Abstract
The lactose utilization genes of Staphylococcus xylosus have been isolated and characterized. The system is comprised of two structural genes, lacP and lacH, encoding the lactose permease and the β-galactosidase proteins, respectively, and a regulatory gene, lacR, coding for an activator of the AraC/XylS family. The lactose utilization genes are divergently arranged, the lacPH genes being opposite to lacR. The lacPH genes are cotranscribed from one promoter in front of lacP, whereas lacR is transcribed from two promoters of different strengths. Lactose transport as well as β-galactosidase activity are inducible by the addition of lactose to the growth medium. Primer extension experiments demonstrated that regulation is achieved at the level of lacPH transcription initiation. Inducibility and efficient lacPH transcription are dependent on a functional lacR gene. Inactivation of lacR resulted in low and constitutive lacPH expression. Expression of lacR itself is practically constitutive, since transcription initiated at the major lacR promoter does not respond to the availability of lactose. Only the minor lacR promoter is lactose inducible. Apart from lactose-specific, LacR-dependent control, the lacPH promoter is also subject to carbon catabolite repression mediated by the catabolite control protein CcpA. When glucose is present in the growth medium, lacPH transcription initiation is reduced. Upon ccpA inactivation, repression at the lacPH promoter is relieved. Despite this loss of transcriptional regulation in the ccpA mutant strain, β-galactosidase activity is still reduced by glucose, suggesting another level of control.
The lactose operon of Escherichia coli is a paradigm for gene regulation (for a review, see reference 33). Studying lac regulation led to fundamental concepts of how a set of genes may be coordinately regulated depending upon the concentration of metabolizable compounds in the growth medium. Soon after repression of the lac operon was established, the universality of that regulatory mode was challenged by the analysis of the arabinose and maltose systems in E. coli, where positive control was realized (for a review, see reference 43). Molecular characterization of sugar utilization systems has also provided valuable knowledge on gene regulation in bacteria other than E. coli. Examples include the complex sucrose metabolism of Bacillus subtilis, lac systems in several AT-rich gram-positive bacteria, and global control by carbon catabolite repression (reviewed in references 9, 41, and 46).
In Staphylococcus xylosus (44), an AT-rich gram-positive bacterium used in meat fermentations (18), the regulation of maltose, sucrose, and xylose catabolic genes has been studied in some detail (10, 13, 45). In addition, two genes encoding proteins involved in carbon catabolite repression in this organism have been isolated (11, 51). One of the genes, the glucose kinase gene glkA, has been detected by transposon mutagenesis and screening for altered β-galactosidase activity in the presence of glucose. It was of interest, therefore, to clone the β-galactosidase gene of S. xylosus and to analyze its regulation.
In this communication, we report on the isolation and characterization of the lactose utilization genes of S. xylosus and their transcriptional regulation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage DNAs.
The staphylococcal strains used in this study are listed in Table 1. E. coli TG1 [supE hsdΔ5 thi Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] was used to screen the S. xylosus library (8) for phage M13 and plasmid cloning. The S. xylosus library had been constructed in pBR322. Genes to be introduced into S. xylosus were cloned in E. coli TG1 by using the shuttle vector pRB473 (8), which is a derivative of pRB373 (6). Plasmid pTV1Ts harboring transposon Tn917 (55) served for transposon mutagenesis.
TABLE 1.
S. xylosus strains used
Transposon mutagenesis.
The transposon mutagenesis was performed as described previously (51). Cells were plated on agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal [100 μg/ml]), erythromycin (2.5 μg/ml), and glucose (1%) and incubated at 37°C for about 48 h.
DNA manipulations, sequencing, transformation, and transfection.
DNA manipulations, plasmid DNA isolation and sequencing, Southern blot analysis, transformation and transfection of E. coli, and preparation of media and agar plates for bacterial growth were done by standard procedures (42). Isolation of chromosomal DNA from S. xylosus and the construction of the genomic library of S. xylosus in E. coli have been described previously (8). Plasmids were introduced into S. xylosus by electroporation (7). PCR was carried out with Vent DNA polymerase (New England Biolabs) or rTth DNA polymerase XL (Perkin-Elmer) in accordance with the instructions of the suppliers.
