Abstract
Recent studies revealed the relationship among homologous recombination repair (HRR), androgen receptor (AR), and poly(adenosine diphosphate-ribose) polymerase (PARP); however, the synergy between anti-androgen enzalutamide (ENZ) and PARP inhibitor olaparib (OLA) remains unclear. Here, we showed that the synergistic effect of ENZ and OLA significantly reduced proliferation and induced apoptosis in AR-positive prostate cancer cell lines. Next-generation sequencing followed by Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses revealed the significant effects of ENZ plus OLA on nonhomologous end joining (NHEJ) and apoptosis pathways. ENZ combined with OLA synergistically inhibited the NHEJ pathway by repressing DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and X-ray repair cross complementing 4 (XRCC4). Moreover, our data showed that ENZ could enhance the response of prostate cancer cells to the combination therapy by reversing the anti-apoptotic effect of OLA through the downregulation of anti-apoptotic gene insulin-like growth factor 1 receptor (IGF1R) and the upregulation of pro-apoptotic gene death-associated protein kinase 1 (DAPK1). Collectively, our results suggested that ENZ combined with OLA can promote prostate cancer cell apoptosis by multiple pathways other than inducing HRR defects, providing evidence for the combined use of ENZ and OLA in prostate cancer regardless of HRR gene mutation status.
Keywords: apoptosis, enzalutamide, nonhomologous end joining, olaparib, prostate cancer
INTRODUCTION
Prostate cancer remains the second leading cause of cancer deaths in the United States, accounting for an estimated 27% of new cases among men.1 Androgen receptor (AR) is an important factor in the progression of prostate cancer; hence, androgen deprivation therapy (ADT) is the treatment of choice for patients with inoperable or recurrent prostate cancer, and the initial response rate is approximately 80% for metastatic prostate cancer.2 Although ADT is effective in the first few years, patients eventually become resistant to the drug and progress to castration-resistant prostate cancer (CRPC). The median survival after progression to CRPC is only 14.5 months.3 Although the overall incidence of prostate cancer in the United States has declined, the diagnosis of patients with distant metastases has grown, so the drug treatment of prostate cancer has become crucial.1,4
The new generation anti-androgen drug enzalutamide (ENZ) was approved for marketing in 2012 and has since been successfully used as a first-line treatment for metastatic CRPC (mCRPC). ENZ can reduce the rate of CRPC metastasis/death by 71% according to several phase 3 clinical trials.5,6 Although ENZ is initially effective, patients eventually develop resistance and the drug fails in most CRPCs.
DNA damage repair (DDR), including single-strand break (SSB) and double-strand break (DSB), is an essential mechanism for cell survival by repairing damaged cells. Poly(adenosine diphosphate-ribose) polymerase (PARP) is a family of proteins involved in DDR and is mainly responsible for SSB repair; homologous recombination repair (HRR) is mainly responsible for DSB repair.7 PARP inhibitors can inhibit the SSB repair of cells and lead to synthetic lethality in HRR-deficient cells. PARP inhibitors have become a research hotspot in the treatment of prostate cancer because of their success in breast and ovarian cancer and their low toxicity. The PARP inhibitor olaparib (OLA) has recently been approved for patients with HRR gene-mutated prostate cancer. However, HRR gene mutations are not common in primary prostate cancer and CRPC, and many patients without HRR gene defects do not have the opportunity to use PARP inhibitors.8,9
Given that AR promotes HRR in prostate cancer, the combined treatment of ADT and PARP inhibitor causes synthetic lethality.10,11 However, the synergistic inhibitory effects of ENZ plus OLA in prostate cancer have not been investigated. In this study, the molecular mechanism of the combination treatment of ENZ and OLA to inhibit the proliferation of prostate cancer cells and promote their apoptosis was explored through cell experiments combined with bioinformatic analysis.
MATERIALS AND METHODS
Antibodies and reagents
OLA (AZD2881) was purchased from Selleck Chemicals (Houston, TX, USA). ENZ (MDV3100) was acquired from MedChemExpress (Monmouth Junction, NJ, USA). Dimethyl sulfoxide (DMSO) was obtained from Sigma-Aldrich (St. Louis, MO, USA). The primary antibodies used in the experiments are listed in Supplementary Table 1.
Supplementary Table 1.
Primary antibodies used in the experiments
| Name | Brand | Applications and dilutions | Article number |
|---|---|---|---|
| β-actin | Beyotime | WB, 1:2000 | AF0003 |
| Cleaved-PARP | Selleck | WB, 1:1000 | A5034 |
| DNA-PKcs | CST | WB, 1:1000 | 4602T |
| GAPDH | Abcam | WB, 1:5000 | AB8245 |
| IGF1R | Beyotime | WB, 1:1000 | AF7182 |
| XRCC4 | Beyotime | WB, 1:1000 | AG3577 |
| γH2AX | Beyotime | WB, 1:500; IF, 1:50 | AF5836 |
WB: Western blot; PARP: poly (ADP-ribose) polymerase; IF: immunofluorescence; DNA-PKcs: DNA-dependent protein kinase catalytic subunit; IGF1R: insulin-like growth factor 1 receptor; XRCC4: X-ray repair cross complementing; γH2AX: gamma H2A histone family member X
Cell culture and transfection
C4-2 and LNCaP prostate cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA), grown in 10% fetal bovine serum-supplemented RPMI-1640 medium (GIBCO, Gaithersburg, MD, USA), and incubated at 37°C in 5% CO2 environment. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect prostate cancer cells with small-interfering RNA (siRNA) in accordance with the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from C4-2 and LNCaP was extracted with TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions. Reverse transcription reactions were carried out with the cDNA archive kit (TaKaRa, Tokyo, Japan) following the manufacturer’s protocol. The reaction conditions for mRNA detection were as follows: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. The primers used for qRT-PCR are listed in Supplementary Table 2. The relative quantity of mRNA was determined by the ΔΔCT method. All PCRs were performed in triplicate.
Supplementary Table 2.
