Abstract
Perilipin 2 (Plin2) binds to the surface of hepatic lipid droplets (LDs) with expression levels that correlate with triacylglyceride (TAG) content. We investigated if Plin2 is important for hepatic LD storage in fasted or high-fat diet-induced obese Plin2+/+ and Plin2−/− mice. Plin2−/− mice had comparable body weights, metabolic phenotype, glucose tolerance, and circulating TAG and total cholesterol levels compared with Plin2+/+ mice, regardless of the dietary regime. Both fasted and high-fat fed Plin2−/− mice stored reduced levels of hepatic TAG compared with Plin2+/+ mice. Fasted Plin2−/− mice stored fewer but larger hepatic LDs compared with Plin2+/+ mice. Detailed hepatic lipid analysis showed substantial reductions in accumulated TAG species in fasted Plin2−/− mice compared with Plin2+/+ mice, whereas cholesteryl esters and phosphatidylcholines were increased. RNA-Seq revealed minor differences in hepatic gene expression between fed Plin2+/+ and Plin2−/− mice, in contrast to marked differences in gene expression between fasted Plin2+/+ and Plin2−/− mice. Our findings demonstrate that Plin2 is required to regulate hepatic LD size and storage of neutral lipid species in the fasted state, while its role in obesity-induced steatosis is less clear.
Supplementary key words: Plin2, liver, triglyceride, cholesteryl ester, lipid droplets, fasting
Lipids constitute a diverse group of organic molecules that are important for cellular membrane synthesis, cell signaling, organelle interactions, energy metabolism, and energy storage (1, 2). Because of their hydrophobic properties, most lipids are poorly soluble in the cytosol and are either incorporated into cellular membranes or stored in lipid droplet (LD) organelles. In mammalian cells, the central LD core mainly consists of neutral lipids such as triacylglycerols (TAGs) and/or cholesteryl esters (CEs), although certain specialized cell types may store other esterified lipids, such as retinyl esters (1). The LD surface is surrounded by a monolayer of phospholipids and a cell-specific population of LD-associated proteins, such as the perilipins. Perilipins exhibit very high sequence similarity and have evolutionary and functional relationships that can be traced for more than 500 million years (2, 3). The perilipin family comprises five genes (Plin1-5) in mammals, and their alternative splicing can generate at least 10 different protein isoforms in mice (4). A standardized Perilipin/Plin nomenclature has been introduced (5) for these tissue-specific proteins that interact with lipases and the lipophagy machinery at the LD surface (6).
One of the first mammalian proteins detected at the surface of cytosolic LDs is perilipin 1 (Plin1) (7). Expression of Plin1 is restricted to adipocytes and steroid cells (7). Perilipin 2 (Plin2), initially named adipose differentiation-related protein/ADRP (8), adipophilin (9), and ADFP (10), is the second family member confirmed to bind to LDs (11). Plin2 is unstable and rapidly ubiquitinylated and degraded by the proteasome when not bound to LDs (12, 13). Plin1 seems able to displace Plin2 from the LD surface. Consequently, Plin2 proteins are not detected in adipocytes expressing Plin1 despite the high presence of Plin2 mRNA (11), perhaps except in lipolytically stimulated adipocytes where Plin2 proteins may be stabilized upon phosphorylation of Plin1 (14). Plin2 is ubiquitously expressed (11, 15), but its expression can be highly elevated when cells are exposed to FAs (16, 17). Plin3 is ubiquitously expressed (15) and the protein is stable in the cytosol independent of intracellular LD levels (18). Plin4 contains a divergent extended central region compared with other Plins and is the least studied family member (19). Plin3 and Plin4 may coat LDs during LD growth when the amounts of other Plin proteins are insufficient to coat expanding phospholipid surfaces (18, 20). Plin5 is primarily expressed in oxidative cells, for example, heart and muscle (19, 21), and is involved in the shuttling of FAs from LDs to mitochondria to maintain mitochondrial integrity (22, 23).
Neutral lipids stored in LDs can be mobilized via lipolytic degradation by cytosolic neutral lipases, alternatively by lipophagy in a process where LDs are engulfed by phagophores and degraded in lysosomes (24). Plin proteins are involved in both processes, but their role in lipolysis is better characterized. In the basal state, nonphosphorylated Plin1 protects TAG stores from lipolytic degradation in white adipose tissue (25), whereas Plin5 has a similar role in tissues with high oxidative capacity (e.g., heart and brown adipose tissue) (26, 27). Upon lipolytic stimulation, Plin1 is phosphorylated (2, 7, 28) and enhances lipolysis by releasing abhydrolase domain containing 5 (ABHD5), enabling this lipase coactivator to bind and activate adipose triglyceride lipase (ATGL) at the LD surface. Phosphorylation of Plin5 will also stimulate lipolysis (29), but since Plin5 binds to both ATGL (29) and Abhd5 (30), Plin5 is likely to regulate ATGL activity differently than Plin1. The first and rate-limiting step of TAG lipolysis in adipocytes is activated by ATGL hydrolyzing TAG into diacylglycerol (DAG) and FFA. Subsequently, DAG is cleaved by hormone-sensitive lipase (HSL), followed by cleavage of monoacylglycerol by monoacylglycerol lipase into FFAs and glycerol (31). The importance of Plin proteins and the lipolytic pathway are less defined in the liver. The liver expresses predominantly Plin2 and Plin3, with lower expression of several key lipolytic factors, such as Plin4, Plin5, and HSL (6, 32). Removal of Plin2 depletes TAG stores in the liver (10), but it remains to be determined if Plin2 and Plin3 bind and immobilize ATGL at the LD surface similar to Plin1 and Plin5.
Accumulation of LDs in the liver and other metabolically active tissues is strongly correlated to obesity-mediated metabolic disorders and insulin resistance (33). It is not fully clarified whether liver steatosis protects against or promotes the development of insulin resistance or if LDs accumulate as a consequence of insulin resistance. Hepatic expression of Plin2 is enhanced in obese mice (34, 35) and humans with fatty liver disease (35). In line with these observations, hepatic overexpression of Plin2 in mice increases lipid accumulation (36, 37), whereas global (10, 38, 39) or liver-specific removal of Plin2 (38, 40) reduces hepatic lipid storage. Removal of Plin2 may also protect against the development of hepatic insulin resistance (38, 39, 41). In contrast, muscular overexpression of Plin2 seems to enhance insulin sensitivity despite increased lipid accumulation (42), implying that the consequences of LD deposition can vary between tissues.
Although obesity and insulin resistance promote TAG accumulation and hepatic steatosis, prolonged energy deprivation has a stronger impact on hepatic TAG levels. Fasting triggers a massive release of FAs from adipose tissue into the circulation, promoting FA uptake into multiple nonadipocyte organs, including the liver. In response to overnight fasting, hepatic expression of Plin2 and Plin5 mRNAs and proteins increase substantially (17, 43). While Plin5 may help to limit hepatic inflammation during fasting (43), the role of hepatic Plin2 during fasting is unclear. In this study, we examined the role of Plin2 in the coating of hepatic LDs by analyzing livers of fasted and diet-induced obese Plin2+/+ and Plin2−/− mice.
Materials and Methods
Ethics and animal housing conditions
All experimental uses of animals were approved and registered by the Norwegian Animal Research Authority (Mattilsynet, approval FOTS IDs: #10901, #10902, and #24912) and conformed to the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and ethical guidelines in Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. All mice were housed with a stable light/dark cycle (07 AM–07 PM), with 55 ± 5% relative humidity at 22 ± 2°C in a specific pathogen-free animal unit. Mice had free access to water and rodent chow or custom-made diets. The presence of pathogens was monitored quarterly in accordance with FELASA (Federation of European Laboratory Animal Science Associations) recommendations without detection of any screened pathogens.
Mice diet interventions
Generation of Plin2−/− mice with disruption of Plin2 exons 4–6 has been described previously (44). All Plin2−/− mice were back-crossed into C57BL/6N for >10 generations prior to experiments. Plin2+/+ and Plin2−/− mice were euthanized by cervical dislocation, followed by a rapid collection of tissues, which were snap-frozen in liquid nitrogen. In total, 116 mice were used.
Fasting intervention
Female Plin2+/+ and Plin2−/− mice (15 weeks of age) from the same breeding colony were housed with free access to standard chow (58 E% carbohydrate, 18 E% fat, 24 E% protein; catalog no.: 2018SX, Envigo, IN) or fasted for 24 h. Chow-fed mice remained in their original cages, whereas mice under fasting were transferred in pairs to clean cages equipped with a water bottle and a plastic shelter but no bedding material or eatable items. In total, 38 mice were included in the study (20 Plin2+/+ and 18 Plin2−/− mice). None of the included animals were excluded from the analyses.
High-fat diet intervention
Female Plin2+/+ and Plin2−/− animals were reproduced and housed with free access to rodent chow (62 E% carbohydrate, 11 E% fat, 27 E% protein; catalog no.: RM3A/SDS RM3, Scanbur, Denmark). Mice were given either a low-fat diet (LFD; 3.85 kcal/g, 70 E% from carbohydrates, 10 E% from fat, and 20 E% from protein, catalog no.: D12450J, Research Diets, New Brunswick, NJ) or a high-fat diet (HFD; 5.24 kcal/g, 20 E% from carbohydrates, 60 E% from fat, and 20 E% from protein, catalog no.: D12492J, Research Diets) from 8 weeks of age until 18 or 28 weeks of age (10 or 20 weeks of diet intervention). The two diets differed only in calorie density and their content of carbohydrates and saturated fat. Plin2+/+ and Plin2−/− animals were randomly allocated in groups of 4 ± 1 mice per cage (both genotypes represented in each cage) to minimize microbiota variations. Body weights were measured weekly during the midportion of the light cycle. In total, 78 mice were included in the study (41 Plin2+/+ and 37 Plin2−/− mice). None of the included animals were excluded from the analyses.
Mice phenotyping procedures
Body composition
Body composition was determined at 18 weeks of age (10 weeks of diet) and 28 weeks of age (20 weeks of diet) with a Minispec LF90II magnetic resonance imaging machine (Bruker, Billerica, MA).
Oral glucose tolerance test
Oral glucose tolerance test was performed at 17 weeks of age (9 weeks of diet) and 27 weeks of age (19 weeks of diet). Briefly, after a fasting period of 5 h (9–14 AM), glucose (d-(+)-glucose, catalog no.: G7021; Sigma, Darmstadt, Germany) was administered via oral gavage dosed at 1.5 g/kg lean mass. Blood glucose was measured with a glucose meter (FreeStyle Precision Neo, Abbot, The Netherlands) in whole blood collected from the tip of the tail at 0, 15, 30, 60, 90, and 120 min time points. Insulin levels during the glucose tolerance test were determined in serum collected at 0, 15, and 30 min time points using an Ultra Sensitive Mouse Insulin ELISA kit (catalog no.: 90080, Crystal Chem, Elk Grove Village, IL).
Indirect calorimetry
A week before euthanasia, mice were placed in an indirect calorimetry system consisting of 20 individual cages (PhenoMaster, TSE Systems, Germany). Indirect calorimetric measurements were performed as described previously (45). Gas flow was 0.42 l/min, with gas exchange measured for 10 s per cage in 20 min intervals. Physical activity was recorded as movements in the XY plane. Food intake was calculated for individual mice by measuring diet before and after indirect calorimetry measurements.
Isolation of total RNA and RT-qPCR
Total RNA was isolated with NucleoSpin RNA kit (Macherey-Nagel, GmbH & Co KG, Düren, Germany), with a modified protocol described previously (44). RNA was eluted in 40 μl RNase-free water and stored at −80°C. Concentrations and purity of RNA samples were determined with NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington).
Total RNA (12.5 ng/μl) was reversely transcribed with random hexamers and the High Capacity cDNA Reverse Transcription Kit (catalog no.: 4368814, Thermo Fisher Scientific) on an Eppendorf Mastercycler EP Gradient S (Eppendorf AG, Hamburg, Germany) as described previously (44). Gene-specific regions were amplified from 10 ng/μl complementary DNA with gene-specific assay primers (200 nmol/l each, listed in supplemental Table S1) and Bio-Rad SsoAdvanced™ Universal SYBR® Green Supermix using the Bio-Rad CFX96 system (Bio-Rad, Hercules, CA). Cycling conditions and primer design have been described previously (44). Data are presented as gene expression levels relative to the expression of Tbp (2−ΔCq).
RNA-Seq and read counting
The total RNA quantity and integrity were measured with a Bioanalyzer RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA). The RNA samples had RIN values between 7.6 and 8.9. Total RNA samples were subjected to Strand-Specific TruSeqTM mRNA-Seq library preparation, and 50 bp paired-end reads were sequenced at the Norwegian Sequencing Centre (Oslo University Hospital, Ullevål, Oslo) as described before (45) on an Illumina Novaseq 600 instrument. BBDuk (BBMap v34.56 was used to remove/trim low-quality reads and adapter sequences sourceforge.net/projects/bbmap/). Reads were aligned against the Mus musculus reference genome (ENSEMBL release 101, GRCh38.101) using HiSat2, version 1.2.1 (46). FeatureCounts v1.4.6-p1 (http://subread.sourceforge.net) was used for read counting (47), with an average read of 43 millions/sample. Raw sequencing data and normalized counts are available in the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE214064).
