Abstract
Cancer cells often metastasize by undergoing an epithelial-mesenchymal transition (EMT). Although abundance of CD8+ T-cells in the tumor microenvironment correlates with improved survival, mesenchymal cancer cells acquire greater resistance to antitumor immunity in some cancers. We hypothesized the EMT modulates the immune response to ovarian cancer. Here we show that cancer cells from infiltrated/inflamed tumors possess more mesenchymal cells, than excluded and desert tumors. We also noted high expression of LGALS3 is associated with EMT in vivo, a finding validated with in vitro EMT models. Dissecting the cellular communications among populations in the tumor revealed that mesenchymal cancer cells in infiltrated tumors communicate through LGALS3 to LAG3 receptor expressed by CD8+ T cells. We found CD8+ T cells express high levels of LAG3, a marker of T cell exhaustion. The results indicate that EMT in ovarian cancer cells promotes interactions between cancer cells and T cells through the LGALS3 - LAG3 axis, which could increase T cell exhaustion in infiltrated tumors, dampening antitumor immunity.
Subject terms: Computational biology and bioinformatics, Cancer, Immunology, Regulatory networks
Introduction
The epithelial-to-mesenchymal transition (EMT) refers to the plastic ability of epithelial cells to undergo a conversion to a mesenchymal state by gradually shedding epithelial features in response to environmental signals1. In tissue microenvironments, cells undergoing EMT exist on a continuum where cells are either in a complete epithelial state, complete mesenchymal cells, or a gradient of intermediate states. In cancer, EMT has been associated with metastatic spread and immunosuppression2–4. In the case of metastasis, cancer cells responsible for invasiveness and migration appear to be at least in a partial EMT (pEMT), if not a fully mesenchymal, state5–7. Mesenchymal and pEMT states also highly contribute to the immunosuppressive burden in the ovarian tumor microenvironment (TME)7–9.
Immune infiltration and inflammation play an important role in tumor development in a variety of cancers10,11. Chronic inflammation established by immune infiltration has been associated with tumor progression, particularly with enhanced metastasis and the EMT process through secreted factors such as transforming growth factor beta 1 (TGF-β1) and tumor necrosis alpha (TNF-α) derived from myeloid cells12,13. Human tumors can be classified according to their level of immune infiltration: an ‘infiltrated-inflamed’ (infiltrated) tumor is characterized by a TME containing tumor infiltrating lymphocytes (TILs) in intraepithelial locations and displaying a cytotoxic transcriptional signature14. This immune phenotype along with the presence of CD8+ cytotoxic T lymphocytes (CTLs) has been associated with better prognosis and overall survival in high-grade serous (HGSOC)15,16. ‘Immune-excluded’ (excluded) tumors are distinguished by TILs found mainly in the stromal portion of the tumor. In ‘immune-desert’ (desert) tumors, immune cells are largely absent from the TME14,17. The differences between excluded and desert tumors may be due to immune exclusion because of the density of the extracellular matrix18 or through a chemorepellent gradient as has been reported for triple-negative breast cancer19.
Even though in ovarian cancer, particularly in HGSOC, immune infiltration by CD8+CD103+ CTLs has been associated with increased progression-free survival, recent findings indicate that a large portion of this population are positive for T-cell immunoglobulin and mucin-domain containing-3 (TIM3)20,21 and Lymphocyte Activation Gene-3 (LAG3)21,22 in epithelial ovarian cancer (EOC), suggesting they are prone to exhaustion and attenuated effector functions. Both TIM3 and LAG3 act as co-inhibitory molecules together with programmed cell death protein-1 (PD1) to dampen CTL mediated antitumor immunity22,23. LAG3 encodes a cell surface protein that is expressed on T-cells, natural killer (NK) cells, plasmacytoid dendritic cells (pDCs) and B cells, and associates with the T-Cell Receptor complex (TCR, CD3) on both CD4+ and CD8+ T cells acting as an immune inhibitory checkpoint. LAG3 binds to Galectin-3 (GAL-3) with strong affinity and chronic LAG3 engagement on CD8+ tumor antigen-specific T cells has been implicated in exhaustion of TILs and reduction in their cytolytic capacity24,25.
EMT in cancer cells has been linked to chemoresistance and immunosuppression in HGSOC. TGF-β/SMAD signaling drives resistance to paclitaxel, BMP9 activates EMT through TGF-β1 and promotes platinum resistance, and expression of lysyl oxidase (LOX) induces EMT through SLUG and TWIST1 and contributes to chemoresistance in activating PI3K/AKT9,26. Recent attempts in breast cancer to elucidate the role of EMT in cancer cells’ ability to facilitate an immunosuppressive TME have shown that mesenchymal cancer cells express low levels of MHC I and are associated with infiltration of regulatory T cells (Treg)27 and resistance to anti-CTLA4 therapy3. In HGSOC, lower abundance of CD8+ TILs is associated with the worst prognosis and a high expression of EMT-related gene signatures in cancer cells28. Despite the interest in how cancer cells affect CD8+ T-cell exhaustion in HGSOC, the role of the EMT has yet to be elucidated.
To better understand the signaling pathways underlying CD8+ T-cell function in HGSOC driven by EMT, we accessed a publicly available single-cell RNA-seq (scRNA-seq) dataset of 16 HGSOC samples29 categorized by their tumor immune phenotypes: infiltrated/inflamed (infiltrated), immune-excluded (excluded), or immune-desert (desert). We first discovered that infiltrated ovarian tumors have a higher EMT signature than either excluded or desert tumors. Pathway analysis revealed that EOC cancer cells in a pEMT or mesenchymal state upregulate pathways related to chemotaxis of immune cells, particularly in infiltrated TMEs. In-depth analysis of cell communication networks of cancer cells signaling to the CD8+ T cell population revealed that a large portion of the communication is facilitated by the mesenchymal cells and is mediated by HLA II molecules and LGALS3 communicating with the LAG3 receptor complex. Our findings suggest that direct signaling between mesenchymal cancer cells and CD8+ T cells trigger LAG3 and other T cell exhaustion pathways, disrupting anti-tumor immunity and supporting tumor development.
Results
Comparing single-cell profiles of distinct immune phenotypes of HGSOC
To assess how EMT contributes to the activity, immunosuppression and/or exhaustion of CD8+ T cells, we began by accessing a scRNA-Seq dataset of 16 human ovarian cancers29. The plan for this data analysis is summarized in Fig. 1a. We chose these datasets due to their inclusion of immune phenotyping metadata confirmed by histology. We assessed for quality-control and clustered every sample individually by merging the respective CD45+, tumor, and stromal fractions of each individual dataset before using our semi-supervised labeling method to identify the individual cell types comprising each tumor sample, as described in the Methods. The tumor samples analyzed are from a single study where each tumor was processed with an identical protocol29,30. Although this protocol may introduce cell type biases, the assumption is that they are consistent among samples.
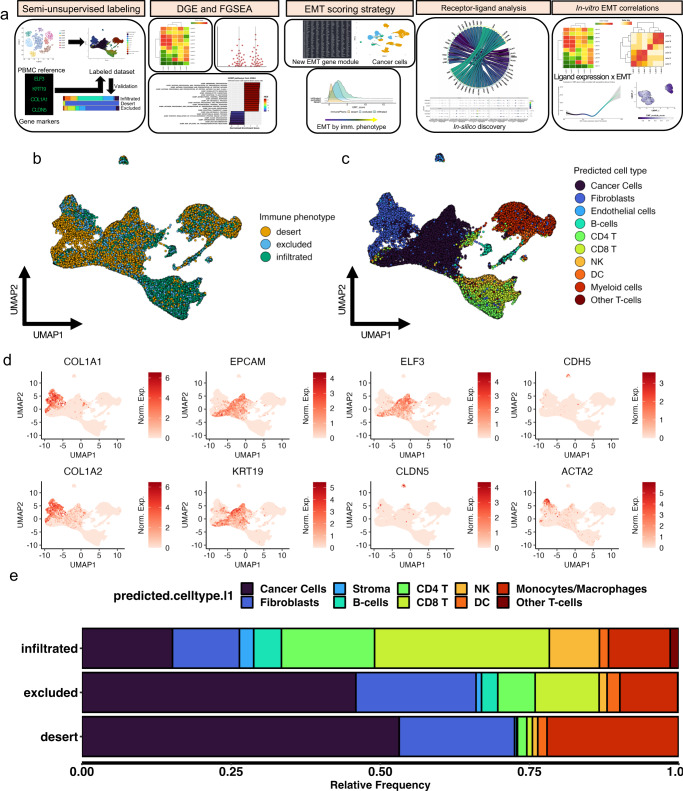
Fig. 1. Identification of cell populations from patient-derived tumor cells in silico confirms greater immune cell presence in infiltrated tumors compared to excluded and desert tumors.
a Overview of analysis and experimental validation procedures. b Uniform manifold and approximation projection (UMAP) of patient derived tumor cells colored by tumor immune phenotype. c Labeled UMAP from patient derived tumor cells colored by cell population. d Series of UMAPs colored by individual gene expression levels in various cell populations from patient derived tumors: KRT19, ELF3, EPCAM for cancer cells; COL1A1, COL1A2 for fibroblasts; CDH5, CLDN5 for endothelial cells; ACTA2 for smooth muscle cells. e Relative frequency of overall cell populations in patient derived tumor samples separated by immune phenotype and colored by cell population.
First, we overlaid the immune phenotypes of the samples in a UMAP to visualize the grouping of all the cells (Fig. 1b). Next, we overlaid the results from our semi-supervised labeling method (Fig. 1c, d) to label the cell types onto the UMAP. While there is some intracellular heterogeneity, most populations group together irrespective of the tumor immune phenotype. This suggests that infiltration status phenotypes minimally contributes to dimensionality reduction and clustering of the cell populations. By examining the population proportion breakdown in the TME by immune phenotype metadata (Fig. 1e, Supplemental Fig. 1), we found that semi-supervised labeling of the 16 tumors matches the originally published findings with respect to immune phenotype, where in infiltrated tumors there are most CD45+ cells relative to cancer cells, fewer CD45+ cells relative to cancer cells in excluded tumors, and fewest CD45+ cells relative to cancer cells in desert tumors. This confirms that our cell labeling method was accurate in correctly identifying CD45+ and CD45- cells.