The sequences of the primers used in this study
| Gene | Sequence forward (5’–3’) | Sequence reverse (5’–3’) |
|---|---|---|
| PRKDC | CATGGAAGAAGATCCCCAGA | TGGGCACACCACTTTAACAA |
| XRCC4 | TCACCCAAGAACTCTTACCA | CAGCCGTAGAAGCCATAA |
| DAPK1 | ACGTGGTCCGGTATCTCTGTCTG | TGCTCGTGCTGTTCCGATCTA |
| IGF1R | TTCAGTTCGTGGTGGACCGAG | TCCACAATGCCTGTCTGAGGTG |
| β-actin | ACTCTTCCAGCCTTCCTTCC | TGTTGGCGTACAGGTCTTTG |
PRKDC: protein kinase DNA-activated catalytic subunit; IGF1R: insulin-like growth factor 1 receptor; XRCC4: X-ray repair cross complementing; DAPK1: death-associated protein kinase 1
RNA sequencing and analysis
C4-2 cells were treated with DSMO, ENZ (16 μmol l−1), OLA (32 μmol l−1), and ENZ (16 μmol l−1) combined with OLA (32 μmol l−1) for 48 h, and total RNA was extracted as described above. Next-generation sequencing (NGS) provided by Allwegene (Beijing, China) was applied to determine the sequence of mRNA. Each sample (three replicates per group) was validated for RNA purity (optical density 260 nm/280 nm [OD 260/280], OD 260/230) by NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and for RNA integrity by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). TruSeq Library Prep Kit (Illumina, San Diego, CA, USA) was used for library preparation, and sequencing was performed on HiSeq2500 sequencer (Illumina). Gene expression was calculated by counting the mapped reads using HTSeq (Python Software Foundation, Fredericksburg, VA, USA). Expression level was measured as fragments per kilobase of exon model per million mapped reads (FPKM).
Western blot
Protein was extracted using radioimmunoprecipitation assay (RIPA) buffer supplemented with phosphatase and protease inhibitors (Selleck Chemicals, Houston, TX, USA) and separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After being transferred to polyvinylidene fluoride membranes, the proteins were probed with indicated antibodies.
Colony-forming assay
C4-2 and LNCaP cells were seeded at a density of 200–1000 cells per well in six-well plates for approximately 24–72 h under usual conditions and then treated with ENZ and OLA alone or in combination. Media and drugs were replenished every 5 days. After 10 days (C4-2) and 20 days (LNCaP) of incubation, the cells were fixed in methanol and stained with 0.5% crystal violet in absolute ethanol. These experiments were performed at least three times.
Apoptosis measurement
Apoptosis in prostate cancer cells was analyzed with Annexin V-FITC Apoptosis Detection Kit (Multisciences Biotech, Hangzhou, China) in accordance with the manufacturer’s protocol. After staining, the cells were examined using flow cytometry on a BD Accuri™ C6 (BD Biosciences, San Jose, CA, USA). Apoptotic cell profiles and quantitative data were generated with FlowJo software (Tree Star, Ashland, OR, USA). These experiments were repeated three times.
Growth inhibition assay and drug combination analysis
Cell counting was performed using CCK-8 (Dojindo Molecular Technologies, Kumamoto, Japan) in accordance with the manufacturer’s protocol. Half maximal inhibitory concentration (IC50) was calculated from sigmoidal dose-response curves by Prism 8 (GraphPad Software, San Diego, CA, USA). Combination index (CI) was calculated with CompuSyn software (ComboSyn, New York, NY, USA) to estimate the combined effect. CI values less than 1 showed synergism, CI values equal to 1 indicated simple additivity, and CI values greater than 1 indicated antagonism.12 A minimum of three tests were conducted.
Immunofluorescence
C4-2 and LNCaP cells were grown for 48 h on coverslips in 12-well plates and treated with ENZ and OLA alone or in combination. The cells were fixed with 4% paraformaldehyde, blocked with immunol staining blocking buffer (Beyotime, Shanghai, China), and treated overnight at 4°C with rabbit anti-γH2AX (Ser139) polyclonal antibody (Beyotime). Unbound antibodies were eliminated through phosphate-buffered saline (PBS) washing. The cells were treated at room temperature for 2 h with second antibody Alexa Fluor 488 (Servicebio, Wuhan, China), and nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). Images were captured and quantified using an immunofluorescence microscope (Nikon, Tokyo, Japan).
Statistical analyses
Analysis of variance (ANOVA) was used for data from more than two groups, including colony assay, qRT-PCR, cell apoptosis analysis, and immunofluorescence analyses. P value less than 0.05 was considered significant. P values of NGS data were corrected to control for false discovery rates (FDRs) using the Benjamini–Hochberg method. Raw read counts were normalized using DESeq to correct for sequencing depth. Given the three biological replicates and that DESeq was used to control experimental error, genes with an adjusted P < 0.05 were considered differentially expressed.
RESULTS
Combination of ENZ and OLA synergistically inhibits the proliferation of AR-positive prostate cancer cells
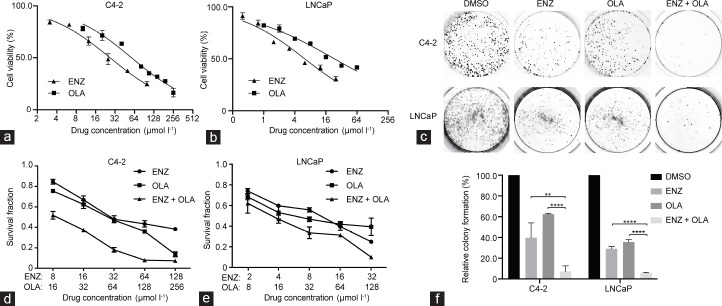
In prostate cancer, AR signaling induces DSB repair by HRR and nonhomologous end joining (NHEJ).11,13 The synthetic lethality caused by the combined use of ADT and PARP inhibitor has been reported, but their synergistic effect remains unclear. Considering that ENZ, a drug affecting AR signaling, may impair HRR and NHEJ, we hypothesized that the combined use of ENZ and OLA might result in a substantial therapeutic impact on AR-positive prostate cancer cell lines. We incubated prostate cancer cell lines with growing concentrations of ENZ and OLA alone and in combined for 48 h to verify this theory. The IC50 of ENZ was 26.85 μmol l−1 and 6.21 μmol l−1 in C4-2 and LNCaP cells, respectively, and the IC50 of OLA was 57.82 μmol l−1 and 37.07 μmol l−1 in C4-2 and LNCaP cells, respectively (Figure 1a and 1b). We evaluated the long-term impact of medication combination on prostate cancer cell lines by performing colony formation assays. The combination of ENZ and OLA essentially eradicated the growth of C4-2 and LNCaP cells compared with the use of either drug alone (Figure 1c and 1f). To further explore the synergistic effect of ENZ combined with OLA, CI values were determined by CCK-8 assay. The results showed that the combination of ENZ and OLA inhibited cell proliferation with a synergistic effect at 0.5 fraction affected (FA) and CI values of less than 1 in C4-2 and LNCaP cells (0.55 and 0.77, respectively; Figure 1d and 1e). These results revealed that the combination of ENZ and OLA has therapeutic efficacy for AR-positive prostate cancer cells.
