Abstract
We show that ribosomal protein S1 specifically binds the boxA transcriptional antiterminator RNAs of bacteriophage λ and the Escherichia coli ribosomal RNA operons. Although S1 competes with the NusB-S10 antitermination complex for binding to boxA, it does not affect antitermination by the λ N protein in vitro, and its role, if any, in rRNA synthesis is still unknown.
Translation of mRNA in Escherichia coli suppresses the transcriptional termination activity of termination factor Rho because the translating ribosomes prevent Rho from binding the nascent RNA (reviewed in reference 5). Untranslated transcripts, such as those synthesized from the ribosomal RNA (rrn) operons, are more readily accessible to Rho. The rrn operons are, however, transcribed without premature termination of transcription. The observation that insertion of strong Rho-dependent terminators within rrnC caused only a small reduction in the transcription of downstream sequences indicated that a mechanism exists that renders the rrn operons resistant to the action of Rho (17). Later experiments suggested that this mechanism is transcriptional antitermination (1).
Transcriptional antitermination has been best characterized in bacteriophage λ (for a review, see reference 8). The λ N protein is able to modify RNA polymerase so that it becomes resistant to both Rho-dependent and Rho-independent terminators (15, 30). A cis-acting element called the nut site (22) must be transcribed into RNA for N to function (11, 19). The nut site consists of two functional elements, boxA and boxB. boxB RNA is able to form a 15-nucleotide stem-loop that binds N (4, 16, 19). boxA RNA is a 12-nucleotide sequence 5′ to boxB that interacts with host factors (16, 20). The host factors involved in N-mediated antitermination are NusA, NusB, NusG, and ribosomal protein S10 (NusE) (6, 27). It is thought that N, the Nus factors, and the nut site form a ribonucleoprotein complex that stays associated with elongating RNA polymerase and directs the enzyme to transcribe through termination signals (19).
Antitermination in the rrn operons depends on boxA sequences that are closely related to the boxA elements of λ nut sites (13). Moreover, rrn boxA RNA has been shown to bind a heterodimer of NusB and S10 (14, 20). NusB was further implicated in rrn antitermination by experiments showing that NusB is important for rRNA synthesis in vivo (23) and that a NusB-depleted extract is unable to support antitermination in vitro (25). However, antitermination in the rrn operons is known to differ from N-mediated antitermination in three major ways: first, the bacteriophage λ N protein is not involved in rrn antitermination; second, rrn boxA is capable of supporting antitermination in the absence of boxB or any other RNA sequence (2); third, an unidentified factor(s) is required for antitermination in vitro in the rrn system but not in the λ system (25). The present study was initiated to attempt to identify this missing factor(s) and other proteins that interact with boxA RNA.
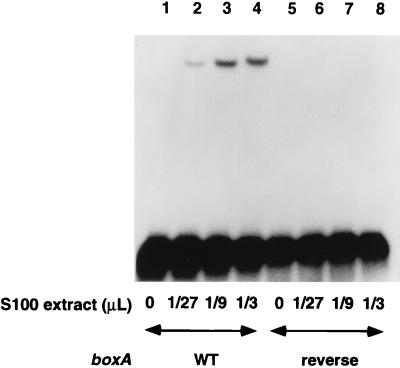
To detect E. coli proteins that bind boxA, S100 extract (7) was incubated in buffer (40 mM HEPES [pH 7.3], 10 mM ammonium sulfate, 15 mM potassium chloride, 0.5 mg of bovine serum albumin/ml, 50 μg of tRNA/ml) with a 35-nucleotide radiolabeled RNA (AGGGAAAGUUCACUGCUCUUUAACAAUUUAGUCGA) containing the 12-nucleotide rrn boxA element (underlined), or boxA inserted in the reverse orientation as a control, and was run on a nondenaturing 10% polyacrylamide gel (60:1 acrylamide/bisacrylamide ratio, 2% glycerol, 0.5× Tris-borate-EDTA). The electrophoretic mobility of the RNA probe containing boxA was reduced in a concentration-dependent manner when it was incubated with various amounts of extract, whereas the mobility of the control probe containing reversed boxA was not (Fig. 1, compare lanes 1 to 4 with lanes 5 to 8). This shifted band was not a consequence of NusB and S10 binding the probe, as this band still formed when antibody against NusB was used to deplete the extract of NusB (data not shown). Furthermore, the band had a mobility different from that of the band that appeared when purified NusB and S10 were incubated with the probe (see Fig. 5).
