Abstract
H-NS is a major constituent of the Escherichia coli nucleoid, whereas ςS is a stress-induced sigma factor. An hns null mutation affects the cellular content of ςS in such a way that a remarkable accumulation of ςS is observed in the logarithmic growth phase, which results in enhanced expression of a number of ςS-dependent genes, including the katE gene. We isolated an extragenic mutation that affects the expression of the katE-lacZ fusion gene in the Δhns background. The relevant gene was identified as yhhP, which encodes a small polypeptide of 81 amino acids. Lesion of this gene seemed to affect the stability of ςS. A deletion analysis of yhhP revealed that this small protein plays a fundamental role in the general physiology of E. coli. The yhhP-deficient cell is not capable of growing in standard laboratory rich medium (i.e., Luria broth), resulting in the formation of filamentous cells. Homologs of this intriguing protein occur in a wide variety of bacterial species, including archaeal species.
H-NS is a major constituent of the Escherichia coli nucleoid (1), whereas ςS is a stress-induced sigma factor (10, 12). The former influences the transcription of a number of apparently unlinked genes on the chromosome (6, 19), whereas the latter modulates the transcription of a certain subset of genes whose expression is enhanced at the stationary phase (5, 13). Although intensive studies of these two proteins have been done separately, it is known that there is a clear link between their cellular functions. We and others previously showed that a mutational lesion of the hns gene (i.e., the hns::neo allele) affects the cellular content of ςS in such a way that a remarkable accumulation of ςS is observed in the logarithmic growth phase (2, 18). This accumulation was shown to be the result of an elevated translational efficiency of rpoS mRNA and also of an increased stability of newly synthesized ςS in the Δhns cells. Consequently, such an event results in enhanced expression of a number of ςS-dependent genes, including the katE gene, which encodes a stationary-phase-specific catalase. However, the mechanism by which H-NS controls the cellular content of ςS has been the subject of debate (2, 18). To gain insight into the underlying molecular mechanism, we attempted to search for chromosomal mutations that affect (or decrease) the expression of the katE-lacZ fusion gene in the Δhns background in the hope of finding a gene(s) that can be implicated in the presumed H-NS–ςS–katE regulatory circuit. During the course of such experiments, we identified a new gene which encodes a small polypeptide of 81 amino acids. Although this protein may be implicated indirectly in the H-NS–ςS regulatory circuit, we found that it plays a fundamental role in the general physiology of E. coli growing under standard laboratory conditions.
Isolation of mutants that affect ςS production in an hns deletion mutant.
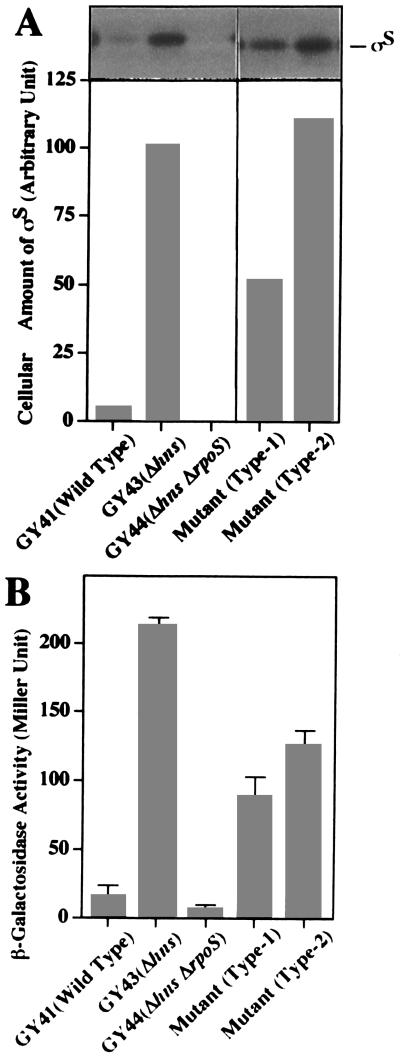
As shown in Fig. 1A, a mutational lesion of the hns gene (i.e., the hns::neo allele) affects the cellular content of ςS in such a way that a great accumulation of ςS is observed in the logarithmic growth phase (GY41, wild type; GY43, Δhns). Consequently, remarkably enhanced expression of katE-lacZ (or β-galactosidase) in GY43 was strictly ςS dependent, although the katE-lacZ fusion gene was not significantly expressed in the wild-type strain grown to the logarithmic growth phase in M9 glucose medium (Fig. 1B). In this study, we attempted to search for chromosomal mutations that affect (or decrease) the expression of katE-lacZ in the Δhns background. GY43 carrying both the Δhns and katE-lacZ alleles gave dark blue colonies on M9 glucose agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The cells were mutagenized with 2% ethyl methanesulfonate (20% viability) and then spread on the selection plate. After an extensive screening, 36 candidates showing white or pale-blue color were isolated, each of which seemed to express a lower level of β-galactosidase activity. Among them, 15 were found to be due to mutations in the katE-lacZ fusion gene itself while 16 had mutations that mapped in the rpoS gene itself. The remaining five candidates were assumed to have extragenic mutations that somehow affect the H-NS–ςS–katE regulatory circuit. Levels of ςS in these mutants were directly measured by means of immunoblotting with a ςS antiserum (Fig. 1A), whereas levels of katE-lacZ expression were measured by monitoring β-galactosidase activities (Fig. 1B). Only one mutant (type 1) exhibited a lower level of ςS (about 50%) than that of the parental strain, while the other four mutants (type 2) had more or less the same level of ςS as the parental strain (Fig. 1A). However, all of them exhibited decreased levels of β-galactosidase activity (40 to 60% of that of the parental strain) (Fig. 1B). We then focused our attention on the type 1 mutant. Although its observed phenotype is not striking, its putative mutation was assumed to affect the content of ςS in the Δhns background, thereby resulting in a reduction of katE expression.