Differential RNA expression analyses were performed in R, version 4.0.3 software with the EdgeR package (48). Graphs were generated using the EnhancedVolcano package (https://github.com/kevinblighe/EnhancedVolcano). A false discovery rate (FDR) of 5% was used as the threshold for statistical significance. Gene Ontology analyses were performed with the clusterProfiler package (49) with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations using the default parameters: organism(mmu), keyType(keg), pvalueCutoff(0.05), pAdjustMethod(BH), minGSSize(20), maxGSSize(500), and qvalueCutoff(0.05).
Immunoblotting
Liver tissue was lysed in radioimmunoprecipitation assay buffer containing cOmplete Proteinase Inhibitor Cocktail (catalog no.: 11836170001; Roche, Basel, Switzerland) and phosphatase inhibitors (catalog no.: P0044; Sigma-Aldrich, St Louis, MO). Samples were homogenized with glass beads in a Precellys 24 tissue homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France) and sonicated on a Bioruptor® Plus device (Diagenode, Liege, Belgium). Lysates were mixed with Laemmli buffer, separated on Criterion™ TGX™ 4–20% gels (Bio-Rad, Hercules, CA), and transferred to nitrocellulose membranes using the RTA transfer kit and Trans-Blot Turbo transfer system (Bio-Rad, Hercules, CA). Membranes were stained with Ponceau S to determine the total protein loaded in each lane. Primary antibodies used were guinea pig anti-Plin2 (catalog no.: GP-40; Progen, Heidelberg, Germany), rabbit anti-Lamp1 (catalog no.: 3243; Cell Signaling, Danvers, MA), rabbit anti-light chain 3 B (LC3B) (catalog no.: 2775; Cell Signaling), rabbit anti-SQSTM1/p62 (catalog no.: 5114; Cell Signaling), rabbit anti-PEX14 (catalog no.: ABC142; MilliporeSigma, MA), rat anti-Lamp2 (catalog no.: ab13524; Abcam, Cambridge, UK), guinea anti-Plin3 (catalog no.: GP-40; Progen), guinea anti-Plin5 (catalog no.: GP-44; Progen), anti-rabbit HSL (catalog no.: 4107; Cell Signaling), anti-rabbit ATGL (catalog no.: 2439S; Cell Signaling), and anti-rabbit phosphorylated HSL (p-HSL) Ser565 (catalog no.: 4137S; Cell Signaling). Secondary antibodies used were goat anti-rabbit IgG (catalog no.: 111-035-144), donkey anti-guinea pig IgG (catalog no.: 706-035-148; Jackson ImmunoResearch, West Grove, PA), and goat anti-rat IgG (catalog no.: AP136P; Sigma-Aldrich, Steinheim, Germany). Chemiluminescence detection was done on a ChemiDoc™ Touch Imaging System (Bio-Rad, Hercules, CA). Band intensities were quantified using ImageJ, version 1.52 software (National Institutes of Health, Bethesda, MD), with protein abundances given relative to total protein levels (Ponceau signal) in the corresponding lane.
Liver histology
Liver tissues were fixed for ∼24 h at 4°C in 4% paraformaldehyde in 0.1 mol/l phosphate buffer (PB; pH 7.4) (4% paraformaldehyde-PB) and subsequently stored at 4°C in 0.4% paraformaldehyde-PB until processing. Liver tissues were cryoprotected with a gradient of sucrose solved in 0.1 mol/l PB and then frozen in optimal cutting temperature embedding matrix (KMA-0100-00A; CellPath, Newtown, UK) over nitrogen vapor in embedding molds (E4140-1EA; Sigma-Aldrich, Steinheim, Germany) as described previously (44). Sections (20 μm) were prepared from optimal cutting temperature blocks with a Leica CM3050S cryostat (Leica Biosystems, Deer Park, IL) at −20°C.
Sections were washed in PB and stained for 25 min while floating in PB containing 1 μmol/l Bodipy 493/503 (Thermo Fisher Scientific, Waltham, MA), 5 μmol/l Hoechst-33342 (Sigma-Aldrich, St Louis, MO), and 1 U/ml CF568 conjugated phalloidin (Biotium, Fremont, CA). After staining, sections were washed two times in PB and then mounted on SuperFrost Plus slides (VWR, Radnor, PA) with ProLong mounting media (Thermo Fisher Scientific, Waltham, MA) and coverslips. The slides were allowed to harden overnight at room temperature and stored at 4°C in the dark. All steps from sectioning until confocal imaging were completed within 2 weeks.
Confocal microscopy
High-resolution confocal images were taken under a 40× oil immersion objective mounted on an LSM 710 confocal microscope (Zeiss) as described previously (44). Laser and detection settings: nucleus staining (excited with a 405 nm laser/fluorescence detected at 414–465 nm, LD staining [488 nm/497–545 nm], and cell skeleton staining [561 nm/563–632 nm]). Signal intensities were optimized for each channel with a representative section and reused for the remaining sections.
Quantification of LDs
Confocal images were used for LD quantification using ImageJ, version 1.54 software. The diameters of individual LDs were manually measured for one representative image per mouse (n = 5 mice in each group). The number of LDs measured on each image ranged from 121 to 336 LDs (average number = ∼200 LDs per image). The corresponding numbers of nuclei were counted on the same images and used for normalization.
Hepatic and plasma TAG, total cholesterol, and ketone bodies content
Lipids were measured in plasma and homogenates of liver tissue using colorimetric enzymatic detection kits for detection of total TAG (catalog no.: T75321L; Pointe Scientific, Canton, MI), total cholesterol (Chol; catalog no.: C75101L; Pointe Scientific, Canton, MI), and ketone bodies (catalog no.: MAK134; Sigma-Aldrich, St Louis, MO) according to the manufacturer's instructions.
Detailed lipid analysis
Around 100 mg of liver tissue was used for lipid analysis using HPLC coupled with time-of-flight mass spectrometry as previously described (44). The following lipid species were measured: cardiolipins (CLs), ceramides (Cers), Chol, CEs, DAG, ether-linked phosphatidylcholine (PCO), FFAs, hexosylceramide (HexCer), lysophosphatidylcholine, lysophosphatidylethanolamine (LPE), bis(monoacylglycerol)phosphate, phosphatidylcholine (PC), phosphatidylcholine plasmalogen, phosphatidylethanolamine (PE), phosphatidylethanolamine plasmalogen, phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), sphingomyelin (SM), and TAG. Detected lipid signals were normalized against liver weights and internal standards. Internal standards used were CL (C56:0), Cer (C35:1), Chold47, CE (C19:0), DAG (C30:0), FFA (C17:1), HexCer (C35:1), lysophosphatidylcholine (C17:1), LPE (C17:1), PC (C28:0), PE (C28:0), PG (C30:0), PI (C31:1), PS (C28:0), SM (C35:1), and TAG (C39:0), all from Sigma-Aldrich (St Louis, MO).
Statistics
Data were analyzed with Prism 5 (GraphPad Software, CA). Two-way ANOVA with Tukey post hoc tests and multiple t-tests were used where P < 0.05 were considered significant. Data in graphs are shown as means ± 95% confidence interval.
Results
Plin2−/− mice have normal whole body and organ weights when fasted
Parallel studies in alternatively generated Plin2-null models have established that loss of Plin2 results in reduced hepatic lipid levels in fed mice (10, 50). We showed previously that fasting in mice initiates massive accumulation of hepatic LDs, accompanied by upregulation of Plin2 mRNA and protein levels (17). To investigate the importance of Plin2 for hepatic LD formation and lipid metabolism during fasting, 15-week-old female and male Plin2+/+ and Plin2−/− mice were given free access to a chow diet (fed) or were fasted for 24 h (fasted). As expected, body weights, and liver, gonadal, and subcutaneous white adipose tissue weights were reduced in both Plin2+/+ and Plin2−/− mice after 24 h of fasting (Table 1). Heart and kidney weights were unaffected by fasting. Except for slightly higher liver weights in chow-fed female Plin2−/− mice and lower heart weight in fasted male Plin2+/+ mice, body weights and organ weights were essentially similar in Plin2+/+ and Plin2−/− mice.
Table 1.
Body weights and organ weights of 15-week-old Plin2+/+ and Plin2−/− mice with ad libitum access to chow diet or after 24 h of fasting
| Organs | Fed |
Fasted (24 h) |
||
|---|---|---|---|---|
| Plin2+/+ | Plin2−/− | Plin2+/+ | Plin2−/− | |
| 15 weeks—males | ||||
| Body weight | 28.1 ± 1.4 | 28.1 ± 2.4 | 22.7 ± 2.1d | 24.5 ± 2.5d |
| Liver (g) | 1.45 ± 0.09 | 1.37 ± 0.16 | 0.97 ± 0.09d | 0.97 ± 0.12d |
| Heart (g) | 0.128 ± 0.011 | 0.129 ± 0.008 | 0.117 ± 0.009 | 0.132 ± 0.015a |
| Kidney (g) | 0.34 ± 0.05 | 0.34 ± 0.04 | 0.31 ± 0.04 | 0.33 ± 0.06 |
| White adipose tissue epididymal (g) | 0.25 ± 0.05 | 0.21 ± 0.05 | 0.14 ± 0.11c | 0.16 ± 0.07 |
| White adipose tissue subcutaneous (g) | 0.16 ± 0.03 | 0.16 ± 0.03 | 0.09 ± 0.06c | 0.08 ± 0.04c |
| 15 weeks—females | ||||
| Body weight (g) | 21.5 ± 0.9 | 22.4 ± 1.2 | 18.0 ± 1.1d | 18.7 ± 1.4d |
| Liver (g) | 0.92 ± 0.08 | 1.03 ± 0.10a | 0.82 ± 0.08b | 0.76 ± 0.1d |
| Heart (g) | 0.094 ± 0.009 | 0.093 ± 0.007 | 0.096 ± 0.006 | 0.092 ± 0.009 |
| Kidney (g) | 0.23 ± 0.02 | 0.23 ± 0.02 | 0.24 ± 0.02 | 0.23 ± 0.03 |
| White adipose tissue gonadal (g) | 0.21 ± 0.10 | 0.19 ± 0.09 | 0.09 ± 0.05b | 0.11 ± 0.06 |
| White adipose tissue subcutaneous (g) | 0.17 ± 0.04 | 0.17 ± 0.05 | 0.08 ± 0.02d | 0.09 ± 0.03d |
Data are presented as means ± SD, n = 8–10.
P < 0.05 indicates difference between Plin2+/+ and Plin2−/− mice with the same feeding condition.
P < 0.05,
P < 0.01, and
P < 0.001 indicates difference between fed and fasted mice of the same genotype.
Plin3 and Plin5 proteins increase in the liver of fasted Plin2−/− mice
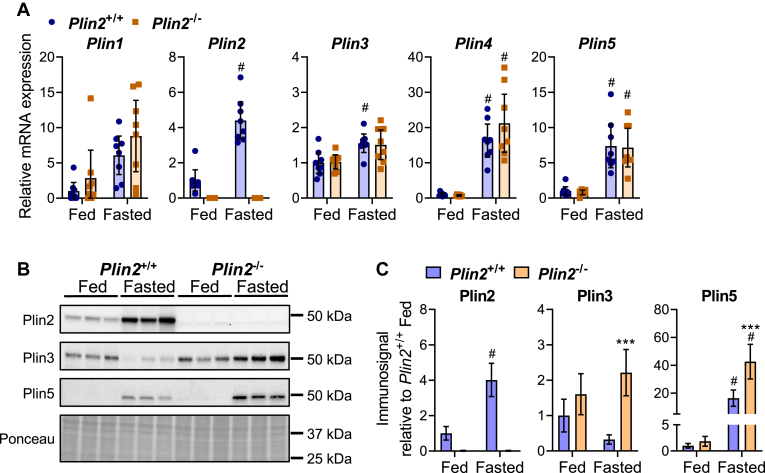
Upon loss of a Plin, other Plin proteins are known to compensate and bind to the LD surface (2). To determine if deletion of Plin2 altered the expression of other Plins, we measured Plin1-5 mRNAs in the liver. Fasting increased hepatic expression of Plin2, Plin4, and Plin5 mRNAs substantially (>5-fold), whereas expression of Plin3 mRNA was modestly increased (Fig. 1A). Expression of Plin1 was low and exhibited higher variation between individuals compared with Plin2-Plin5 mRNAs. Except for the anticipated absence of Plin2 mRNA in Plin2−/− mice, the remaining Plin mRNAs were expressed at similar levels in Plin2+/+ and Plin2−/− mice. Several Plins are subjected to post-translational regulation (2), and their mRNA levels do not reliably predict their protein expression levels. We therefore measured protein levels of the abundantly expressed perilipin proteins; Plin2, Plin3, and Plin5 with immunoblotting. Consistent with previous findings, Plin2 protein levels increased in the livers of Plin2+/+ mice upon fasting (17), whereas Plin2 protein was not detected in fed or fasted Plin2−/− mice (Fig. 1B). Whole-cell Plin3 protein levels were similar in fed Plin2+/+ and Plin2−/− mice but increased in fasted Plin2−/− mice and tended to decrease in fasted Plin2+/+ mice, resulting in higher Plin3 protein levels in fasted Plin2−/− mice compared with Plin2+/+ mice (Fig. 1C). Plin5 protein levels were similar in fed Plin2+/+ and Plin2−/− mice but substantially increased with fasting, reaching significantly higher levels in fasted Plin2−/− mice compared with Plin2+/+ mice. Hence, both Plin3 and Plin5 proteins were increased in fasted Plin2−/− mice, suggesting they compensate for the hepatic loss of Plin2.