FIG. 1.
A factor in a crude E. coli extract binds rrn boxA RNA. The indicated amount of S100 extract was incubated with radiolabeled boxA RNA (lanes 1 to 4) or reverse boxA RNA (lanes 5 to 8) and electrophoresed on a nondenaturing polyacrylamide gel. WT, wild type.
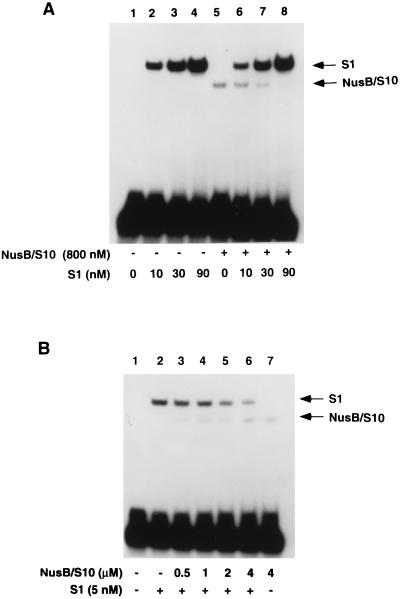
FIG. 5.
S1 competes with NusB-S10 for binding of boxA. S1, NusB, and S10 (as indicated) were incubated with radiolabeled boxA RNA. The reaction mixtures were electrophoresed on a nondenaturing polyacrylamide gel. +, present; −, absent.
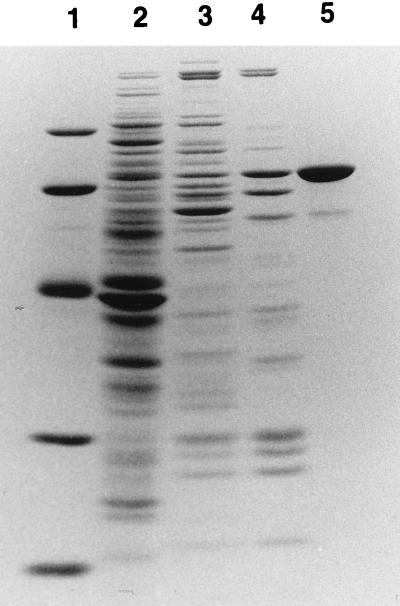
Column chromatography was used to purify and identify the protein that was retarding the mobility of bound boxA in the mobility shift experiment (Fig. 2). S100 extract was first passed over a DEAE-cellulose column (Whatman DE52), and the column was washed with buffer (10 mM Tris-acetate [pH 7.8], 14 mM magnesium acetate, 1 mM dithiothreitol) containing 0.14 M KCl. The protein with boxA-binding activity was then eluted with buffer containing 0.25 M KCl. Fractions containing the protein were pooled (Fig. 2, lane 3) and loaded onto a phenyl-Sepharose column. The protein was eluted with buffer containing 0.05 M KCl, and fractions containing the protein were again pooled (Fig. 2, lane 4) and loaded onto a poly(U)-agarose column (Pharmacia Biotech). The protein remained bound when this column was washed with 1 M KCl and was eluted with 6 M urea. After these purification steps, only one major polypeptide with an apparent molecular mass of 70 kDa and a minor, 60-kDa polypeptide were evident on a sodium dodecyl sulfate-polyacrylamide gel stained with Coomassie blue (Fig. 2, lane 5). Gel purification and renaturation (10) of the 70-kDa protein confirmed that it was responsible for the boxA-binding activity (data not shown).
FIG. 2.
Purification of a 70-kDa protein that binds boxA RNA. Fractions containing boxA-binding activity were pooled after DE52 (lane 3), phenyl-Sepharose (lane 4), and poly(U)-agarose (lane 5) column chromatography and were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue. Lane 1 contains protein molecular mass standards (97.4, 66.2, 45.0, 31.0, and 21.5 kDa from top to bottom), and lane 2 contains E. coli S100 extract.