Cloning of the DNA region upstream of lacP.
Since plasmid pBG303, a representative of the β-galactosidase plasmids from the S. xylosus library (11), contained a truncated putative lactose transporter gene upstream of the β-galactosidase gene, this region was isolated from the chromosome. Southern blot analysis with a lacH-specific probe revealed a second SstI restriction site about 8 kb upstream of the one within lacH (Fig. 1). Chromosomal DNA of S. xylosus was digested with SstI, ligated, and used for inverse long-range PCR with the following primers: 5′-CCAATTCGTAATATCCCCGCTCC (positions 2982 to 2960 at the 3′ end of lacP) and 5′-CACTAACGGTCCCATCGGTTTGG (positions 3590 to 3612 at the 5′ end of lacH). Restriction of the PCR product with HpaI produced two HpaI fragments of 2.5 and 2.8 kb in size. The 2.8-kb fragment, located next to lacH (Fig. 1), was cloned into pUC18, generating plasmid pBG304 (Fig. 1), which was used for further analysis.
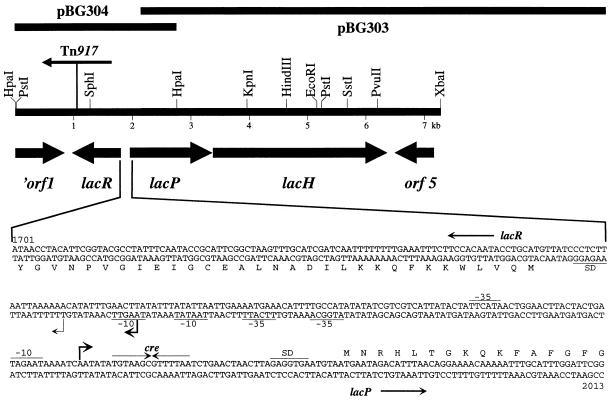
FIG. 1.
Genetic organization of the lactose utilization genes of S. xylosus and nucleotide sequence of the lacR-lacPH promoter region. The lac region that has been sequenced is shown. The size and orientation of the genes were deduced from the nucleotide sequence. The position and orientation of Tn917 in the lacR mutant strain TX258 are marked. The sequence of the lacR-lacP intergenic region is shown. Numbering refers to the complete lac sequence available from the EMBL database under accession no. Y14599. The transcriptional start sites of the lac promoters and an inverted repeat resembling cre are indicated by arrows. The major transcriptional start site of the lacR promoter is symbolized by a boldface arrow. Putative RNA polymerase- and ribosome-binding sites are underlined.
Construction of a lacP deletion mutant of S. xylosus by gene replacement.
To construct a lacP deletion mutant, two PCR fragments were produced. The first, a 1.1-kb fragment, included the whole lacR gene and the lacP region encoding the first 20 amino acids of LacP. The second fragment, 3.5 kb in length, contained the last 20 codons of lacP together with the complete lacH gene. The primers for these amplifications were as follows. For the lacR-lacP′ fragment, 5′-GACGGATCCGGGCAGAAAACCAATGGAAG (positions 897 to 916; BamHI restriction site underlined) and 5′-GCAGTCGACCTTACCGATGGCACCGAATCC (positions 2027 to 2006; SalI restriction site underlined); for the ′lacP-lacH fragment, 5′-GCAGTCGACACGGATATTGAAAAGACATTACAG (positions 3293 to 3316; SalI restriction site underlined) and 5′-GACGCTAGCGTAGGTATTGGAGGCGCAGG (positions 6763 to 6744; NheI restriction site underlined). After restriction with the appropriate restriction enzymes, both PCR fragments were cloned in one step as a BamHI-SalI fragment and a SalI-NheI fragment in the BamHI-NheI-restricted temperature-sensitive shuttle vector pBT2 (7), generating plasmid pBT2RΔPH2. On this plasmid, about 92% of lacP was removed. The plasmid was introduced into S. xylosus C2a, and appropriate dilutions of an overnight culture in B medium with chloramphenicol (20 μg/ml), incubated at 30°C, were plated on lactose utilization test plates (32) and incubated at 37°C for about 48 h. White colonies (lactose negative) that integrated the ΔlacP gene into their chromosome by double crossover and that were chloramphenicol sensitive could be detected. One representative colony, designated S. xylosus TX260 (ΔlacP), was shown to carry the expected ΔlacP mutation based on PCR analysis (data not shown).