Fig. 1.
Hepatic expression of Plins in fed and fasted Plin2+/+ and Plin2−/− mice. Female Plin2+/+ and Plin2−/− mice (15 weeks) were housed with ad libitum access to a chow diet (fed) or were fasted for 24 h (fasted) prior to euthanasia at 8–10 AM (n = 8 per group). A: Relative expression of Plin1, Plin2, Plin3, Plin4, and Plin5 mRNAs in liver. B: Representative immunoblots showing expression of Plin2, Plin3, and Plin5 proteins in three individual animals. C: Relative quantification of hepatic Plin protein expression levels (n = 6). Statistical testing was performed with two-way ANOVA and Tukey test for multiple comparisons. #P < 0.05 indicates the statistical difference between fed and fasted conditions for the same genotype; ∗∗∗P < 0.001 indicates the difference between Plin2+/+ and Plin2−/− mice with the same feeding condition. Data in graphs are shown as means ± 95% confidence interval.
Fasted Plin2−/− mice have decreased hepatic TAG content but store LDs with increased size
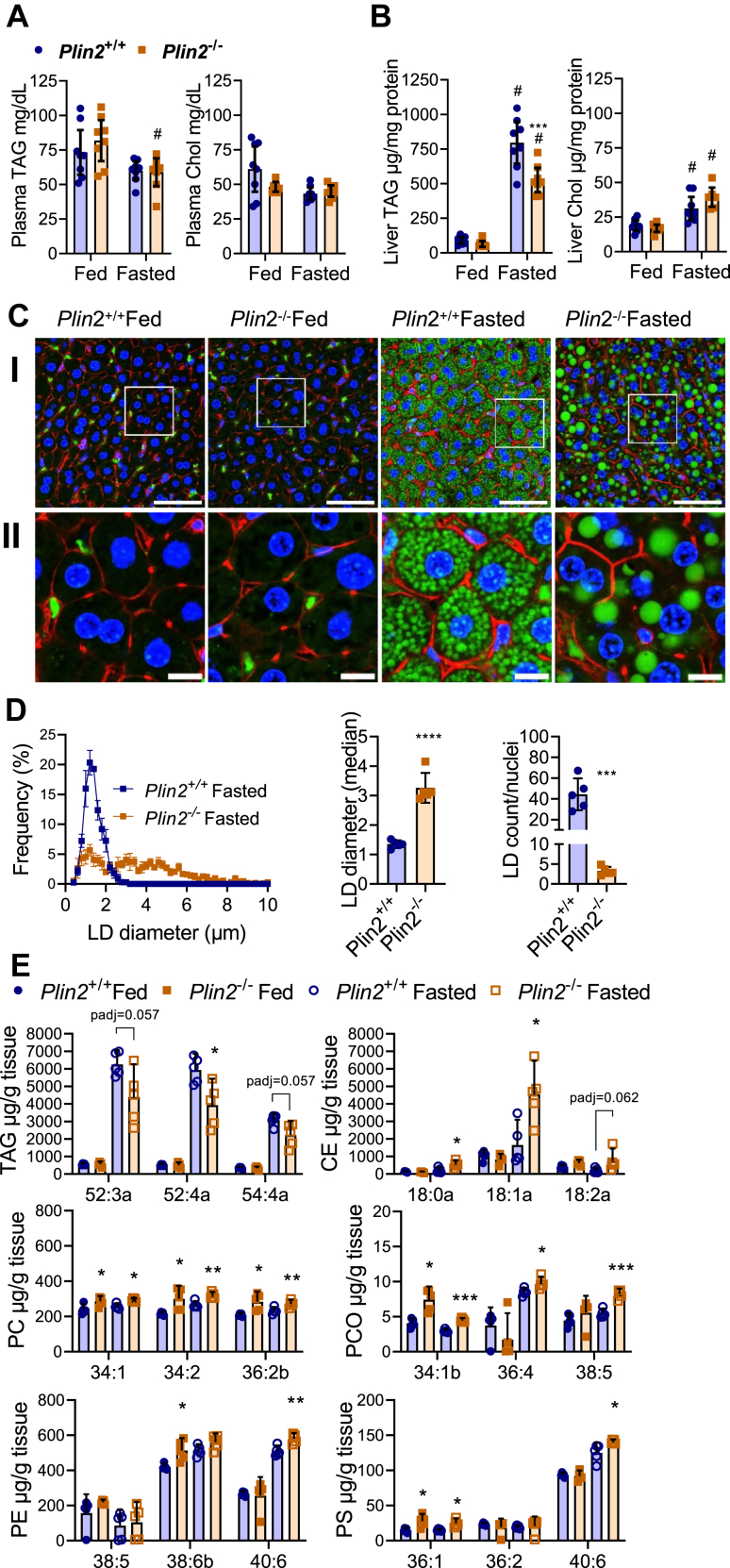
Several studies have reported a reduction in hepatic lipids upon loss of Plin2 in fed conditions (10, 38, 39). To compare lipid levels in fed and fasted conditions, we examined plasma and hepatic TAG and total Chol content in Plin2+/+ and Plin2−/− mice. Plasma TAG and total Chol levels were similar in Plin2+/+ and Plin2−/− mice and remained essentially unaltered by fasting (Fig. 2A). Hepatic TAG levels were similar in fed Plin2+/+ and Plin2−/− mice, whereas levels were massively increased by fasting but less in fasted Plin2−/− mice compared with fasted Plin2+/+ mice (Fig. 2B). Hepatic total Chol increased with fasting with a trend for higher levels in fasted Plin2−/− mice compared with Plin2+/+ mice (P = 0.056).
Fig. 2.
Lipids abundance in fed and fasted Plin2+/+ and Plin2−/− mice. Female Plin2+/+ and Plin2−/− mice (15 weeks) were housed with ad libitum access to a chow diet (fed) or were fasted for 24 h (fasted) prior to euthanasia at 8–10 AM (n = 8 per group). A: Plasma TAGs and total Chol levels. B: Liver TAG and Chol levels. Statistical testing was performed with two-way ANOVA and Tukey test for multiple comparisons. #P < 0.05 indicates statistical difference between fed and fasted condition; ∗∗∗P < 0.001 difference between Plin2+/+ and Plin2−/− mice with the same feeding condition. C: Representative images of liver cryosections. Cryosections (20 μm thick) were stained to visualize LDs (Bodipy 493/503, green), nuclei (Hoechst 33342, blue), and plasma membrane-located F-actin (Phalloidin-CF568, red). I: Confocal images of liver sections taken with a 40× objective. II: Pictures zoomed in from I to visualize individual cells (inserted boxes indicate enlarged areas). Scale bars represent 50 μm (I) and 10 μm (II). D: Size distribution of hepatic LDs, LD diameter, and LD numbers in fasted Plin2+/+ and Plin2−/− mice (n = 5 in each group). LD numbers were normalized against nuclei. E: Analysis of selected hepatic lipid species with HPLC-qTOF/MS: TAG, CE, PC, PCO, PE, and PS species (n = 5 per group). Statistical testing was performed with multiple t-tests between Plin2+/+ and Plin2−/− mice in fed or fasted states. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Data in graphs are shown as means ± 95% confidence interval.
To follow up on differences in hepatic TAG content in fasted mice, we examined hepatic LD content and histological structures in fed and fasted Plin2+/+ and Plin2−/− mice. Sections were triple stained to visualize LDs (Bodipy 493/503, green), nuclei (Hoechst 33342, blue), and plasma membrane-located F-actin (Phalloidin-CF568, red). In consistency with measured TAG levels, LD levels (green staining) were comparable at low levels in fed Plin2+/+ and Plin2−/− mice (Fig. 2C). In contrast, a massive increase in LD levels was observed in fasted mice (Fig. 2C) but with LDs that were distinctly different between Plin2+/+ and Plin2−/− mice. Hepatic LDs in fasted Plin2+/+ mice were small, numerous, and dispersed in the cytoplasm of the lipid-containing hepatocytes, whereas a large majority of LDs in fasted Plin2−/− mice were typically much larger in size compared with LDs observed in fasted Plin2+/+ mice (Fig. 2D). Quantification of LDs in the fasted state demonstrated a clear reduction in the LD number and an increase in the LD size in fasted Plin2−/− mice compared with fasted Plin2+/+ mice (Fig. 2D). Because of the considerably smaller size of LDs in the fed state, we were not able to accurately quantify the size and number of those LDs.
To gain insights into differences in hepatic lipid species between Plin2+/+ and Plin2−/− mice, we performed detailed lipid analysis with HPLC-qTOF/MS. Various species of TAG (number of carbons:double bounds ranging from TAG48:1a to TAG58:10a), DAG species (DAG32:0 to DAG40:6), and FFAs (16:0 to 22:6) tended consistently to be less abundant in livers of fed Plin2−/− mice compared with fed Plin2+/+ mice (supplemental Fig. S1). Upon fasting, various species of TAG, DAG, and PE, as well as most FFAs, were increased, whereof TAG and DAG seem to be robustly increased and represent the major hepatic lipids accumulating in the liver upon fasting (supplemental Fig. S1). The three most abundant TAG species (Fig. 2E) and the majority of other TAG species were less abundant in the livers of fasted Plin2−/− mice compared with fasted Plin2+/+ mice (supplemental Fig. S1). All measured CE species were elevated in the livers of fasted Plin2−/− mice compared with fasted Plin2+/+ mice (Fig. 2C and supplemental Fig. S1), and CE species were differently affected by fasting based on their FA chain length. CE species consisting of saturated/monounsaturated FAs (CE16:0 to CE18:1) increased by fasting, whereas CE species consisting of longer polyunsaturated FAs (CE18:2 to CE22:6) decreased by fasting (supplemental Fig. S1). For the other measured lipids, the abundance of PC, PCO, and PS species tended to be higher in fed or fasted Plin2−/− compared with Plin2+/+ mice (Fig. 2D, and supplemental Fig. S1). Levels of some PE species were also elevated in Plin2−/− mice (Fig. 2E and supplemental Fig. S2). Bis(monoacylglycerol)phosphate and HexCer seemed to be higher in fed Plin2−/− mice compared with Plin2+/+ mice, though these lipid species were found at low levels affecting the accuracy of their quantification. The abundance of other lipids like Cer, LCP, LPE, PG, PI, SM, and CL in the liver was essentially similar between Plin2+/+ and Plin2−/− mice (supplemental Fig. S1).
Hepatic gene expression differs between fasted Plin2+/+ and Plin2−/− mice
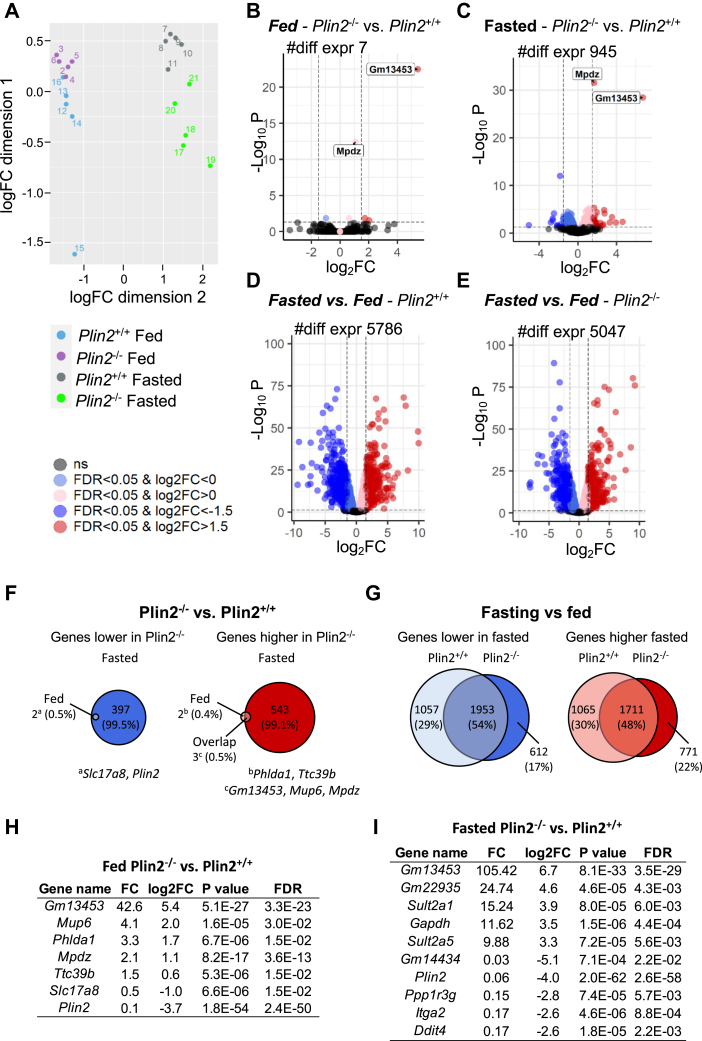
Next, we determined if lack of Plin2 expression influenced hepatic gene expression in the fed or fasted state. Livers from five representative individuals of fed or fasted Plin2+/+ and Plin2−/− mice were subjected to RNA-Seq. Principal component analysis revealed substantial hepatic transcriptomic changes with fasting of Plin2−/− and Plin2+/+ mice (Fig. 3A), with groups that clustered well together except for one distinctively different individual amongst fed Plin2+/+ mice. Differential gene expression analyses with this outlier included or excluded were similar, and the presented data include all individuals.