To identify the 70-kDa protein, its N-terminal sequence was determined for 19 amino acids, providing the identity of the amino acids at all positions except 14 and 16. This sequence was identical to that predicted for ribosomal protein S1 (Fig. 3). S1 is a component of the 30S subunit of the ribosome, where it is thought to interact nonspecifically with the nascent mRNA (26). S1 contains six copies of an approximately 70-amino-acid motif that has been implicated in binding RNA (9). S1 motifs have been found in a variety of proteins, including NusA (3). Considering that NusA has also been implicated in binding boxA RNA (16), it is possible that this interaction is mediated through the S1 domain of NusA. Since the S1 gene is essential for the growth of E. coli (12), presumably because of the role of S1 in translation, participation by S1 in other processes might easily have escaped attention. S1 is an essential subunit of the replicase of bacteriophage Qβ (29). Recently, S1 has also been shown to be complexed with NusA and recombination protein β of phage λ, although the significance of this complex is unclear (28). Nevertheless, we have found that NusA alone does not bind rrn boxA or nut site RNA in a gel mobility shift assay (16, 20) and does not affect the binding of S1 to boxA RNA (data not shown).
FIG. 3.
Comparison of the N-terminal sequence of the 70-kDa protein with the known sequence of ribosomal protein S1. xxx, unknown amino acid.
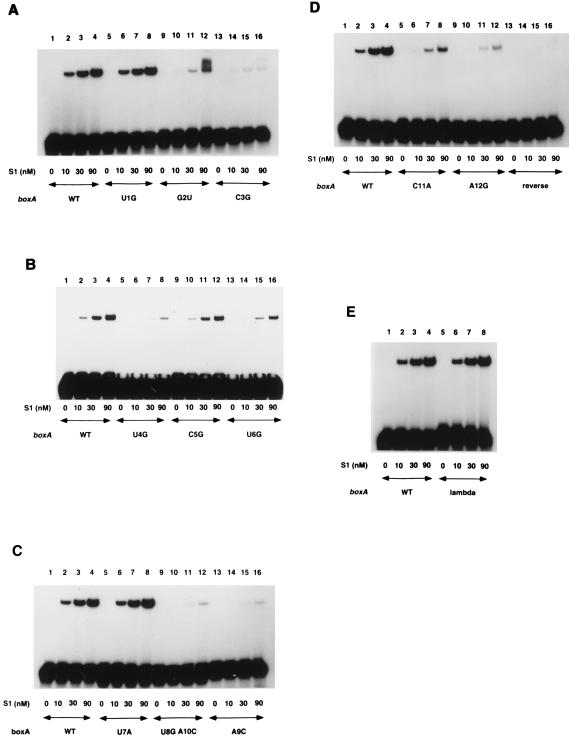
In order to characterize further the specificity of the interaction between S1 and rrn boxA, gel mobility shift experiments were performed with mutant boxA probes (Fig. 4 and Table 1). Certain point mutations in boxA at positions 1, 5, and 7 did not affect the amount of S1 required to shift the mutant probe (Table 1; compare, e.g., Fig. 4A lanes 1 to 4 with lanes 5 to 8). A second class of mutations at positions 2, 4, 6, and 11 substantially affected the interaction between S1 and boxA so that approximately nine times more S1 was required to shift the mutant probe (Table 1; compare, e.g., Fig. 4A lanes 1 to 4 with lanes 9 to 12). A third class of mutations at positions 3, 8 plus 10, 9, and 12 affected the interaction between S1 and boxA so that at least 30 times as much S1 was required to shift the mutant probe (Table 1; compare, e.g., Fig. 4A lanes 1 to 4 with lanes 13 to 16). An almost undetectable amount of probe containing reverse boxA was shifted even at the highest concentration of S1 used (Fig. 4D, lane 16). Approximately equal amounts of S1 were needed to shift probes containing the λ nutR boxA sequence and wild-type rrn boxA (Fig. 4E, compare lanes 1 to 4 with lanes 5 to 8). Thus, the interaction of S1 with λ or rrn boxA RNA is highly specific and could potentially play a role in λ and/or rrn antitermination.