Figure 1.
Responses of prostate cancer cells to ENZ and OLA as single agents and in combination. IC50 values of (a) C4-2 and (b) LNCaP cells treated with ENZ or OLA for 48 h were determined using the CCK-8 assay. (c) Representative images of the colony formation after treatment with DMSO, ENZ (2 µmol l−1 for C4-2 and 0.5 µmol l−1 for LNCaP), OLA (4 µmol l−1 for C4-2 and 2 µmol l−1 for LNCaP), or ENZ in combination with OLA. (d) C4-2 and (e) LNCaP cells were treated with ENZ and OLA as single agents or in combination for 48 h and then subjected to CCK-8 assay. (f) Quantification of colony numbers of C4-2 and LNCaP cells subjected to the indicated treatments. Mean with s.d. for three independent experiments is shown. **P < 0.01; ****P < 0.0001 (ANOVA). IC50: half maximal inhibitory concentration; ENZ: enzalutamide; OLA: olaparib; s.d.: standard deviation; ANOVA: analysis of variance; DMSO: dimethyl sulfoxide.
Combination of ENZ and OLA promotes apoptosis in AR-positive prostate cancer cells
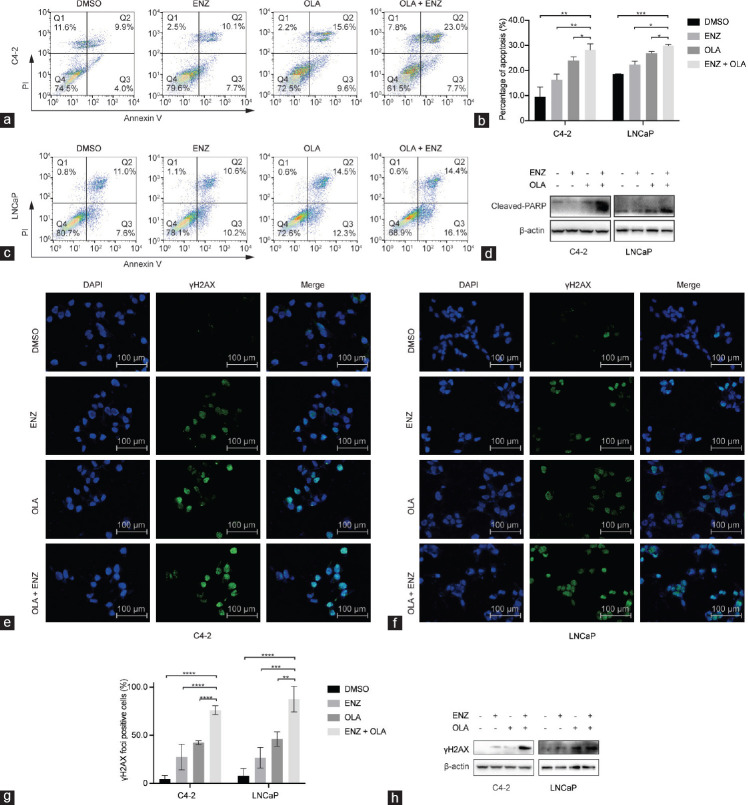
We then evaluated whether the combination of ENZ and OLA affects apoptotic cell death using annexin V-FITC/PI dual staining assays. The use of ENZ or OLA alone resulted in a moderated increase in the proportion of apoptotic cells, and their combination significantly increased apoptosis in C4-2 and LNCaP cell lines (Figure 2a–2c). In agreement with this finding, the combination of ENZ and OLA significantly increased the expression of cleaved-PARP, an active apoptotic marker, in C4-2 and LNCaP cells (Figure 2d). We next determined whether the combined treatment would induce DNA damage by measuring gamma H2A histone family member X (γH2AX), which is generated when DSB appears. In both cell lines, immunofluorescence staining of γH2AX showed that the combination of ENZ and OLA greatly improved the number and fluorescence intensity of foci compared with the use of either drug alone (Figure 2e–2g). Western blot analysis also revealed that the combination treatment of ENZ and OLA increased the expression of γH2AX (Figure 2h). All these results demonstrated that ENZ combined with OLA could induce apoptosis by causing DSB, suggesting that the combination has a strong synergistic effect on inhibiting the growth of prostate cancer cells.
Figure 2.
Combined treatment of ENZ and OLA induces apoptosis by increasing DNA damage in AR-positive prostate cancer cell lines. (a) The representative dot plots of C4-2 cells treated with DMSO, ENZ (25 µmol l−1), OLA (50 µmol l−1), or their combination for 48 h. (b) The histogram of apoptotic rates was shown after Annexin V-FITC/PI staining. (c) The representative dot plots of LNCaP cells treated with DMSO, ENZ (5 µmol l−1), OLA (30 µmol l−1), or their combination for 48 h. (d) The expression of cleaved-PARP after indicated treatments monitored by Western blot. Representative images of immunofluorescence staining of γH2AX in (e) C4-2 and (f) LNCaP cells treated as indicated for 48 h, and cell nuclei were stained with DAPI. (g) The percentage of cells with γH2AX foci. (h) The expression of γH2AX after indicated treatments monitored by Western blot. Mean with s.d. for three independent experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (ANOVA). AR: androgen receptor; ENZ: enzalutamide; OLA: olaparib; cleaved-PARP: cleaved-poly(adenosine diphosphate-ribose) polymerase; γH2AX: gamma H2A histone family member X; s.d.: standard deviation; Q1–4: quadrant 1–4; ANOVA: analysis of variance; DAPI: 4′,6-diamidino-2-phenylindole; DMSO: dimethyl sulfoxide; PI: prodium iodide.
