Abstract
Bacillus subtilis is unable to grow by consuming galactose because it is unable to transport it into the cell. The transcription of galE is not influenced by galactose but is repressed by glucose. Galactose is toxic for galE-negative bacteria because it results in elevated levels of metabolic intermediates. These negative effects are reduced in galK and galT mutants. Glucose is also toxic for galE-negative strains.
Bacillus subtilis 168 is unable to use sugars such as lactose, galactose, and xylose (11, 18) as its sole carbon source. This is surprising given the fact that genes which encode the proteins necessary for the degradation of lactose and xylose are present in B. subtilis and that these genes are expressed under inducing conditions (2, 4). For xylose it has been shown that this sugar is not transported inside the cell (14).
We cloned and sequenced a B. subtilis homolog of the Escherichia coli galE gene (16). Meanwhile, the complete B. subtilis genome has been sequenced (9). Besides the galE homolog, putative galK and galT genes have been identified (5). Again, the genetic equipment of the bacterium should allow the utilization of galactose as the sole carbon source, but nevertheless galactose does not support growth. We were therefore interested in whether the B. subtilis galE gene serves any function. We were able to prove that the gene is expressed in B. subtilis 168. The transcription is not influenced by galactose but is repressed in the presence of glucose. Galactose is not actively transported into B. subtilis 168. Even minor amounts of internal UDP-galactose are toxic for galE-negative B. subtilis strains. Glucose is able to alleviate the effects of galactose but causes cell death during late log phase. One task of the GalE protein is therefore to protect the cell from the toxic effects of galactose and glucose or derivatives of both sugars which accumulate in its absence.
Identification of the galE gene.
We cloned and sequenced a B. subtilis galE homolog (16), which encodes a protein that is 57% identical to the product of the E. coli gene. The structure of the protein from E. coli has been solved recently (19). Fourteen of 15 amino acids known to interact with NADH are conserved between E. coli and B. subtilis; the only difference is at position 59 where the E. coli protein has isoleucine and the B. subtilis protein has leucine. Of eight amino acids known to interact with either UDP-glucose or UDP-galactose, seven are conserved, with alanine 216 in E. coli versus serine 215 in B. subtilis. This residue is known to interact via the backbone of the protein; therefore, the side chain of the residue is of minor importance.
The strains and plasmids used in this study are listed in Table 1. To test whether the galE homolog gene is functional in vivo we transformed plasmid pB8 into galE-negative E. coli MK30-3. This plasmid was constructed by ExoIII digestion of the galE-containing HindIII fragment described in reference 16 (EMBL gene bank accession no. X99339). The resulting 1,677-bp fragment (nucleotide numbers 1 to 1677, as described by Schrögel et al. [16]) was cloned into pBluescriptII SK+ (Stratagene, La Jolla, Calif.). The transformed bacterium was able to grow on minimal media with galactose as the sole carbon source (data not shown). This proves that the cloned gene indeed encodes a UDP-glucose epimerase.
TABLE 1.