Fig. 3.
Global hepatic gene expression in fed and fasted Plin2+/+ and Plin2−/− mice. Liver gene expression in female Plin2+/+ and Plin2−/− mice (15 weeks) housed with ad libitum access to chow diet (fed) or fasted for 24 h (fasted). RNA-Seq was performed on five representative individuals per group (n = 5). A: Principal component analysis (PCA) plot of fed and fasted Plin2+/+ and Plin2−/− mice. B: Volcano plot illustrating the number of individual transcripts with induced (red Log2FC >0.585 [50% increase] or light red Log2FC >0) or repressed (blue Log 2FC <−0.585 or light blue Log 2FC <0) expression levels in fed Plin2−/− mice compared with fed Plin2+/+ mice. FDR (false discovery rate) <0.05. C: Transcripts with induced (red) or repressed (blue) expression levels in fasted Plin2−/− mice compared with fasted Plin2+/+ mice. FDR <0.05. D: Transcripts with induced (red) or repressed (blue) expression levels in fasted compared with fed Plin2+/+ mice. FDR <0.05. E: Transcripts with induced (red) or repressed (blue) expression levels in fasted compared with fed Plin2−/− mice. FDR <0.05. F: Diagram of genes that were lower (blue) or higher (red) expressed in Plin2−/− mice compared with Plin2+/+ mice in the fed (light color) and fasted (dark color) states. G: Diagram of genes that were lower (blue) or higher (red) expressed in fasted versus fed Plin2+/+ mice (light color) and Plin2−/− mice (dark color). H: List of transcripts that were differentially expressed in fed Plin2−/− mice compared with fed Plin2+/+ mice. I: The top altered transcripts (five upregulated and five downregulated) that were differentially expressed in fasted Plin2−/− and Plin2+/+ mice.
Detailed analyses of individual genes are illustrated in volcano plots, comparing Plin2+/+ and Plin2−/− mice in fed and fasted states. Comparison of gene expression in fed Plin2−/− versus Plin2+/+ mice (Fig. 3B, F and supplemental Table S2B) revealed only seven genes (Fig. 3H) that were differentially expressed (FDR <0.05). Two genes were downregulated (Plin2 and Slc17a8), and five genes were upregulated (Gm13453 [pseudogene], Mpdz, Phlda1, Ttc39b, and Mup6). As expected, fasting initiated a profound switch in hepatic gene expression. In total, 5,786 genes (3,010 down and 2,776 up) were differentially expressed (FDR <0.05) in fasted versus fed Plin2+/+ mice (Fig. 3D and supplemental Table S2D). A lower number of genes (5,047 genes, 2,565 down, and 2,482 up) were differentially expressed in fasted versus fed Plin2−/− mice (Fig. 3E and supplemental Table S2E). Of note, of the total number of transcripts altered by fasting compared with feeding in Plin2+/+ and Plin2−/− mice, around two-thirds were common, whereas around one-third were uniquely regulated for each genotype (Fig. 3G). These results suggest a moderately compromised fasting response in Plin2−/− mice. Comparison of gene expression in fasted Plin2−/− mice versus Plin2+/+ mice (Fig. 3C and supplemental Table S2C) revealed 945 differentially expressed genes (FDR <0.05; 399 genes were lower expressed and 546 genes higher expressed in Plin2−/−). The five genes that were strongest downregulated or upregulated in fasted Plin2−/− mice versus Plin2+/+ mice are listed (Fig. 3I).
To identify pathways that were differently regulated upon fasting, we performed a KEGG pathway overrepresentation analysis with the 945 genes that were significantly increased or decreased in fasted Plin2−/− mice compared with fasted Plin2+/+ mice. A majority of pathways that were increased in fasted Plin2−/− mice were related to the immune system, in addition to cell adhesion and steroid biosynthesis (Table 2). Pathways identified as lower expressed in fasted Plin2−/− mice were related to autophagy, insulin signaling, longevity, and apoptosis.
Table 2.
KEGG pathway overrepresentation analysis on genes that were significantly increased or decreased in fasted Plin2−/− mice compared with fasted Plin2+/+ mice (FDR < 0.05)
| KEGG pathway | GeneRatio | Count | P | FDR (Adjusted P) |
|---|---|---|---|---|
| Genes that were higher in Plin2−/− (top 15 pathways) | ||||
| Cell adhesion molecules | 19/275 | 19 | 2.10E-06 | 2.97E-04 |
| Viral myocarditis | 13/275 | 13 | 1.82E-06 | 2.97E-04 |
| Antigen processing and presentation | 12/275 | 12 | 1.35E-05 | 1.27E-03 |
| Intestinal immune network for IgA production | 8/275 | 8 | 3.32E-05 | 2.35E-03 |
| Asthma | 6/275 | 6 | 7.34E-05 | 4.15E-03 |
| Toxoplasmosis | 12/275 | 12 | 1.02E-04 | 4.80E-03 |
| Human T-cell leukemia virus 1 infection | 19/275 | 19 | 1.70E-04 | 6.86E-03 |
| Influenza A | 15/275 | 15 | 2.01E-04 | 7.09E-03 |
| Rheumatoid arthritis | 10/275 | 10 | 2.50E-04 | 7.09E-03 |
| Steroid biosynthesis | 5/275 | 5 | 2.45E-04 | 7.09E-03 |
| Th17 cell differentiation | 11/275 | 11 | 2.83E-04 | 7.28E-03 |
| Phagosome | 15/275 | 15 | 3.49E-04 | 8.22E-03 |
| Epstein-Barr virus infection | 17/275 | 17 | 5.41E-04 | 9.00E-03 |
| Hematopoietic cell lineage | 10/275 | 10 | 4.71E-04 | 9.00E-03 |
| Inflammatory bowel disease | 8/275 | 8 | 4.74E-04 | 9.00E-03 |
| Genes that were lower in Plin2−/− | ||||
| Autophagy—animal | 13/132 | 13 | 1.07E-07 | 2.56E-05 |
| Insulin signaling pathway | 9/132 | 9 | 1.64E-04 | 1.97E-02 |
| Longevity regulating pathway | 7/132 | 7 | 2.93E-04 | 2.35E-02 |
| Apoptosis | 8/132 | 8 | 7.26E-04 | 4.36E-02 |
Altered expression of immune-related genes in fasted Plin2−/− mice
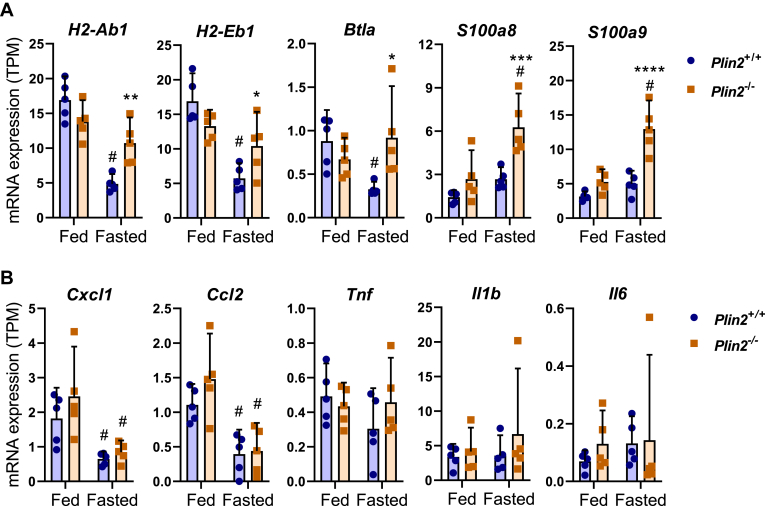
Hepatic expression of the family member Plin5 has been shown to dampen inflammation during fasting (43). Since hepatic Plin5 expression is elevated (Fig. 1) and KEGG analysis revealed that immune-related pathways are differently regulated in fasted Plin2−/− mice, we investigated the expression of immune-related genes in fed and fasted Plin2+/+ and Plin2−/− mice. Several histocompatibility 2 genes (e.g., H2-Ab1 and H2-Eb1) and the B and T lymphocyte-associated gene (Btla) were reduced by fasting in Plin2+/+ mice but to a lesser extent in Plin2−/− mice (Fig. 4A). Conversely, the neutrophil-enriched calprotectin/S100 calcium-binding a8/a9 genes (S100a1 and S100a9) were induced in fasted Plin2−/− mice as compared with Plin2+/+ mice. Hepatic inflammatory markers such as Cxcl1 and Ccl2 were reduced by fasting but were not different between genotypes, whereas Tnf, Il1b, and Il6 were not affected by fasting or removal of Plin2 (Fig. 4B). These expression patterns demonstrate that typical hepatic inflammation is unaffected in fed or fasted Plin2−/− mice, which suggests that differences in the expression of immune-related genes are linked to alterations in hepatic immune cell populations.
Fig. 4.
Hepatic expression of immune-related transcripts in fed and fasted Plin2+/+ and Plin2−/− mice. Hepatic expression of selected mRNAs in female Plin2+/+ and Plin2−/− mice (15 weeks) housed with ad libitum access to chow diet (fed) or fasted for 24 h (fasted). A: Expression of immune-related genes with altered expression in fasted Plin2−/− mice (H2-Ab1, H2-Eb1, Btla, S100a8, and S100a9). B: Expression of typical hepatic inflammation marker genes (Cxcl1, ccl2, Tnfa, Il1b, and Il6). Statistical testing was done with two-way ANOVA and Tukey test for multiple comparisons. #P < 0.05 indicates statistical difference between fed and fasted conditions; ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001 indicate differences between Plin2+/+ and Plin2−/− mice with the same feeding condition.
Unaltered autophagy but increased lipolysis in fasted Plin2−/− mice
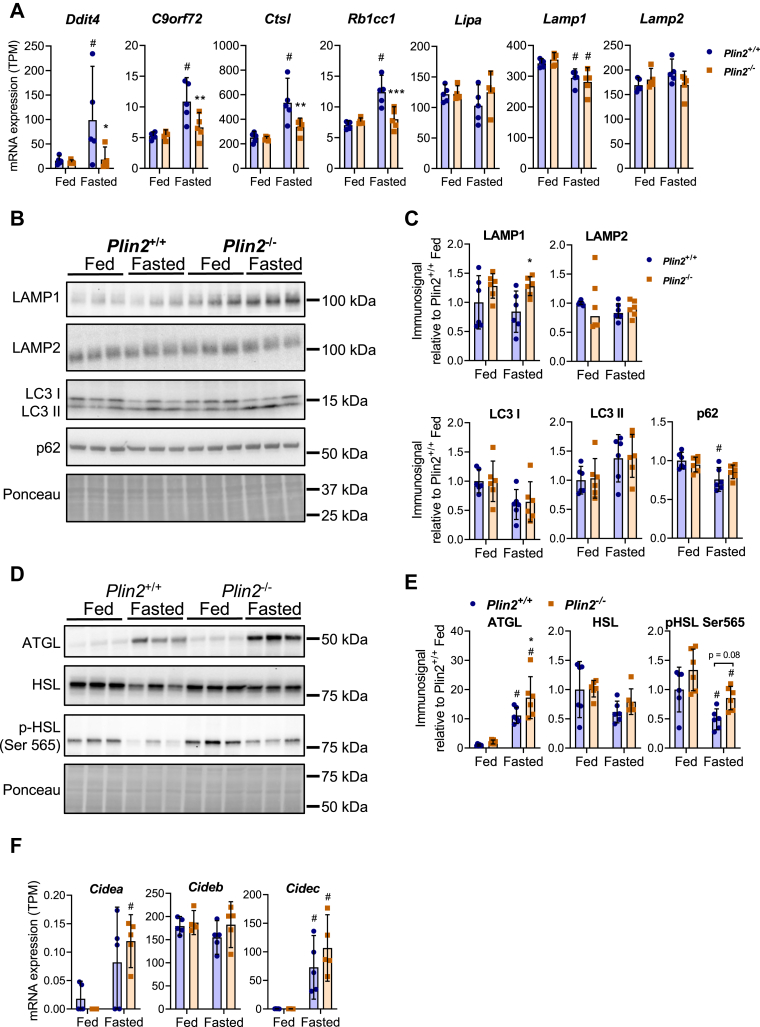
The KEGG pathway analysis (Table 2) revealed that many of the genes that were lower expressed in fasted Plin2−/− mice are involved in autophagy. Because altered lipophagy can potentially explain the depleted TAG stores and enlargement of LDs in fasted Plin2−/− mice, we analyzed the hepatic expression of genes involved in lipophagy. Expression of several autophagy-related genes (Ddit4, C9orf72, Ctsl, and Rb1cc1) was induced by fasting in Plin2+/+ mice but not in fasted Plin2−/− mice (Fig. 5A). Expression of lysosomal acid lipase (Lipa) and the lysosomal-associated membrane proteins 1 and 2 (Lamp1 and Lamp2) were similarly expressed in fasted Plin2+/+ and Plin2−/− mice. To evaluate lipophagy activity, we measured protein expression levels of Lamp1 and Lamp2, microtubule-associated protein 1A/1B- LC3, and the ubiquitin-binding protein p62. LAMP1 protein levels tended to be higher in fed Plin2−/− mice and were higher in fasted Plin2−/− mice compared with Plin2+/+ mice (Fig. 5B–C). Cytosolic LC3 I and membrane-bound and lipidated LC3 II were expressed at similar levels in fed and fasted Plin2+/+ and Plin2−/− mice. Expression of the p62 protein remained unchanged, suggesting that hepatic lipophagy activity is unaffected by the removal of Plin2.