ENZ combined with OLA affects global gene transcription in the C4-2 cell line
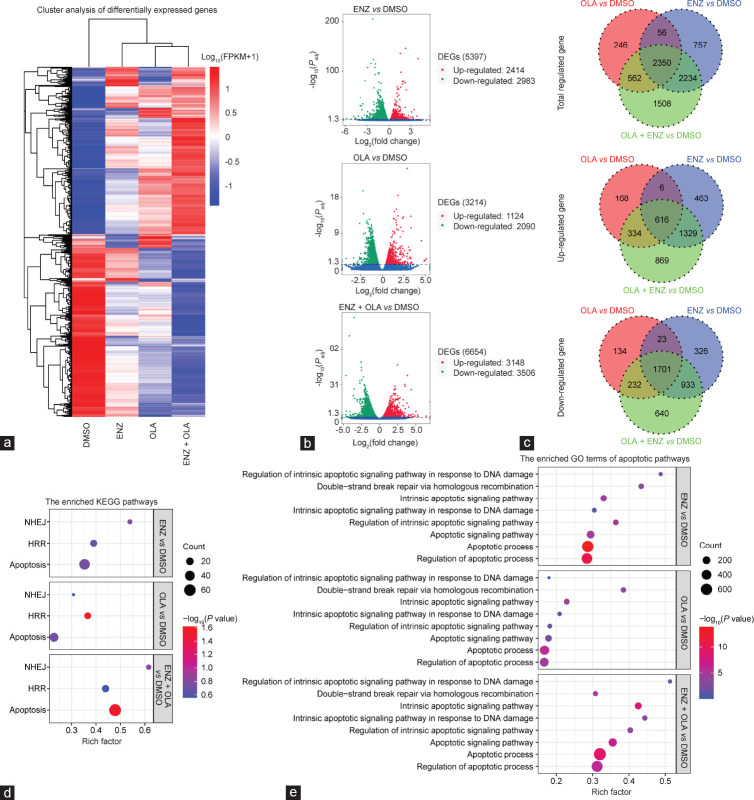
We explored the molecular mechanism by which the two drugs synergistically inhibit the growth of C4-2 cells. We carried out NGS to compare gene expression profiles in the C4-2 cell line following treatment with DMSO, ENZ, OLA, or ENZ combined with OLA. Principal component analysis (PCA) was performed to reduce the dimensionality of the original complex data. The findings revealed that the combination group had the highest degree of variance with the DMSO group compared with ENZ or OLA alone (Supplementary Figure 1a (110.5KB, tif) ). PCA results showed that different drugs caused genes to be expressed in different directions. Box plot, violin plot, and density distribution plot of FPKM suggested that the data from each group were similarly distributed (Supplementary Figure 1b (110.5KB, tif) –1d (110.5KB, tif) ). Hierarchical clustering was conducted to distinguish differential gene expression patterns among different groups (Figure 3a). Volcano plots showed that no matter which group would have a huge impact on the upregulation and downregulation of genes, the effect of the combination group was even more prominent (Figure 3b). Venn diagram of differentially expressed genes showed many intersections with the differential genes in the single-drug group and many differential genes distinct from the single-drug group, indicating the presence of many new differential genes that appeared after the combined treatment of ENZ and OLA (Figure 3c). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of genes with differential expression showed that the apoptotic pathway was significantly enriched in the combination group compared with that in the single-drug group (Supplementary Figure 2a (179.6KB, tif) –2c (179.6KB, tif) ).
Figure 3.
Global gene expression profiles were analyzed by NGS in C4-2 cells. (a) Hierarchical clustering showing gene expression profiles between groups as indicated. (b) The volcano plots showing the amounts of genes significantly influenced after indicated treatment with DMSO. (c) Venn diagram of the overlaps of genes regulated in indicated groups. (d) Results of KEGG analysis of the differentially expressed genes upon NHEJ, HR, and apoptosis in indicated groups. (e) Results of GO analysis of the differentially expressed genes upon apoptotic pathways in indicated groups. NGS: next-generation sequencing; FPKM: fragments per kilobase of exon model per million mapped reads; ENZ: enzalutamide; OLA: olaparib; DEGs: differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; NHEJ: nonhomologous end joining; HR: homologous recombination; GO: Gene Ontology; DMSO: dimethyl sulfoxide; Padj: adjusted P value.
ENZ combined with OLA inhibits NHEJ and induces apoptosis
NHEJ is one of the major pathways in DSB repair besides HRR.14 Considering that AR signaling promotes DSB repair through HRR and NHEJ in prostate cancer,11,13 we performed KEGG enrichment analysis of homologous recombination (HR; hsa03440), NHEJ (hsa03450), and apoptosis (hsa04210). The results showed that in NHEJ and apoptosis pathways, the enrichment degree of the combination group was higher than that of the single-drug group (Figure 3d). To further understand the effect of combination therapy on the specific genes of the NHEJ pathway relative to that of a single drug, we listed the statistically significant NHEJ differential genes in Figure 4a. We found that the key genes protein kinase DNA-activated catalytic subunit (PRKDC) and X-ray repair cross complementing 4 (XRCC4) affecting the NHEJ pathway were significantly downregulated after the combined treatment. qRT-PCR and Western blot analyses validated the results in C4-2 and LNCaP cell lines (Figure 4b and 4c). The PRKDC gene, which encodes DNA-dependent protein kinase catalytic subunit (DNA-PKcs), is highly activated in advanced tumors and affects gene expression in CRPC possibly by selective AR signaling.15,16 To further explore the role of PRKDC, we performed qRT-PCR and Western blot analyses and found that OLA had a significant effect on PRKDC and DNA-PKcs. To determine the role of OLA-downregulated DNA-PKcs in ENZ sensitivity, we detected the expression level of γH2AX that was synthesized from ENZ and DNA-PKcs silencing combination treatment. The results demonstrated that the knockdown of DNA-PKcs combined with ENZ significantly increased the abundance of γH2AX protein, indicating severe DSB (Figure 4d). Although no statistical difference in the expression of PRKDC was observed between tumor tissues and normal tissues according to The Cancer Genome Atlas (TCGA) combined with Genotype-Tissue Expression database, the mutual comparison of tumor tissues revealed that the expression of PRKDC in tumor tissues with Gleason score 6 was statistically different from that in the other groups (Figure 4e and 4f, and Supplementary Table 3 and 4).17 These results suggested that DNA repair impairment mediates the cytotoxic effects of the combination treatment.