Strains and plasmids used during this study
| Strain or plasmid | Genotype or relevant markers | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | hsdR17 endA1 recA1 gyrA96 thi relA1 supE44 φ80dlacZΔM15 Δ(lacZ-argF)U169 | BRL |
| E. coli MK30-3 | Δ(lac-proAB) recA galE strA F′ [proAB lacIqZΔM15] | 8a |
| B. subtilis 168 | trpC2 | BGSCa |
| B. subtilis MD156 | trpC2 ccpA::spec | 1 |
| B. subtilis DH32M | trpC2 amyE::(spoVG-lacZ, cat) | This work |
| B. subtilis EP2 | trpC2 galE::aphA3 | This work |
| B. subtilis EP10 | trpC2 amyE::(P galE-spoVG-lacZ cat) | This work |
| B. subtilis EP10ccpA | trpC2 amyE::(P galE-spoVG-lacZ cat) ccpA::spec | This work |
| B. subtilis GK1 | trpC2 galK-pGK1 | This work |
| B. subtilis GT1 | trpC2 galT-pGT1 | This work |
| B. subtilis EP2GK1 | trpC2 galE::aphA3 galK-pGK1 | This work |
| B. subtilis EP2GT1 | trpC2 galE::aphA3 galT-pGT1 | This work |
| Plasmids | ||
| pBluescriptII SK+ | bla | Stratagene |
| pB8 | bla galE | This work |
| pDH32M | bla amyE::(cat spoVG-lacZ) | 8b |
| pBEST501 | bla aphA3 | 8 |
| pEP2 | bla galE::aphA3 | This work |
| pEP10 | bla amyE::(galE-lacZ cat) | This work |
| pKL6 | bla erm bgaB | 16 |
| pKL9 | bla erm | This work |
| pGK1 | bla erm ′galK′ | This work |
| pGT1 | bla erm ′galT′ | This work |
BGSC, Bacillus Genetic Stock Center.
Regulation of galE transcription and mapping of the galE promoter.
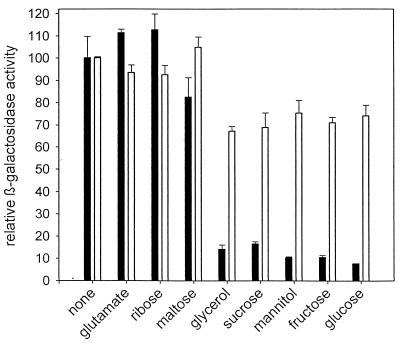
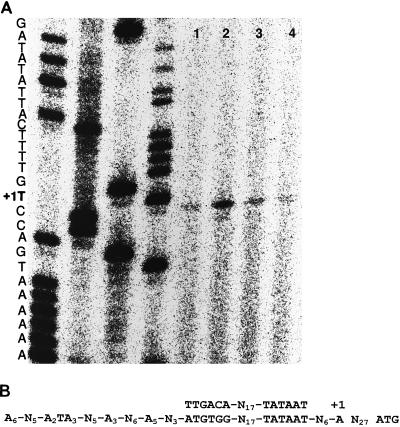
We wanted to test whether galE is actively transcribed in B. subtilis 168. To do so, we constructed plasmid pEP10. This plasmid was constructed by cloning the NciI-PvuII fragment from pB8 (nucleotide numbers 1048 to 1359 [16]) into pDH32M. The resulting plasmid contains a transcriptional fusion of the region upstream of galE to lacZ. pEP10 was integrated into the amyE locus of B. subtilis 168. The β-galactosidase activity of the resulting strain in minimal media with different carbon sources was determined as described previously (4). Fifteen to 16 U of β-galactosidase activity was obtained, irrespective of the presence of galactose. This is significantly higher than background activity (about 1 U under these conditions). lacZ expression by B. subtilis EP10 was reduced to 1.3 U in the presence of glucose, whereas background activity was reduced to 0.2 U. We were interested in whether this effect was specific for glucose or carbon catabolite repression. We tested a set of carbon sources known to mediate catabolite repression (7). As seen in Fig. 1, all carbon sources known to repress xylA transcription had a similar effect on galE transcription. In a ccpA-negative mutant the effect was abolished. This indicates that galE is transcribed in B. subtilis 168 under direct or indirect control of CcpA. To characterize the promoter in detail, we mapped the 5′ end of the galE transcript by primer extension. The primer (5′ GCCAATGTAACCGGCACCGC 3′) was labeled with [γ-32P]dATP and used for the primer extension reaction as described previously (21). It hybridizes to the translated part of the galE transcript. A signal was obtained 27 bp upstream of the translational start codon (Fig. 2A). A sequence with homology to ςA-dependent promoters was found upstream of the putative transcriptional start site (Fig. 2B). Eight of 12 bp are conserved.
FIG. 1.