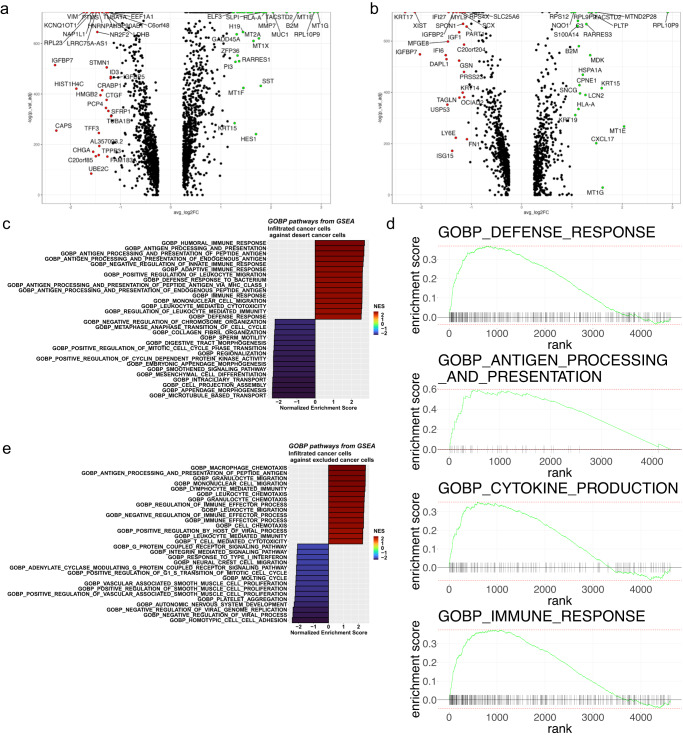
To assess the role of cancer cells in contributing to the unique composition of each immune phenotype, we performed differential gene expression analysis to identify the top and bottom most differentially regulated genes between cancer cells from the infiltrated TME compared to cancer cells from desert TME (Fig. 2a) and excluded TME (Fig. 2b). Cancer cells from infiltrated TMEs differentially express genes related to immune regulation within the context of carcinogenesis, such as: B2M31, SLPI32, HLA-A33, ELF334, MUC135, MDK36, CXCL1737 and others. Based on upregulation of these immune regulatory genes, we performed GSEA first comparing cancer cells from the infiltrated to the desert TMEs. We discovered that many biological pathways (GOBP) related to immune regulation were upregulated in cancer cells from infiltrated TMEs (Fig. 2c, d). Among the main findings, some pathways were related to chemotaxis and enrichment for processes related to antigen presentation and cytokine activity, suggesting that cancer cells in infiltrated tumors actively participate in modulating the immune response and shape the tumor immune phenotype. Similar results were obtained when investigating the pathway enrichment analyses comparing cancer cells from infiltrated versus excluded TMEs (Fig. 2e) and excluded to desert TMEs (Supplemental Fig. 2) suggesting that pathways related to immune regulation could be “turned on” either as a response to immune infiltration or as a precursor to the arrival of immune cells promoted by cancer cells themselves. These results may be indicative of EMT-related cell-cell signaling driving chemotaxis of various CD45+ cells from different populations to the TME.
Fig. 2. Cancer cells from infiltrated tumors upregulate signaling pathways related to immune cell chemotaxis, immune modulation, and antigen presentation compared to cancer cells from excluded or desert tumors.
a Volcano plots of differentially expressed genes (DGE) between cancer cells of desert and infiltrated tumors (left) and excluded and infiltrated tumors (right). b Enrichment plot of infiltrated GOBP terms from GSEA of a selection of significant (*p-adjusted <0.05) pathways between cancer cells of infiltrated (red) and desert tumors (blue). c Enrichment plot of infiltrated GOBP terms from GSEA of a selection of significant (*p-adjusted <0.05) pathways between cancer cells of infiltrated (red) and excluded tumors (blue). d GSEA enrichment ranks of significantly upregulated (*p-adjusted <0.05) pathways related to immune regulation in cancer cells from infiltrated tumors. e Enrichment plot of infiltrated GOBP terms from GSEA of a selection of significant (*p-adjusted <0.05) pathways between cancer cells of infiltrated (red) and desert tumors (blue).
Cancer cells from infiltrated tumors are more mesenchymal compared to cancer cells from other immune phenotypes
Our discovery that cancer cells in infiltrated TMEs activate immune regulatory pathways prompted us to assess the EMT status of cancer cells given that cells in partial or fully mesenchymal states are known for their immunoregulatory capabilities38–40. Hornburg et al., (2021)29 showed that desert TMEs contain malignant cells with higher expression of EMT-associated genes using the MSigDB Hallmark gene set41. Our gene set scoring using the subset of 58 Hallmark genes they found associated with the desert tumors similarly reproduced their findings (Supplemental Fig. 3a, b). However, when we performed gene set scoring on the cells with the complete 200-gene Hallmark gene set, no difference was found between tumor immune phenotypes (Supplemental Fig. 3c, d). By applying GSEA enrichment analysis of the three different EMT gene modules, we also found their subset enriched in DEGs from desert derived cancer cells, whereas applying both the full Hallmark EMT gene set and our cancer-specific EMT gene signature yielded negative enrichment scores (Supplemental Fig. 3e; Supplemental Data 1). Based on this analysis, it is unclear if scores calculated using the Hallmark EMT gene set are sufficient to indicate a mesenchymal phenotype. Signature activity among malignant cells from different tumor immune phenotypes revealed some key differences in the cancer cell population (Fig. 3a). The score distribution was then evaluated across the malignant cells derived from the 16 tumors (Fig. 3b) revealing a notably higher EMT activity score of infiltrated tumors compared to excluded and desert tumors (Fig. 3c, d; Supplemental Fig. 4). This result was surprising as desert TMEs have been previously linked to higher EMT signatures in the cancer cells of HGSOC29,42,43. Consistent with our finding, individual cancer-specific EMT signature genes such as PDIA3, HLA-A, BCAM, B2M, LGALS3BP, and HLA-C were highly expressed in cancer cells from infiltrated and excluded TMEs (Supplemental Fig. 5). Importantly, none of the tumor samples was found as an obvious outlier for cancer-specific EMT signature scores (Supplemental Fig. 4). Interestingly, cancer cells from infiltrated TMEs appeared to be more heterogeneous in their EMT scoring and skewed towards mesenchymal phenotypes.
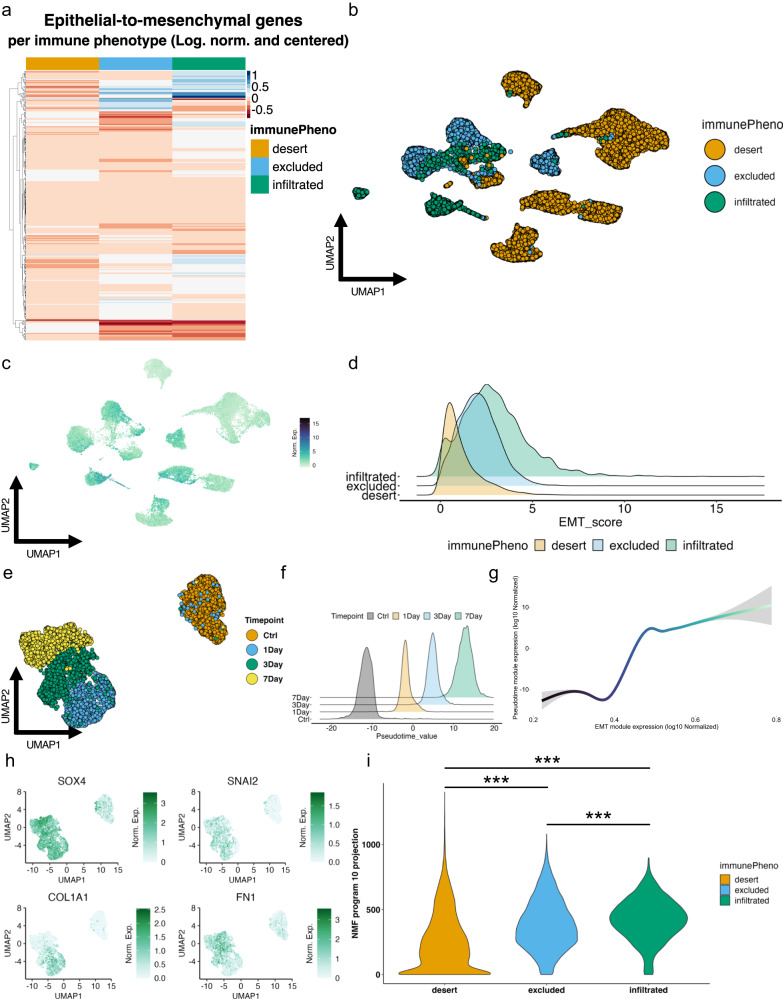
Fig. 3. Cancer cells from infiltrated tumors have higher cancer-specific EMT signature scores compared to cancer cells from excluded and desert tumors and are more progressed on the EMT axis.
a Quantile scaled heatmap of cancer-specific EMT signature genes arrayed by immune phenotype. b UMAP clustering of cancer cell compartment of scRNA-Seq dataset of 16 HGSOC tumors colored by the immune phenotype of the specific source tumor. c Enrichment UMAP plot for the cancer-specific EMT signature scores in the clustered cancer cell compartment. d Ridge plot of individual cancer-specific EMT signature module scores in the cancer cell compartment of each tumor’s immune phenotype. e UMAP clustering of OVCA420 cells treated with TGF-β1 for 4 different timepoints: Ctrl (no treatment), 1-day, 3-day, and 7-day. f Pseudotime value density for each individual treatment timepoint in OVCA420 cells treated with TGF-β1. g Generalized additive model (GAM) fitted line correlating pseudotime values and cancer-specific EMT signature scores. h Enrichment UMAP plot of classical EMT markers Sox4, Snai2, Col1a1, and FN1 in OVCA420 cells treated with TGF-β1. (i) Violin plot of NMF program 10 ‘h’ coefficient values for cancer cells in vivo in 16 HGSOC arrayed by immune phenotype. A one-way ANOVA test with Tukey’s multiple comparisons performed on program 10 ‘h’ coefficient values reveals significant differences among all immune subtypes (***p < 0.001).
To validate the cancer-specific EMT signature’s fidelity in determining EMT, we leveraged an EOC cell line that we have previously shown to undergo transcriptional changes associated with EMT when treated with TGF-β144. OVCA420 cells were treated with TGF-β1 to induce EMT and the cells were collected at three different time-points to perform scRNA-seq (Fig. 3e). We modeled a continuous pseudotemporal EMT trajectory from the data (Fig. 3f) and found that scores from the EMT signature increase throughout EMT progression as expected, alongside pseudotime values (Fig. 3g). Furthermore, the OVCA420 in vitro model of EMT involved activation of classical EMT genes such SOX4, SNAI2, COL1A1, and FN1 (Fig. 3h). Finally, we leveraged non-negative matrix factorization (NMF), a technique which enables the investigation of coordinated gene expression sources of heterogeneity in the data in a semi-supervised manner with machine learning. We therefore generated a list of 10 possible cell state programs (Supplemental Fig. 6) representing the inherent expression of unique sub-groups of cells throughout the time course (Fig. 3i). Of all the NMF-derived programs, programs 2 and 10 were most consistent with an EMT program. Program 10 (Supplemental Fig. 6) enriches the cell cluster located at the final timepoint of 7-days after TGF-β1 treatment, where we expect a ‘maximal’ mesenchymal state. To further explore program 10, we took the top 500 most-weighted genes in the program and ran an Enrichr analysis (Supplemental Fig. 7a). We discovered that program 10 enriches for Hallmark gene modules related to immunoregulatory pathways such as ‘TNF-alpha signaling via NF-kB’ (***p < 0.001), and others to ‘p53 pathway’ (***p < 0.001), and ‘mTORC1 signaling’ (***p < 0.001) while also enriching the Hallmark EMT gene module. When we performed the same analysis for program 2 (Supplemental Fig. 6), we found reduced enrichment for these same pathways (Supplemental Fig. 7b). These results suggest that a distinct immunoregulatory program is activated upon TGF-β1 induced-EMT and may not be well represented by the Hallmark EMT gene set.
