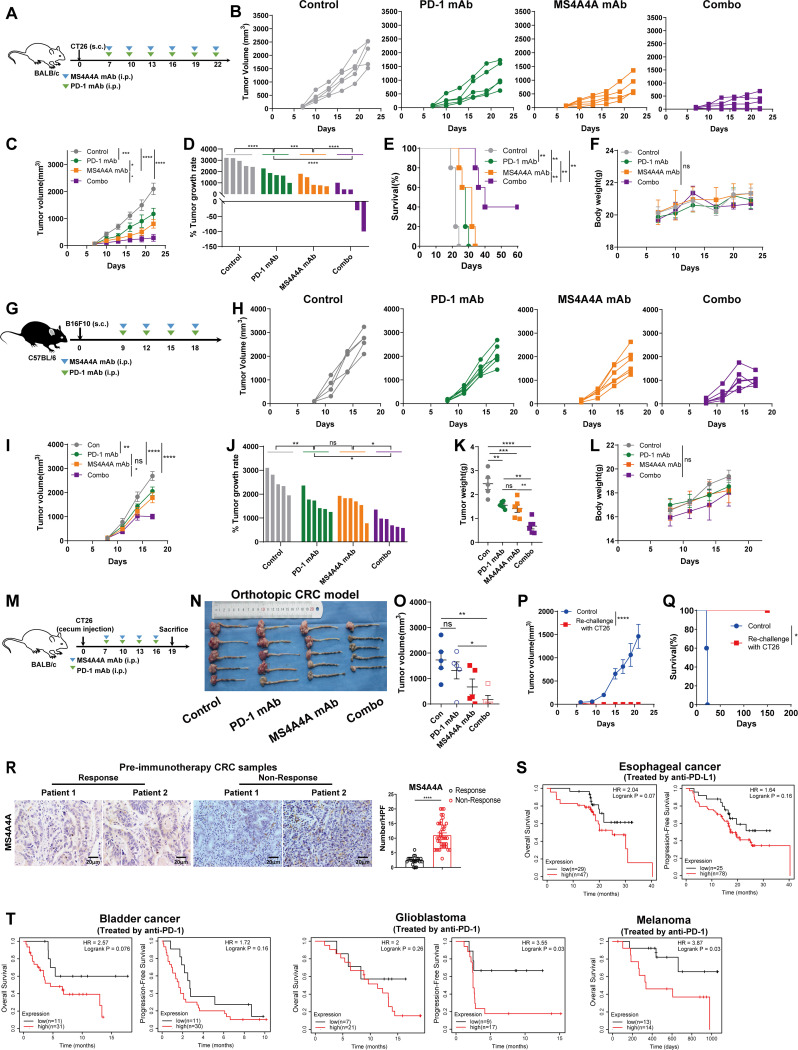
Similarly, we used the anti-MS4A4A mAb combined with the anti-PD-1 mAb to treat CT26 tumour-bearing mice. Consistent with the effect of siMs4a4a, we observed that the combination therapy group had slower CT26 tumour growth compared with the monotherapy groups, and the OS of this group was significantly extended (figure 6A–E). Further examination revealed no differences in body weight among groups (figure 6F), suggesting that anti-MS4A4A mAb may have limited general toxicity in mouse models. Similar results were observed in the B16F10 model and CT26 orthotopic implant model (figure 6G–O). To test whether the combination therapy could elicit a long-lived memory T-cell response that protected mice from tumour relapse, we rechallenged survivor mice using CT26 cells and observed complete tumour regression in the rechallenged survivor mice (figure 6P,Q). Collectively, these in vivo animal results suggest that MS4A4A blockade treatment produces potent T-cell-mediated antitumour effects and prevents tumour recurrence through memory T-cell responses.
Figure 6.