FIG. 1.
Immunoblotting analyses showing the cellular content of ςS and the β-galactosidase activities expressed by the cells carrying the katE-lacZ fusion gene. The E. coli cells with each indicated relevant genetic background and two types of mutant cells from GY43 were grown in M9 minimal medium containing glucose (0.2%) at 30°C. (A) Total proteins were prepared by precipitation with trichloroacetic acid from the cells grown to the mid-logarithmic growth phase (after a 2-h cultivation). Samples (10 μg of protein) were subjected to immunoblotting analysis with an anti-ςS antiserum, as described previously (18). Above the graph is an immunostained membrane filter, whereas the graph shows the quantitative representation of the stained bands corresponding to ςS. (B) The same harvested cells used for panel A were subjected to measurement of β-galactosidase activity. The procedures were essentially the same as those described by Miller (14).
Identification of the mutant gene.
To transfer this putative mutation into a fresh genetic background and to map its position on the chromosome, a rough mapping was first carried out by means of F-factor-mediated conjugation with a set of Hfr strains (KL series) (17). Then, a fine mapping was done by means of P1 transduction with a large set of Tn10 insertions in the chromosome (zxx series::Tn10) (3). The mutation was mapped to a position between 67 and 82 min on the E. coli chromosome and then to a position between 76.9 and 77.8 min. This result indicated that the phenotypic alteration observed for the putative mutant resulted from a single mutational event (the mutant allele was tentatively designated sirA1, for ςS regulation). As a result of these experiments, we have established the following two strains: GY47, which carries only the sirA1 allele, and GY49, which carries the sirA1 allele as well as the Δhns allele (note that without these modifications, these strains have isogenic backgrounds derived from MC4100, which carries the katE-lacZ fusion gene).
The sirA1 mutation affects the stability of ςS.
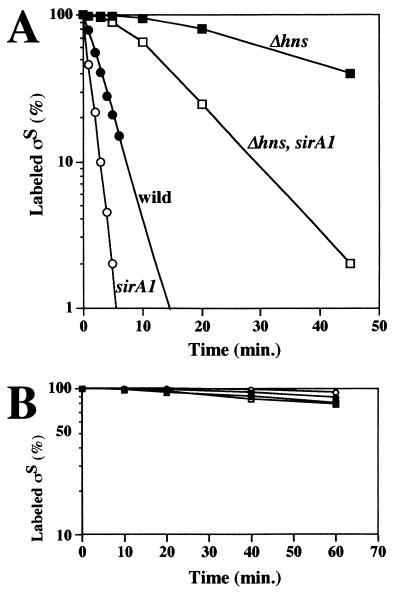
ςS production was examined in GY47 (sirA1) and GY49 (sirA1 Δhns). It is known that the regulation of ςS production in E. coli is modulated during transcription and translation and that it affects stability (11). For both GY47 and GY49, the possible transcriptional control of rpoS was examined by employing an rpoS-lacZ transcriptional fusion gene whereas the possible translational control of rpoS mRNA was examined by means of pulse-labeling with [35S]methionine for a short period (2 min), as described previously (18). The results showed that neither the transcriptional nor the translational efficiency of rpoS mRNA was affected by the sirA1 mutation in both GY47 and GY49 (data not shown). However, it was found that the sirA1 mutation affects significantly the stability of ςS, particularly during the logarithmic growth phase (Fig. 2A). As demonstrated previously (2, 18), ςS becomes more stable in the Δhns background than in the wild-type background. The sirA1 mutation caused a destabilization of ςS in both the Δhns and wild-type backgrounds. Based on this finding, the phenotypes shown in Fig. 1 can be reasonably explained. However, it should be noted that this effect of the sirA1 lesion was not observed for the cells grown to the stationary phase (Fig. 2B). In any case, from these results, we concluded that the sirA1 mutation affects, albeit moderately, ςS stability at the logarithmic growth phase.