Analysis of the enrichment of cancer cells in the dataset of 16 HGSOC for genes encapsulated in our in vitro-derived NMF program 10 (Supplemental Data 2; Fig. 3i) revealed a greater overall presence of mesenchymal cells in the cancer cell population derived from the infiltrated tumors, followed by cancer cells from excluded and then desert tumors. Scoring the tumors with this defined NMF signature showed enrichment in infiltrated tumors consistent with the distribution of scores from the cancer-specific EMT signature. These findings suggest that program 10 identifies genes related to an immunoregulatory program activated by EMT in HGSOC cancer cells, that is most prevalent in mesenchymal cancer cells from infiltrated tumors. We therefore sought next to determine whether the EMT program could have a direct role in shaping the tumor immune phenotypes.
EMT is linked to CD8+ T cell activity and exhaustion through the LAG3 receptor
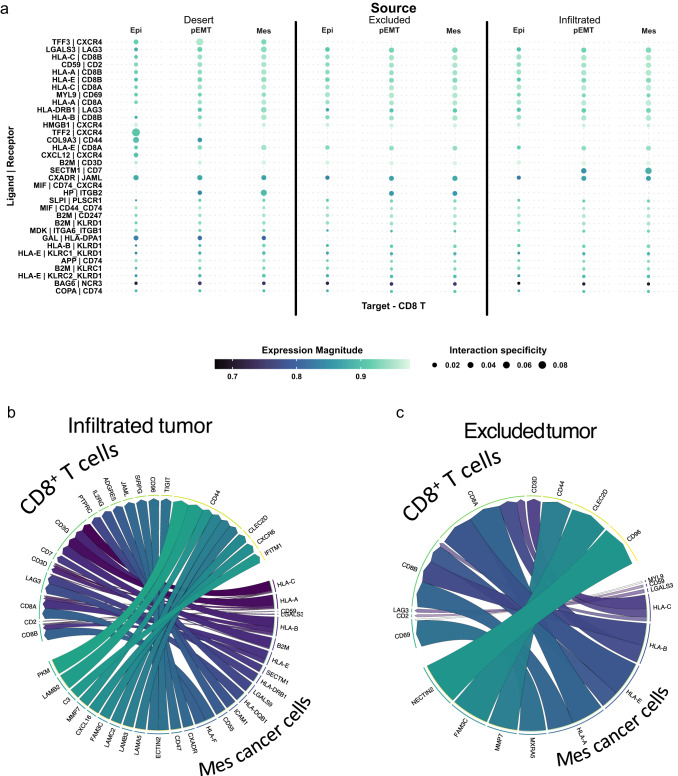
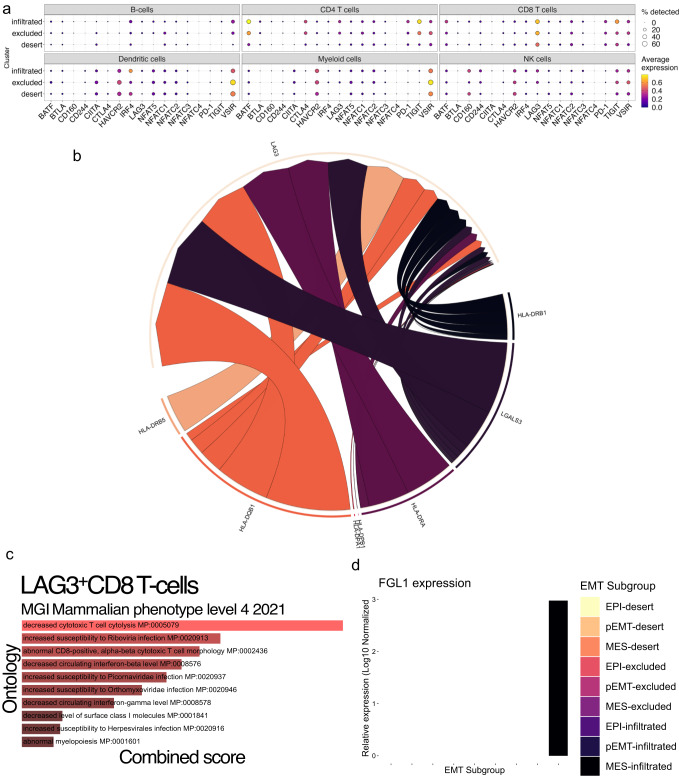
Since our findings indicate that EMT in cancer cells correlates with the presence of immune cells (Fig. 3d; Supplemental Fig. 5a), we dissected the inter-cellular communications in each tumor immune phenotype by evaluating the unique expression of ligands in malignant cells and their cognate receptors in CD8+ T cells. CD8+ T cells are very abundant in infiltrated tumors (Fig. 1e, Supplemental Fig. 1), and we were interested in assessing whether EMT influences antitumor immunity in infiltrated and excluded cancers. The ligand-receptor interactome analysis of communication between cancer cells from infiltrated and excluded tumors to CD8+ T-cell receptors revealed that the majority of interactions are borne from mesenchymal cancer cells towards CD8+ T cells in both immune phenotypes (Supplemental Fig. 8a, b). We then assessed the top cancer cell to CD8+ T cell receptor-ligand interactions with the former arrayed by EMT status (Fig. 4a). Heterogeneity in signaling between different immune phenotypes is evident, where receptor-ligand pairs such as SECTM1 - CD7 appear to be unique to infiltrated tumors (Fig. 4a). The top five ligands targeting CD8+ T-cells by mesenchymal cancer cells from infiltrated tumors were HLA-C, HLA-A, CD59, LGALS3, and B2M (Fig. 4a, b). We found similar gene patterns in mesenchymal cells’ ligands in excluded tumors with CD59, LGALS3, HLA-C, HLA-B, and HLA-E (Fig. 4a, c). The finding of classic MHC I molecule expression by cancer cells was not surprising as HLA I expressing tumors have been previously linked to TIL frequency, with TILs eventually inducing HLA allotype selection, consequently enabling cancer cells to evade antitumoral immunity45,46. Nonetheless, we were expecting to find other dominant inhibitory ligands such as PD-L1 promoting immunosuppression, but LGALS3 (Galectin-3, Gal-3) emerged as a top ligand, highlighting its potential role in the EMT driven immunosuppression in EOC9,47–50.
Fig. 4. Most ligands targeting CD8+ T-cells in the TME originate from the mesenchymal cancer cells.
a Cell-cell communication analysis of top-most ligand-receptor pairs from cancer cells from 16 HGSOC samples arrayed by infiltration phenotype of tumors and EMT to CD8+ T-cells. ‘Expression magnitude’ is a measure of expression levels of a certain interaction. ‘Specificity’ is a measure of the specificity of interactions across all cell types. Interactions are ordered by p-value where top-most receptor-ligand pair has lowest p-value. Circle plot of ligands originating from mesenchymal cancer cells in infiltrated tumors (b) and excluded tumors (c), and the receptors targeted on recipient CD8+ T-cells. Order and color of ligands corresponds to p-value aggregate rank reflecting specificity of interactions (most specific – purple color, clockwise). The number of tracks per ligand corresponds to the number of overall interactions the ligand has with different receptors. Track width corresponds to p-value aggregate rank within a ligand if the ligand has multiple receptor targets.
Further investigation of the specific receptors targeted by LGALS3 showed that the LAG3 complex on CD8+ T cells is the primary recipient of LGALS3, as well as MHC II molecules, matching previous literature findings51–53 (Fig. 4a−c; Supplemental Fig. 8c, d). In fact, LAG3 appears to be one of the most targeted receptor complexes by rank in both infiltrated and excluded tumors, suggesting the possible exhaustion of these cells. The observation linking the LGALS3-LAG3 interaction to EMT prompted us to further investigate CD8+ T cell exhaustion as a possible consequence of the EMT process in primary HGSOC.
Common exhaustion markers or checkpoint inhibitors of CD8+ T-cells include BTLA, CD160, CD244, CTLA4, HAVCR2, LAG3, TIGIT, and PD1, the NFAT family (NFAT5, NFATC1-4) of transcription factors involved in promoting T cell exhaustion, and IRF4, BATF, VSIR, and CIITA, a master regulator of MHC II expression which is known to interact with LAG3. Of these markers, LAG3 and TIGIT are most active in CD8+ T cells in infiltrated tumors (Fig. 5a). In fact, high LAG3 and TIGIT expression in infiltrated tumors could be indicative of a specific inflammatory milieu triggering LAG3 upon CD8+ T cell infiltration and sensitizing these cells to exhaustion signals from mesenchymal cancer cells. A breakdown of all the ligands targeting the LAG3 receptor on CD8+ T cells, regardless of cancer cell EMT status or tumor immune phenotype, showed that most ligands for LAG3 are HLA II molecules, with LGALS3 being the only non-MHC related ligand (Fig. 5b). Despite HLA II ligands making up the bulk of interactions between cancer cells and LAG3 on CD8+ T-cells, we did not find any associations between EMT and HLA II expression (data not shown). Additionally, NECTINs 2, 3, and 4 are the major interacting partner with TIGIT between cancer cells and CD8+ T-cells, suggesting another possible vector for T cell exhaustion by mesenchymal cancer cells.
Fig. 5. Most CD8+ T-cells in HGSOC express LAG3, a marker of T cell exhaustion.
a Dot plot showing expression levels and frequency of common markers of T-cell exhaustion in the entire CD45+ cell compartment. b Circle plot of CD8+ T-cell LAG3 receptor complex and all the ligands that target it. c Analysis of enriched pathways from a DEG analysis of LAG3+ CD8 T-cells compared to LAG3- T-cells in the Mouse Genome Informatics (MGI) database. d Log-normalized expression of FGL1 in the EMT-delineated compartment of cancer cells from each tumor immune phenotype.
GSEA of the DEGs between LAG3+ and LAG3- CD8 T cells revealed enrichment for molecular pathways related to decreased cytotoxic T cell cytolysis (MP:0005079) and abnormal CD8+ T cell morphology (MP:0002436) (Fig. 5c), suggesting that there is a significant contingent of CD8+ T cells in infiltrated tumors that are cytolytically non-functional as a result of LAG3-related signaling. Fibrinogen-like protein 1 (FGL1) has been shown recently to be a ligand of LAG3, also potentially influencing T cell exhaustion54. Interestingly, we found FGL1 is expressed only by mesenchymal cancer cells in infiltrated tumors (Fig. 5d). As our findings show that there is a significant proportion of LAG3+ CD8+ T-cell targeted by ligand LGALS3+ originating from mesenchymal cancer cells, we sought to further explore the link between LGALS3 and EMT to better elucidate how EMT could modulate CD8+ T-cell exhaustion.