Fig. 5.
Hepatic expression of lipophagy markers and lipolytic enzymes in fed and fasted Plin2+/+ and Plin2−/− mice. Expression of selected mRNAs and proteins in female Plin2+/+ and Plin2−/− mice (15 weeks) housed with ad libitum access to chow diet (fed) or fasted for 24 h (fasted). A: Expression of autophagy-related genes with altered expression in fasted Plin2−/− mice (Ddit4, C9orf72, Ctsl, Rb1cc1), lysosomal acid lipase (Lipa), and lysosomal markers (Lamp1 and Lamp2). B: Expression of proteins involved in lipophagy and lysosomal function. Representative immunosignals of Lamp1 and Lamp2 (lysosomal), cytosolic (LC3-I) and lipidated (LC3-II) LC3 proteins, and the autophagy receptor (p62) in individual mice. Six representative animals were selected for each group (n = 6 per group). C: Relative quantification of hepatic Lamp1, Lamp2, LC3-I, LC3-II, and p62 protein immunosignals relative to expression in fed Plin2+/+ mice. D: Expression of proteins involved in lipolysis. Representative immunosignals of ATGL, HSL, and Ser 565 p-HSL (Ser 565) in individual mice. E: Relative quantification of hepatic ATGL, HSL, p-HSL (Ser 565) protein immunosignals relative to expression in fed Plin2+/+ mice. F: Expression of Cidea. Cideb and Cidec mRNAs in fed and fasted Plin2+/+ and Plin2−/− mice. Statistical testing was done with two-way ANOVA and Tukey test for multiple comparisons. #P = 0.05 indicates the statistical difference between fed and fasted conditions. Data in graphs are shown as means ± 95% confidence interval.
Plin2 is known to affect lipolysis and ATGL activity (51, 52). To determine if lipolysis is altered in Plin2−/− mice, we analyzed the protein abundance of the lipases ATGL and HSL. ATGL protein levels increased with fasting, with higher expression levels in fasted Plin2−/− mice compared with Plin2+/+ mice (Fig. 5D, E). Expression of HSL tended to be lower with fasting, whereas p-HSL (Ser 565) was reduced. There was an insignificant trend for higher levels of p-HSL (Ser 565) in fasted Plin2−/− mice compared with fasted Plin2+/+ mice. Overall, increased levels of ATGL and a trend for higher levels of p-HSL (Ser 565) in Plin2−/− mice compared with Plin2+/+ mice suggests that lipolytic activity is increased in fasted Plin2−/− mice.
We then considered if differences in LD size morphology could be related to expression of cell death-inducing DNA fragmentation factor alpha-like effector (Cide) proteins, which are known regulators of LD fusion (53). Gene expression of Cidea, Cideb, and Cidec was unaltered between Plin2+/+ mice and Plin2−/− mice, in the fed and fasted states (Fig. 5F). Expression levels were confirmed with RT-qPCR analysis (data not shown). Expression of Cidea was low, and while expression of Cidea and Cideb was unaffected by fasting, expression of Cidec mRNA was highly induced by fasting. These data do not suggest a simple mechanistic regulation of altered LD morphology in Plin2−/− livers caused by differences in the expression of Cide proteins.
Minor alterations in the expression of genes involved in FA oxidation and endoplasmic reticulum stress in fasted Plin2−/− mice
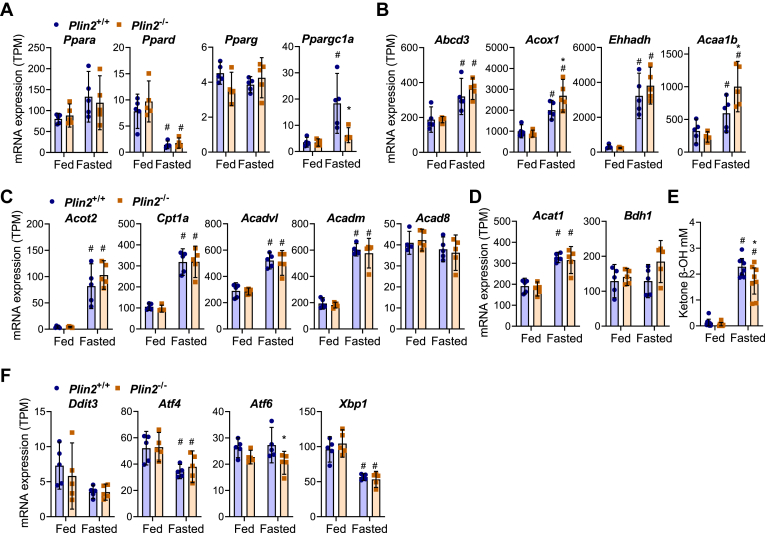
We have shown previously that loss of Plin2 increases lipolysis and FA oxidation in myotubes (52). To determine if the same occurs in the liver of fasted Plin2−/− mice, we analyzed expression of FA sensors, β-oxidation genes, and endoplasmic reticulum (ER) stress markers in our RNA-Seq dataset. As reported previously (54), fasting increased expression of peroxisome proliferator-activated receptor alpha (Ppara), an important hepatic FA sensor during fasting (Fig. 6A). Expression of Ppara and the family related Ppard and Pparg was similar in Plin2+/+ mice and Plin2−/− mice, whereas expression of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Ppargc1a) was induced in fasted Plin2+/+ mice but not in fasted Plin2−/− mice. Several mRNAs involved in peroxisomal β-oxidation (Acox1 and Acaa1b) were expressed at somewhat higher levels in fasted Plin2−/− mice compared with fasted Plin2+/+ mice (Fig. 6B). Expression of Pex14 protein, a peroxisomal marker (55), was unchanged in fasted Plin2+/+ compared with fasted Plin2−/− mice (data not shown), suggesting a similar content of peroxisomes. Expression levels of mRNAs encoding enzymes involved in mitochondrial FA transport (Acot2, Cpt1) and mitochondrial β-oxidation (Acadv1, Acadm) increased at similar levels with fasting in Plin2+/+ and Plin2−/− mice, whereas expression of Acad8 mRNA, encoding an enzyme involved in the oxidation of both FAs and branch-chained amino acids, remained unaltered (Fig. 6C). Expression of Acat1 and Bdh1 mRNAs, which encode enzymes catalyzing the conversion of acetyl-CoA into acetoacetate and β-hydroxybutyrate, respectively, was similarly expressed in fasted Plin2+/+ and Plin2−/− mice. Plasma levels of β-hydroxybutyrate were significantly increased with fasting but were less increased in fasted Plin2−/− mice compared with Plin2+/+ mice (Fig. 6D).
Fig. 6.
Hepatic expression of β-oxidation enzymes, ER stress markers, and inflammation in fed and fasted Plin2+/+ and Plin2−/− mice. Hepatic gene expression and levels of plasma ketone bodies in female Plin2+/+ and Plin2−/− mice (15 weeks) housed with ad libitum access to chow diet (fed) or fasted for 24 h (fasted). For all analyses (n = 8 per group). A: Expression of transcription factors activated by FAs (Ppara, Ppard, Pparg) and coactivator (Ppargc1a). B: Expression of peroxisomal β-oxidation genes (Abcd3, Acox1, Ehhadh, and Acaa1b). C: Expression of mitochondrial β-oxidation genes (Acot2, Cpt1, Acadvl, Acadm, and Acad8). D: Expression of ketogenic genes (Acat1 and Bdh1). E: Plasma levels of the ketone body beta-hydroxybutyrate (BOH). F: Expression of the ER stress markers (Ddit3, Atf4, Atf6, and Xbp1). Statistical testing was done with two-way ANOVA and Tukey test for multiple comparisons. #P < 0.05 indicates statistical difference between fed and fasted conditions; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 indicate difference between Plin2+/+ and Plin2−/− mice with the same feeding condition.
Peroxisomal β-oxidation is known to generate free radicals, which potentially induce oxidative stress (55). Expression of mRNAs encoding proteins activated by ER stress (Ddit3, Aft4, Atf6, and Xbp1) was essentially similarly expressed in Plin2+/+ and Plin2−/− mice, although some minor differences in expression levels were noted for some of these genes between fed and fasted mice (Fig. 6E). Based on these expression profiles, it seems unlikely that free radicals are induced sufficiently to cause ER stress in fasted Plin2−/− mice.
Comparable body weights, metabolic phenotypes, and glucose clearance in Plin2+/+ and Plin2−/− mice
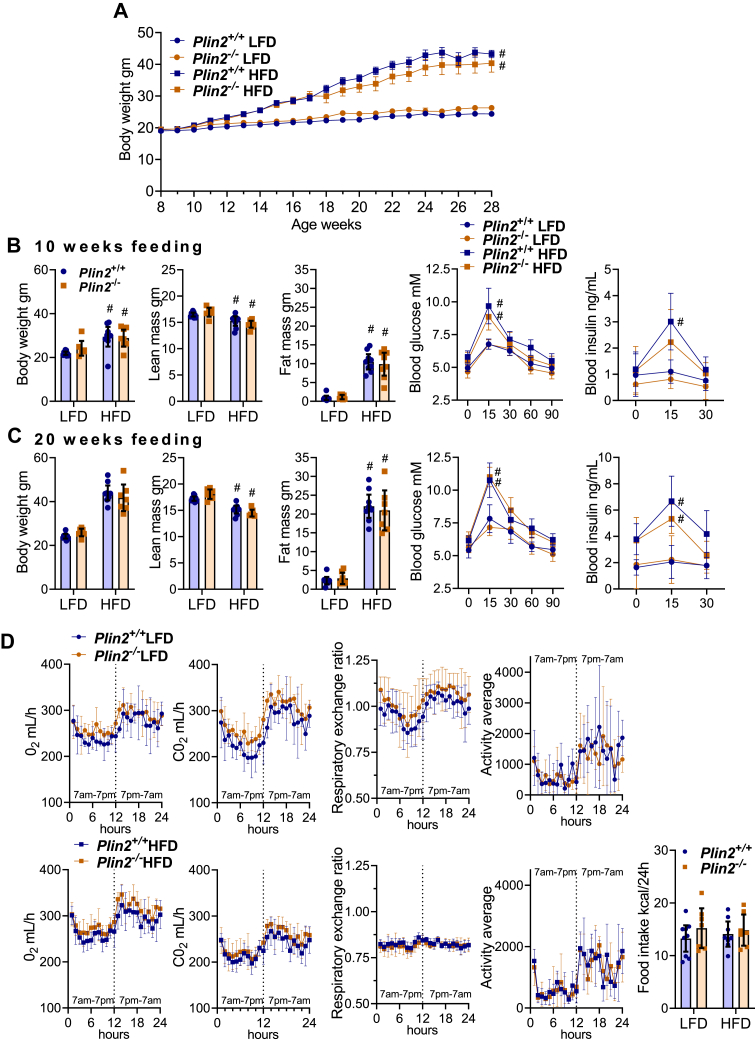
The previous analyses demonstrate that hepatic Plin2 is required for normal lipid storage and lipid metabolism during fasting (metabolically; whole-body energy deficiency). Hepatic lipids are also increased with obesity (metabolically; whole body energy excess) (56). To investigate the role of Plin2 for hepatic LD formation and lipid metabolism in diet-induced obese mice, we exposed female Plin2+/+ and Plin2−/− mice to LFD or HFD for up to 20 weeks. From around 7 weeks on the diet (15 weeks of age), the body weights of HFD-fed mice separated from the body weights of LFD-fed mice (Fig. 7A), but there were no differences in body weight curves between Plin2+/+ and Plin2−/− mice on the same diet. After 10 weeks (Fig. 7B) and 20 weeks (Fig. 7C) of feeding, body composition measurements revealed that lean mass was slightly decreased with HFD compared with LFD, whereas fat mass was increased ∼10 g and ∼20 g with HFD, respectively. Hence, weight gain obtained with HFD was caused by fat accumulation. There were no differences in lean or fat mass between Plin2+/+ and Plin2−/− mice (Fig. 7B–C). To evaluate glucose clearance, mice were exposed to an oral glucose bolus (1.5 g/kg lean mass). HFD-fed mice had increased blood glucose and insulin levels compared with LFD-fed mice, but there were no differences in glucose response or plasma insulin levels between Plin2+/+ and Plin2−/− mice fed the diets for 10 weeks (Fig. 7B) or 20 weeks (Fig. 7C).
Fig. 7.