FIG. 2.
Characterization of the stability of ςS. The E. coli cells with each indicated relevant genetic background were grown to the mid-logarithmic growth phase (after a 2 h cultivation) (A) or the stationary phase (after a 6 h cultivation) (B). A portion of each culture (1.2 ml) was labeled with [35S]methionine for 1 min (A) or 2 min (B) and then chased with nonradioactive methionine. At the intervals indicated, portions (0.2 ml) were taken and the amounts of labeled ςS were determined, as described previously (18). The amounts of labeled ςS were expressed (percentages) relative to that determined for the sample without chase.
Cloning of the sirA gene.
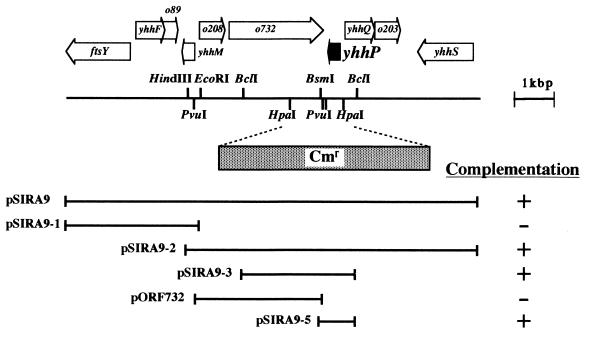
We then wanted to clarify the nature of the sirA gene. To this end, we attempted to clone this putative gene. Based on the finding that the sirA gene is located at between 76.7 and 77.8 min, we employed the two Kohara λ phage clones (named 5F2 and 7H7) (9), which should carry the chromosomal DNA segments covering this region. The DNA inserts from these λ phages were digested partially with Sau3AI and then cloned randomly onto a plasmid vector. GY49, which carries the sirA1 allele, was transformed with the plasmid mixture, and then SirA+ transformants were screened by spreading them on M9 glucose agar plates containing X-Gal, based on the rationale described above. One of the positive clones showing dark-blue color was found to harbor a recombinant plasmid (named pSIRA9), shown in Fig. 3A. Based on the latest version of the E. coli genome structure, a number of open reading frames (ORFs) were predicted within this region. Most of them, except for that of the ftsY gene, are hypothetical. We further constructed several subclones from the original plasmid and found that pSIRA9-5 carrying the short BsmI-BclI fragment is able to complement the sirA1 mutation (Fig. 3A). According to sequences in the E. coli databases, this segment should encompass only a single complete ORF (or gene), named yhhP. We determined the entire nucleotide sequence for the cloned BsmI-BclI fragment from pSIRA9-5, and the sequence was confirmed to be identical to those in the databases. The yhhP gene is predicted to encode a protein of only 81 amino acids. To determine whether the yhhP gene corresponds to the sirA gene, we sequenced the yhhP gene from the chromosome of the sirA1 mutant with appropriate oligonucleotide PCR primers. The sirA1 mutant allele was found to have a single base substitution (G to A), which results in an amino acid substitution (Glu to Lys) at amino acid position 18 in the yhhP coding sequence. Therefore, we conclude that the sirA gene corresponds to the yhhP gene (hereafter, sirA will be referred to as yhhP).
FIG. 3.
Schematic representation of the E. coli chromosomal region surrounding the yhhP gene. The genome organization is schematically shown, based on the latest version of the E. coli entire genome sequence (4). Below the picture, the plasmids used in this study are listed. These plasmids are derivative of pBR322, and each carries the indicated chromosomal region (horizontal lines). They were introduced into GY49, which carries the sirA1 allele as well as the katE-lacZ fusion gene. Then, the abilities to complement the sirA1 lesion were examined and are indicated by signs (+, positive; −, negative). The HpaI-HpaI region on the chromosome was replaced by a cassette carrying the chloramphenicol resistance gene (Cmr), as indicated, to yield GY51 (a derivative of MC4100).