LGALS3 is linked with EMT in both in vivo and in vitro contexts
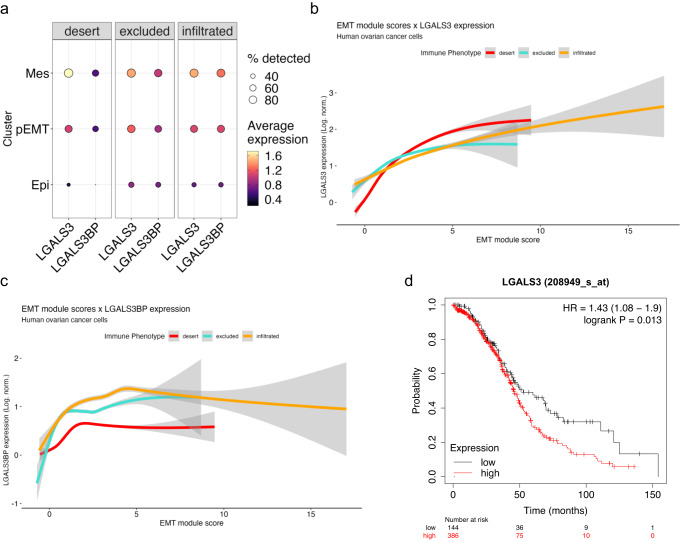
To determine whether LGALS3 has any association with EMT, we examined its expression across cancer cells arrayed by both tumor immune phenotype and EMT status. We also assessed LGALS3 Binding Protein (LGALS3BP) as it is known to synergize with LGALS3 in certain contexts55. Both LGALS3 and LGALS3BP appear to have higher expression in mesenchymal cancer cells, regardless of the tumor immune phenotype (Fig. 6a). We fitted expression of LGALS3 (Fig. 6b) and LGALS3BP (Fig. 6c) against EMT score and arrayed by tumor immune phenotype to find that LGALS3 expression increases alongside the cancer specific EMT signature expression in every tumor immune phenotype and every delineated EMT phase (i.e., EPI, pEMT, and MES). By contrast, LGALS3BP increases most in earlier, epithelial phases in all tumor immune phenotypes, and then plateaus, suggesting it is more important in earlier phases of the EM program. To determine whether LGALS3 correlates with survival in HGSOC, we analyzed the TCGA data and found that low LGALS3 expression in bulk RNA-Seq of HGSOC is associated with longer survival (*p = 0.013) (Fig. 6d). These findings suggest that LGALS3 expression correlates with EMT in cancer cells and contributes to reduced survival of ovarian cancer patients.
Fig. 6. LGALS3 and LGALS3BP expression in cancer cells correlates with EMT in vivo.
a Dot plot showing LGALS3 and LGALS3BP in the cancer cell compartment arrayed by EMT status and the percentage of cells that express these genes in each immune phenotype. Log-normalized LGALS3 (b) and LGALS3BP (c) expression fitted via generalized linear model against cancer-specific EMT signature scores in cancer cells, segregated by tumor immune phenotype. d Kaplan−Meier curve comparing patient overall survival in LGALS3-high and LGALS3-low expressing ovarian tumors from TCGA data.
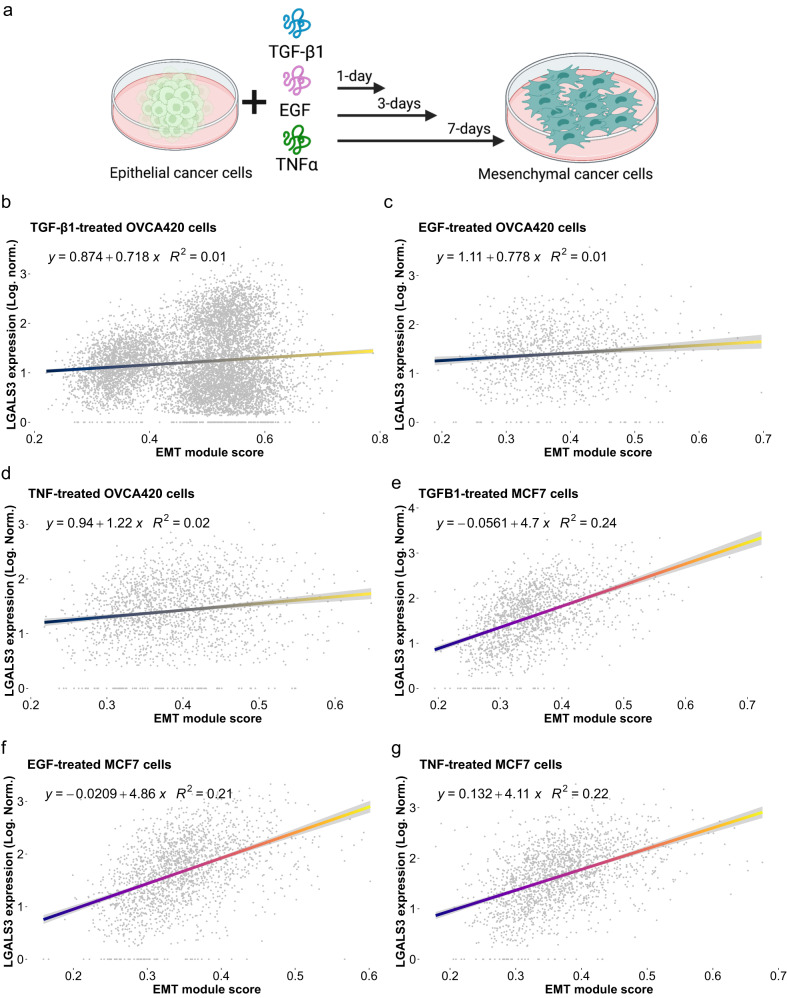
To elucidate if LGALS3 is associated with the EMT in other model systems and under different conditions, we referred to our TGF-β1 treated OVCA420 cancer cells and correlated expression of LGALS3 with the cancer specific EMT signature scores (Fig. 7a). We fitted a generalized linear model and noted a modest positive association (R2 = 0.01) between LGALS3 and cancer specific EMT signature scores (Fig. 7b). We noted that it appeared as though there are two populations of mesenchymal cells where LGALS3 is expressed in one but not the other. To determine if LGALS3 is consistently implicated in EMT rather than simply a consequence of TGF-β1 treatment, we accessed our previously published data44 where multiple cell lines of different cancer types (ovarian, OVCA420; breast, MCF7; prostate, DU145; and lung, A549) were each treated with EMT inducers (TGF-β1, TNFα, EGF). While these proteins were chosen for their ability to induce EMT through different receptors and pathways, in certain cancers they can be secreted by immune cells, such as TNFα by CD8+ T cells56, thus mimicking the immune response to cancer cells. We found a similar positive correlation between LGALS3 and cancer-specific EMT signature scores in OVCA420 cells treated with either EGF (R2 = 0.01) or TNF-α (R2 = 0.02) (Fig. 7c, d). Other cancer cell types also showed positive correlations: MCF7 cells (TGF-β1, R2 = 0.24; TNF-α, R2 = 0.21, EGF, R2 = 0.22) (Fig. 7e−g), DU145 (TGF-β1, R2 = 0.1; TNF-α, R2 = 0.11, EGF, R2 = 0.11) (Supplemental Fig. 9a−c), and A549 (TGF-β1, R2 = 0.05; TNF-α, R2 = 0.1, EGF, R2 = 0.12) (Supplemental Fig. 9d−f), with breast cancer cells showing the strongest correlations between LGALS3 expression and EMT. Taken together, our findings indicate that LGALS3 expression is correlated with the EMT of cancer cells in vivo and in vitro, and that EMT drives expression of LGALS3 in HGSOC cells. Additionally, LGALS3 expression is a “core” gene in the EMT program that is activated under a variety of EMT inducers through TGF-β1-, or TNF-α-, or EGF-induced signaling. Mesenchymal cancer cells in turn exert inhibitory antitumoral signaling on CD8+ T cell promoting a dysfunctional cytotoxic state and exhaustion by binding to the LAG3 receptor complex.
Fig. 7. LGALS3 is directly correlated with the EMT in vitro in ovarian and breast cancer cell lines.
a Schematic of cell line treatment (b−d) Log-normalized expression of LGALS3 as generalized linear model against cancer-specific EMT signature scores in OVCA420 ovarian cancer cells treated with TGF-β1 (b), TNFα (c), and EGF (d). Log-normalized expression of LGALS3 GLM-correlated against cancer-specific EMT signature scores in breast cancer MCF7 cells treated with TGF-β1 (e), TNFα (f), and EGF (g).
GSK3 and Aurora-A kinase inhibitors attenuate LGALS3 expression
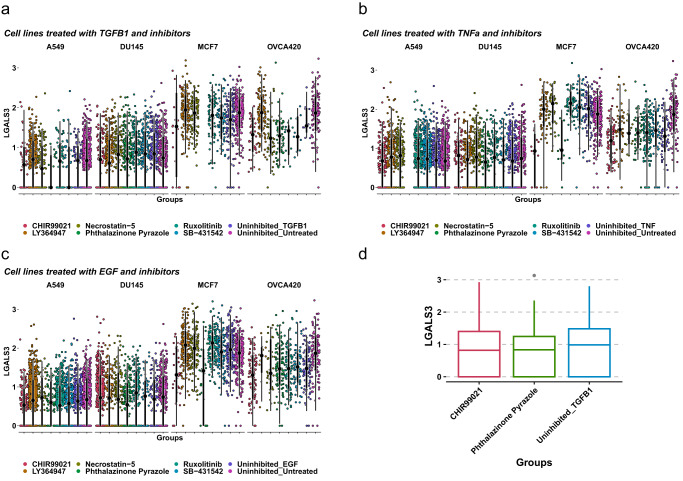
To determine the possible pathways linking EMT and LGALS3 expression we referred to a previously published kinase inhibitor screen that elucidated some of the signaling dependencies of the EMT44. Particularly, the modularity of the EMT spectrum was revealed to be a core part of the process where some TGF-β1R-independent kinases attenuated the EMT despite treatment with various inducers (TGF-β1, TNFα, and EGF). In the original screen it was discovered that both RIP1 kinase inhibitor Necrostatin-5 and TGF-β1R inhibitor LY364947 abrogated EMT in all treatment conditions. When we explored this screen further (Fig. 8a−c), we noted that two kinase inhibitors produced the most consistent reduction in LGALS3 expression across cell lines and treatments: CHIR99021, a GSK3 kinase inhibitor and phthalazinone pyrazole, an Aurora kinase A inhibitor (Fig. 8d), suggesting LGALS3 expression is potentially dependent on activation of the Wnt and NF-kB pathways, or the β-catenin pathway during EMT. Finally, when we explored the effects of these two kinase inhibitors on pseudotime values as a measure of EMT and LGALS3 expression, we found that neither CHIR99021 nor phthalazinone pyrazole appear to attenuate the EMT, suggesting that blocking LGALS3 upregulation is not sufficient to prevent EMT (Supplemental Fig. 10).
Fig. 8. Kinase inhibitor screen shows GSK3 and Aurora-A kinase inhibitors attenuate EMT and LGALS3 expression.