Body weight and composition, energy metabolism, and glucose clearance in Plin2+/+ and Plin2−/− mice fed LFD or HFD. Female Plin2+/+ and Plin2−/− mice were fed an LFD or an HFD from 8 weeks of age until 28 weeks of age. Mice were euthanized in the morning (8–10 AM) after 10 weeks of diet intervention (18 weeks of age) or 20 weeks of diet intervention (28 weeks of age). A: Body weights from the start of diet intervention (8 weeks of age) until the end of diet intervention (28 weeks of age). Number of individuals included in data points up to 10/20 weeks of diet intervention: Plin2+/+ LFD (n = 17/8), Plin2+/+ HFD (n = 19/9), Plin2−/− LFD (n = 13/7), Plin2−/− HFD (n = 17/7). B: Metabolic measurements after 10 weeks of diet intervention (18-week-old mice). Total body weights, lean mass, and fat mass at euthanasia. Whole blood glucose (0–120 min) and plasma insulin (0–30 min) levels after oral gavage of 1.5 g glucose/kg lean mass (OGTT) in 17-week-old mice. C: Metabolic measurements after 20 weeks of diet intervention (28-week-old mice). Total body weights, lean mass, and fat mass at euthanasia. Whole blood glucose (0–120 min) and plasma insulin (0–30 min) levels after oral gavage of 1.5 g glucose/kg lean mass (OGTT) in 27-week-old mice. D: Metabolic phenotyping of Plin2+/+ and Plin2−/− mice fed LFD (upper raw) or HFD (lower raw) after 18 weeks of diet intervention. Oxygen consumption (O2, ml/h), carbon dioxide production (CO2, ml/h), respiratory exchange ratio (RER), activity levels (in the XY plane), mean activity levels in the light (7 AM–7 PM) and dark (7 PM–7 AM) phases, and daily calorie intake (kcal/24 h). Data in graphs are shown as means ± 95% confidence interval. Statistical testing was performed with two-way ANOVA and Tukey or Dunnett’s test for multiple comparisons. #P < 0.05 indicates the statistical difference between LFD and HFD for the same genotype.
To investigate whole-body energy expenditure, Plin2+/+ and Plin2−/− mice were monitored with indirect calorimetry after 9 and 18 weeks of diet intervention (data from 19 weeks of diet intervention are shown). O2 consumption was relatively similar in LFD- or HFD-fed mice. CO2 production and respiratory exchange ratio were different between LFD- and HFD-fed mice because of the consumption of primarily carbohydrates versus fat as the main energy substrates, respectively (Fig. 7D). There were no differences in O2 consumption, CO2 production, respiratory exchange ratio, activity levels, or food intake between Plin2+/+ and Plin2−/− mice. We also measured the tissue weights of various organs in animals terminated after 10 and 20 weeks of diet intervention (18 and 28 weeks of age). Total body weights, as well as weights of epididymal and inguinal subcutaneous white adipose tissue depots, were elevated in HFD- compared with LFD-fed mice (Table 3). Liver, heart, and kidney weights were not affected by diets. There were no differences in organ weights between Plin2+/+ and Plin2−/− mice (Table 3).
Table 3.
Body weights and organ weights of Plin2+/+ and Plin2−/− mice on LFD or HFD at 18 weeks (10 weeks on diet) and 28 weeks of age (20 weeks on diet)
| Organs | LFD |
HFD |
||
|---|---|---|---|---|
| Plin2+/+ | Plin2−/− | Plin2+/+ | Plin2−/− | |
| 18 weeks—females | ||||
| Body weight (g) | 22.1 ± 1.0 | 23.8 ± 1.7 | 29.6 ± 4.5a | 31.7 ± 6.0a |
| Liver (g) | 0.99 ± 0.08 | 0.99 ± 0.08 | 1.12 ± 0.126 | 1.09 ± 0.15 |
| Heart (g) | 0.108 ± 0.024 | 0.119 ± 0.041 | 0.111 ± 0.033 | 0.118 ± 0.027 |
| Kidney (g) | 0.22 ± 0.02 | 0.24 ± 0.04 | 0.28 ± 0.06 | 0.28 ± 0.04 |
| White adipose tissue gonadal (g) | 0.35 ± 0.09 | 0.55 ± 0.27 | 1.38 ± 0.63a | 1.7 ± 0.89a |
| White adipose tissue subcutaneous (g) | 0.27 ± 0.09 | 0.36 ± 0.15 | 1.02 ± 0.49a | 1.02 ± 0.50a |
| 28 weeks—females | ||||
| Body weight (g) | 24.4 ± 1.3 | 26.3 ± 2.1 | 43.3 ± 3.6a | 40.4 ± 8.0a |
| Liver (g) | 1.04 ± 0.08 | 1.11 ± 0.17 | 1.14 ± 0.41 | 1.13 ± 0.18 |
| Heart (g) | 0.100 ± 0.006 | 0.109 ± 0.008 | 0.118 ± 0.007 | 0.118 ± 0.022 |
| Kidney (g) | 0.23 ± 0.02 | 0.25 ± 0.02 | 0.28 ± 0.02 | 0.28 ± 0.03 |
| White adipose tissue gonadal (g) | 0.047 ± 0.11 | 0.73 ± 0.24 | 3.41 ± 0.39a | 3.31 ± 1.02a |
| White adipose tissue subcutaneous (g) | 0.36 ± 0.13 | 0.53 ± 0.20 | 2.30 ± 0.38a | 2.10 ± 0.75a |
Data are presented as means ± SD, n = 8–10.
Indicates difference between LFD and HFD of the same genotype (P < 0.001).
Plin2−/− mice fed HFD were more resistant to hepatic TAG accumulation
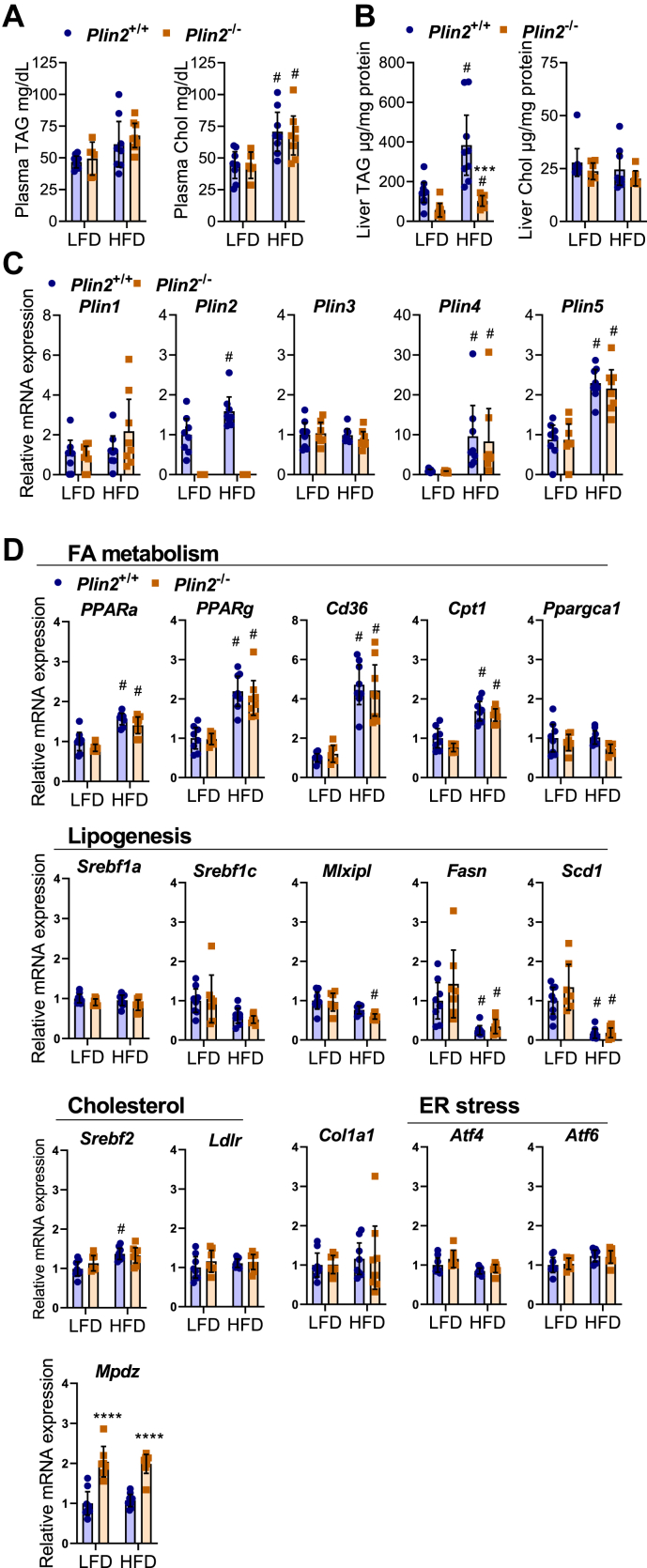
Although Plin2−/− mice showed no distinguished phenotype with regard to adiposity, glucose clearance, or whole-body energy expenditure, we investigated if LFD- or HFD-fed Plin2−/− mice had altered plasma or hepatic lipid content. Plasma TAG and total Chol levels were similar in Plin2+/+ and Plin2−/− mice after 10 weeks (supplemental Fig. S3A) and 20 weeks of diet intervention (Fig. 8A), in line with no differences in these lipids between fed or fasted Plin2+/+ and Plin2−/− mice. Hepatic TAG levels were similarly low with LFD feeding in Plin2+/+ and Plin2−/− mice but were less induced in HFD-fed Plin2−/− mice compared with Plin2+/+ mice after 10 weeks (supplemental Fig. S3) and 20 weeks of diet intervention (Fig. 8B). Hepatic total Chol levels were similar in LFD- or HFD-fed Plin2+/+ and Plin2−/− mice. Next, we performed a detailed lipid analysis of liver tissue from five representative individuals per group of mice fed LFD and HFD for 10 weeks (18-week-old mice). The majority of TAG and DAG species measured were decreased in LFD- or HFD-fed Plin2−/− mice compared with Plin2+/+ mice (supplemental Fig. S2). CE and other lipid species measured were essentially similar in Plin2+/+ and Plin2−/− mice fed the same diet (supplemental Fig. S2).
Fig. 8.
Lipid levels and hepatic mRNA expression in Plin2+/+ and Plin2−/− mice fed LFD or HFD. Female Plin2+/+ and Plin2−/− mice were fed an LFD or an HFD for 20 weeks (from 8 weeks of age until 28 weeks of age). Number of individuals: Plin2+/+ LFD (n = 8), Plin2+/+ HFD (n = 9), Plin2−/− LFD (n = 7), and Plin2−/− HFD (n = 7). A: Plasma TAGs and total Chol levels. B: Hepatic TAG and Chol levels. C: Hepatic expression of Plin1, Plin2, Plin3, Plin4, and Plin5 mRNAs. D: Hepatic expression of mRNAs involved in FA metabolism (Ppara, Pparg, Cd36, Cpt1, Ppargc1a), lipogenesis (Srebf1a, Srebf1c, Mlxipl, Fasn, Scd1), Chol metabolism (Srebf2, Ldlr), fibrosis (Col1a1), ER stress (Atf4 and Atf6), and expression of Mpdz. Statistical testing was done with two-way ANOVA and Tukey test for multiple comparisons. #P < 0.05 indicates the statistical difference between LFD and HFD for the same genotype. ∗∗∗P < 0.001 indicates difference between Plin2+/+ and Plin2−/− mice receiving the same diet. Data in graphs are shown as means ± 95% confidence interval.
Hepatic expression of metabolic genes is unaltered in HFD-fed Plin2−/− mice
Others have reported differences in weight gain and altered hepatic expression of metabolic genes in Plin2−/− mice fed obesogenic diets (38, 39, 57). We therefore measured the hepatic expression of Plins and metabolic genes in LFD- and HFD-fed Plin2+/+ and Plin2−/− mice. Expression of Plin2, Plin4, and Plin5 mRNAs increased with HFD feeding compared with LFD after 20 weeks of diet intervention (Fig. 8C). This dietary effect was less pronounced after 10 weeks of feeding (supplemental Fig. S3B). Except for loss of Plin2 expression in Plin2−/− mice, expression levels of other Plins were similar in Plin2+/+ and Plin2−/− mice with both diets, suggesting there were no transcriptional compensations in Plin expression upon loss of Plin2. Furthermore, we observed no differences in expression levels between Plin2+/+ and Plin2−/− mice for genes involved in FA metabolism (Ppara, Pparg, Cd36, Cpt1, Ppargc1a), lipogenesis (Srebf1, Mlxipl, Fasn, Scd1), Chol metabolism and uptake (Srebf2 and Ldlr), fibrosis (Col1a1), or ER stress (Atf4 and Atf6) (Fig. 8D). Mpdz mRNA was upregulated ∼2-fold in Plin2−/− mice compared with Plin2+/+ mice but was unaffected by diet (Fig. 8D).
Discussion
Studies to characterize the role of hepatic Plin2 have primarily focused on its role in obesity (38, 39) and the development of nonalcoholic fatty liver disease caused by surplus energy intake (10, 39, 58, 59). In the current study, we compared the significance of Plin2 expression for hepatic lipid storage in mice subjected to energy deficiency (fasting) or energy surplus (HFD feeding). Fasting for 24 h resulted in a >10-fold increase in hepatic TAG levels in contrast to a ∼3-fold increase in hepatic TAG levels with 20 weeks of HFD feeding. Hepatic TAG levels in Plin2−/− mice were reduced by both fasting and HFD feeding, yet with different consequences upon loss of Plin2. Hepatic LD morphology (result not shown) and expression of glucose and FA-oxidizing genes were unaltered in Plin2−/− mice given an HFD. In contrast, LD morphology was drastically altered upon fasting, accompanied by somewhat elevated expression of immune-related genes and genes involved in autophagy and peroxisomal FA oxidative pathways in Plin2−/− mice compared with Plin2+/+ mice. Taken together, our results show that hepatic expression of Plin2 is more important for coating of hepatic LDs formed upon fasting compared with LDs accumulated as a consequence of diet-induced hepatic steatosis.