FIG. 4.
Mutations in boxA affect its ability to bind ribosomal protein S1. Various concentrations of S1 were incubated with radiolabeled RNAs containing wild-type (WT) or mutant boxA as indicated, and the reaction mixtures were electrophoresed on nondenaturing polyacrylamide gels.
TABLE 1.
Binding of ribosomal protein S1 to mutant boxA RNAs
| Probe | boxA sequencea | Binding to S1b | Binding to NusB-S10c |
|---|---|---|---|
| rrn boxA | UGCUCUUUAACA | +++ | + |
| boxAU1G | GGCUCUUUAACA | +++ | +/− |
| boxAG2U | UUCUCUUUAACA | ++ | − |
| boxAC3G | UGGUCUUUAACA | + | − |
| boxAU4G | UGCGCUUUAACA | ++ | − |
| boxAC5G | UGCUGUUUAACA | +++ | − |
| boxAU6G | UGCUCGUUAACA | ++ | − |
| boxAU7A | UGCUCUAUAACA | +++ | +/− |
| boxAU8GA10C | UGCUCUUGACCA | + | − |
| boxAA9C | UGCUCUUUCACA | + | − |
| boxAC11A | UGCUCUUUAAAA | ++ | + |
| boxAA12G | UGCUCUUUAACG | + | +/− |
| λ boxA | CGCUCUUACACA | +++ | − |
| boxA (reversed) | ACAAUUUCUCGU | +/− | − |
Boldface indicates substitutions.
+++, wild-type ability to bind S1; ++, apparent ninefold decrease in affinity; +, apparent 30-fold decrease in affinity; +/−, almost undetectable level of binding.
+, wild-type ability to bind NusB-S10; +/−, apparent ninefold decrease in affinity; −, no detectable binding (20).
To determine if S1 could enter a ribonucleoprotein complex with NusB, S10, and rrn boxA RNA, a gel mobility shift experiment was performed in which purified NusB, S10, and S1 were incubated at various concentrations with RNA containing boxA (Fig. 5). Probe shifted by S1 had a lower electrophoretic mobility than probe shifted by the NusB-S10 complex (Fig. 5A, compare lanes 2 and 5). As S1 was added in increasing amounts to reaction mixtures containing NusB and S10, the NusB-S10-RNA complex disappeared and no supershifted band was observed (Fig. 5A, lanes 5 to 8). Thus, S1 apparently competes with NusB and S10 for binding to boxA RNA. This result is consistent with our observations that the nucleotides most important for binding S1 (Table 1) are also important for binding NusB and S10 (20) (Table 1). In a similar experiment, addition of increasing amounts of NusB-S10 to reaction mixtures containing S1 decreased the amount of S1-RNA complex without producing a supershifted complex (Fig. 5B, lanes 2 to 6), again demonstrating that NusB-S10 and S1 cannot simultaneously bind boxA RNA.
Our binding data (Fig. 5) indicated that the affinity of rrn boxA RNA for S1 is at least 200 times greater than its affinity for the NusB-S10 complex. This raised the possibility that S1 might be an inhibitor of antitermination. Nevertheless, adding purified S1 did not inhibit rrn boxA-mediated antitermination in reactions containing crude E. coli extract (25), nor did it make possible rrn antitermination in vitro when it was added to reactions containing purified Nus factors (24). Therefore, the significance of the specific interaction we have described between rrn boxA and ribosomal protein S1 is still unclear.