Growth of S. xylosus to monitor expression of the lactose utilization genes.
S. xylosus was grown in B medium, which consisted of 1% peptone (Gibco BRL), 0.5% yeast extract, 0.5% NaCl, and 0.1% K2HPO4 · 3H2O. Carbohydrates were added to a final concentration of 25 mM, if required. The complex growth medium was used since no suitable minimal medium is available for S. xylosus. Fermentation of carbohydrates by S. xylosus was monitored on agar plates (32) containing 0.5% of the respective sugar.
For the determination of lactose transport and β-galactosidase activity, and to prepare RNA for primer extension analysis, the following growing conditions were applied. To test for inducibility, cells were grown in B medium without additional carbohydrate to an optical density at 578 nm (OD578) of 1.2. Sugars (25 mM) were added, and the cultures were incubated for 1 additional hour and harvested (OD578, 2 to 2.5). The culture without added carbohydrate grown for the same time period served as the uninduced control. To measure glucose repression, lactose and glucose were added concomitantly to cultures grown as described above.
Measurements of lactose uptake.
Transport of lactose was measured by using whole cells. The harvested cells were washed with ice-cold MT buffer (100 mM MOPS [morpholinepropanesulfonic acid; pH 7.0], 0.5 mM MgSO4, 10 mM NaCl) and resuspended in the same buffer to yield an OD578 of 3.0. These cells were kept on ice until use. One milliliter of the cell suspension was preincubated for 2 min at 30°C, and then 200 μM lactose (25 μM [14C]lactose [57.0 mCi/mmol]) was added. Samples 0.15 ml in size were taken after 1, 2, 4, 8, and 15 min, collected on cellulose nitrate disks (pore size, 0.45 μm), and washed with 5 ml of MT buffer. Filters were dried at 80°C for 30 min, and the radioactivity was determined by liquid scintillation counting. Uptake rates are expressed in picomoles of lactose accumulated per minute per milligram of cell protein. The amount of protein was determined by the method of Bradford (3).
Determination of β-galactosidase activity in cell extracts.
Crude extracts were prepared by vortexing cells repeatedly with glass beads in β-galactosidase buffer (T4) containing 0.1 M Tris (pH 8.0), 0.5 M KCl, 1 mM MgSO4, 0.4 mM MnCl2, and 4 mM dithiothreitol. The assays were performed at 30°C with p-nitrophenyl-β-d-galactopyranoside (7.5 mM) as the substrate and 15 to 400 μg of cellular protein. The release of nitrophenol was monitored at 405 nm. Specific β-galactosidase activity is expressed in nanomoles of nitrophenol released per minute per milligram of protein. Protein concentrations in the cell extracts were determined by the method of Bradford (3).
RNA preparation and primer extension analysis.
Isolation of total RNA (except 5S RNA and tRNAs) was performed with the RNeasy Midi Kit (Qiagen). Five to 8 milliliters of the culture, harvested at an OD578 of 2.0 to 2.5, were washed with 5 ml of ice-cold EDTA solution (0.5 M, pH 8.0), and the pellet was resuspended for cell disruption in a mixture of 1 ml of lysostaphin solution (0.5 mg/ml of H2O) and 10 μl of 100× TE buffer (1 M Tris [pH 8.0], 0.1 M EDTA) and incubated at 37°C. By using large amounts of lysostaphin, the cells lysed rather quickly, within about 2 min. After cell lysis occurred, preparation of the RNA was continued in accordance with the RNeasy protocol for isolation of total RNA from bacteria, with the larger volumes of buffers and solutions given in the protocol always used. The final RNA solution was concentrated to a volume of 25 μl, which contained 2 to 10 μg of RNA/μl. Primer extension experiments were performed with avian myeloblastosis virus reverse transcriptase (Stratagene). The following primers yielded reverse transcripts: for lacR, 5′-CCTACATTCGGTACGCC (positions 1705 to 1721); and for lacPH, 5′-CATCCTTACCGATGGCAC (positions 2030 to 2013). The 5′-end 32P-labeled oligonucleotides were used in primer extension reactions with 15 μg of cellular RNA. Reverse transcripts were resolved on 5% urea-containing polyacrylamide gels. DNA sequencing reactions using the same oligonucleotide were used for sizing the primer extension products.