A prominent phenotype in our studies was reduced hepatic steatosis in both fasted or HFD-fed Plin2−/− mice. The first generated Plin2-null model (with disruption of Plin2 exons 2–3) also showed reduced hepatic TAG levels (10), but this model expresses a shortened Plin2 isoform that might confound this observation (60). The same hepatic phenotype has later been reported in a different Plin2-null model (disrupting Plin2 exon 5) (39), with siRNA silencing of Plin2 in diet-induced obese mice (41) or genetically obese leptin-deficient (ob/ob) mice (61), or in mice with liver-specific loss of Plin2 fed a methionine-choline deficient diet (40). Similarly, we also observed reduced hepatic TAG in our Plin2 model (disruption of exons 4–6, (44)), albeit not in mice fed diets with low-fat content, which had low hepatic TAG levels. A less pronounced hepatic phenotype in nonsteatotic livers may be explained by the cellular location of Plin2 and distinct TAG reservoirs in hepatocytes. Hepatocytes can incorporate TAG in cytoplasmic LDs and membrane-enclosed VLDL particles. Plin2 binds to cytoplasmic LDs (11) and is actively degraded by proteasomes when not bound to LDs (12, 13). Although the presence of Plin2 may balance transfer of TAG toward LD versus VLDL pools (37, 59), VLDL is synthesized in the ER lumen and the Golgi apparatus and has no direct contact with Plin2 or LDs. Hepatocytes with low TAG levels are therefore expected to direct most TAG toward VLDL, with low dependence on Plin2 expression. In contrast, HFD or fasting elevate TAG incorporation into cytosolic LD in hepatocytes, opening for more manifested effects upon Plin2 removal. Our findings that lack of Plin2 has a limited effect on hepatic gene transcription in fed hepatocytes (low TAG) versus considerable effect in fasted hepatocytes (elevated TAG) imply that phenotypical alterations of Plin2 removal are primarily linked to the cellular need for storage of TAG.
It is well established that Plin2 overexpression increases LD accumulation (37, 42) and that loss of Plin2 reduces LD accumulation in various cells (10, 40, 41, 59, 61). It is, however, unclear if loss of Plin2 primarily affects LD size or LD numbers, and if loss of Plin2 enhances LD degradation primarily by altering rates of lipolysis or lipophagy. The first study demonstrating that removal of nonadipose Plin proteins could affect LD morphology was performed in hepatocyte-like cells (AML12), where siRNA silencing of Plin3 reduced LD size (Plin2 overexpression), silencing of Plin2 had no effect, whereas Plin2 and Plin3 silencing resulted in few but enlarged LDs (62). Studies in animals have reported enlarged hepatic LDs in Plin2−/− mice crossed into obese Lep−/− deficient mice (59), and in adrenals of Plin2−/− mice (44), yet with diverse effects on LD size and total LD mass. Removal of Plin2 results in increased adrenal CE levels and LD size (44), whereas hepatic TAG levels and total LD content are reduced despite minor elevations in hepatic CE levels in fasted or HFD-fed Plin2−/− mice. Reduced LD content has also been reported in foam cells of Plin2−/− mice in an atherogenic ApoE−/− background (63), but lipid species and LD sizes were not evaluated. We now demonstrate that perhaps the most radical increase in hepatic LD size occurs in fasted Plin2−/− mice, although the mechanisms involved in this phenotype are unclear and may be beyond the role of Plin2 as a protector against lipases. We observed increased levels of Plin3 and Plin5 proteins in fasted Plin2−/− mice, implying that these Plin proteins compensate at the LD surface in the lack of Plin2. Such a shift in LD protein coating is likely to alter the recruitment of lipases and other proteins. Attempts to isolate LDs and reproducibly compare the hepatic LD proteome and lipidome in fasted Plin2+/+ and Plin2−/− mice have proven difficult because of the extreme differences in size and buoyancy for those LD populations. However, whole hepatic cell lysates of fed Plin2−/− mice had increased levels of the most abundant PC and PCO lipid species as well as increased levels of some of the PE and PS lipids upon fasting. It has been suggested that the cylindrical shape of PC facilitates good coverage of the LD surface and promotes LD stability (64, 65), whereas PE forms cone-shaped structures resulting in LD packing defects when present in excess (64). It is possible that increased hepatic levels of PC and PCO species in fed Plin2−/− mice accumulate on LDs and help to stabilize the LD surface, whereas the increased levels of PE species in fasted Plin2−/− mice cause LD packaging defects. Detailed experiments on isolated LDs will be needed to determine if the lack of Plin2 affects the LD phospholipid layer and contributes to the abnormal LD morphology in fasted Plin2−/− mice.
The ability of various Plins to protect LDs seems influenced by the type of neutral lipids stored in the core (66). Emerging data suggest that TAG reservoirs are more strongly affected in Plin2−/− mice compared with CE reservoirs that tend to be maintained or may even be enhanced (44). ATGL is the key regulator of lipolytic TAG degradation in many metabolic tissues, with activity levels influenced by expressed Plins (67). Overexpression of Plin2 in fibroblasts reduces ATGL activity and preserves TAG stores (52), whereas ATGL activity is enhanced and TAG stores depleted in Plin2−/− myotubes (53). A limitation with these in vitro cell culture studies is the insignificant Plin5 expression (53, 68) which does not fully recapitulate the LD proteomes of the same cell types in vivo. Such limitations are especially relevant when analyzing Plin2 function in the liver, given that Plin5 binds and regulates the activity of ATGL (69), steatosis is exacerbated in Pnpla2 (ATGL)−/− mice (70), and as shown in this study, Plin5 is overexpressed in Plin2−/− livers. The current literature supports the notion that hepatic Plin2 protects LDs against ATGL activity. Of note, we observed increased hepatic expression of Plin5 and ATGL in fasted Plin2−/− mice, which suggests that elevated lipolysis is involved in the depletion of hepatic TAG in Plin2−/− mice. Another possible mechanism involved is lipophagy, a form of autophagy where whole or partial LDs are engulfed by phagosomes and neutral lipids are degraded by lysosomal lipases and other enzymes (71). We observed slightly increased hepatic LAMP1 expression in fasted Plin2−/− mice compared with the Plin2+/+ mice but similar levels of LC isoforms and p62, suggesting that altered lipophagy activity likely not contribute to reduced hepatic TAG levels in female Plin2−/− mice. These results differ from a previous study reporting increased hepatic autophagy in fed male Plin2−/− mice (51). It is unclear if these discrepancies are caused by different experimental conditions or gender-specific consequences of Plin2 loss. Of note, we previously observed increased content of the lipidated LC3 II and LAMP1/2 proteins in the adrenals of fed female Plin2−/− mice (44), but these alterations may occur because of the accumulation of ceroid-like structures in some apoptotic/dead cortical cells and may not necessarily be a sign of altered autophagy in functional cortical cells. In summary, although ATGL seems involved, additional studies will be needed to fully define mechanisms involved in the depletion of hepatic TAG reservoirs in Plin2−/− mice upon fasting.
Secondary systemic consequences of reduced hepatic LD accumulation in Plin2−/− mice have been reported by independent laboratories. Improved insulin sensitivity has been observed with hepatic siRNA silencing of Plin2 in HFD-fed (61) and alcohol-fed mice (72), in Plin2−/− mice crossed into obese Lep−/− deficient mice (59), and with HFD or Western diet feeding of mice with global or hepatic-specific Plin2 deletion (38, 39, 58). In contrast to these reports, the initial characterization of Plin2−/− mice did not report effects on insulin sensitivity (10). In addition, overexpression of Plin2 in muscle seems to increase insulin sensitivity in parallel with enhanced LD accumulation (42), which opens for tissue-specific effects upon Plin2 deletion. Contradictory to most studies mentioned previously performed in males, we observed similar body weights, organ weights, body composition, glucose tolerance, energy expenditure, and hepatic expression of metabolic genes between female Plin2+/+ mice and Plin2−/− mice fed an HFD for up to 20 weeks. Although gender differences could potentially explain divergent effects on insulin sensitivity in this case, we have not been able to replicate gross differences in weight gain between male Plin2+/+ and Plin2−/− mice given an obesogenic Western-like diet (result not shown). Whole-body insulin sensitivity is determined by several organs, and inconsistent effects on glucose clearance between studies can potentially be caused by the usage of models with global or hepatocyte-specific deletion of Plin2. However, effects on body weight gain are inconsistently reported in many of the dietary studies performed in Plin2 models (38, 39, 58), suggesting reproducibility issues. Importantly, parameters reported to differ between Plin2+/+ and Plin2−/− mice with obesogenic diet feeding in previous studies, inducing improved insulin sensitivity, are paralleled by a lack of weight gain in Plin2−/− mice (38, 39, 58). The same metabolic differences will in large be observed between lean versus obese mice independent of Plin2 removal (72). Hence, it will be necessary to conduct new diet interventions to elucidate why effects on weight gain differ between laboratories and Plin2-null models used, and whether phenotypic differences observed in diet interventions are caused by Plin2 deletion in the liver or systemic or unknown confounding factors affecting body weight gain under certain conditions. Such confounding factors could perhaps be related to microflora or the immune system. Our diet intervention was performed in healthy mice absent from all pathogens according to FELASA guidelines. Details on animal health were not described in the other studies. Another possibility is differences in back-crossing between the models examined. We observed ∼2-fold elevated expression of multiple PDZ domain crumbs cell polarity complex component (Mpdz) mRNA in Plin2−/− mice independent of dietary conditions. Our Plin2 model was generated in ES cells of a 129 background and has been backcrossed into C57BL/6N for at least 10 generations. The Mpdz loci is located ∼5 Mbp downstream of Plin2 on mouse chromosome 4 (National Center for Biotechnology Information, assembly #NC_000070.7), and this whole chromosomal region is therefore unlikely to be backcrossed but remain as a 129 genome. Hence, it cannot be determined if the altered expression of the Mpdz gene in our model is caused by Plin2 loss or SNP variability in the Mpdz gene region between the 129 genome and C57BL/6 strains that affects expression levels of the Mpdz gene.
To conclude, our study examined hepatic effects upon deletion of Plin2 in female mice. Fasted female Plin2−/− mice had altered hepatic TAG and CE levels, abnormal LD morphology, increased protein levels of Plin3, Plin5, and ATGL, and grossly altered hepatic gene expression compared with fasted Plin2+/+ mice. In contrast, female Plin2−/− mice fed an HFD for up to 20 weeks had reduced hepatic TAG, but otherwise, no differences in body weight gain, tissue weights, body composition, glucose tolerance, energy expenditure, or hepatic expression of metabolic genes compared with Plin2+/+ mice were observed. Our analysis suggests that Plin2 is more important for coating of hepatic LDs during fasting as compared with obesity-induced hepatic steatosis.
Data availability
Data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank engineers Ingunn Jermstad and Shaista Khan at the Norwegian Transgenic Centre and Pratibha Shrikant Kolan for technical assistance. Members of the Lipid Droplet group and the Molecular Nutrition Unit at the Department of Nutrition are acknowledged for scientific discussions.
Author contributions
A. I. D., Y. L., P. K., and K. T. D. conceptualization; A. R. K. and K. T. D. methodology; A. I. D. and K. T. D. validation; A. I. D., Y. L., M. H., and K. T. D. formal analysis; A. I. D., Y. L., P. K., M. H., and K. T. D. investigation; A. I. D. and K. T. D. writing–original draft; A. I. D., Y. L., P. K., M. H., S. O. K., F. A. N., A. R. K., and K. T. D. writing–review & editing; A. I. D., Y. L., and M. H. visualization; K. T. D. supervision; S. O. K., F. A. N., A. R. K., and K. T. D. project administration; S. O. K., A. R. K., and K. T. D. funding acquisition.