Figure 4.
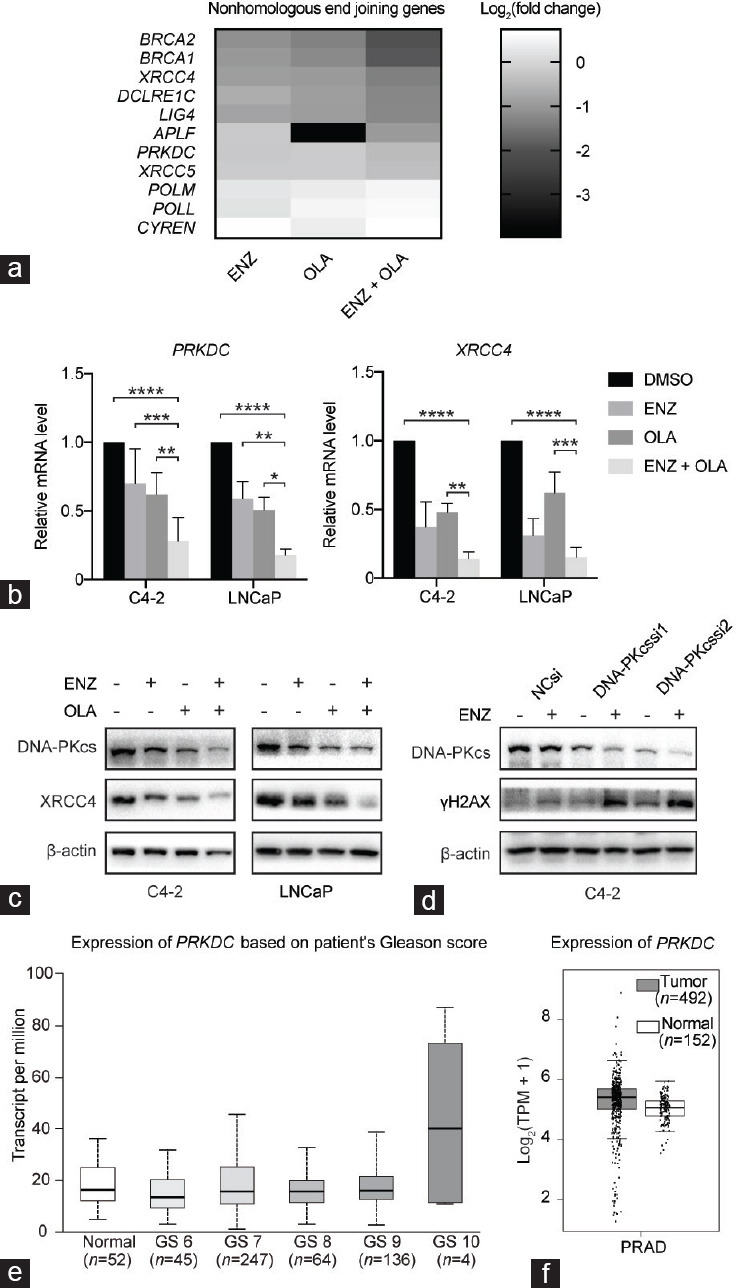
ENZ combined with OLA inhibits NHEJ and induces apoptosis. (a) Heatmap of the expression levels of some NHEJ genes following indicated treatment. (b) The mRNA expression level of PRKDC and XRCC4 in C4-2 and LNCaP cells with indicated treatments. (c) The expression of DNA-PKcs and XRCC4 in C4-2 and LNCaP after indicated treatments monitored by Western blot. (d) The expression of DNA-PKcs and γH2AX after the knockdown of DNA-PKcs monitored by Western blot. (e) Expression of PRKDC in TCGA database according to Gleason score. (f) PRKDC expression of tumor and normal tissues in TCGA database. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (ANOVA). ENZ: enzalutamide; OLA: olaparib; NHEJ: nonhomologous end joining; PRKDC: protein kinase DNA-activated catalytic subunit; XRCC4: X-ray repair cross complementing 4; DNA-PKcs: DNA-dependent protein kinase catalytic subunit; γH2AX: gamma H2A histone family member X; si: small interfering; TCGA: The Cancer Genome Atlas; GS: Gleason score; TPM: transcripts per million; PRAD: prostate adenocarcinoma; T: tumor; N: normal; ANOVA: analysis of variance; NC: normal control.
Supplementary Table 3.
Protein kinase DNA-activated catalytic subunit expression level in different stages in prostate of The Cancer Genome Atlas
| TCGA samples | Series 1 (low) | Series 1 (q1) | Series 1 (median) | Series 1 (q3) | Series 1 (high) |
|---|---|---|---|---|---|
| Normal<br> (n=52) | 4.943 | 11.927 | 16.286 | 24.771 | 36.051 |
| Gleason score 6<br> (n=45) | 2.965 | 9.118 | 13.631 | 20.092 | 31.97 |
| Gleason score 7<br> (n=247) | 1.081 | 10.76 | 15.767 | 25.096 | 45.471 |
| Gleason score 8<br> (n=64) | 2.978 | 11.534 | 15.595 | 19.864 | 32.708 |
| Gleason score 9<br> (n=136) | 2.707 | 12.595 | 16.076 | 21.488 | 38.708 |
| Gleason score 10<br> (n=4) | 10.859 | 11.519 | 40.25 | 73.287 | 86.869 |
TCGA: The Cancer Genome Atlas
Supplementary Table 4.