Nucleotide sequence accession number.
The nucleotide sequence is available from the EMBL database under accession no. Y14599.
RESULTS
Cloning of the lactose utilization genes of S. xylosus.
To clone the β-galactosidase gene, aliquots of an amplified S. xylosus library that was stored as plasmid pools (11) were introduced into E. coli TG1 and the transformants were checked for β-galactosidase activity on X-Gal-containing agar plates. Blue colonies were obtained from six plasmid pools. Subsequently, the plasmid contents of four representative transformants of each pool were analyzed. Plasmids originating from one pool were found to be identical. In addition, two plasmids from different pools showed the same restriction patterns. Therefore, five distinct β-galactosidase-expressing plasmids were isolated from the amplified library. Comparative restriction analysis of these plasmids revealed a common region of about 4 kb. Plasmid pBG303 containing an insertion of about 8 kb was chosen for further analysis. By Southern blot analysis of chromosomal S. xylosus DNA, it was verified that the insertion represented a continuous segment of the genome. Partial sequencing of the cloned DNA identified an open reading frame with high sequence similarity to various β-galactosidases. In addition, an incomplete open reading frame that resembled sugar transporters of the GPH family was detected upstream of the β-galactosidase gene (Fig. 1) (37). Since none of the plasmids from the library contained the complete putative lactose transporter gene, the gene was isolated from the S. xylosus genome by a different approach.
The missing information was cloned on plasmid pBG304 by inverse PCR as described in Materials and Methods. Several attempts to obtain the whole lactose utilization region, as depicted in Fig. 1, in E. coli or Staphylococcus carnosus (15) failed. Subsequently, the lactose utilization region of S. xylosus was sequenced by using the plasmids pBG303 and pBG304.
Nucleotide sequence of the lactose utilization genes.
The nucleotide sequence was determined for both strands from the HpaI restriction site within orf1 to the unique XbaI restriction site (Fig. 1). It comprises 7,206 bp and contains 5 open reading frames, one of which (orf1) is truncated at the 5′ end. The largest open reading frame, encoding a protein of 994 amino acids with a calculated molecular mass of 115.246 kDa, constitutes the β-galactosidase gene and is designated lacH. The β-galactosidase protein of S. xylosus has the highest degree of similarity to the β-galactosidase protein of Actinobacillus pleuropneumoniae (2), with almost 40% identical residues. With the β-galactosidase protein from E. coli (27), LacH has 33% amino acids in common. Several conserved regions, which appear to be important for the hydrolytic activity in β-galactosidases (26), are also present in the S. xylosus enzyme.
The lacP gene, located upstream of lacH, encodes a protein of 462 amino acids (51.667 kDa) that shows significant similarity to members of the GPH protein family that transport galactosides, pentoses, and hexunorides (37). Within this family, the melibiose permeases of enteric bacteria (17, 31, 54) are most similar to the S. xylosus protein (37 to 39% identical residues). Hydropathy analysis (28) and structural predictions (22) have suggested that LacP is a membrane protein with 11 transmembrane segments. Therefore, lacP appears to encode the lactose permease of S. xylosus.
Upstream of lacP and on the opposite strand of the DNA, an open reading frame, named lacR, with a coding capacity of 279 amino acids is found. The deduced LacR protein has a molecular mass of 32.339 kDa and resembles regulatory proteins of the AraC/XylS family (12). LacR of S. xylosus has 31% identical residues to the raffinose operon activator from Pediococcus pentosaceus (Swiss-Prot no. P43465) and 24% to both AraC from E. coli (52) and XylS from Pseudomonas putida (25). The helix-turn-helix DNA binding motif of the regulator family, which is located in the C-terminal portion of the proteins, is present at the appropriate position in LacR (amino acids 190 to 209). Therefore, lacR probably encodes the regulator of the lactose utilization genes.