Characterization of a yhhP deletion mutant.
To gain further insight into the function of the yhhP gene product, the HpaI-HpaI chromosomal region of E. coli MC4100 was replaced by the chloramphenicol cassette (Cmr) to yield GY51 (o732-yhhP::Cmr) (Fig. 3A). This substitution was carried out by the established method of a linear DNA transformation in a recD mutant background (16). This construct should lack both the functional yhhP and o732 genes on the chromosome. Keeping this fact in mind, we characterized this strain in terms of yhhP’s function. We first examined GY51 with special reference to ςS stability. Essentially the same experiment as that conducted for the sirA1 mutant shown in Fig. 2A was done for GY51, and essentially the same result as that observed for GY49 (sirA1) was obtained (data not shown). It should be emphasized that this altered property of GY51 was completely complemented by the introduction of pSIRA9-5 carrying only the yhhP gene. This result fully supported our previous notion that this gene is implicated in ςS stability.
The yhhP gene product is crucial for cell viability.
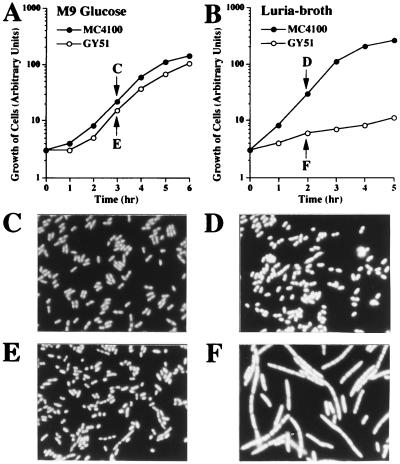
Surprisingly, we were unable to isolate the o732-yhhP::Cmr construct on Luria agar rich medium (i.e., we used M9 glucose minimal medium). In addition, although we wanted to construct the o732-yhhP::Cmr deletion in the Δhns background, such a stable construct was never generated. We interpreted these results by assuming that either the o732 or the yhhP gene plays a crucial physiological role under standard conditions for growth. Typical growth curves for MC4100 (wild type) and GY51 (o732-yhhP::Cmr) in M9 glucose minimal medium and in Luria broth rich medium at 37°C are shown in Fig. 4A and B, respectively. It was found that when GY51 cells grown in M9 glucose medium were inoculated into the rich medium, cell growth completely ceased almost immediately. Essentially the same result was obtained with agar plates containing rich medium. This phenomenon was seen even when the cells were grown at various temperatures. This typical phenotype is the so-called rich medium sensitivity for growth. We then examined cell morphology for GY51, at the growth points indicated in Fig. 4A. When the GY51 cells were incubated for several hours in the rich medium, they started to show an extremely elongated cell morphology, probably due to a defect in cell division (Fig. 4F) (4′,6-diamidino-2-phenylindole [DAPI]-stained cells). Note that chromosome segregation appeared to occur normally and that each filamentous cell contained multiple nuclei positioned regularly in the compartment. In any case, the rich medium’s sensitivity for growth and filamentous-cell formation were strikingly evident in GY51. The yhhP gene should be responsible for these particular phenotypes, because pSIRA9-5 carrying the yhhP gene could completely complement these lesions, but another plasmid carrying the o732 gene could not do so (data not shown). Furthermore, when the original mutant carrying the sirA1 allele was examined with regard to these particular aspects, it also exhibited the same properties, though to a slightly more moderate extent (i.e., very slow growth on the rich medium). It was thus revealed that the yhhP gene, which encodes a small polypeptide, plays a crucial role in E. coli cell growth under standard laboratory conditions.
FIG. 4.
The E. coli mutant carrying a deletion of the yhhP gene shows characteristic phenotypes. GY51 mutant cells, along with wild-type MC4100 cells, were grown either in M9 glucose medium (A) or Luria broth (B) at 37°C. The growth curves were monitored by measuring turbidity. At the times indicated, the cells were stained with DAPI and then examined by fluorescence microscopy. (C) MC4100 grown in M9 glucose medium; (D) MC4100 grown in Luria-broth; (E) GY51 grown in M9 glucose medium; (F) GY51 grown in Luria broth. Note that scale bars are not indicated because the wild-type cells (C and D) were found to be of average sizes (around 2 μm in length), and all micrographs were prepared with the same magnification as that used for the wild type.
Implications.