Targeted MS4A4A treatment synergises with anti-PD-1 mAb treatment. (A–F) Tumour growth of CT26 tumour-bearing mice treated with an isotype control, an anti-PD-1 mAb, an anti-MS4A4A mAb or the anti-PD-1 mAb combined with the anti-MS4A4A mAb (n=5/group). (A) Schematic showing the treatment plan. (B–D) Tumour growth. (E) Survival of CT26 tumour-bearing mice. (F) Body weight of mice in each group. (G–L) Tumour growth of B16F10 tumour-bearing mice treated with an isotype control, an anti-PD-1 mAb, an anti-MS4A4A mAb or the anti-PD-1 mAb combined with the anti-MS4A4A mAb (n=5–6/group). (G) Schematic showing the treatment plan. (H–J) Tumour growth. (K) Tumour weight. (L) Body weight of mice in each group. (M–O) Mouse CT26 colon cancer cells were injected in situ into the cecum wall of mice and then treated with an isotype control, an anti-PD-1 mAb, an anti-MS4A4A mAb or the anti-PD-1 mAb combined with the anti-MS4A4A mAb (n=5/group). (M) Schematic showing the treatment plan. (N) Macroscopic appearance of orthotopic CRC tumours for each indicated treatment. (O) Tumour volume. (P) Tumour growth curves of mice rechallenged s.c. as in (E) (survivor) (n=3) with MC38 cells and age-matched tumour-naive mice (control) (n=5). (Q) Overall survival of rechallenged mice depicted with a Kaplan-Meier curve (n=3–5/group). (R) Representative images and statistical analysis of immunohistochemical staining for MS4A4A in tumour samples from patients with CRC (n=12) who underwent surgical resection or colonoscopic biopsy prior to immunotherapy. Five random high-power fields were selected for analysis on each slide. (S) Kaplan-Meier plots showing the relationships between the level of MS4A4A gene expression in tumours and the overall survival or progression-free survival of patients with cancer treated with anti-PD-L1 therapy in a cohort of patients with oesophageal cancer. (T) Kaplan-Meier plots showing the relationships between the level of MS4A4A gene expression in tumours and the overall survival or progression-free survival of patients with cancer treated with anti-PD-1 therapy in a cohort of patients with bladder cancer, patients with glioblastoma or patients with melanoma. CRC, colorectal cancer; i.p., intraperitoneal injection; mAb, monoclonal antibody; PD-1, programmed cell death protein 1; s.c., subcutaneous.
Further, we investigated the clinical correlation between the MS4A4A protein level and immunotherapy response in patients with CRC. We obtained 12 CRC samples prior to immunotherapy for IHC analysis. The results revealed that high MS4A4A expression was strongly associated with no response to immunotherapy, while responders tended to have significantly lower MS4A4A protein expression (figure 6R). Using public databases, we found that in several cancer types, patients with high MS4A4A expression tended to have lower overall and progression-free survival rates after immunotherapy (figure 6S,T). In conclusion, these data suggest that high MS4A4A expression may be a marker of non-response to immunotherapy in patients with cancer.
Anti-MS4A4A mAb treatment enhances the therapeutic efficacy against large established murine CRC tumours
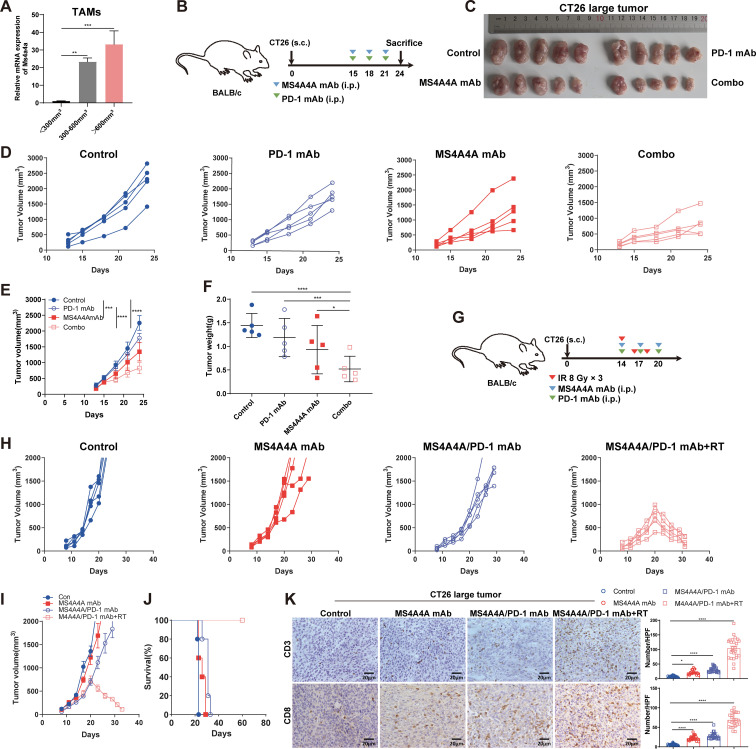
The effect of anti-PD-1 therapy is related to pretreatment tumour size.33 Moreover, large established tumours tend to develop complex immunosuppressive networks that are resistant to immunotherapy.34 In this study, TAMs in tumours of different sizes were sorted by flow cytometry, and qRT-PCR data showed that higher Ms4a4a mRNA expression levels were significantly associated with larger tumour sizes, suggesting that increased MS4A4A expression levels were correlated with tumour progression (figure 7A).