Log-normalized expression of LGALS3 in A549, DU145, MCF7, or OVCA420 cell lines either treated either with EMT inducers TGF-β1 (a), TNFα (b), EGF (c) or EMT inducers and kinase inhibitors: RIP1 kinase inhibitor Necrostatin5, TGFβR1 kinase inhibitor LY364947, JAK1/2 kinase inhibitor Ruxolitinib, TGFβ/ALK kinase inhibitor SB-431542, GSK3 kinase inhibitor CHIR99021, or TGF-β1 and Aurora kinase A inhibitor phthalazinone pyrazole. d LGALS3 expression in OVCA420 cells treated either with TGF-β1, TGF-β1 and GSK3 kinase inhibitor CHIR99021, or TGF-β1 and Aurora kinase A inhibitor phthalazinone pyrazole.
Discussion
In this study, we show that the EMT, a complex cellular process underpinning metastasis, associates with LGALS3 expression, which may act as ligands for LAG3 in CD8+ T cells, promoting T cell exhaustion and dampening antitumor immunity. Additionally, we show that infiltrated TMEs in HGSOC have cancer cells that are, on average, more mesenchymal than those found in the excluded or desert phenotypes, with the potential implication that a greater chance for metastasis and immunosuppression is possible in those tumors. Given that infiltrated and excluded tumors have been correlated with better survival among HGSOC patients compared to desert tumors, and the presence of CD8+ T cells in these tumors, there is great potential for treating these patients with LAG3 checkpoint blockade therapy combined with other immune checkpoint inhibitors as it has been demonstrated in mouse IE9mp1 EOC model by Huang et al. (2015)22. Recently, the US Food and Drug administration (FDA) approved second-generation checkpoint inhibitor ‘Opdualag’, an anti-LAG3 and anti-PD1 combination drug that targets metastatic or unresectable melanoma57. Additionally, the development of new peptides, such as C25, that block LAG3 binding to MHC II has been proven to activate CD8+ T cells52,58. LAG3 blockade therapies have been shown to have therapeutic benefit for patients with chronic lymphocytic leukemia, melanoma, and pancreatic adenocarcinoma59,60. In BRCA-mutated HGSOC patients, LAG3 was positively correlated with PD-L1; however, combination immunotherapies in human HGSOC to block the activity of both inhibitory checkpoints were found to have negligible efficacy, suggesting other underlying mechanisms governing the immunosuppressive TME of HGSOC in the context of LAG3 expression47.
Recent scRNA-Seq investigations on BRCA1/2 mutated HGSOCs have showed a potential link between EMT and CD8+ T-cell exhaustion61. As well, EMT and T cells found in malignant ascites from EOC express high levels of LAG3 and PD-162,63. This latter observation is of particular interest as it has been previously suggested that EOC ascites contain a significant mesenchymal cancer cell population64,65. As well, mesenchymal and pEMT cancer cell states have been linked to higher PD-L1 expression in breast cancer66 suggesting that even a pEMT state is correlated with immunosuppression. Similar relationships between LGALS3 and T cell exhaustion in TGF-β1 induced fibrotic disease67 have been uncovered, where LGALS3 inhibitors68,69 have proven to be effective in treating the disease. In cancer contexts where TGF-β1 plays a major role in initiation and maintenance of EMT, patients may also benefit from LGALS3 inhibitors. For example, LGALS3 has a strong proinflammatory role when expressed by fibroblasts70, eliciting secretion of IL-6, CXCL8, CCL2, and CCL5 all of which are factors that play a role in carcinogenesis and immunosuppression. Additionally, anti-TIGIT therapy has shown mixed success in treating a variety of solid tumors, including ovarian cancer71 with other clinical trials demonstrating better clinical benefits in tumors with high TIGIT expression72. Curiously, CD8+ T cells infiltrated ovarian tumors have high TIGIT expression as shown in this paper (Fig. 5), which may suggest ovarian tumors are a potential target for combination anti-LAG3 and anti-TIGIT therapy. Moreover, combination therapies including anti-TIGIT and anti-PD-L1 synergize to enhance cytolytic CD8+ T cell activity73,74 which could be used to target ovarian tumors of the infiltrated subtype to alleviate exhaustion and immunosuppression.
Galectin-3 (Gal-3/LGALS3), encoded by the LGALS3 gene, is a lectin that can be both expressed on the cell surface75 and secreted76, and is expressed by the majority of human cells. It exhibits several immune-regulatory functions such as reducing the affinity of TCR for its cognate MHC I-peptide ligand by sequestering the TCR from its CD8+ co-receptor77, causing apoptosis78, and internalization of the TCR79, leading to decreased interferon-gamma (IFN-γ) production upon LAG3 engagement on CD8+ T cells51. LGALS3 has been linked to poor prognosis in EOC80 with the assertion that it may be activating the Wnt/β-Catenin pathway to effect cancer stemness mechanisms81. Additionally, overexpression of Galectin-1 (LGALS1), a protein similar to LGALS3, promotes EMT in fibroblasts through TGF-β signaling pathways82. LGALS3 has also been linked to the EMT previously83 and has been suggested as a T cell-directed immunotherapy to increase efficacy of current immune checkpoint inhibitors. Interestingly, some of our in vitro results demonstrate there could be two different EMT trajectories where LGALS3 is expressed in one but not the other, suggesting that the EMT can lead to heterogenous populations of mesenchymal cells.
The potential to rescue CD8 + T-cells from exhaustion has been shown, notably in studies where anti-PD1 therapy was shown to improve the function of exhausted tumor infiltrating CD8+ T cells in ovarian cancer84. Whether targeting LGALS3-LAG3 axis has similar potential to rescue CD8 + T cell function alone or in combination with other immunotherapies such as anti-PD1, anti-CTLA4, and anti-TIGIT, requires further investigation. The LGALS3 inhibitor GB0139 has shown promise in acute lung injury where its mechanism of action involves reducing IL-6, TNF-α, and MIP-1α69, and thus may also prove efficacious in ovarian cancer where TNF-α plays a role in EMT initiation and maintenance. Galectin-3C, a dominant-negative inhibitor of LGALS3, reduces the metastatic potential of ovarian cancer either in combination with Paclitaxel or alone85.
It should be noted that the suggestion to target infiltrated ovarian tumors for checkpoint therapy is based on our interpretation of data from primary tumors. Indeed, the immune environment in the ascites or metastases may be different where CD8 + T cells may not be exhausted and could clear cancer cells, thus reducing metastatic spread and contributing to the positive survival prognosis of infiltrated tumors. In these cases, checkpoint therapy may be less effective depending on the TME but could still assist the immune system in clearing the primary tumor site.
In our screen of various kinase inhibitors, we found the highly selective and potent GSK3 inhibitor CHIR99021 reduced expression levels of LGALS3 while not affecting EMT signature scores, suggesting it may be suitable for therapeutic investigation due to its possible specificity to LGALS3. GSK3 inhibition or downregulation can potentiate the cytotoxicity of CD8 + T cells against lymphoma cells86, gastric cancer cells87, and melanoma cells, with the latter also showing a blockage of LAG3 because of GSK3 targeting by small molecule inhibitors88.
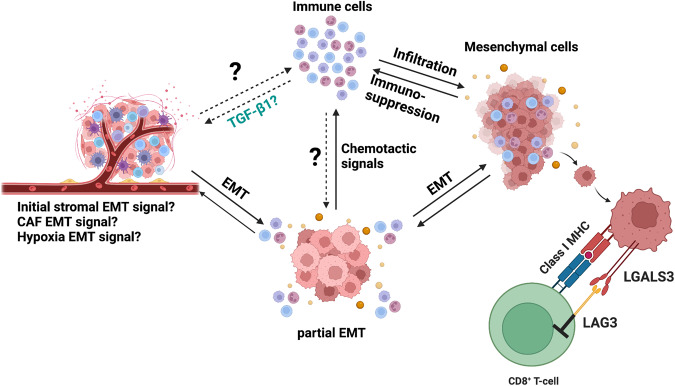
A surprising finding in this study is the correlation between EMT and tumor immune cell infiltration, since previous studies have reported greater EMT in desert tumors. Certain immune cells, such as CD4+ CD25+ Treg cells, tumor-associated macrophages, and myeloid-derived suppressor cells (MDSCs) can induce EMT in cancer cells8 and our study shows upregulation of chemotactic signals during EMT. Consequently, we propose the existence of a positive feedback loop between immune cells and cancer cells, based on their EMT status. In this scenario, initial signals favoring EMT are propagated through the TME, perhaps from the stroma and fibroblasts89–91, or the result of hypoxic conditions92–94. Cancer cells receiving those signals undergo EMT and upregulate pathways related to leucocyte and lymphoid chemotaxis by secreting relevant interleukins and chemokines, as predicted by our findings. Tumor infiltration by activated immune cells promote secretion of factors such as TGF-β1, TNF-α, and ADAM17 intensifying EMT signals and further driving EMT progression in the cancer cell population, resulting in tumor-promoting positive feedback loop. This would ultimately lead to T cell exhaustion through elevated LAG3 stimulation, among other coinhibitory markers, such as TIGIT (Fig. 9). The validity of this feedback loop hypothesis warrants further investigation, for example, by assessing the tumor immune cell composition in mouse models of ovarian cancer that express an inducible EMT signal, such as SNAIL.
Fig. 9. Schematic of hypothesized positive feedback loop between immune cells and cancer cells leading to metastasis and immunosuppression.
In cancer, mesenchymal cancer cells have been shown to be associated with immunosuppression and exhaustion of T-cells in the TME. In our paper, we demonstrate one way in which mesenchymal cells may exhaust CD8+ T-cells in the TME of epithelial ovarian cancer. Additionally, we show there is an association between infiltration of immune cells, particularly T-cells, and EMT. It is currently unknown whether cancer cells instigate the infiltration of T-cells after undergoing EMT, or whether the initial infiltration of T-cells and other immune cells triggers EMT in cancer cells. We propose this model as a potential future direction, where the relationship between immune cell infiltration and EMT can be further investigated.
Our results do not eliminate the possibility that active CD8 + T cells select for survival of pro-exhaustion mesenchymal cells by successfully attacking the less immunosuppressive epithelial cells. However, this still leaves a question about the origin of signals that promote EMT, whether they originate from another population of cells such as fibroblasts secreting TGF-β1. Our results demonstrate that while desert tumors have a large fibroblast population, there are fewer mesenchymal cancer cells in them than infiltrated tumors, further suggesting that EMT signals likely originate from a different cell population, such as immune cells. Therefore, even if CD8 + T cells and other immune cells select for mesenchymal cells, the trigger for EMT may also originate in those cells.
Our results indicating that there are more mesenchymal cells in infiltrated TMEs as opposed to desert TMEs likely differ from Hornburg et al. 29 for a few reasons. First, we used a cancer-specific EMT signature that can capture the intra-tumoral heterogeneity of the epithelial/mesenchymal program and states. The Hallmark EMT gene set, when combined with classical EMT markers (i.e., SNAI1/2, ZEB1/2, TWIST1) captures mostly pEMT states95 and is based on founder gene sets, some of which are not from a cancer context41,96. Second, the Hallmark EMT gene set enriches in stromal compartments97 and also in cancer-associated fibroblasts95 suggesting it may be poorly optimized to capture malignant mesenchymal cells at all, at least on a single-cell RNA level. Only 58 out of 200 genes in the Hallmark EMT gene set were enriched in Hornburg et al.’s analysis of desert tumors and were on the cusp of significance. Third, our scoring strategy to compute EMT scores differs from their enrichment analysis in that our strategy provides valuable single-cell level scoring compared to bulk sample averages. Pseudo-bulking scoring methods have difficulty accounting for cell-to-cell variability98 which could impact EMT calculations where different cell states exist even within the same cell type. We believe that as the cancer-specific EMT signature was derived from scRNA-Seq datasets, it may be best used in single-cell contexts using analysis algorithms best-suited to the technology.