In the current E. coli databases, the yhhP gene is listed as hypothetical and its gene product shows no significant similarity to any other protein of known biological function. However, the above-described findings encouraged us to inspect its amino acid sequence more closely by computer-aided searches (with BLAST and FASTA). Such inspections revealed that similar proteins are predicted to occur in a variety of microorganisms whose entire genomic sequences have recently been completed (e.g., Haemophilus influenzae Rd, Bacillus subtilis, Synechocystis sp., and Methanocccus jannaschii). As listed in Fig. 5, they are significantly similar to each other not only in that a number of conserved amino acids can be aligned but also in that their sizes are relatively small (73 to 84 amino acids). Furthermore, E. coli was predicted to have two other similar proteins (YedF and YeeD) and H. influenzae has two (YhhP homolog and Y242). Thus, the YhhP-like proteins appear to be widespread among microorganisms, including archaeal organisms. Among these putative YhhP homologs (or orthologs), N-terminal sequences in which the existence of the CPxP motif is evident are highly conserved. In fact, this motif has already been documented for the set of E. coli and H. influenzae proteins in the PROSITE database (dictionary of protein sites and patterns; http://expasy.hcuge.ch/; the Geneva University Hospital and the University of Geneva), although its functional significance is unknown. It is worth mentioning that the CPeP sequence in the E. coli YhhP protein has been changed to CPkP in the sirA1 mutation characterized in this study. These facts together are compatible with the idea that this family of proteins has a fundamental role in the physiology of bacterial cells.
FIG. 5.
Alignment of the deduced amino acid sequence from the E. coli yhhP gene with those of putative YhhP paralogs and orthologs in microorganisms. In each line, the abbreviations in parentheses indicate the microorganisms from which the sequences are derived. Ec, E. coli; Hi, H. influenzae; Bs, B. subtilis; Ss, Synechocystis sp.; Mj, M. jannaschii. The proteins are labeled according to their accession numbers in the SwissProt database, except for ssl1707 from Synchocystis sp. (7, 8). The numbers indicate total amino acid residues. Among these aligned sequences, highly conserved amino acids are shaded and conserved CPxP motifs are boxed. The amino acid substitution, identified in the sirA mutant, is also indicated (Glu-18 to Lys).
Although the yhhP gene was originally identified as one whose mutational lesion affected the H-NS–ςS regulatory circuit, the underlying molecular mechanism is not known. As judged by the extent of its effect on ςS stability, this protein may exert its effect on this particular event indirectly. Rather, the YhhP protein seems to play a more fundamental (or broader) physiological function. The observed effect of the yhhP mutant on ςS stability may also be relevant to this presumedly broad function of YhhP. Characterization of the evident phenotype (i.e., rich medium sensitivity for growth and cell elongation) should give us a clue to understand the possible function of this presumably ubiquitous protein. Experiments along this line, including isolation of multicopy suppressor genes for the ΔyhhP mutant, are under way. In this respect, it may be worth mentioning that the complete genome sequence of E. coli revealed that, of 4,288 protein-coding genes annotated, 38% (1,632) have no attributed function (4). Furthermore, of the 4,288 genes, 9% (381) have protein products smaller than 100 amino acids (14). Blattner and his colleagues (4) mentioned that it was difficult in general to assign functions to small ORFs, unless they had been characterized genetically or biochemically. In this sense, our results proved that the small yhhP gene is indeed functional. As recently emphasized by Moxon and Higgins (15), we may not know as much about the biology of E. coli as we believe.
Acknowledgments
We thank K. Ito (Institute for Virus Research, Kyoto University) for kind gifts of E. coli strains (KL series of Hfr strains and those carrying zxx series::Tn10).
This study was supported by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 2.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of ςS and many ςS-dependent genes in Escherichia coli. J Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 6.Higgins C F, Hinton J C, Hulton C S, Owen-Hughes T, Pavitt G D, Seirafi A. Protein H1: a role for chromatin structure in the regulation of bacterial gene expression and virulence? Mol Microbiol. 1990;4:2007–2012. doi: 10.1111/j.1365-2958.1990.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 9.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 10.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 11.Lange R, Hengge-Aronis R. The cellular concentration of the ςS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 12.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 13.McCann M P, Kidwell J P, Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 15.Moxon E R, Higgins C F. E. coli genome sequence. A blueprint for life. Nature. 1997;389:120–121. doi: 10.1038/38107. [DOI] [PubMed] [Google Scholar]
- 16.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, ςS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida T, Ueguchi C, Yamada H, Mizuno T. Function of the Escherichia coli nucleoid protein, H-NS: molecular analysis of a subset of proteins whose expression is enhanced in a hns deletion mutant. Mol Gen Genet. 1993;237:113–122. doi: 10.1007/BF00282791. [DOI] [PubMed] [Google Scholar]