Figure 7.
Treatment of large established CT26 tumours with the anti-MS4A4A mAb improves the therapeutic benefit of anti-PD-1 immunotherapy. (A) qRT-PCR was performed to detect the expression of MS4A4A on TAMs in tumours with different volumes. (B–F) CT26 cells (5×105) were transplanted subcutaneously (s.c.) into BALB/c mice and treated with an isotype control, an anti-PD-1 mAb, an anti-MS4A4A mAb or the anti-PD-1 mAb combined with the anti-MS4A4A mAb when the tumour volume was 300–600 mm3 (n=5/group). (B) Schematic showing the treatment plan. (C) Representative images of CT26 tumours. (D and E) Tumour growth. (F) Tumour weight. (G–K) CT26 cells (5×105) were transplanted s.c. into BALB/c mice and treated with an isotype control, an anti-MS4A4A mAb, anti-PD-1+ anti-MS4A4A mAbs or anti-PD-1+ anti-MS4A4A mAbs combined with RT when the tumour volume was 300–600 mm3 (n=5/group). (G) Schematic showing the treatment plan. (H and I) Tumour growth. (J) Survival of CT26 tumour-bearing mice. (K) Representative images of IHC staining for CD3 and CD8 in tumour sections and statistical analysis. The number of CD3-positive cells and CD8-positive cells per HPF was counted in subcutaneous tumour sections from each group. Five random HPFs were selected for analysis on each slide. HPF, high-power field; IHC, immunohistochemistry; IR, ionizing radiation; i.p., intraperitoneal injection; mAb, monoclonal antibody; mRNA, messenger RNA; PD-1, programmed cell death protein 1; qRT-PCR, quantitative real-time PCR; RT, radiotherapy; TAMs, tumour-associated macrophages.
Therefore, we speculated that anti-MS4A4A/PD-1 treatment may produce a synergistic effect on more advanced and larger CRC tumours. To test this hypothesis, we constructed a large CT26 tumour model (volume >300 mm3) (figure 7B). The results showed that anti-PD-1 treatment alone had no significant effect on the growth of large CT26 tumours, while combination with the anti-MS4A4A mAb significantly inhibited tumour growth (figure 7C–F). These data suggest that anti-MS4A4A therapy re-establishes the sensitivity of large established CRC tumours to anti-PD-1 therapy.
Studies have shown that multiple infiltrating immune cell populations, including CD8+ T-cells, are less frequently found in large tumours than in small tumours.35 36 In this study, anti-MS4A4A/PD-1 treatment slowed the growth of large tumours to some extent, but did not induce tumour regression. We speculated that this may be related to reduced T-cell infiltration within large tumours. Many studies have confirmed that radiotherapy (RT) can increase T-cell infiltration into tumours.37 38 Therefore, we speculated that RT combined with anti-MS4A4A/PD-1 therapy can further inhibit the growth of large CRC tumours. Subsequently, we used the anti-MS4A4A/PD-1 mAbs in combination with RT (8 Gy) to treat large tumours (figure 7G). The results showed that combining the three therapies could promote apparent regression of large tumours and significantly improve survival in mice (figure 7H–J). Furthermore, IHC assays showed that anti-MS4A4A/PD-1 combined with RT significantly increased CD3+ T-cell and CD8+ T-cell infiltration within large tumours (figure 7K). Overall, these results demonstrate that anti-MS4A4A/PD-1 therapy is more effective in combination with RT and has the potential for translation to the clinic.