Catabolite repression of galE transcription. B. subtilis EP10 and B. subtilis EP10ccpA were grown in MOPSO minimal medium containing 0.5% succinate and 0.2% concentrations of the indicated carbon sources. The LacZ activities of the respective cultures were determined. One hundred percent corresponds to 23 U for the wild-type strain (solid bars) and 39 U for the ccpA-negative mutant (open bars).
FIG. 2.
(A) Mapping of the putative transcriptional start site by primer extension. Left side (from left to right): A, C, G, and T lanes of a sequence reaction done with the primer extension primer. Lanes 1 and 2, RNA isolated from two different NB medium-grown cultures; lanes 3 and 4, RNA from cells grown on MOPSO medium without and with galactose, respectively. A signal is obtained 27 bp upstream of the translational start codon. (B) Sequence of the putative galE promoter. The poly(dA) stretches upstream of the promoter, the putative promoter sequence, and the start point of transcription and translation are indicated.
Effects of a galE inactivation.
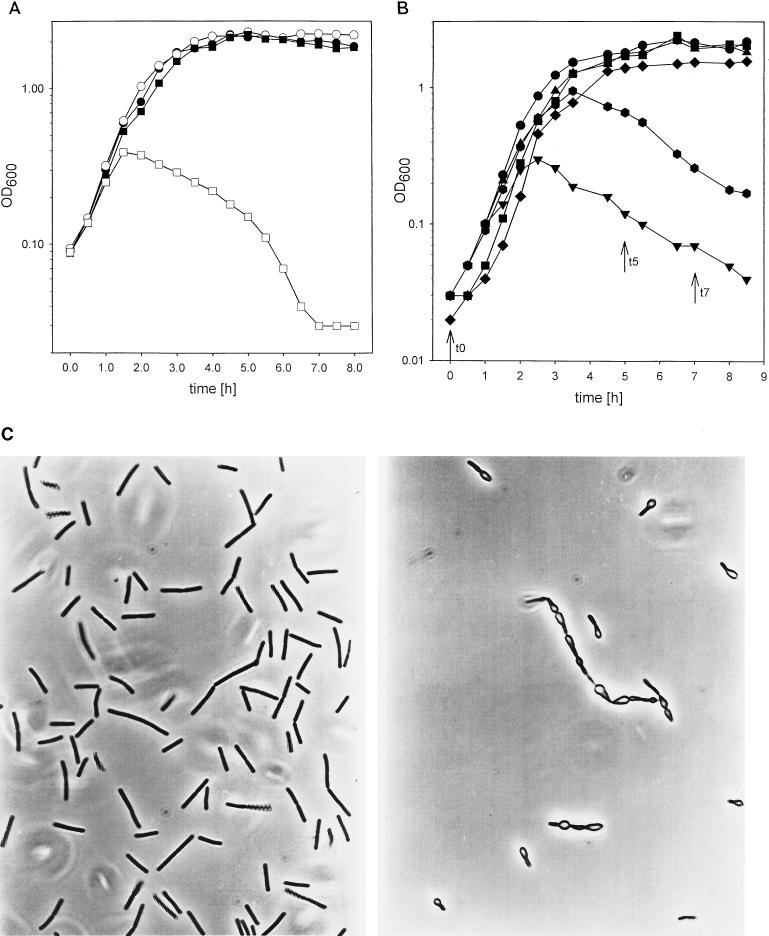
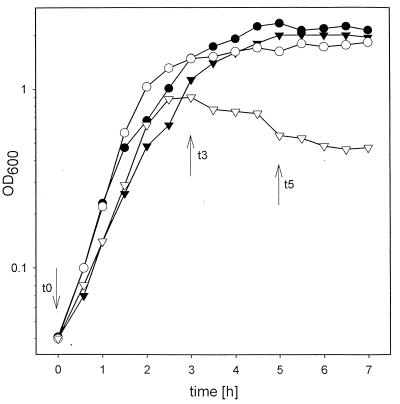
B. subtilis 168 is unable to grow on galactose as the sole carbon source (18). Therefore, lack of growth on galactose is not a specific trait for galE-negative B. subtilis strains. We tested whether the presence of galactose has a negative effect on galE strains. Such effects have been described for gal mutants of E. coli and Salmonella typhimurium (3, 10, 13, 22). To obtain a galE-negative mutant, we constructed plasmid pEP2. This was done by inserting the aphA3-containing SmaI fragment from pBEST501 (8) into StyI-digested pB8. The protruding ends were filled in with Klenow polymerase. The resistance gene was inserted in the direction opposite to that of the galE gene. pEP2 was linearized and transformed into B. subtilis 168 to give B. subtilis EP2. We grew B. subtilis 168 and the isogenic galE-negative derivative in NB medium with and without 10 mM galactose. Growth of the wild type and the mutant was similar in unsupplemented NB medium, indicating that galE expression is not necessary in complex media. The presence of galactose had no influence on the growth of B. subtilis 168 but caused a decrease of the cell density of B. subtilis EP2 (Fig. 3A). In galactose-supplemented medium the mutant cells started to swell and lysed at an optical density at 600 nm (OD600) of about 0.4 (Fig. 3C). Cell titers indicated that galactose is a bactericidal compound for these cells; the decrease in viable-cell number is even more pronounced than the decrease in OD (Table 2). Mutations in galK and galT encoding galactokinase and galactose-1-phosphate uridylyltransferase, respectively, abolish the lytic effects of the galE mutation in S. typhimurium (17). We tested whether the same is true for B. subtilis. To do so, we created plasmid pKL9 by digesting plasmid pKL6 (15) with SalI and by subsequent religation. Plasmid pGK1 was constructed to inactivate the B. subtilis galK gene. Therefore, an internal region of the galK gene was amplified by PCR with chromosomal B. subtilis 168 DNA as the template (primer I: 5′ GCAAACTCAATGTCTCAGGCC 3′; primer II: 5′ GTTCTCTTTGAACATATGGGCCG 3′). The resulting fragment was digested with NruI and AccI (nucleotide numbers 36381 and 36578 [5]) and cloned into NruI- and AccI-digested pKL9. Plasmid pGT1 was constructed to inactivate galT. An internal fragment of galT was amplified (primer I: 5′ CGAGAAACCGTCTTTATGAAGC 3′; primer II: 5′ CACCGTCCAATGAACATTTTTGG 3′), digested with AvaI and EcoRI (nucleotide numbers 37050 and 37282 [5]), and ligated to EcoRI- and AvaI-digested pKL9. We inactivated galK and galT by a single crossover and determined the effects of galactose on the respective mutants and on mutants with a defect in galE and additionally with a defect in galK or galT. As seen in Fig. 3B galactose had no effect on either the galT- or the galK-defective strain. Furthermore, the toxic effect of galactose on galE-negative strains was abolished or at least reduced if galK or galT was also inactivated. This experiment indicates that none of the gal genes is cryptic because inactivation of any of the genes had an effect on the phenotype, at least in media with galactose. The toxic effect of galactose is most likely mainly due to UDP-galactose or a derivative thereof, because it is observed only in the presence of galactose and is reduced or completely abolished if the synthesis of this compound is inhibited by the inactivation of galT or galK. Neither the galT nor the galK mutation alone had a toxic effect on growth in the presence of galactose (Table 2). Therefore, it is unlikely that the amounts of galactose-phosphate which are present in the galT strain are toxic for B. subtilis. In a second set of experiments we tested the influence of glucose on galE-negative mutants. Glucose is able to suppress the effects of galactose on galE-negative enterobacteria because of catabolite repression (17, 22). The toxic effects of galactose on B. subtilis are also reduced by the presence of glucose (data not shown). But in contrast to what is found for enterobacteria, glucose itself is toxic for B. subtilis galE mutants. At the end of the exponential growth phase the cells do not lyse (data not shown) but nevertheless die rapidly in the presence of glucose (Table 3 and Fig. 4). Taken together, these results indicate that GalE is per se not necessary for the growth of B. subtilis 168 in complex media. It is necessary only in the presence of either glucose or galactose. In the presence of both glucose and galactose the galE-negative strain is able to synthesize every compound which is synthesized by the activity of UDP-glucose epimerase in wild-type galE strains. But the addition of both sugars at one time is nevertheless lethal. Therefore, the effects of the mutation are not due to the fact that a compound necessary for survival is missing. Although it is likely that GalE serves in a biosynthetic role in the cell, our results rule out an unanticipated biosynthetic function of GalE which is necessary for the survival of the cells in the absence of galactose or glucose. They strongly argue for the hypothesis that B. subtilis produces compounds in the presence of glucose and galactose which are removed by the activity of GalE. If these compounds are not removed the cell is bound to die.