The deduced polypeptide of the truncated orf1 (Fig. 1) had similarity to regulators of the LysR family (19), whereas the product of orf5 did not have significant similarity to proteins in databases. It appears that neither gene takes part in lactose utilization in S. xylosus.
Isolation of transposon-induced lactose utilization mutants of S. xylosus.
In a previous study intended to isolate S. xylosus mutants altered in global catabolite repression, Tn917 transposon mutagenesis was performed, using β-galactosidase expression for screening. Besides several dark blue colonies, which were catabolite repression mutants (51), one white and one pale blue colony were isolated. Both mutant strains lost the ability to ferment lactose as determined by acid production on lactose-containing indicator plates (32). By PCR analysis using transposon- as well as lac-specific primers, Tn917 was localized to lacR in S. xylosus TX258 (Fig. 1) and to lacH in S. xylosus TX259. Due to the integration of Tn917 into the lacH gene, no β-galactosidase activity was detectable in TX259 (data not shown). Therefore, the strain was not further analyzed. In the mutant, TX258, however, the exact location of Tn917 in lacR was determined by DNA sequencing. Tn917 was found to have integrated 120 bp apart from the end of lacR (Fig. 1). To analyze the consequences of lacR inactivation for lac gene expression, lactose transport and β-galactosidase activity were determined in the wild type and the mutant strain.
Lactose transport and β-galactosidase activity in the wild-type and lacR mutant strains.
To measure activities specified by proteins encoded by the lac genes, strains were grown in complex medium with lactose or galactose or without additional sugar. As summarized in Table 2, lactose transport and β-galactosidase activity in the wild-type strain are induced by the addition of lactose to the growth medium, whereas galactose has no effect. Induction of lactose transport was found to be about 14-fold and β-galactosidase activity was stimulated 11-fold, indicating coordinated expression of both genes. In the lacR mutant strain, TX258, both activities were unregulated and much lower than in the wild type (Table 2). The low noninducible expression of the lac genes in the lacR mutant strongly suggests that LacR functions as an activator.
TABLE 2.
Lactose transport and β-galactosidase activity in the S. xylosus wild type, C2a, and in the lacR mutant, TX258
| Strain | Growth conditionsa | Lactose transportb (pmol of lactose transported/min/mg of protein) | β-Galactosidase activityc (nmol of nitrophenol produced/min/mg of protein) |
|---|---|---|---|
| C2a | B | 371 ± 99 | 9 ± 2 |
| B + lactose | 5,143 ± 187 | 98 ± 14 | |
| B + galactose | 578 ± 94 | 7 ± 2 | |
| TX258 (lacR) | B | 78 ± 8 | <1 |
| B + lactose | 83 ± 8 | <1 |
The strains were grown in B medium (B) to an OD578 of 1.2. The indicated sugar was added at 25 mM. After 1 h of growth, the cells were harvested. Cultures without additional sugar were grown and harvested accordingly.
Lactose transport was determined by using 200 μM [14C]lactose (7.12 mCi/mmol) and 1 ml of washed cells at an OD578 of 3. The values (means ± standard deviations) represent the initial uptake rates within the first 4 min of the experiment. They were taken from two measurements, each from three cultures.
The values (means ± standard deviations) represent two determinations, each with bacterial extracts from three cultures.
Inducibility of β-galactosidase activity in a lacP mutant strain.
To determine whether a functional lactose permease is required for induction of the system, a lacP mutant strain was constructed (see Materials and Methods). In that strain, TX260, an in-frame deletion within lacP removed the coding region for 422 amino acids, leaving the lacR-lacP as well as the lacP-lacH intergenic region intact. The lacP deletion strain was lactose negative, as determined on utilization test plates, transported background levels of lactose (50 pmol lactose/min/mg of protein), and showed a noninducible β-galactosidase activity of about 5 U. Therefore, LacP cannot be replaced by another transporter and is required for induction. In the sucrose utilization system of S. xylosus, inactivation of the sucrose permease gene did not result in the loss of sucrose-mediated induction of gene expression (50).
Glucose-mediated repression of β-galactosidase activity in the wild type and in catabolite repression mutants.