Discussion
This study shows that in vivo inhibition of MS4A4A or anti-MS4A4A treatment can curb tumour growth and synergise with anti-PD-1 immunotherapy. mAbs that inhibit CTLA-4 or PD-1 have been shown to unleash anti-tumour T-cell effector functions in mice and some patients with cancer.39 40 However, checkpoint blockade is not fully effective against some tumours, which can escape by multiple mechanisms, including the generation of myeloid-rich TME with potent immunosuppressive properties.41 Therefore, efforts are underway to complement ICI therapies with treatments that target myeloid cells.42
Given the importance of TAMs in regulating antitumour immunity, many cancer treatment strategies targeting TAMs have been developed. These strategies can be broadly classified into (1) TAM removal and (2) alteration of TAM immunosuppressive activity. The first category is divided into strategies targeting TAM recruitment and survival, including (1) blocking the CCL2-CCR2 axis to inhibit TAM recruitment43 and (2) blocking the CSF1-CSF1R axis to induce TAM apoptosis.44 However, an inherent drawback of eliminating TAMs is losing the natural immunostimulatory role of macrophages as primary phagocytes and professional antigen-presenting cells in solid tumours.45 Therefore, reprogramming or repolarising immunosuppressive TAMs into immunostimulatory TAMs is an attractive research direction. The second class of strategies aimed at altering TAM immunosuppressive activity includes (1) blocking CD24-Siglec1046 and CD47-SIRPa signalling47 to enhance macrophage phagocytosis; (2) reprogramming TAMs with CD40 agonists; and (3) targeting macrophage IRs such as TREM241 to restore antitumour immunity. The anti-MS4A4A treatment introduced in this study provides a new therapeutic approach that broadens the arsenal of myeloid cell targets in tumours.
Currently, immunotherapy for CRC still has bottlenecks. Only MSI-H-type CRC benefits from ICI therapy; the immune efficacy in microsatellite-stable-type CRC is less satisfactory. Developing ways to expand the population that will benefit from CRC immunotherapy is an urgent challenge in current basic immune research. Because preclinical evidence largely supports the need for combination strategies to achieve significant antitumour efficacy, most targeted TAM strategies currently in clinical trials are tested in combination with chemotherapy, RT or ICIs.24 Here, we found a synergistic effect for anti-MS4A4A treatment combined with anti-PD-1 treatment in inhibiting CRC growth in mice. Moreover, we found that this combination treatment modality significantly retarded tumour growth even when the tumour was large. Furthermore, when anti-MS4A4A/PD-1 therapy was combined with RT, we found that large CRC tumours showed significant regression. Our findings suggest that targeting macrophage MS4A4A has potential as a novel target for combination immunotherapy in CRC.
CD20/MS4A1 belongs to the same family as MS4A4A and is maintained during abnormal human B-cell proliferation.48 An mAb against CD20/MS4A1 has shown efficacy in treating relapsed B-cell lymphoma.49 Similar to CD20/MS4A1 in B cells, MS4A4A is highly expressed by human TAMs,14 and its expression has been associated with multiple cancers.15 50 MS4A4A expression has also been associated with autoimmune diseases, such as polyangiitis.48 Therefore, MS4A4A is considered a suitable target for developing therapeutic mAbs. Here, we developed and designed an antibody recognising the mouse MS4A4A protein and explored the feasibility of this anti-MS4A4A mAb in the treatment of CRC in animal experiments. Notably, anti-MS4A4A treatment did not induce significant toxicity in mice, suggesting that MS4A4A-targeted therapy is safe for cancer treatment. Additionally, we found that MS4A4A was specifically highly expressed within TAMs in multiple cancers, implying that anti-MS4A4A therapies could be applied to a wide range of tumours.
Collectively, our results reveal the critical role of MS4A4A+ TAMs in regulating tumour immune evasion, suggesting that anti-MS4A4A treatment may effectively improve the efficacy of anti-PD-1 therapy. These findings provide a new perspective for understanding the role of TAMs in regulating antitumour immunity and new directions for developing effective immunotherapeutic strategies for CRC.