FIG. 3.
(A) Growth of different B. subtilis strains in NB medium and NB medium supplemented with galactose. B. subtilis 168 and B. subtilis EP2 were diluted in NB medium to an OD600 of 0.1. The cell suspensions were divided into two parts, and galactose was added to one part. Solid symbols, growth in NB medium; open symbols, growth in NB medium supplemented with galactose; circles, B. subtilis 168; squares, B. subtilis EP2. (B) Growth of B. subtilis with mutations in different gal genes on NB medium with galactose. Bacteria were resuspended in media containing galactose to an OD600 of 0.02. Circles, B. subtilis 168; inverted triangles, B. subtilis EP2; squares, B. subtilis GK1; diamonds, B. subtilis GT1; hexagons, B. subtilis EP2GT1; triangles, B. subtilis EP2GK1. The numbers of CFU were determined at the indicated times (arrows; see Table 2). (C) Cell morphology of bacteria grown on NB medium containing galactose. Left panel, B. subtilis 168; right panel, B. subtilis EP2.
TABLE 2.
Toxic effect of galactose on B. subtilis EP2
| B. subtilis strain | Growth of bacteria in indicated medium at timea:
|
|||||
|---|---|---|---|---|---|---|
|
t0 (107 CFU)
|
t5 (109 CFU)
|
t7 (109 CFU)
|
||||
| NB | NB + gal | NB | NB + gal | NB | NB + gal | |
| 168 | 1.2 | 0.8 | 0.6 | 0.9 | 3.1 | 1.4 |
| EP2 | 0.6 | 0.5 | 1.3 | 0.005 | 2.7 | 0.002 |
| GK1 | 0.7 | 0.6 | 0.4 | 0.6 | 1.1 | 1.3 |
| EP2GK1 | 0.2 | 0.3 | 0.6 | 0.7 | 1.5 | 1.4 |
| GT1 | 0.4 | 0.4 | 0.8 | 0.2 | 1.2 | 0.9 |
| EP2GT1 | 0.2 | 0.1 | 1.2 | 0.2 | 2.3 | 0.026 |
t0, t5, and t7, 0, 5, and 7 h, respectively; see Fig. 3B. gal, galactose.
TABLE 3.
Toxic effect of glucose on B. subtilis EP2
| B. subtilis strain | CFU (107) in indicated medium ata:
|
|||||
|---|---|---|---|---|---|---|
|
t0
|
t3
|
t5
|
||||
| NB | NB + glucose | NB | NB + glucose | NB | NB + glucose | |
| 168 | 0.4 | 0.3 | 30 | 8 | 150 | 15 |
| EP2 | 0.4 | 0.3 | 3 | 0.05 | 43 | 0.009 |
t0, t3, and t5, 0, 3, and 5 h, respectively; see Fig. 4.
FIG. 4.