Funding and additional information
The studies were supported by grants from the Medical Faculty at the University of Oslo (to A. I. D., S. O. K., and K. T. D.), the South-Eastern Norway Regional Health Authority (to Y. L. and K. T. D.), Direktør Throne Holsts fond for ernæringsforskning (to K. T. D)., Henning and Johan Throne-Holst Foundation (to K. T. D.), Aktieselskabet Freia Chocolade Fabrik’s Medical Foundation, Anders Jahre’s Foundation (to K. T. D.), and the Intramural Research Program of the National Institutes of Health, and the National Institute of Diabetes and Digestive and Kidney Diseases (to A. R. K. and K. T. D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplemental Data
References
- 1.Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimmel A.R., Sztalryd C. The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu. Rev. Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- 3.Afonso M.S., Machado R.M., Lavrador M.S., Quintao E.C.R., Moore K.J., Lottenberg A.M. Molecular pathways underlying cholesterol homeostasis. Nutrients. 2018;10:760. doi: 10.3390/nu10060760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X., Gruia-Gray J., Copeland N.G., Gilbert D.J., Jenkins N.A., Londos C., et al. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome. 2001;12:741–749. doi: 10.1007/s00335-01-2055-5. [DOI] [PubMed] [Google Scholar]
- 5.Kimmel A.R., Brasaemle D.L., McAndrews-Hill M., Sztalryd C., Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sztalryd C., Brasaemle D.L. The perilipin family of lipid droplet proteins: gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:1221–1232. doi: 10.1016/j.bbalip.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg A.S., Egan J.J., Wek S.A., Garty N.B., Blanchette-Mackie E.J., Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 8.Jiang H.P., Serrero G. Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7856–7860. doi: 10.1073/pnas.89.17.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heid H.W., Moll R., Schwetlick I., Rackwitz H.R., Keenan T.W. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res. 1998;294:309–321. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- 10.Chang B.H., Li L., Paul A., Taniguchi S., Nannegari V., Heird W.C., et al. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell Biol. 2006;26:1063–1076. doi: 10.1128/MCB.26.3.1063-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasaemle D.L., Barber T., Wolins N.E., Serrero G., Blanchette-Mackie E.J., Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 12.Xu G., Sztalryd C., Lu X., Tansey J.T., Gan J., Dorward H., et al. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 2005;280:42841–42847. doi: 10.1074/jbc.M506569200. [DOI] [PubMed] [Google Scholar]
- 13.Masuda Y., Itabe H., Odaki M., Hama K., Fujimoto Y., Mori M., et al. ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J. Lipid Res. 2006;47:87–98. doi: 10.1194/jlr.M500170-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y., Shinoda A., Kamada H., Shimizu M., Inoue J., Sato R. Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci. Rep. 2016;6 doi: 10.1038/srep20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalen K.T., Schoonjans K., Ulven S.M., Weedon-Fekjaer M.S., Bentzen T.G., Koutnikova H., et al. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes. 2004;53:1243–1252. doi: 10.2337/diabetes.53.5.1243. [DOI] [PubMed] [Google Scholar]
- 16.Gao J., Ye H., Serrero G. Stimulation of adipose differentiation related protein (ADRP) expression in adipocyte precursors by long-chain fatty acids. J. Cell Physiol. 2000;182:297–302. doi: 10.1002/(SICI)1097-4652(200002)182:2<297::AID-JCP19>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Dalen K.T., Ulven S.M., Arntsen B.M., Solaas K., Nebb H.I. PPARalpha activators and fasting induce the expression of adipose differentiation-related protein in liver. J. Lipid Res. 2006;47:931–943. doi: 10.1194/jlr.M500459-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Wolins N.E., Rubin B., Brasaemle D.L. TIP47 associates with lipid droplets. J. Biol. Chem. 2001;276:5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 19.Dalen K.T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., et al. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta. 2007;1771:210–227. doi: 10.1016/j.bbalip.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Wolins N.E., Quaynor B.K., Skinner J.R., Schoenfish M.J., Tzekov A., Bickel P.E. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 2005;280:19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 2006;281:14232–14240. doi: 10.1074/jbc.M601682200. [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Sreenivasan U., Hu H., Saladino A., Polster B.M., Lund L.M., et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 2011;52:2159–2168. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kien B., Kolleritsch S., Kunowska N., Heier C., Chalhoub G., Tilp A., et al. Lipid droplet-mitochondria coupling via perilipin 5 augments respiratory capacity but is dispensable for FA oxidation. J. Lipid Res. 2022;63 doi: 10.1016/j.jlr.2022.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia A., Subramanian V., Sekowski A., Bhattacharyya S., Love M.W., Brasaemle D.L. The amino and carboxyl termini of perilipin a facilitate the storage of triacylglycerols. J. Biol. Chem. 2004;279:8409–8416. doi: 10.1074/jbc.M311198200. [DOI] [PubMed] [Google Scholar]
- 26.Kuramoto K., Okamura T., Yamaguchi T., Nakamura T.Y., Wakabayashi S., Morinaga H., et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 2012;287:23852–23863. doi: 10.1074/jbc.M111.328708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollak N.M., Schweiger M., Jaeger D., Kolb D., Kumari M., Schreiber R., et al. Cardiac-specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J. Lipid Res. 2013;54:1092–1102. doi: 10.1194/jlr.M034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granneman J.G., Moore H.P., Krishnamoorthy R., Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) J. Biol. Chem. 2009;284:34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollak N.M., Jaeger D., Kolleritsch S., Zimmermann R., Zechner R., Lass A., et al. The interplay of protein kinase A and perilipin 5 regulates cardiac lipolysis. J. Biol. Chem. 2015;290:1295–1306. doi: 10.1074/jbc.M114.604744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granneman J.G., Moore H.P., Mottillo E.P., Zhu Z., Zhou L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 2011;286:5126–5135. doi: 10.1074/jbc.M110.180711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quiroga A.D., Lehner R. Liver triacylglycerol lipases. Biochim. Biophys. Acta. 2012;1821:762–769. doi: 10.1016/j.bbalip.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Holm C., Belfrage P., Fredrikson G. Immunological evidence for the presence of hormone-sensitive lipase in rat tissues other than adipose tissue. Biochem. Biophys. Res. Commun. 1987;148:99–105. doi: 10.1016/0006-291x(87)91081-3. [DOI] [PubMed] [Google Scholar]
- 33.Shulman G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 2014;371:1131–1141. doi: 10.1056/NEJMra1011035. [DOI] [PubMed] [Google Scholar]
- 34.Inoue M., Ohtake T., Motomura W., Takahashi N., Hosoki Y., Miyoshi S., et al. Increased expression of PPARgamma in high fat diet-induced liver steatosis in mice. Biochem. Biophys. Res. Commun. 2005;336:215–222. doi: 10.1016/j.bbrc.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 35.Motomura W., Inoue M., Ohtake T., Takahashi N., Nagamine M., Tanno S., et al. Up-regulation of ADRP in fatty liver in human and liver steatosis in mice fed with high fat diet. Biochem. Biophys. Res. Commun. 2006;340:1111–1118. doi: 10.1016/j.bbrc.2005.12.121. [DOI] [PubMed] [Google Scholar]
- 36.Schadinger S.E., Bucher N.L., Schreiber B.M., Farmer S.R. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1195–1205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 37.Magnusson B., Asp L., Bostrom P., Ruiz M., Stillemark-Billton P., Linden D., et al. Adipocyte differentiation-related protein promotes fatty acid storage in cytosolic triglycerides and inhibits secretion of very low-density lipoproteins. Arterioscler. Thromb. Vasc. Biol. 2006;26:1566–1571. doi: 10.1161/01.ATV.0000223345.11820.da. [DOI] [PubMed] [Google Scholar]
- 38.Orlicky D.J., Libby A.E., Bales E.S., McMahan R.H., Monks J., La Rosa F.G., et al. Perilipin-2 promotes obesity and progressive fatty liver disease in mice through mechanistically distinct hepatocyte and extra-hepatocyte actions. J. Physiol. 2019;597:1565–1584. doi: 10.1113/JP277140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McManaman J.L., Bales E.S., Orlicky D.J., Jackman M., MacLean P.S., Cain S., et al. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J. Lipid Res. 2013;54:1346–1359. doi: 10.1194/jlr.M035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Najt C.P., Senthivinayagam S., Aljazi M.B., Fader K.A., Olenic S.D., Brock J.R., et al. Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G726–738. doi: 10.1152/ajpgi.00436.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai Y., Varela G.M., Jackson M.B., Graham M.J., Crooke R.M., Ahima R.S. Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology. 2007;132:1947–1954. doi: 10.1053/j.gastro.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 42.Bosma M., Hesselink M.K., Sparks L.M., Timmers S., Ferraz M.J., Mattijssen F., et al. Perilipin 2 improves insulin sensitivity in skeletal muscle despite elevated intramuscular lipid levels. Diabetes. 2012;61:2679–2690. doi: 10.2337/db11-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang E., Cui W., Lopresti M., Mashek M.T., Najt C.P., Hu H., et al. Hepatic PLIN5 signals via SIRT1 to promote autophagy and prevent inflammation during fasting. J. Lipid Res. 2020;61:338–350. doi: 10.1194/jlr.RA119000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Khanal P., Norheim F., Hjorth M., Bjellaas T., Drevon C.A., et al. Plin2 deletion increases cholesteryl ester lipid droplet content and disturbs cholesterol balance in adrenal cortex. J. Lipid Res. 2021;62 doi: 10.1016/j.jlr.2021.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hjorth M., Doncheva A., Norheim F., Ulven S.M., Holven K.B., Saether T., et al. Consumption of salmon fishmeal increases hepatic cholesterol content in obese C57BL/6 J mice. Eur. J. Nutr. 2022;61:4027–4043. doi: 10.1007/s00394-022-02930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 48.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai T.H., Chen E., Li L., Saha P., Lee H.J., Huang L.S., et al. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy. 2017;13:1130–1144. doi: 10.1080/15548627.2017.1319544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Listenberger L.L., Ostermeyer-Fay A.G., Goldberg E.B., Brown W.J., Brown D.A. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 2007;48:2751–2761. doi: 10.1194/jlr.M700359-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Feng Y.Z., Lund J., Li Y., Knabenes I.K., Bakke S.S., Kase E.T., et al. Loss of perilipin 2 in cultured myotubes enhances lipolysis and redirects the metabolic energy balance from glucose oxidation towards fatty acid oxidation. J. Lipid Res. 2017;58:2147–2161. doi: 10.1194/jlr.M079764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen F.J., Yin Y., Chua B.T., Li P. CIDE family proteins control lipid homeostasis and the development of metabolic diseases. Traffic. 2020;21:94–105. doi: 10.1111/tra.12717. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Gao T., Deng S., Shang L., Chen X., Chen K., et al. Fasting induces hepatic lipid accumulation by stimulating peroxisomal dicarboxylic acid oxidation. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandalio L.M., Rodríguez-Serrano M., Romero-Puertas M.C., del Río L.A. In: Peroxisomes and their Key Role in Cellular Signaling and Metabolism. del Río L.A., editor. Springer Netherlands; Dordrecht: 2013. Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules; pp. 231–255. [DOI] [PubMed] [Google Scholar]
- 56.Speakman J.R. Use of high-fat diets to study rodent obesity as a model of human obesity. Int. J. Obes. (Lond). 2019;43:1491–1492. doi: 10.1038/s41366-019-0363-7. [DOI] [PubMed] [Google Scholar]
- 57.Libby A.E., Bales E., Orlicky D.J., McManaman J.L. Perilipin-2 deletion impairs hepatic lipid accumulation by interfering with sterol regulatory element-binding protein (SREBP) activation and altering the hepatic lipidome. J. Biol. Chem. 2016;291:24231–24246. doi: 10.1074/jbc.M116.759795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang B.H., Li L., Saha P., Chan L. Absence of adipose differentiation related protein upregulates hepatic VLDL secretion, relieves hepatosteatosis, and improves whole body insulin resistance in leptin-deficient mice. J. Lipid Res. 2010;51:2132–2142. doi: 10.1194/jlr.M004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell T.D., Palmer C.A., Orlicky D.J., Bales E.S., Chang B.H., Chan L., et al. Mammary glands of adipophilin-null mice produce an amino-terminally truncated form of adipophilin that mediates milk lipid droplet formation and secretion. J. Lipid Res. 2008;49:206–216. doi: 10.1194/jlr.M700396-JLR200. [DOI] [PubMed] [Google Scholar]
- 60.Varela G.M., Antwi D.A., Dhir R., Yin X., Singhal N.S., Graham M.J., et al. Inhibition of ADRP prevents diet-induced insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;295:G621–628. doi: 10.1152/ajpgi.90204.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bell M., Wang H., Chen H., McLenithan J.C., Gong D.W., Yang R.Z., et al. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57:2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paul A., Chang B.H., Li L., Yechoor V.K., Chan L. Deficiency of adipose differentiation-related protein impairs foam cell formation and protects against atherosclerosis. Circ. Res. 2008;102:1492–1501. doi: 10.1161/CIRCRESAHA.107.168070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seebacher F., Zeigerer A., Kory N., Krahmer N. Hepatic lipid droplet homeostasis and fatty liver disease. Semin. Cell Dev. Biol. 2020;108:72–81. doi: 10.1016/j.semcdb.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 64.Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsieh K., Lee Y.K., Londos C., Raaka B.M., Dalen K.T., Kimmel A.R. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 2012;125:4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 67.Bindesboll C., Berg O., Arntsen B., Nebb H.I., Dalen K.T. Fatty acids regulate perilipin5 in muscle by activating PPARdelta. J. Lipid Res. 2013;54:1949–1963. doi: 10.1194/jlr.M038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H., Bell M., Sreenivasan U., Sreenevasan U., Hu H., Liu J., et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 2011;286:15707–15715. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ong K.T., Mashek M.T., Bu S.Y., Mashek D.G. Hepatic ATGL knockdown uncouples glucose intolerance from liver TAG accumulation. FASEB J. 2013;27:313–321. doi: 10.1096/fj.12-213454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Y., Li J., Kang R., Tang D. Interplay between lipid metabolism and autophagy. Front. Cell Dev. Biol. 2020;8:431. doi: 10.3389/fcell.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carr R.M., Peralta G., Yin X., Ahima R.S. Absence of perilipin 2 prevents hepatic steatosis, glucose intolerance and ceramide accumulation in alcohol-fed mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Small L., Brandon A.E., Turner N., Cooney G.J. Modeling insulin resistance in rodents by alterations in diet: what have high-fat and high-calorie diets revealed? Am. J. Physiol. Endocrinol. Metab. 2018;314:E251–E265. doi: 10.1152/ajpendo.00337.2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available from the corresponding author upon reasonable request.