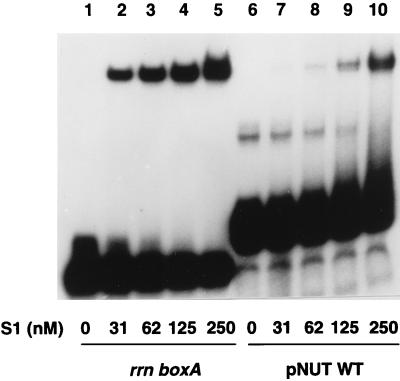
The existence of an E. coli inhibitor of N-mediated antitermination that binds boxA has been predicted by Patterson and colleagues on the basis of genetic experiments with deletion and point mutations in boxA (21). To determine whether S1 could be this inhibitor, we first compared the strength of binding between S1 and a probe containing rrn boxA with that of S1 and a probe containing a λ nut site (boxA + boxB) (Fig. 6). S1 bound the nut site probe, although approximately eight times as much S1 was required to shift the nut site probe as to shift a similar amount of the rrn boxA probe (Fig. 6, compare lanes 2 to 5 with lanes 7 to 10), raising the possibility that boxB might partially hinder the S1-boxA interaction. Even though S1 could bind boxA in the presence of boxB, we found that S1 did not inhibit N-mediated nonprocessive antitermination in vitro in reactions in which NusA was the only E. coli cofactor (30), nor did it inhibit processive antitermination in reactions containing NusA, NusB, NusG, and S10 (15) (data not shown). The inability of S1 to inhibit antitermination even though it binds boxA with an apparently higher affinity than NusB-S10 (which does not bind nut site RNA in the absence of N and other factors [16]) suggests that protein-protein interactions within the N-modified transcription complex involving N, RNA polymerase, NusA, and NusG allow NusB-S10 to outcompete S1 for binding boxA (8). The ability of S1 to compete with NusB-S10 for binding boxA makes it seem unlikely that S1 has a positive role in antitermination. It is conceivable that S1 may be just one component of an antitermination inhibitory complex whose other component(s) remains to be identified.
FIG. 6.
S1 binds RNA containing a nut site. Various amounts of S1 (as indicated) were incubated with probe containing either the rrn boxA or λ (pNUT WT) nut site. The reaction mixtures were electrophoresed on a nondenaturing polyacrylamide gel.
Alternatively, S1 may be involved in some other kind of boxA-mediated process. For example, the processing stalks of the 16S and 23S rRNAs are located near boxA sequences in the leader regions of the rrn operons and in the spacer regions between the 16S and 23S rRNAs. Morgan (18) has suggested that the processing stalks could be juxtaposed for processing by their adjacent boxA sequences. If that is so, one could imagine that boxA and S1 might play a role in the processing of rRNA.
Another possibility is provided by the recent observation that the λ N protein can repress the translation of its own mRNA in a process that requires the nut site but appears to be independent of NusA, NusB, and S10 (31). The boxA16 mutation, which alters the fifth nucleotide (underlined) of λ boxA (CGCUCUUACACA), prevents antitermination of transcription but not repression of translation, whereas the boxA5 mutation, which alters the second nucleotide of λ boxA (CGCUCUUACACA), prevents both (31). Interestingly, nucleotides 2 and 5 of boxA are both important for the binding of NusB and S10 (20) (Table 1), whereas only nucleotide 2 is important for the binding of S1 (Fig. 3 and Table 1). Translational repression may therefore involve a complex containing the ribosome in which the binding of N to boxB and the binding of ribosomal protein S1 to boxA at the nutL site somehow prevent or aid binding of the ribosome to the AUG initiator codon of the N gene located not far downstream of nutL.