Growth of different B. subtilis strains in NB medium and NB medium supplemented with glucose. Solid symbols, growth in NB medium; open symbols, growth in NB medium supplemented with glucose; circles, B. subtilis 168; inverted triangles, B. subtilis EP2. The numbers of CFU were determined at the indicated times (see Table 3).
Metabolic intermediates in galactose-grown B. subtilis EP2.
For enterobacteria it is believed that the toxic effects of galactose are due to the accumulation of galactose-1-phosphate and UDP-galactose (10, 13). We tested whether B. subtilis 168 and B. subtilis EP2 are able to transport galactose. While we were able to demonstrate galactose accumulation by E. coli, only background levels were obtained for B. subtilis in an identical assay (data not shown). Nevertheless, we grew different B. subtilis strains in media containing radioactively labeled galactose.
Cells were grown to an OD600 of 0.2 in NB medium at 37°C. Then 0.2% unlabeled galactose (final concentration) and 5 × 106 cpm of labeled galactose (final concentration, 105 cpm/ml) were added, and the bacteria were incubated for another 60 min. The cells were concentrated by centrifugation and washed thoroughly in β-mercaptoethanol-free Z buffer (12). The cells were lysed by sonication, and the protein concentration of the supernatant was determined. Equal amounts of supernatant were analyzed by thin-layer chromatography. The samples were fractionated on SilicaGel 60 (Merck, Darmstadt, Germany) with a mixture of n-butanol, ethanol, and H2O (5:3:2) in the liquid phase. The plates were dried and exposed against a BAS-III imaging plate (Fuji Photo Film Co., Tokyo, Japan). The sugars were additionally stained with a mixture of p-anisaldehyde, acetic acid, methanol, and H2SO4 (1:20:340:10), which allows both detection of sugars and their differentiation by color (6). To differentiate between sugar acids and phosphorylated sugars, the samples were treated with either trifluoroacetic acid (TFA) or alkaline phosphatase to remove PO43− groups and analyzed as described above.
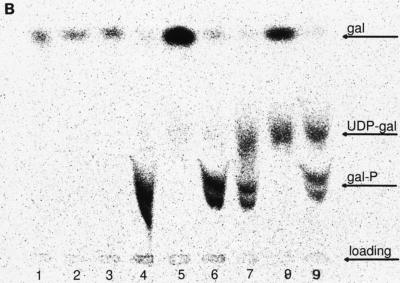
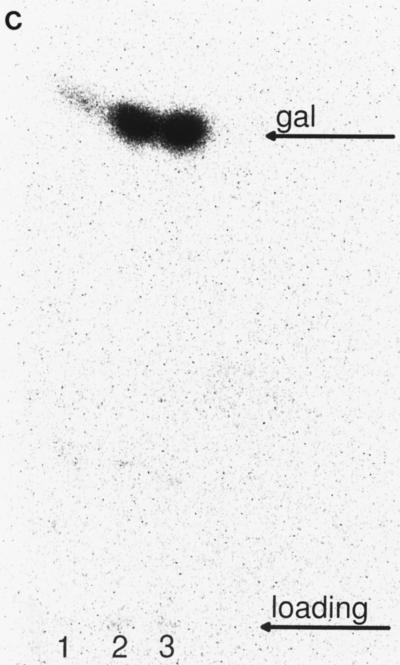
The result of this experiment is depicted in Fig. 5A. Several different galactose derivatives were obtained from the strain with mutant galE and from the strain with mutant galE and galT. One band migrated similarly to UDP-galactose, and one band migrated similarly to galactose-1-phosphate. A third band migrated slower than galactose-1-phosphate (Fig. 5A). To determine the chemical natures of the respective compounds, we treated the samples with phosphatase. After that treatment the two bands which migrated similarly to and slower than galactose-1-phosphate both migrated like galactose, indicating that these bands are galactose phosphates. We do not know whether the two bands are due to different forms of galactose phosphate or indicate different numbers of phosphates linked to one galactose. The band that migrated like UDP-galactose was not impaired by that treatment (Fig. 5B). TFA treatment removed the modification linked to galactose in this band (Fig. 5C), indicating that this modification is linked via an acid-labile ester to galactose. Treatment with UDP-glucose epimerase and subsequent TFA hydrolysis resulted in two spots, galactose and glucose (data not shown). Therefore, the original spot is indeed UDP-galactose.