Statistical significance of protein kinase DNA-activated catalytic subunit expression level in different stages in prostate of The Cancer Genome Atlas
| Comparison | P | Statistical significance |
|---|---|---|
| Normal-vs-Gleason score 6 | 0.027842 | Yes |
| Normal-vs-Gleason score 7 | 0.67012 | No |
| Normal-vs-Gleason score 8 | 0.85906 | No |
| Normal-vs-Gleason score 9 | 0.177278 | No |
| Normal-vs-Gleason score 10 | 0.26868 | No |
| Gleason score 6-vs-Gleason score 7 | 0.0025369 | Yes |
| Gleason score 6-vs-Gleason score 8 | 0.023304 | Yes |
| Gleason score 6-vs-Gleason score 9 | 0.0069567 | Yes |
| Gleason score 6-vs-Gleason score 10 | 0.2228 | No |
| Gleason score 7-vs-Gleason score 8 | 0.86614 | No |
| Gleason score 7-vs-Gleason score 9 | 0.23216 | No |
| Gleason score 7-vs-Gleason score 10 | 0.2767 | No |
| Gleason score 8-vs-Gleason score 9 | 0.22858 | No |
| Gleason score 8-vs-Gleason score 10 | 0.27328 | No |
| Gleason score 9-vs-Gleason score 10 | 0.098277 | Yes |
ENZ combined with OLA induces apoptosis by upregulating pro-apoptotic genes and downregulating anti-apoptotic genes
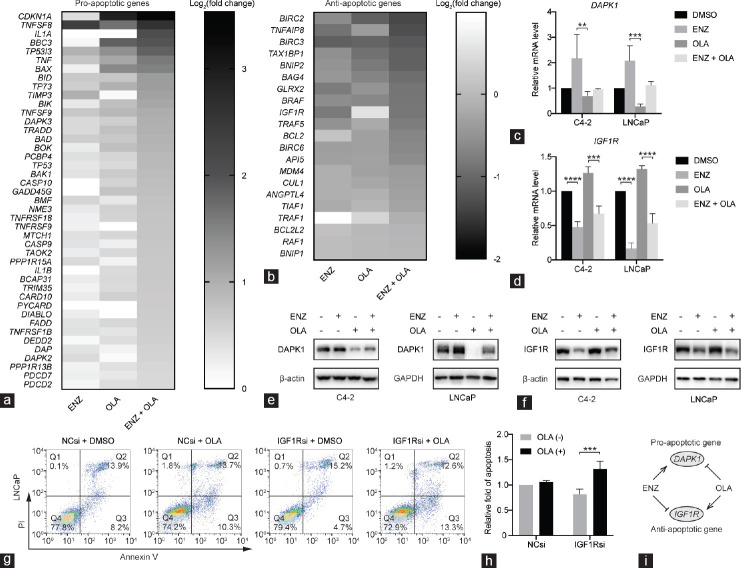
KEGG enrichment analysis showed that the combination group had a higher enrichment degree in apoptosis (hsa04210) than the single-drug group (Figure 3d). To further explore the apoptotic pathways, we performed enrichment analysis for differentially expressed genes in each group according to different apoptotic pathways in Gene Ontology (GO) terms. We found that the enrichment of the combination group in the apoptotic process (GO: 0006915), apoptotic signaling pathway (GO: 0097190), intrinsic apoptotic signaling pathway (GO: 0097193), regulation of apoptotic process (GO: 0042981), regulation of intrinsic apoptotic signaling pathway (GO: 2001242), intrinsic apoptotic signaling pathway in response to DNA damage (GO: 0008630), and regulation of intrinsic apoptotic signaling pathway in response to DNA damage (GO: 1902229) were significantly enhanced compared with those in the single-drug groups (Figure 3e). This finding suggested that ENZ combined with OLA may synergistically enhance the toxicity to prostate cancer cells by promoting intrinsic apoptosis pathways.
To further explore which apoptotic genes are affected by the combined treatment, we produced a heatmap of differentially expressed pro-apoptotic (Figure 5a) and anti-apoptotic genes (Figure 5b) in each group according to the apoptotic process (GO: 0006915). We found that a pro-apoptotic gene, death-associated protein kinase 1 (DAPK1), was upregulated by ENZ but downregulated by OLA (Supplementary Table 5). DAPK1 is considered a tumor suppressor gene in prostate cancer.18 qRT-PCR and Western blot validated that although the expression of DAPK1 was not statistically changed after the combined use of ENZ and OLA, ENZ could reverse the inhibitory function of OLA on DAPK1 (Figure 5c and 5e). In addition, we found that insulin-like growth factor 1 receptor (IGF1R), an anti-apoptotic gene, was upregulated by OLA but downregulated by ENZ (Supplementary Table 5). IGF1R is overexpressed in prostate cancer tissues and plays a crucial role in prostate cancer cell proliferation and ADT resistance.19,20 The anti-apoptotic gene IGF1R was upregulated by OLA but downregulated by ENZ or ENZ+OLA (Figure 5d and 5f). To verify the anti-apoptotic effect of IGF1R, we knocked down IGF1R in LNCaP cells and assessed the proportion of apoptosis using flow cytometry. Knockdown of IGF1R alone resulted in a slight decrease in the proportion of apoptotic cells but a significant increase in the proportion of apoptotic cells when ENZ was combined with OLA (Figure 5g and 5h). Overall, our results demonstrated that ENZ combined with OLA could promote apoptosis by jointly upregulating pro-apoptotic genes and downregulating anti-apoptotic genes. In addition, ENZ can enhance the response of prostate cancer cells to the combination therapy by reversing the anti-apoptotic effect of OLA through the downregulation of anti-apoptotic gene IGF1R and upregulation of pro-apoptotic gene DAPK1 (Figure 1i).
Figure 5.
ENZ combined with OLA induces apoptosis by upregulating pro-apoptotic genes and downregulating anti-apoptotic genes. Heatmap of the expression levels of (a) pro-apoptotic genes and (b) anti-apoptotic genes following indicated treatment. The mRNA expression level of (c) DAPK1 and (d) IGF1R in C4-2 and LNCaP with indicated treatments. The expression of (e) DAPK1 and (f) IGF1R in C4-2 and LNCaP cells after indicated treatments monitored by Western blot. (g) The representative dot plots of LNCaP cells with indicated treatment and (h) the histogram of apoptotic rates after Annexin V-FITC/PI staining. (i) Functional illustration of apoptotic genes DAPK1 and IGF1R. **P < 0.01; ***P < 0.001; ****P < 0.0001 (ANOVA). ENZ: enzalutamide; OLA: olaparib; DAPK1: death-associated protein kinase 1; IGF1R: insulin-like growth factor 1 receptor; NC: normal control; si: small interfering; Q1–4: quadrant 1–4; ANOVA: analysis of variance; DMSO: dimethyl sulfoxide; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; PI: prodium iodide.