With single-cell sequencing technologies still rapidly improving in both capability and fidelity, there is tremendous potential to expand the study of tumor interactomes either through advances in technology or improved algorithm design. For example, technologies such as CITE-Seq can be leveraged to supplement genomic sequencing data with cell surface-level protein expression data combined with established receptor-ligand algorithms to generate a clearer picture of the interactome of the TME99. Future exploration of the interactome that we have generated could reveal novel interactions between cancer cells and other immune cell types, or further delve into subtypes of T cells such as Treg cells, to paint a more complete picture of the receptor-ligand interactions in the TME of HGSOC. To conclude, ovarian mesenchymal cancer cells suppress CD8+ T cell activity through the pro-exhaustion LAG3-LGALS3 pathway. There is a therapeutic opportunity to target HGSOC that are already infiltrated by CD8+ T cells and relieve them of barriers that dampen antitumoral activity, such as T cell exhaustion. Other than targeting the LAG3-LGALS3 pathway itself, it may be possible to attenuate the EMT to prevent pathway activity specifically and remove a possible source of immunosuppression.
Methods
Cell culture
The human ovarian cancer cell line OVCA420 was kindly provided by Dr. Gordon Mills. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/L glucose, L-glutamine, and sodium pyruvate (Corning, 10-013-CV), supplemented with 10% of fetal bovine serum (FBS) and cultured at 37 oC with 5% CO2.
OVCA420 scRNA-seq EMT time course experiment
OVCA420 cells (10,000/well) were plated into in 6-well plates. Cells were treated with 10 ng/mL TGF-β1 (R&D Systems, #240-B-010), with treatment timed in such a way that all time-points were synchronized at the time of collection. Cells were passaged as needed to avoid confluence, and fresh TGF-β1 was added every two days with refreshed media. Cells were not passaged in the 2 days prior to final collection to avoid artifacts during sequencing. Single-cell suspensions were processed using the 10x Genomics Single Cell 3’ RNA-seq kit v3. Final libraries were sequenced on an Illumina HiSeq 4000 after gene expression libraries were prepared according to the manufacturer’s protocol. Raw sequencing reads were processed using CellRanger v2.0.1 using the GRCh38 build of the human genome and default parameters. Graphics were created using ‘ggplot2’ and ‘SCpubr’100.
Data quality control and processing
Quality control was first performed independently on each 10x Genomic library, and all main processing steps were performed with Seurat V3101 for the OVCA420 cells and Seurat v4102 for the ovarian cancer datasets. Expression matrices for the OVCA420 cells were imported into R as Seurat objects. Only cells with more than 200 genes detected were retained and cells with a high percentage of mitochondrial gene expression were also removed. For the OVCA420 treatment time course, an independent Seurat object was made combining all timepoints, followed by a standard workflow by first removing genes detected in fewer than 1% of the cells for each timepoint. We then obtained the top 2000 most variable genes using the ‘vst’ selection method in Seurat, scaled RNA expression values and regressed out mitochondrial reads, total UMI count, and cell cycle scoring. Cell cycle regression was handled by ‘SCTransform’103,104, which was also used to normalize the RNA matrices for each sample using regularized negative binomial regression. After this, PCA was run on the variable genes and all UMAP embeddings were calculated from the first 30 principal components.
For the ovarian cancer datasets42, each individual tumor sample matrix was obtained as tables divided into stroma, CD45+, and tumor cell files that we first made into Seurat objects with a minimum of 200 genes per cell and then merged using the ‘base::merge()’ function prior to processing, yielding 16 individual samples (i.e., 16 tumors). Each sample was processed independently similarly to the OVCA420 cells. Briefly, cells with high percentage mitochondrial genes and low feature number were subset out and cell cycle genes were regressed out for each sample using ‘SCTransform’103,104. ‘SCTransform’ was also used to normalize the RNA matrices for each sample using regularized negative binomial regression. PCA was then applied to each individual sample and UMAP embeddings were calculated from the first 30 principal components. We also added metadata such as immune phenotype and patient-ID in each samples’ Seurat metadata slot.
Semi-supervised cell labeling and integration
Cell type labeling was first performed using common markers for cancer cells (KRT19, AMHR2, ELF3, EPCAM) and fibroblasts (COL1A1, COL1A2). We also labeled endothelial cells (CDH5, CLDN5) and smooth muscle cells (ACTA2) to find very small populations (<100 cells) for each, so we removed these populations. To label the CD45+ population we applied Seurat’s multimodal reference mapping method102 to each individual sample with their published CITE-Seq reference object of 162,000 PBMCs measured with 228 antibodies. We used all default settings provided by Seurat for this part of the processing pipeline. After mapping the cell populations by the reference object, we merged this with our labels for cancer cells and fibroblasts to finalize the individual sample objects with labels for all clusters.
Seurat’s integration101 was used to align and combine shared populations across the 16 tumors. Briefly, Seurat matches pairs of cells across datasets that share certain biological states, or anchors, based on bulk RNA expression. The corrected data was then scaled and UMAP embeddings were applied to it based on 30 principal components from a PCA run.
EMT scoring strategy
For EMT scoring, we subset the cancer cell population from the integrated object and used the ‘AddModuleScore()’ Seurat function together with our previously published cancer-specific EMT signature96 to assign an EMT score to each cancer cell. This EMT signature reflects the most consistent expression patterns associated with epithelial-mesenchymal plasticity in cancer96. EMT scoring was similarly performed for the OVCA420 object. We then calculated the mean of each individual samples’ EMT scores and labeled every cell that fell under a threshold of ‘mean–1 standard deviation’ an epithelial cell and every cell above a threshold of ‘mean+1 standard deviation’ a mesenchymal cell, with all cells in-between labeled as partial-EMT (pEMT).
Differential gene expression analysis
The Wilcoxon rank sum test implemented in the ‘FindMarkers()’ or ‘FindAllMarkers()’ functions of Seurat were used to calculate all differentially expressed genes between the input populations. For volcano plots and analysis of most differentially expressed genes we used a cutoff of p-adjusted < 0.05 and for log2 fold-change (log2fc) the mean of the log2fc ± 2*standard deviation of log2fc.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using the ‘fgsea’ R package105. Input genes were ranked by their log2 fold-change values. Reference gene sets were collected from the Molecular Signatures Database (MSigDB) v6.2. For gene set enrichment of LAG3+-high CD8+ T-cells, we used the Enrichr106–108 online tool (https://maayanlab.cloud/Enrichr/).
Pseudotemporal ordering of cells
For a detailed explanation of the pseudotime pipeline, refer to Cook and Vanderhyden (2020)44. Briefly, we used R package ‘psupertime’109 v0.2.6 (https://github.com/wmacnair/psupertime) to calculate pseudotime scores on the top 3000 most variable genes for the OVCA420 Seurat object. Psupertime requires scRNA-seq data with ordinal labels to build a linear combination of genes that vary consistently over the time course and are used to assign a pseudotemporal value to individual cells. Individual cells with pseudotemporal values were correlated with other genes or modules (eg. EMT signature scores).
Non-negative matrix factorization
Non-negative matrix factorization (NMF) was performed using the ‘RcppML’ R package110. Briefly, ‘RcppML’ leverages NMF as a machine learning strategy to learn coordinated gene activity in sparse data and present summaries of biological processes as broken into individual vectors of weighted values that contribute to the overall dimensionality in the data.
Cell-cell communication analysis
For cell-cell communication analysis, we used LIANA111, a tool that integrates multiple methods for cell-cell communication inference in single-cell data. LIANA provides a consensus-based rank aggregate for receptor-ligand pairs from the results of multiple cell-cell communication algorithms through ‘robust rank aggregation’ (RRA). Briefly, we chose the default settings of LIANA for our analysis that use methods from SCA, NATMI, Connectome, CellPhoneDB, and CytoTalk to evaluate receptor-ligand pairs. We ran LIANA using the function ‘liana_wrap()’ on our integrated object. The cancer cell population was divided into unique identities based on immune phenotype and EMT score, resulting in 9 identities: Infiltrated-epithelial, Infiltrated-pEMT, infiltrated-mesenchymal, excluded-epithelial, excluded-pEMT, excluded-mesenchymal, desert-epithelial, desert-pEMT, desert-mesenchymal. We then aggregated all the methods into a single matrix using ‘liana_aggregate()’ to construct maps of cell-cell communications with ‘Circlize’112 based on the top ‘aggregate_rank’ of receptor-ligand pairs.
Kaplan-Meier plots
For KM plots, https://kmplot.com/113 was accessed to use bulk RNA-Seq TCGA and microarray data to construct KM survival plots. For LAG3 and LGALS3, we plotted overall survival of optimally debulked patients with high-grade (3 + 4), later stage (2 + 3 + 4) tumors with TP53 mutation. We chose these settings because they match well with the patient data from Hornburg et al. (2021)29 and represent a subset of some of the worst HGSOC samples based on expected patient outcomes.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Dr. Michele Ardolino for his helpful feedback, and the rest of the Vanderhyden lab for their helpful discussion and feedback. We acknowledge Genome Quebec for their technical support, and assistance with scRNA-seq experiments. This work was supported in part by a grant from the National Science and Engineering Research Council (NSERC). We acknowledge BioRender® clip art used for Figs. 1a and 9.
Author contributions
Conceptualization, E.Y. and G.M.R.; Data curation, D.P.C.; Formal analysis, E.Y.; Funding acquisition, B.C.V.; Investigation, E.Y.; Methodology, E.Y. and D.P.C.; Project administration, B.C.V.; Resources, D.P.C.; Software, E.Y.; Supervision, B.C.V.; Visualization, E.Y.; Writing – original draft, E.Y.; Writing – review & editing, D.P.C., G.M.R. and B.C.V.
Data availability
Sixteen high-grade serous ovarian cancer datasets were obtained with permission from European Genome-Phenom Archive (EGAD00001006974). For kinase inhibitor-treated time-course experiment, raw sequencing files and processed UMI count matrices have been obtained from the NCBI Gene Expression Omnibus under the accession GSE147405. For OVCA420 time course treated with TGF-β1 experiment, raw sequencing files and processed UMI count matrices have been deposited in the NCBI Gene Expression Omnibus under the accession GSE247098.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41540-023-00322-4.