FIG. 5.
(A) Analysis of galactose derivatives accumulated in B. subtilis galE-negative strains. The respective strains were fed with 14C-labeled galactose. A cell suspension with an OD600 of 1 was sonicated, and the supernatant was analyzed by thin-layer chromatography. The chromatograms were developed and compared to chromatograms of pure reagents. Lane 1, B. subtilis EP2GK1; lane 2, B. subtilis EP2; lane 3, B. subtilis EP2GT1; lane 4, B. subtilis 168. (B) Analysis of galactose derivatives after treatment with phosphatase. Lanes 1, 4, and 7, untreated samples; lanes 2, 5, and 8, phosphatase-treated samples; lanes 3, 6, and 9, samples in phosphatase buffer without phosphatase. Lanes 1 to 3, B. subtilis EP2GK1; lanes 4 to 6, B. subtilis EP2GT1; lanes 7 to 9, B. subtilis EP2. The two lower bands are converted to galactose by the treatment. (C) Analysis of galactose derivatives after treatment with TFA. Lane 1, B. subtilis EP2GK1; lane 2, B. subtilis EP2GT1; lane 3, B. subtilis EP2; All bands are converted to galactose by that treatment.
We quantified the amounts of the respective compounds. To do so, we determined the amount of radioactivity in each single spot of the thin-layer chromatography plate. For this determination, 104 cpm corresponded to 1 μmol of galactose. The amount of radioactivity was divided by the number of cells loaded on the respective lane. To obtain galactose concentrations, it was assumed that a single cell has a volume of 1 μm3 (20). Whereas we were unable to detect any momomeric galactose derivatives in wild-type B. subtilis, B. subtilis EP2 had internal concentrations of 0.2 μM galactose-phosphate and 0.2 μM UDP-galactose. B. subtilis EP2GT1, whose growth is only slightly impaired by the presence of galactose, accumulated 0.4 μM galactose phosphate. The concentration of these compounds within the cell is therefore less than 1 μM. This is low compared to the external galactose concentration of 10 mM and is in accordance with the fact that there is no active import of galactose in the cell. This concentration is comparable to the bacteriotoxic concentrations of antibiotics. The antibiotic tetracycline, which is able to cross the membrane, accumulates to a concentration of about 1 μM inside the cell (14a). This concentration is lethal for the cell. Therefore UDP-galactose is as toxic as the antibiotic tetracycline.
Conclusion.
The results presented in this paper prove that the galE gene of B. subtilis is essential for growth in media containing either glucose or galactose. This is not due to the fact that metabolic intermediates are not produced in the absence of GalE, because this effect is not abolished in the presence of both sugars. Therefore, the presence and not the absence of such an intermediate is the reason for the toxic effect. In the presence of galactose this compound is most likely UDP-galactose. Even very small amounts of UDP-galactose seem to be toxic for B. subtilis. The molecular reason for this toxicity is unknown.
Acknowledgments
We thank W. Hillen for support and discussion, M. Dahl for B. subtilis MD156, and E. Krüger for critically reading the manuscript.
This work was supported by the DFG (via Schwerpunkt Molekulare Analyse von Regulationsnetzwerken in Bakterien).