DISCUSSION
The new-generation anti-androgen drug ENZ has been used for metastatic castration-sensitive prostate cancer and CRPC with or without metastases but eventually fails in most patients.21 The incidence of germline mutations in HRR genes was significantly higher in men with metastatic prostate cancer than that in men with localized prostate cancer.22 Given that such gene alterations induce a dependency on PARP function, PARP inhibitor OLA could be used to treat prostate cancer. BRCAness is a characteristic feature of HRR defects with breast cancer gene mutation. ADT could induce BRCAness through AR signaling, thereby increasing prostate cancer cells’ susceptibility to PARP inhibitors.10,11 Using NGS to compare gene expression profiles in prostate cancer cells following treatment with DMSO, ENZ, OLA, or ENZ combined with OLA, we found that ENZ combined with OLA induces apoptosis by affecting NHEJ and jointly upregulating pro-apoptotic genes and downregulating anti-apoptotic genes.
Given that the rationale for using OLA in prostate cancer is mainly based on synthetic lethality, most research focused on pharmaceutically impairing HRR by inducing BRCAness. We first demonstrated that the combination of ENZ and OLA inhibited the proliferation of prostate cancer cell lines in a synergistic manner. Immunofluorescence showed that ENZ combined with OLA significantly increased the number and enhanced fluorescence intensity of foci of γH2AX, indicating DSB accumulation. Apart from HRR, NHEJ is one of the major pathways in DSB repair in human cells.14 Through NGS, we found that the combined use of ENZ and OLA affected NHEJ in addition to HRR. NHEJ occurs through several steps that mainly include recognition, end processing, and ligation. DNA-PKcs, encoded by PRKDC, participates in the recognition and end processing of NHEJ, and XRCC4 is involved in the NHEJ ligation step.23 DNA-PKcs also promotes tumor progression and metastasis and makes prostate cancer cells resistant to docetaxel.16,24 High expression of DNA-PKcs is a predictor of poor biochemical recurrence survival in patients with prostate cancer treated with radiotherapy.25,26 Our data suggested that ENZ combined with OLA corporately downregulated DNA-PKcs and XRCC4. Meanwhile, OLA-downregulated DNA-PKcs combined with ENZ-downregulated XRCC4 contributed to DSB, perhaps through their roles in NHEJ pathway.
We also demonstrated that ENZ combined with OLA promoted apoptosis via upregulating pro-apoptotic genes and downregulating anti-apoptotic genes. Furthermore, we found that the inhibitory function of OLA on the pro-apoptotic gene DAPK1 could be reversed by ENZ. DAPK1 mediates pro-apoptotic activity through tumor necrosis factor-alpha (TNF-α),27 which is consistent with our results. DAPK1 also mediates the inhibition of the IκB kinase β/signalosome 5/programmed death receptor-ligand 1 (PD-L1) axis,28 providing a direction for the future combined use of PD-L1 inhibitors. Although the high expression of IGF1R has a clear relationship with prostate cancer development and progression,29 IGF1R inhibitor has failed in mCRPC clinical trial because of the complexity of the IGF1R system.30 However, IGF1R inhibitor plus mitoxantrone and prednisone has shown improvement in composite progression-free survival among men with mCRPC.31 In our study, IGF1R exhibited an anti-apoptotic effect in the combination group and was upregulated by OLA and downregulated by ENZ. Knockdown of IGF1R showed a synergistic effect with OLA, providing a new strategy for the combined use of IGF1R inhibitors.
Several clinical trials of drug combination are underway to further expand the adaption of OLA, reduce side effects, and slow drug resistance. A recent phase 2 clinical trial using OLA in combination with abiraterone showed a statistically significant radiographic progression-free survival in mCRPC regardless of HRR gene mutation status (13.8 months in combination vs 8.2 months in control, P = 0.034, hazard ratio = 0.65, 95% confidence interval: 0.44–0.97), and a phase 3 clinical trial is now ongoing.32 Our results suggested that ENZ combined with OLA can promote prostate cancer cell apoptosis by multiple pathways other than inducing BRCAness, providing evidence for future clinical trials of ENZ plus OLA in CRPC regardless of HRR gene mutation status.
AUTHOR CONTRIBUTIONS
JL and CL conceived the study. HYD and CL designed the experiments. HYD and PZ performed the experiments and drafted the initial manuscript. MLB, TRZ, and CBN helped to perform data analysis. MLB, LD, and XSZ provided technical assistance. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
Global gene expression profiles analyzed by NGS. (a) Principal component analysis of C4-2 cells in indicated groups. (b) Box and whisker plots of FPKM levels in indicated groups. (c) Violin plots of FPKM distribution in indicated groups. (d) Distribution of FPKM density in indicated groups. NGS: next-generation sequencing; ENZ: enzalutamide; OLA: olaparib; PC: principal component; FPKM: fragments per kilobase of exon model per million mapped reads.
GO enrichment analyses of the differentially expressed genes. Top 20 terms of the differentially expressed genes analyzed by GO enrichment analyses in (a) ENZ vs DMSO group, (b) OLA vs DMSO group, and (c) ENZ plus OLA group. ENZ: enzalutamide; OLA: olaparib; GO: Gene Ontology.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (82002718), the Jiangsu Natural Science Foundation (BK20191077), and Jiangsu Province Hospital (the First Affiliated Hospital of Nanjing Medical University) Clinical Capacity Enhancement Project (JSPH-MC-2021-12).