Materials and methods
Details regarding the materials and methods are described in the online supplemental methods. Reagents used in this study are listed in online supplemental table 1 and online supplemental table 2.
gutjnl-2022-329147supp017.pdf (151.4KB, pdf)
gutjnl-2022-329147supp018.pdf (72.6KB, pdf)
gutjnl-2022-329147supp019.pdf (187.7KB, pdf)
Footnotes
Twitter: @I have no twitter
YLi, ZS, ZC and YZhan contributed equally.
Contributors: HD, YF and GL were responsible for the concept and experimental design. YLi, ZC, YZhan and ZLiu carried out the experiments. YZhang, ML, YLiu, WL and YD contributed to clinical sample collection. ZLi, ZZ, JZ and JQ helped with the animal study. YLi, ZC and SG performed statistical analysis. YLi and ZS wrote and edited the manuscript. HD is the guarantor for this paper. All authors read and approved the final manuscript.
Funding: This study was supported by grants from the National Natural Science Foundation of China (82073063, 81972631, 82103595, 82273039); The Natural Science Foundation of Guangdong Province (2022A1515010298, 2021A1515010989, 2023A1515010274, 2023A1515010980); Guangdong Provincial Regional Joint Fund-Youth Fund Project (2020A1515110006); The Foundation of President of Nanfang Hospital (2020B012); Outstanding Youth Development Scheme of Nanfang Hospital, Southern Medical University (2020J010); The Guangdong Provincial Major Talents Project (2019JC05Y361); The Guangdong Provincial Key Laboratory of Precision Medicine for Gastrointestinal Cancer (2020B121201004).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Medical Ethics Committee of NanFang Hospital of Southern Medical University (NFEC-2021-396). Participants gave informed consent to participate in the study before taking part.
References
- 1. Page DB, Postow MA, Callahan MK, et al. Immune modulation in cancer with antibodies. Annu Rev Med 2014;65:185–202. 10.1146/annurev-med-092012-112807 [DOI] [PubMed] [Google Scholar]
- 2. Chen EX, Jonker DJ, Loree JM, et al. Effect of combined immune checkpoint inhibition vs best supportive care alone in patients with advanced colorectal cancer: the Canadian cancer trials group CO.26 study. JAMA Oncol 2020;6:831–8. 10.1001/jamaoncol.2020.0910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467–80. 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- 4. Marigo I, Zilio S, Desantis G, et al. T cell cancer therapy requires CD40-CD40L activation of tumor necrosis factor and inducible nitric-oxide-synthase-producing dendritic cells cancer cell. Cancer Cell 2016;30:377–90. 10.1016/j.ccell.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Panni RZ, Herndon JM, Zuo C, et al. Agonism of CD11B reprograms innate immunity to sensitize pancreatic cancer to immunotherapies. Sci Transl Med 2019;11:eaau9240. 10.1126/scitranslmed.aau9240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Johnson KCC, Gatti-Mays ME, et al. Emerging strategies in targeting tumor-resident myeloid cells for cancer immunotherapy. J Hematol Oncol 2022;15:118. 10.1186/s13045-022-01335-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938–45. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H, Tian T, Zhang J. Tumor-associated Macrophages (TAMs) in colorectal cancer (CRC): from mechanism to therapy and prognosis. Int J Mol Sci 2021;22:8470. 10.3390/ijms22168470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forssell J, Oberg A, Henriksson ML, et al. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer Clin cancer Res. Clin Cancer Res 2007;13:1472–9. 10.1158/1078-0432.CCR-06-2073 [DOI] [PubMed] [Google Scholar]
- 10. Feng Q, Chang W, Mao Y, et al. Tumor-associated macrophages as prognostic and predictive biomarkers for postoperative adjuvant chemotherapy in patients with stage II colon cancer. Clin Cancer Res 2019;25:3896–907. 10.1158/1078-0432.CCR-18-2076 [DOI] [PubMed] [Google Scholar]
- 11. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012;122:787–95. 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Long SA, Thorpe J, DeBerg HA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol 2016;1:eaai7793. 10.1126/sciimmunol.aai7793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanyal R, Polyak MJ, Zuccolo J, et al. MS4A4A: a novel cell surface marker for M2 macrophages and plasma cells. Immunol Cell Biol 2017;95:611–9. 10.1038/icb.2017.18 [DOI] [PubMed] [Google Scholar]