REFERENCES
- 1.Dahl M K, Hillen W. Contributions of XylR, CcpA and Hpr to catabolite repression of the xyl operon in Bacillus subtilis. FEMS Microbiol Lett. 1995;132:79–83. [Google Scholar]
- 2.Errington J, Vogt C H. Isolation and characterization of mutations in the gene encoding an endogenous Bacillus subtilis beta-galactosidase and its regulator. J Bacteriol. 1990;172:488–490. doi: 10.1128/jb.172.1.488-490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukasawa T, Nikaido H. Galactose-sensitive mutants of Salmonella. II. Bacteriolysis induced by galactose. Biochim Biophys Acta. 1961;48:470–483. doi: 10.1016/0006-3002(61)90045-2. [DOI] [PubMed] [Google Scholar]
- 4.Gärtner D, Geissendorfer M, Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988;170:3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser P, Kunst F, Arnaud M, Coudart M P, Gonzales W, Hullo M F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweizer J, Vertes A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97 kb region from 325 degrees to 333 degrees. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 6.Haußmann W. Ph.D. thesis. Tübingen, Germany: Universität Tübingen; 1994. [Google Scholar]
- 7.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 8.Itaya M, Kondo K, Tanaka T. A neomycin resistance cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Kramer W, Drutsa V, Jansen H W, Kramer B, Pflugfelder M, Fritz H J. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984;12:9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b.Kraus A, Hueck C, Gartner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 10.Kurahashi K, Wahba A J. Interference of growth with certain Escherichia coli mutants by galactose. Biochim Biophys Acta. 1958;30:298–302. doi: 10.1016/0006-3002(58)90054-4. [DOI] [PubMed] [Google Scholar]
- 11.Lindner C, Stülke J, Hecker M. Regulation of xylanolytic enzymes in Bacillus subtilis. Microbiology. 1994;140:753–757. doi: 10.1099/00221287-140-4-753. [DOI] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 13.Nikaido H. Galactose-sensitive mutants of Salmonella. I. Metabolism of galactose. Biochim Biophys Acta. 1961;48:460–469. doi: 10.1016/0006-3002(61)90044-0. [DOI] [PubMed] [Google Scholar]
- 14.Schmiedel D, Hillen W. A Bacillus subtilis 168 mutant with increased xylose uptake can utilize xylose as sole carbon source. FEMS Microbiol Lett. 1996;135:175–178. [Google Scholar]
- 14a.Scholz, O. Personal communication.
- 15.Schrögel O, Allmansberger R. Optimisation of the BgaB reporter system: determination of transcriptional regulation of stress responsive genes in Bacillus subtilis. FEMS Microbiol Lett. 1997;153:237–243. doi: 10.1111/j.1574-6968.1997.tb10488.x. [DOI] [PubMed] [Google Scholar]
- 16.Schrögel O, Krispin O, Allmansberger R. Expression of a pepT homologue from Bacillus subtilis. FEMS Microbiol Lett. 1996;145:341–348. doi: 10.1111/j.1574-6968.1996.tb08598.x. [DOI] [PubMed] [Google Scholar]
- 17.Shuster C W, Rundell K. Resistance of Salmonella typhimurium mutants to galactose death. J Bacteriol. 1969;100:103–109. doi: 10.1128/jb.100.1.103-109.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinmetz M. Carbohydrate metabolism: pathways, enzymes, genetic regulation, and evolution. In: Sonenshein A, Hoch J, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 157–170. [Google Scholar]
- 19.Thoden J B, Frey P A, Holden H M. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 20.Whatmore A M, Reed R H. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J Gen Microbiol. 1990;136:2521–2526. doi: 10.1099/00221287-136-12-2521. [DOI] [PubMed] [Google Scholar]
- 21.Williams J G, Mason P J. Hybridisation in the analysis of RNA. In: Hames B D, Higgins S J, editors. Nucleic acid hybridisation, a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 139–160. [Google Scholar]
- 22.Yarmolinsky M B, Wiesmeyer H, Kalckar H M, Jordan E. Hereditary defects in galactose metabolism in Escherichia coli mutants. II. Galactose-induced sensitivity. Proc Natl Acad Sci USA. 1959;45:1786–1791. doi: 10.1073/pnas.45.12.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]