In previous studies on catabolite repression in S. xylosus, we have characterized two genes which are involved in this global regulatory process. glkA, the first gene that was isolated, encoded a glucose kinase (51). Inactivation of glkA resulted in a partial loss of glucose-specific repression of several catabolic enzymes, including β-galactosidase. The second gene encoded the catabolite control protein CcpA (11). Disruption of ccpA relieved some catabolic enzymes completely from repression by sugars, such as glucose, sucrose, or fructose. However, part of the glucose-mediated repression of β-galactosidase activity persisted. Since growing conditions used in the previous studies were slightly different from those in the lac induction experiments reported above, glucose repression of β-galactosidase activity was reexamined.
In the wild type, the addition of glucose prevents lactose-mediated induction, reducing β-galactosidase activity about 20-fold (Table 3). As expected, glucose repression of β-galactosidase activity was partially relieved in the mutants. The residual reduction was about fourfold in the glucose kinase mutant, TX140, and in the ccpA mutant strain, TX154 (Table 3). In both cases, β-galactosidase activity in the presence of lactose appeared to be slightly different than that in the wild type. In TX140, it reached about 85% of the wild-type level, whereas the wild-type value was exceeded by about 20% in the ccpA mutant strain. The new determinations are in good agreement with the β-galactosidase activities obtained earlier (11, 51). Obviously, neither mutations in glkA nor those in ccpA lead to a complete loss of glucose repression of β-galactosidase activity.
TABLE 3.
β-Galactosidase activity in the S. xylosus wild type, C2a, and in the catabolite repression mutants TX140 (glkA) and TX154 (ccpA)
| Strain | Growth conditionsa | β-Galactosidase activityb (nmol of nitrophenol produced/min/mg of protein) |
|---|---|---|
| C2a | B | 9 ± 2 |
| B + lactose | 98 ± 14 | |
| B + lactose + glucose | 5 ± 1 | |
| TX140 (glkA) | B | 9 ± 1 |
| B + lactose | 83 ± 10 | |
| B + lactose + glucose | 20 ± 5 | |
| TX154 (ccpA) | B | 8 ± 2 |
| B + lactose | 119 ± 31 | |
| B + lactose + glucose | 28 ± 3 |
The strains were grown in B medium (B) to an OD578 of 1.2. The indicated sugars were added at 25 mM. After 1 h of growth, the cells were harvested. Cultures without additional sugar were grown and harvested accordingly.
The values (means ± standard deviations) represent two determinations, each with bacterial extracts from three cultures.
Analysis of lacPH transcription in the wild type, C2a.
To localize the transcriptional start site(s) of the lac genes and to analyze lac regulation at the transcriptional level, RNA was isolated from induced and noninduced S. xylosus cells and reverse transcription experiments were performed with lacH- and lacP-specific primers. As shown in Fig. 2A, reverse transcripts were obtained with a lacP primer. No transcriptional start site between lacP and lacH was detected. This result is consistent with lacH subcloning experiments, which indicated that lacH does not possess its own promoter (data not shown). Therefore, lacP and lacH are cotranscribed, forming a bicistronic operon.
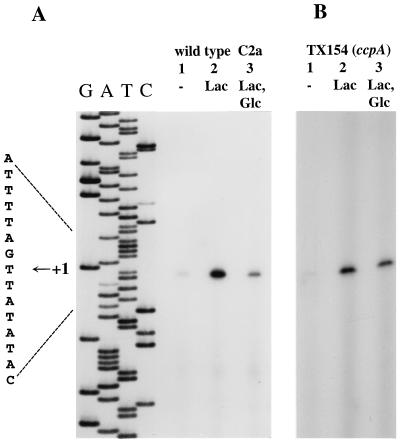
FIG. 2.