REFERENCES
- 1.Aksoy S, Squires C L, Squires C. Evidence for antitermination in Escherichia coli rRNA transcription. J Bacteriol. 1984;159:260–264. doi: 10.1128/jb.159.1.260-264.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg K L, Squires C, Squires C L. Ribosomal RNA operon anti-termination. Function of leader and spacer region boxB-boxA sequences and their conservation in diverse micro-organisms. J Mol Biol. 1989;209:345–358. doi: 10.1016/0022-2836(89)90002-8. [DOI] [PubMed] [Google Scholar]
- 3.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay S, Garcia-Mena J, DeVito J, Wolska K, Das A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage λ. Proc Natl Acad Sci USA. 1995;92:4061–4065. doi: 10.1073/pnas.92.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman D I. Regulation of phage gene expression by termination and antitermination of transcription. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 263–319. [Google Scholar]
- 7.Goda Y, Greenblatt J. Efficient modification of E. coli RNA polymerase in vitro by the N gene transcription antitermination protein of bacteriophage lambda. Nucleic Acids Res. 1985;13:2569–2582. doi: 10.1093/nar/13.7.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenblatt J, Nodwell J R, Mason S W. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 9.Gribskov M. Translational initiation factor IF1 and factor eIF2a share a RNA binding motif with prokaryotic ribosomal protein S1 and polynucleotide phosphorylase. Gene. 1992;119:107–111. doi: 10.1016/0378-1119(92)90073-x. [DOI] [PubMed] [Google Scholar]
- 10.Hager D A, Burgess R A. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ topoisomerase, and other enzymes. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 11.Horwitz R J, Li J, Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987;51:631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- 12.Kitakawa M, Isono K. An amber mutation in the gene rpsA for ribosomal protein S1 in Escherichia coli. Mol Gen Genet. 1982;185:445–447. doi: 10.1007/BF00334137. [DOI] [PubMed] [Google Scholar]
- 13.Li S C, Squires C L, Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 14.Mason S W, Li J, Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J Mol Biol. 1992;223:55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- 15.Mason S W, Li J, Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage λ. J Biol Chem. 1992;267:19418–19426. [PubMed] [Google Scholar]
- 16.Mogridge J, Mah T-F, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the λ N protein. Genes Dev. 1995;9:2831–2844. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 17.Morgan E A. Insertions of Tn10 into an E. coli ribosomal RNA operon are incompletely polar. Cell. 1980;21:257–265. doi: 10.1016/0092-8674(80)90133-6. [DOI] [PubMed] [Google Scholar]
- 18.Morgan E A. Antitermination mechanisms in rRNA operons of Escherichia coli. J Bacteriol. 1986;168:1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nodwell J R, Greenblatt J. The nut site of bacteriophage λ is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 1991;5:2141–2151. doi: 10.1101/gad.5.11.2141. [DOI] [PubMed] [Google Scholar]
- 20.Nodwell J R, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 21.Patterson T A, Zhang Z, Baker T, Johnson L L, Friedman D I, Court D L. Bacteriophage lambda N-dependent transcription antitermination: competition for an RNA site may regulate antitermination. J Mol Biol. 1994;236:217–228. doi: 10.1006/jmbi.1994.1131. [DOI] [PubMed] [Google Scholar]
- 22.Salstrom J S, Szybalski W. Coliphage λ nutL−: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978;124:195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- 23.Sharrock R A, Gourse R L, Nomura M. Inhibitory effect of high-level transcription of the bacteriophage λ nutL region on transcription of rRNA in Escherichia coli. J Bacteriol. 1985;163:704–708. doi: 10.1128/jb.163.2.704-708.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squires, C. L. Personal communication.
- 25.Squires C L, Greenblatt J, Li J, Condon C, Squires C L. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci USA. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A R. Structure and functions of the largest Escherichia coli ribosomal protein. Trends Biochem. 1984;9:491–494. [Google Scholar]
- 27.Sullivan S L, Ward D F, Gottesman M E. Effect of Escherichia coli nusG function on λ N-mediated transcription antitermination. J Bacteriol. 1992;174:1339–1344. doi: 10.1128/jb.174.4.1339-1344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesh T V, Radding C M. Ribosomal protein S1 and NusA protein complexed to recombination protein β of phage λ. J Bacteriol. 1993;175:1844–1846. doi: 10.1128/jb.175.6.1844-1846.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahba A J, Miller M J, Niveleau A, Landers T A, Carmichael G G, Weber K, Hawley D A, Slobin L I. Subunit I of Qβ replicase and 30S ribosomal protein S1 of Escherichia coli: evidence for the identity of the two proteins. J Biol Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- 30.Whalen W, Ghosh B, Das A. NusA protein is necessary and sufficient in vitro for phage λ N gene product to suppress a ρ-independent terminator placed downstream of nutL. Proc Natl Acad Sci USA. 1988;85:2494–2498. doi: 10.1073/pnas.85.8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson H R, Kameyama L, Zhou J, Guarneros G, Court D L. Translational repression by a transcriptional elongation factor. Genes Dev. 1997;11:2204–2213. doi: 10.1101/gad.11.17.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]