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, et al. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal RR, Feng FY, Small EJ. Emerging categories of disease in advanced prostate cancer and their therapeutic implications. Oncology (Williston Park) 2017;31:467–74. [PubMed] [Google Scholar]
- 4.Negoita S, Feuer EJ, Mariotto A, Cronin KA, Petkov VI, et al. Annual report to the nation on the status of cancer, part ii:recent changes in prostate cancer trends and disease characteristics. Cancer. 2018;124:2801–14. doi: 10.1002/cncr.31549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong AJ, Szmulewitz RZ, Petrylak DP, Holzbeierlein J, Villers A, et al. Arches:a randomized, phase iii study of androgen deprivation therapy with enzalutamide or placebo in men with metastatic hormone-sensitive prostate cancer. J Clin Oncol. 2019;37:2974–86. doi: 10.1200/JCO.19.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain M, Fizazi K, Saad F, Rathenborg P, Shore N, et al. Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer. N Engl J Med. 2018;378:2465–74. doi: 10.1056/NEJMoa1800536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bono J, Mateo J, Fizazi K, Saad F, Shore N, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Xu C, Lee HJ, Ren S, Zi X, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580:93–9. doi: 10.1038/s41586-020-2135-x. [DOI] [PubMed] [Google Scholar]
- 9.Golan T, Kindler HL, Park JO, Reni M, Macarulla T, et al. Geographic and ethnic heterogeneity of germline BRCA1 or BRCA2 mutation prevalence among patients with metastatic pancreatic cancer screened for entry into the polo trial. J Clin Oncol. 2020;38:1442–54. doi: 10.1200/JCO.19.01890. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Karanika S, Yang G, Wang J, Park S, et al. Androgen receptor inhibitor-induced “BRCAness”and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci Signal. 2017;10:eaam7479. doi: 10.1126/scisignal.aam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asim M, Tarish F, Zecchini HI, Sanjiv K, Gelali E, et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat Commun. 2017;8:374. doi: 10.1038/s41467-017-00393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 13.Tarish FL, Schultz N, Tanoglidi A, Hamberg H, Letocha H, et al. Castration radiosensitizes prostate cancer tissue by impairing DNA double-strand break repair. Sci Transl Med. 2015;7:312re11. doi: 10.1126/scitranslmed.aac5671. [DOI] [PubMed] [Google Scholar]
- 14.Radhakrishnan SK, Jette N, Lees-Miller SP. Non-homologous end joining:emerging themes and unanswered questions. DNA Repair (Amst) 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, et al. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell. 2015;28:97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giguère V. DNA-PK, nuclear mTOR, and the androgen pathway in prostate cancer. Trends Cancer. 2020;6:337–47. doi: 10.1016/j.trecan.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, et al. Ualcan:an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong Y, Song Y, Deng S. Combined analysis and validation for DNA methylation and gene expression profiles associated with prostate cancer. Cancer Cell Int. 2019;19:50. doi: 10.1186/s12935-019-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueckl SL, Sikes RA, Edlund NM, Bell RH, Hurtado-Coll A, et al. Increased insulin-like growth factor I receptor expression and signaling are components of androgen-independent progression in a lineage-derived prostate cancer progression model. Cancer Res. 2004;64:8620–9. doi: 10.1158/0008-5472.CAN-04-2446. [DOI] [PubMed] [Google Scholar]
- 20.Baures M, Puig Lombardi E, Di Martino D, Zeitouni W, Pacreau E, et al. Transcriptomic signature and growth factor regulation of castration-tolerant prostate luminal progenitor cells. Cancers (Basel) 2022;14:3775. doi: 10.3390/cancers14153775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai K, McManus JM, Sharifi N. Hormonal therapy for prostate cancer. Endocr Rev. 2021;42:354–73. doi: 10.1210/endrev/bnab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–53. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao OS, Goodman OB., Jr DNA-PKc inhibition overcomes taxane resistance by promoting taxane-induced DNA damage in prostate cancer cells. Prostate. 2021;81:1032–48. doi: 10.1002/pros.24200. [DOI] [PubMed] [Google Scholar]
- 25.Bouchaert P, Guerif S, Debiais C, Irani J, Fromont G. DNA-PKcs expression predicts response to radiotherapy in prostate cancer. Int J Radiat Oncol Biol Phys. 2012;84:1179–85. doi: 10.1016/j.ijrobp.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Molina S, Guerif S, Garcia A, Debiais C, Irani J, et al. DNA-PKcs expression is a predictor of biochemical recurrence after permanent iodine 125 interstitial brachytherapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2016;95:965–72. doi: 10.1016/j.ijrobp.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Yoo HJ, Byun HJ, Kim BR, Lee KH, Park SY, et al. DAPK1 inhibits NF-κB activation through TNF-αand INF-γ-induced apoptosis. Cell Signal. 2012;24:1471–7. doi: 10.1016/j.cellsig.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Guo Z, Zhou C, Zhou L, Wang Z, Zhu X, et al. Overexpression of DAPK1-mediated inhibition of IKKβ/CSN5/PD-L1 axis enhances natural killer cell killing ability and inhibits tumor immune evasion in gastric cancer. Cell Immunol. 2022;372:104469. doi: 10.1016/j.cellimm.2021.104469. [DOI] [PubMed] [Google Scholar]
- 29.Baserga R. The insulin receptor substrate-1:a biomarker for cancer? Exp Cell Res. 2009;315:727–32. doi: 10.1016/j.yexcr.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Barata P, Cooney M, Tyler A, Wright J, Dreicer R, et al. A phase 2 study of OSI-906 (linsitinib, an insulin-like growth factor receptor-1 inhibitor) in patients with asymptomatic or mildly symptomatic (non-opioid requiring) metastatic castrate resistant prostate cancer (CRPC) Invest New Drugs. 2018;36:451–7. doi: 10.1007/s10637-018-0574-0. [DOI] [PubMed] [Google Scholar]
- 31.Hussain M, Rathkopf D, Liu G, Armstrong A, Kelly WK, et al. A randomised non-comparative phase II trial of cixutumumab (IMC-A12) or ramucirumab (IMC-|y1121B) plus mitoxantrone and prednisone in men with metastatic docetaxel-pretreated castration-resistant prostate cancer. Eur J Cancer. 2015;51:1714–24. doi: 10.1016/j.ejca.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saad F, Thiery-Vuillemin A, Wiechno P, Alekseev B, Sala N, et al. Patient-reported outcomes with olaparib plus abiraterone versus placebo plus abiraterone for metastatic castration-resistant prostate cancer:a randomised, double-blind, phase 2 trial. Lancet Oncol. 2022;23:1297–307. doi: 10.1016/S1470-2045(22)00498-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global gene expression profiles analyzed by NGS. (a) Principal component analysis of C4-2 cells in indicated groups. (b) Box and whisker plots of FPKM levels in indicated groups. (c) Violin plots of FPKM distribution in indicated groups. (d) Distribution of FPKM density in indicated groups. NGS: next-generation sequencing; ENZ: enzalutamide; OLA: olaparib; PC: principal component; FPKM: fragments per kilobase of exon model per million mapped reads.
GO enrichment analyses of the differentially expressed genes. Top 20 terms of the differentially expressed genes analyzed by GO enrichment analyses in (a) ENZ vs DMSO group, (b) OLA vs DMSO group, and (c) ENZ plus OLA group. ENZ: enzalutamide; OLA: olaparib; GO: Gene Ontology.