- 14. Mattiola I, Tomay F, De Pizzol M, et al. The macrophage tetraspan MS4A4A enhances dectin-1-dependent NK cell-mediated resistance to metastasis. Nat Immunol 2019;20:1012–22. 10.1038/s41590-019-0417-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang H, Wu X, Chen Y. Stromal-immune score-based gene signature: a prognosis stratification tool in gastric cancer. Front Oncol 2019;9:1212. 10.3389/fonc.2019.01212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hakimi AA, Voss MH, Kuo F, et al. Transcriptomic profiling of the tumor microenvironment reveals distinct subgroups of clear cell renal cell cancer: data from a randomized phase III trial. Cancer Discov 2019;9:510–25. 10.1158/2159-8290.CD-18-0957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao Y, Zhang W, Huo M, et al. Xbp1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer signal. Signal Transduct Target Ther 2021;6:357. 10.1038/s41392-021-00761-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takenaka MC, Gabriely G, Rothhammer V, et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and Cd39. Nat Neurosci 2019;22:729–40.:1533. 10.1038/s41593-019-0446-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petty AJ, Li A, Wang X, et al. Hedgehog signaling promotes tumor-associated macrophage polarization to suppress Intratumoral Cd8+. J Clin Invest 2019;129:5151–62. 10.1172/JCI128644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Henau O, Rausch M, Winkler D, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγin myeloid cells. Nature 2016;539:443–7. 10.1038/nature20554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pu J, Xu Z, Nian J, et al. M2 macrophage-derived extracellular Vesicles facilitate CD8+T cell exhaustion in hepatocellular carcinoma via the miR-21-5p/YOD1/YAP/Β-Catenin pathway cell death Discov. Cell Death Discov 2021;7:182. 10.1038/s41420-021-00556-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L, Li Z, Skrzypczynska KM, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer cell. Cell 2020;181:442–459. 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 23. Revel M, Sautès-Fridman C, Fridman W-H, et al. C1Q+ macrophages: passengers or drivers of cancer progression. Trends Cancer 2022;8:517–26. 10.1016/j.trecan.2022.02.006 [DOI] [PubMed] [Google Scholar]
- 24. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and Immunotherapy. Nat Rev Immunol 2019;19:369–82. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao S, Mi Y, Guan B, et al. Tumor-derived Exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol 2020;13:156. 10.1186/s13045-020-00991-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goswami S, Anandhan S, Raychaudhuri D, et al. Myeloid cell-targeted therapies for solid tumours. Nat Rev Immunol 2023;23:106–20. 10.1038/s41577-022-00737-w [DOI] [PubMed] [Google Scholar]
- 27. Gong B, Kiyotani K, Sakata S, et al. Secreted PD-L1 variants mediate resistance to PD-L1 blockade therapy in non-small cell lung cancer. J Exp Med 2019;216:982–1000. 10.1084/jem.20180870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275–87. 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim TK, Vandsemb EN, Herbst RS, et al. Adaptive immune resistance at the tumour site: mechanisms and therapeutic opportunities. Nat Rev Drug Discov 2022;21:529–40. 10.1038/s41573-022-00493-5 [DOI] [PubMed] [Google Scholar]
- 30. Hicks KC, Chariou PL, Ozawa Y, et al. Tumour-targeted Interleukin-12 and Entinostat combination therapy improves cancer survival by reprogramming the tumour immune cell landscape. Nat Commun 2021;12:5151. 10.1038/s41467-021-25393-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gurbatri CR, Lia I, Vincent R, et al. Engineered Probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med 2020;12:eaax0876. 10.1126/scitranslmed.aax0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Momin N, Mehta NK, Bennett NR, et al. Anchoring of intratumorally administered cytokines to collagen safely potentiates systemic cancer Immunotherapy. Sci Transl Med 2019;11:eaaw2614. 10.1126/scitranslmed.aaw2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–5. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X, Liu Z, Zhang A, et al. NQO1 targeting prodrug triggers innate sensing to overcome checkpoint blockade resistance. Nat Commun 2019;10:3251. 10.1038/s41467-019-11238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang M, He M, Guo Y, et al. The influence of immune heterogeneity on the effectiveness of immune checkpoint inhibitors in multifocal hepatocellular Carcinomas. Clin Cancer Res 2020;26:4947–57. 10.1158/1078-0432.CCR-19-3840 [DOI] [PubMed] [Google Scholar]