References
- 1.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front. Med. 2018;12:361–373. doi: 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dongre A, et al. Direct and indirect regulators of epithelial–mesenchymal transition–mediated immunosuppression in breast carcinomas. Cancer Discov. 2021;11:1286–1305. doi: 10.1158/2159-8290.CD-20-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricciardi M, et al. Epithelial-to-mesenchymal transition (EMT) induced by inflammatory priming elicits mesenchymal stromal cell-like immune-modulatory properties in cancer cells. Br. J. Cancer. 2015;112:1067–1075. doi: 10.1038/bjc.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxena K, Jolly MK, Balamurugan K. Hypoxia, partial EMT and collective migration: Emerging culprits in metastasis. Transl. Oncol. 2020;13:100845. doi: 10.1016/j.tranon.2020.100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simeonov KP, et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell. 2021;39:1150–1162.e9. doi: 10.1016/j.ccell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latifi A, et al. Isolation and characterization of tumor cells from the ascites of ovarian cancer patients: molecular phenotype of chemoresistant ovarian tumors. PLoS ONE. 2012;7:e46858. doi: 10.1371/journal.pone.0046858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taki M, et al. Tumor immune microenvironment during epithelial–mesenchymal transition. Clin. Cancer Res. 2021;27:4669–4679. doi: 10.1158/1078-0432.CCR-20-4459. [DOI] [PubMed] [Google Scholar]
- 9.Loret N, Denys H, Tummers P, Berx G. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers. 2019;11:838. doi: 10.3390/cancers11060838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuelten CH, et al. Acute wounds accelerate tumorigenesis by a T-Cell dependent mechanism. Cancer Res. 2008;68:7278–7282. doi: 10.1158/0008-5472.CAN-08-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson J, et al. Acute inflammation induced by the biopsy of mouse mammary tumors promotes the development of metastasis. Breast Cancer Res. Treat. 2013;139:391–401. doi: 10.1007/s10549-013-2575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L. TGFβ and cancer metastasis: an inflammation link. Cancer Metastasis Rev. 2010;29:263–271. doi: 10.1007/s10555-010-9226-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bates RC, Mercurio AM. Tumor Necrosis Factor-α stimulates the Epithelial-to-mesenchymal transition of human colonic organoids. Mol. Biol. Cell. 2003;14:1790–1800. doi: 10.1091/mbc.e02-09-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandalaft, L. E., Dangaj Laniti, D. & Coukos, G. Immunobiology of high-grade serous ovarian cancer: lessons for clinical translation. Nat. Rev. Cancer 1–17 10.1038/s41568-022-00503-z (2022) [DOI] [PubMed]
- 15.Ovarian Tumor Tissue Analysis (OTTA) Consortium. Dose-response association of CD8+ tumor-infiltrating lymphocytes and survival time in high-grade serous ovarian cancer. JAMA Oncol. 2017;3:e173290. doi: 10.1001/jamaoncol.2017.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachmayr-Heyda A, et al. Prognostic impact of tumor infiltrating CD8+ T cells in association with cell proliferation in ovarian cancer patients - a study of the OVCAD consortium. BMC Cancer. 2013;13:422. doi: 10.1186/1471-2407-13-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woude LL, Van Der, Gorris MAJ, Halilovic A, Figdor CG, Vries IJMde. Migrating into the Tumor: a Roadmap for T Cells. Trends Cancer. 2017;3:797–808. doi: 10.1016/j.trecan.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, et al. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature. 2021;599:673–678. doi: 10.1038/s41586-021-04057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, et al. Infiltration of CD8+ T cells into tumor cell clusters in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA. 2019;116:3678–3687. doi: 10.1073/pnas.1817652116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fucikova J, et al. TIM-3 dictates functional orientation of the immune infiltrate in ovarian cancer. Clin. Cancer Res. 2019;25:4820–4831. doi: 10.1158/1078-0432.CCR-18-4175. [DOI] [PubMed] [Google Scholar]
- 21.Fucikova J, et al. Immunological configuration of ovarian carcinoma: features and impact on disease outcome. J. Immunother. Cancer. 2021;9:e002873. doi: 10.1136/jitc-2021-002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang R-Y, et al. LAG3 and PD1 co-inhibitory molecules collaborate to limit CD8+ T cell signaling and dampen antitumor immunity in a murine ovarian cancer model. Oncotarget. 2015;6:27359–27377. doi: 10.18632/oncotarget.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chihara N, et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 2018;558:454–459. doi: 10.1038/s41586-018-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruffo E, Wu R, Bruno TC, Workman CJ, Vignali DAA. Lymphocyte-Activation Gene 3 (LAG3): the next immune checkpoint receptor. Semin. Immunol. 2019;42:101305. doi: 10.1016/j.smim.2019.101305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews LP, Marciscano AE, Drake CG, Vignali DAA. LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 2017;276:80–96. doi: 10.1111/imr.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shayesteh L, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat. Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 27.Dongre A, et al. Epithelial-to-mesenchymal Transition contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami R, et al. Establishment of a novel histopathological classification of high-grade serous ovarian carcinoma correlated with prognostically distinct gene expression subtypes. Am. J. Pathol. 2016;186:1103–1113. doi: 10.1016/j.ajpath.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 29.Hornburg M, et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell. 2021;39:928–944.e6. doi: 10.1016/j.ccell.2021.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Desbois M, et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat. Commun. 2020;11:5583. doi: 10.1038/s41467-020-19408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, et al. B2M overexpression correlates with malignancy and immune signatures in human gliomas. Sci. Rep. 2021;11:5045. doi: 10.1038/s41598-021-84465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nugteren S, Samsom JN. Secretory Leukocyte Protease Inhibitor (SLPI) in mucosal tissues: protects against inflammation, but promotes cancer. Cytokine Growth Factor Rev. 2021;59:22–35. doi: 10.1016/j.cytogfr.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Sabbatino F, et al. Role of human leukocyte antigen system as a predictive biomarker for checkpoint-based immunotherapy in cancer patients. Int. J. Mol. Sci. 2020;21:7295. doi: 10.3390/ijms21197295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Wang H, Li G, Jin X, Chen B. The immune-related Gene ELF3 is a novel biomarker for the prognosis of ovarian cancer. Int. J. Gen. Med. 2021;14:5537. doi: 10.2147/IJGM.S332320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David JM, Hamilton DH, Palena C. MUC1 upregulation promotes immune resistance in tumor cells undergoing brachyury-mediated epithelial-mesenchymal transition. OncoImmunology. 2016;5:e1117738. doi: 10.1080/2162402X.2015.1117738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippou PS, Karagiannis GS, Constantinidou A, Midkine MDK. growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene. 2020;39:2040–2054. doi: 10.1038/s41388-019-1124-8. [DOI] [PubMed] [Google Scholar]
- 37.MacGregor HL, et al. Tumor cell expression of B7-H4 correlates with higher frequencies of tumor-infiltrating APCs and higher CXCL17 expression in human epithelial ovarian cancer. Oncoimmunology. 2019;8:e1665460. doi: 10.1080/2162402X.2019.1665460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera-Cruz CM, Shearer JJ, Figueiredo Neto M, Figueiredo ML. The immunomodulatory effects of mesenchymal stem cell polarization within the tumor microenvironment niche. Stem Cells Int. 2017;2017:4015039. doi: 10.1155/2017/4015039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trivanović D, et al. The roles of mesenchymal stromal/stem cells in tumor microenvironment associated with inflammation. Mediators Inflamm. 2016;2016:7314016. doi: 10.1155/2016/7314016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss, A. R. R. & Dahlke, M. H. Immunomodulation by Mesenchymal Stem Cells (MSCs): Mechanisms of Action of Living, Apoptotic, and Dead MSCs. Front. Immunol. 10, 1191 (2019). [DOI] [PMC free article] [PubMed]
- 41.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, et al. Intratumoral T Cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 43.Bou-Tayeh B, Miller ML. Ovarian tumors orchestrate distinct cellular compositions. Immunity. 2021;54:1107–1109. doi: 10.1016/j.immuni.2021.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Cook DP, Vanderhyden BC. Context specificity of the EMT transcriptional response. Nat. Commun. 2020;11:2142. doi: 10.1038/s41467-020-16066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kooi S, et al. HLA class I expression on human ovarian carcinoma cells correlates with T-cell infiltration in vivo and T-cell expansion in vitro in low concentrations of recombinant interleukin-2. Cell. Immunol. 1996;174:116–128. doi: 10.1006/cimm.1996.0301. [DOI] [PubMed] [Google Scholar]
- 46.Aptsiauri N, Ruiz-Cabello F, Garrido F. The transition from HLA-I positive to HLA-I negative primary tumors: the road to escape from T-cell responses. Curr. Opin. Immunol. 2018;51:123–132. doi: 10.1016/j.coi.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Whitehair R, Peres LC, Mills AM. Expression of the Immune Checkpoints LAG-3 and PD-L1 in High-grade Serous Ovarian Carcinoma: Relationship to Tumor-associated Lymphocytes and Germline BRCA Status. Int. J. Gynecol. Pathol. 2020;39:558–566. doi: 10.1097/PGP.0000000000000657. [DOI] [PubMed] [Google Scholar]
- 48.Alsuliman A, et al. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol. Cancer. 2015;14:149. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nhokaew W, Kleebkaow P, Chaisuriya N, Kietpeerakool C. Programmed Death Ligand 1 (PD-L1) expression in epithelial ovarian cancer: a comparison of type i and type ii tumors. Asian Pac. J. Cancer Prev. APJCP. 2019;20:1161–1169. doi: 10.31557/APJCP.2019.20.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fei Z, et al. PD-L1 induces epithelial–mesenchymal transition in nasopharyngeal carcinoma cells through activation of the PI3K/AKT pathway. Oncol. Res. 2019;27:801–807. doi: 10.3727/096504018X15446984186056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kouo T, et al. Galectin-3 shapes antitumor immune responses by suppressing CD8+ T cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol. Res. 2015;3:412–423. doi: 10.1158/2326-6066.CIR-14-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhai W, et al. A novel cyclic peptide targeting LAG-3 for cancer immunotherapy by activating antigen-specific CD8+ T cell responses. Acta Pharm. Sin. B. 2020;10:1047–1060. doi: 10.1016/j.apsb.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graydon, C. G., Mohideen, S. & Fowke, K. R. LAG3’s Enigmatic Mechanism of Action. Front. Immunol. 11, 615317 (2021). [DOI] [PMC free article] [PubMed]
- 54.Wang J, et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell. 2019;176:334–347.e12. doi: 10.1016/j.cell.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He X, Zhang S, Chen J, Li D. Increased LGALS3 expression independently predicts shorter overall survival in patients with the proneural subtype of glioblastoma. Cancer Med. 2019;8:2031–2040. doi: 10.1002/cam4.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kearney CJ, et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 2018;3:eaar3451. doi: 10.1126/sciimmunol.aar3451. [DOI] [PubMed] [Google Scholar]