- 36. von Itzstein MS, Khan S, Popat V, et al. “Statin intolerance, anti-HMGCR antibodies, and immune checkpoint inhibitor-associated myositis: a "two-hit" autoimmune toxicity or clinical predisposition?” Oncologist 2020;25:e1242–5. 10.1634/theoncologist.2019-0911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta A, Probst HC, Vuong V, et al. Radiotherapy promotes tumor-specific effector Cd8+ T cells via dendritic cell activation. J Immunol 2012;189:558–66. 10.4049/jimmunol.1200563 [DOI] [PubMed] [Google Scholar]
- 38. Matsumura S, Wang B, Kawashima N, et al. Radiation-induced CXCl16 release by breast cancer cells attracts effector T cells. J Immunol 2008;181:3099–107. 10.4049/jimmunol.181.5.3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348:56–61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 40. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450–61. 10.1016/j.ccell.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Molgora M, Esaulova E, Vermi W, et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy cell. Cell 2020;182:886–900. 10.1016/j.cell.2020.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399–416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lesokhin AM, Hohl TM, Kitano S, et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res 2012;72:876–86. 10.1158/0008-5472.CAN-11-1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cannarile MA, Weisser M, Jacob W, et al. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 2017;5:53. 10.1186/s40425-017-0257-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y-C, Wang X, Yu J, et al. Targeting monoamine oxidase A-regulated tumor-associated macrophage polarization for cancer Immunotherapy. Nat Commun 2021;12:3530. 10.1038/s41467-021-23164-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barkal AA, Brewer RE, Markovic M, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer Immunotherapy. Nature 2019;572:392–6. 10.1038/s41586-019-1456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Logtenberg MEW, Scheeren FA, Schumacher TN. The CD47-SIRPα immune checkpoint. Immunity 2020;52:742–52. 10.1016/j.immuni.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mattiola I, Mantovani A, Locati M. The Tetraspan MS4A family in homeostasis, immunity, and disease. Trends Immunol 2021;42:764–81. 10.1016/j.it.2021.07.002 [DOI] [PubMed] [Google Scholar]
- 49. Maloney DG, Liles TM, Czerwinski DK, et al. Phase I clinical trial using escalating single-dose infusion of chimeric anti-CD20 monoclonal antibody (IDEC-C2B8) in patients with recurrent B-cell lymphoma. Blood 1994;84:2457–66. 10.1182/blood.V84.8.2457.bloodjournal8482457 [DOI] [PubMed] [Google Scholar]
- 50. Pan X, Chen Y, Gao S. Four genes relevant to pathological grade and prognosis in ovarian cancer. Cancer Biomark 2020;29:169–78. 10.3233/CBM-191162 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-329147supp001.pdf (1,012.9KB, pdf)
gutjnl-2022-329147supp002.pdf (1.1MB, pdf)
gutjnl-2022-329147supp003.pdf (437KB, pdf)
gutjnl-2022-329147supp004.pdf (505.4KB, pdf)
gutjnl-2022-329147supp005.pdf (625.8KB, pdf)
gutjnl-2022-329147supp006.pdf (199.8KB, pdf)
gutjnl-2022-329147supp007.pdf (1.8MB, pdf)
gutjnl-2022-329147supp008.pdf (1.2MB, pdf)
gutjnl-2022-329147supp009.pdf (1.3MB, pdf)
gutjnl-2022-329147supp010.pdf (901.4KB, pdf)
gutjnl-2022-329147supp011.pdf (416.2KB, pdf)
gutjnl-2022-329147supp012.pdf (568KB, pdf)
gutjnl-2022-329147supp013.pdf (2.1MB, pdf)
gutjnl-2022-329147supp014.pdf (1.8MB, pdf)
gutjnl-2022-329147supp015.pdf (1.2MB, pdf)
gutjnl-2022-329147supp016.pdf (1.1MB, pdf)
gutjnl-2022-329147supp017.pdf (151.4KB, pdf)
gutjnl-2022-329147supp018.pdf (72.6KB, pdf)
gutjnl-2022-329147supp019.pdf (187.7KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.