- 57.FDA approves anti-LAG3 checkpoint. Nat. Biotechnol. 2022;40:625–625. doi: 10.1038/s41587-022-01331-0. [DOI] [PubMed] [Google Scholar]
- 58.Zhao L, Wang H, Xu K, Liu X, He Y. Update on lymphocyte-activation gene 3 (LAG-3) in cancers: from biological properties to clinical applications. Chin. Med. J. 2022;135:1203–1212. doi: 10.1097/CM9.0000000000001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sordo-Bahamonde C, et al. LAG-3 Blockade with Relatlimab (BMS-986016) Restores Anti-Leukemic Responses in Chronic Lymphocytic Leukemia. Cancers. 2021;13:2112. doi: 10.3390/cancers13092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chocarro, L. et al. Clinical landscape of LAG-3-targeted therapy. Immuno-Oncol. Technol. 14, 100079 (2022). [DOI] [PMC free article] [PubMed]
- 61.Launonen I-M, et al. Single-cell tumor-immune microenvironment of BRCA1/2 mutated high-grade serous ovarian cancer. Nat. Commun. 2022;13:835. doi: 10.1038/s41467-022-28389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imai Y, et al. Expression of multiple immune checkpoint molecules on T cells in malignant ascites from epithelial ovarian carcinoma. Oncol. Lett. 2018;15:6457–6468. doi: 10.3892/ol.2018.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rådestad E, et al. Immune profiling and identification of prognostic immune-related risk factors in human ovarian cancer. Oncoimmunology. 2018;8:e1535730. doi: 10.1080/2162402X.2018.1535730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rafehi S, et al. TGFβ signaling regulates epithelial-mesenchymal plasticity in ovarian cancer ascites-derived spheroids. Endocr. Relat. Cancer. 2016;23:147–159. doi: 10.1530/ERC-15-0383. [DOI] [PubMed] [Google Scholar]
- 65.Capellero S, et al. Ovarian cancer cells in ascites form aggregates that display a hybrid epithelial-mesenchymal phenotype and allows survival and proliferation of metastasizing cells. Int. J. Mol. Sci. 2022;23:833. doi: 10.3390/ijms23020833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahoo S, et al. Immunosuppressive Traits of the Hybrid Epithelial/Mesenchymal Phenotype. Front. Immunol. 2021;12:797261. doi: 10.3389/fimmu.2021.797261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacKinnon AC, et al. Regulation of transforming growth Factor-β1–driven Lung Fibrosis by Galectin-3. Am. J. Respir. Crit. Care Med. 2012;185:537–546. doi: 10.1164/rccm.201106-0965OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slack RJ, Mills R, Mackinnon AC. The therapeutic potential of galectin-3 inhibition in fibrotic disease. Int. J. Biochem. Cell Biol. 2021;130:105881. doi: 10.1016/j.biocel.2020.105881. [DOI] [PubMed] [Google Scholar]
- 69.Humphries, D. C. et al. Galectin-3 inhibitor GB0139 protects against acute lung injury by inhibiting neutrophil recruitment and activation. Front. Pharmacol. 13, 949264 (2022). [DOI] [PMC free article] [PubMed]
- 70.Filer A, et al. Galectin 3 induces a distinctive pattern of cytokine and chemokine production in rheumatoid synovial fibroblasts via selective signaling pathways. Arthritis Rheum. 2009;60:1604–1614. doi: 10.1002/art.24574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mettu NB, et al. A Phase 1a/b Open-Label, Dose-Escalation Study of Etigilimab Alone or in Combination with Nivolumab in Patients with Locally Advanced or Metastatic Solid Tumors. Clin. Cancer Res. 2022;28:882–892. doi: 10.1158/1078-0432.CCR-21-2780. [DOI] [PubMed] [Google Scholar]
- 72.Sarikonda G, et al. 111P Interim biomarker analysis of a phase Ib/II study of anti-TIGIT etigilimab (MPH313) and nivolumab in subjects with select locally advanced or metastatic solid tumors (ACTIVATE) Ann. Oncol. 2022;33:S589. doi: 10.1016/j.annonc.2022.07.143. [DOI] [Google Scholar]
- 73.Johnston RJ, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 74.Chauvin J-M, et al. TIGIT and PD-1 impair tumor antigen-specific CD8+ T cells in melanoma patients. J. Clin. Investig. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farhad M, Rolig AS, Redmond WL. The role of Galectin-3 in modulating tumor growth and immunosuppression within the tumor microenvironment. OncoImmunology. 2018;7:e1434467. doi: 10.1080/2162402X.2018.1434467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krześlak A, Lipińska A. Galectin-3 as a multifunctional protein. Cell. Mol. Biol. Lett. 2004;9:305–328. [PubMed] [Google Scholar]
- 77.Demotte N, et al. A galectin-3 ligand corrects the impaired function of human CD4 and CD8 tumor-infiltrating lymphocytes and favors tumor rejection in mice. Cancer Res. 2010;70:7476–7488. doi: 10.1158/0008-5472.CAN-10-0761. [DOI] [PubMed] [Google Scholar]
- 78.Fukumori T, et al. CD29 and CD7 Mediate Galectin-3-Induced Type II T-Cell Apoptosis. Cancer Res. 2003;63:8302–8311. [PubMed] [Google Scholar]
- 79.Chen H-Y, et al. Galectin-3 negatively regulates TCR-mediated CD4+ T-cell activation at the immunological synapse. Proc. Natl. Acad. Sci. USA. 2009;106:14496–14501. doi: 10.1073/pnas.0903497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luk H-M, et al. Expression and clinical significance of Gal-3 and NFκB pathway-related factors in epithelial ovarian carcinoma. Int. J. Clin. Exp. Pathol. 2020;13:1197–1205. [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, et al. Galectin-3 and β-catenin are associated with a poor prognosis in serous epithelial ovarian cancer. Cancer Manag. Res. 2018;10:3963–3971. doi: 10.2147/CMAR.S171146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu T, Liu T, Xing L, Ji G. Baicalin and puerarin reverse epithelial-mesenchymal transition via the TGF-β1/Smad3 pathway in vitro. Exp. Ther. Med. 2018;16:1968–1974. doi: 10.3892/etm.2018.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Priglinger CS, et al. Epithelial-to-Mesenchymal Transition of RPE Cells In Vitro Confers Increased β1,6-N-Glycosylation and Increased Susceptibility to Galectin-3 Binding. PLOS ONE. 2016;11:e0146887. doi: 10.1371/journal.pone.0146887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leem G, et al. 4-1BB co-stimulation further enhances anti-PD-1-mediated reinvigoration of exhausted CD39+ CD8 T cells from primary and metastatic sites of epithelial ovarian cancers. J. Immunother. Cancer. 2020;8:e001650. doi: 10.1136/jitc-2020-001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mirandola L, et al. Galectin-3 inhibition suppresses drug resistance, motility, invasion and angiogenic potential in ovarian cancer. Gynecol. Oncol. 2014;135:573–579. doi: 10.1016/j.ygyno.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 86.Taylor A, Rudd CE. Glycogen Synthase Kinase 3 Inactivation Compensates for the Lack of CD28 in the Priming of CD8+ Cytotoxic T-Cells: Implications for anti-PD-1 Immunotherapy. Front. Immunol. 2017;8:1653. doi: 10.3389/fimmu.2017.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J-Y, et al. Modulation of CD8+ memory stem T cell activity and glycogen synthase kinase 3β inhibition enhances anti-tumoral immunity in gastric cancer. Oncoimmunology. 2018;7:e1412900. doi: 10.1080/2162402X.2017.1412900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudd CE, Chanthong K, Taylor A. Small molecule inhibition of GSK-3 specifically inhibits the transcription of inhibitory Co-receptor LAG-3 for enhanced anti-tumor immunity. Cell Rep. 2020;30:2075–2082.e4. doi: 10.1016/j.celrep.2020.01.076. [DOI] [PubMed] [Google Scholar]
- 89.Hussain, S. et al. The roles of stroma-derived chemokine in different stages of cancer metastases. Front. Immunol. 11, 598532 (2020). [DOI] [PMC free article] [PubMed]
- 90.Zvaifler NJ. Relevance of the stroma and epithelial-mesenchymal transition (EMT) for the rheumatic diseases. Arthritis Res. Ther. 2006;8:210. doi: 10.1186/ar1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bulle A, Lim K-H. Beyond just a tight fortress: contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target. Ther. 2020;5:1–12. doi: 10.1038/s41392-020-00341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hapke RY, Haake SM. Hypoxia-induced epithelial to mesenchymal transition in cancer. Cancer Lett. 2020;487:10–20. doi: 10.1016/j.canlet.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Misra A, Pandey C, Sze SK, Thanabalu T. Hypoxia activated EGFR signaling induces Epithelial to Mesenchymal Transition (EMT) PLOS ONE. 2012;7:e49766. doi: 10.1371/journal.pone.0049766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tam SY, Wu VWC, Law HKW. Hypoxia-induced epithelial-mesenchymal transition in cancers: HIF-1α and Beyond. Front. Oncol. 2020;10:486. doi: 10.3389/fonc.2020.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tyler M, Tirosh I. Decoupling epithelial-mesenchymal transitions from stromal profiles by integrative expression analysis. Nat. Commun. 2021;12:2592. doi: 10.1038/s41467-021-22800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cook DP, Vanderhyden BC. Transcriptional census of epithelial-mesenchymal plasticity in cancer. Sci. Adv. 2022;8:eabi7640. doi: 10.1126/sciadv.abi7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCorry AM, Loughrey MB, Longley DB, Lawler M, Dunne PD. Epithelial-to-mesenchymal transition signature assessment in colorectal cancer quantifies tumour stromal content rather than true transition. J. Pathol. 2018;246:422–426. doi: 10.1002/path.5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hou W, Ji Z, Ji H, Hicks SC. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol. 2020;21:218. doi: 10.1186/s13059-020-02132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim HJ, Lin Y, Geddes TA, Yang JYH, Yang P. CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics. 2020;36:4137–4143. doi: 10.1093/bioinformatics/btaa282. [DOI] [PubMed] [Google Scholar]
- 100.Blanco-Carmona, E. Generating publication ready visualizations for Single Cell transcriptomics using SCpubr. 2022.02.28.482303 Preprint at 10.1101/2022.02.28.482303 (2022).
- 101.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choudhary S, Satija R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 2022;23:27. doi: 10.1186/s13059-021-02584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Korotkevich, G. et al. Fast gene set enrichment analysis. 060012 Preprint at 10.1101/060012 (2021).
- 106.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xie Z, et al. Gene set knowledge discovery with enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macnair W, Gupta R, Claassen M. psupertime: supervised pseudotime analysis for time-series single-cell RNA-seq data. Bioinformatics. 2022;38:i290–i298. doi: 10.1093/bioinformatics/btac227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.DeBruine, Z. J., Melcher, K. & Triche, T. J. Fast and robust non-negative matrix factorization for single-cell experiments. 2021.09.01.458620 Preprint at 10.1101/2021.09.01.458620 (2021).
- 111.Dimitrov D, et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 2022;13:3224. doi: 10.1038/s41467-022-30755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 113.Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): development and implementation. J. Med. Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sixteen high-grade serous ovarian cancer datasets were obtained with permission from European Genome-Phenom Archive (EGAD00001006974). For kinase inhibitor-treated time-course experiment, raw sequencing files and processed UMI count matrices have been obtained from the NCBI Gene Expression Omnibus under the accession GSE147405. For OVCA420 time course treated with TGF-β1 experiment, raw sequencing files and processed UMI count matrices have been deposited in the NCBI Gene Expression Omnibus under the accession GSE247098.