Primer extension analysis of lacPH transcription. (A) Analysis of lacPH transcription in the S. xylosus wild type, C2a. Total RNA was prepared from S. xylosus wild-type cells grown in B medium containing no additional sugar (lane 1), 25 mM lactose (lane 2), or 25 mM each lactose and glucose (lane 3). Fifteen micrograms of RNA and a 32P-labeled lacP-specific primer were used for the primer extension reactions. One-fourth of each reaction mixture was separated on a 5% polyacrylamide–urea gel, together with a sequencing reaction mixture obtained with the same primer. The autoradiograph and the sequence interpretation around the +1 site are shown. (B) Analysis of lacPH transcription in the S. xylosus ccpA mutant, TX154. Total RNA was prepared from S. xylosus cells grown in B medium containing no additional sugar (lane 1), 25 mM lactose (lane 2), or 25 mM each glucose and lactose (lane 3). Fifteen micrograms of RNA and a 32P-labeled lacP-specific primer were used in the primer extension reactions. One-fourth of each reaction mixture was separated on a 5% polyacrylamide–urea gel. The autoradiograph of the gel is shown.
The transcriptional start site of the lacPH promoter was found 46 bp upstream of the lacP start codon (Fig. 1). The amount of reverse transcript obtained with RNA isolated from lactose-grown cells (Fig. 2A, lane 2) is much larger than that obtained with RNA from uninduced cultures (Fig. 2A, lane 1). The results of the primer extension experiments are in accordance with the lactose uptake and β-galactosidase activities measured under these conditions. They demonstrate that initiation of lacPH transcription is regulated in a lactose-dependent manner. Induction of lacPH transcription is dependent on LacR, since no lacPH transcript was detectable in the lacR mutant strain TX258 (data not shown).
Primer extension experiments with RNA isolated from cells that were grown with lactose and glucose yielded a less intense band than those with RNA from lactose-induced cells (Fig. 2A, lane 3). Therefore, glucose prevents efficient initiation of transcription at the lacPH promoter.
Analysis of lacPH transcription in catabolite repression mutants.
A palindromic sequence that could serve as an operator for the catabolite control protein CcpA is located in the lacPH promoter region (Fig. 1) (11, 20, 24). It resembles catabolite responsive elements (cres) (23), which have been found to be essential for CcpA-mediated catabolite repression in a number of AT-rich gram-positive bacteria and which constitute the binding sites for CcpA. It was therefore of interest to determine the consequences of ccpA inactivation on glucose-mediated repression of lacPH transcription.
As shown in Fig. 2B, lactose induced lacPH transcription initiation in the ccpA mutant as it did in the wild type. The reverse transcript obtained from RNA prepared from glucose-grown cells (Fig. 2B, lane 3) had nearly the same intensity as the band from induced cells (Fig. 2B, lane 2). Therefore, glucose repression of lacPH transcription initiation is mainly, but not exclusively, due to the action of CcpA.
The same primer extension experiments were performed with RNA isolated from the glucose kinase mutant, TX140. Inducibility of lacPH transcription by lactose was the same as that in the other tested strains (data not shown). In contrast to the reduced repression in the ccpA mutant, the primer extension product from glucose-repressed TX140 cells had about the same intensity as that from the wild type grown under the same conditions (data not shown). Apparently, glucose-mediated regulation of transcription initiation at the lacPH promoter is not significantly altered by the glkA mutation.
Transcriptional analysis of lacR.
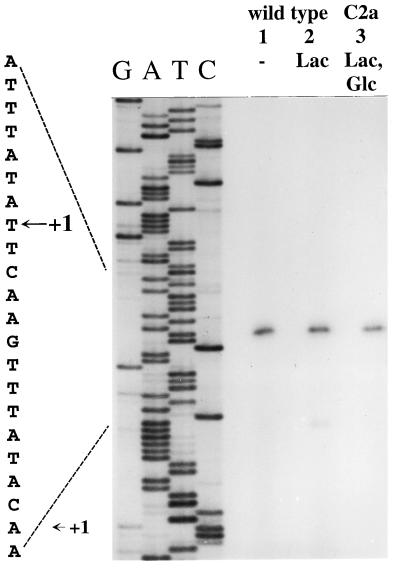
To determine the transcriptional start site of lacR, the same RNAs as those for the lacPH analysis were used in reverse transcription experiments with a lacR-specific primer. As shown in Fig. 3, a strong reverse transcript was observed, localizing the site of initiation 35 bp from the lacR start codon (Fig. 1). The bands were equally strong with RNAs from cells grown in the presence or absence of lactose. Therefore, initiation at this site is not inducible by lactose. However, a smaller, less intense primer extension product appeared when RNA from lactose-grown cells was used (Fig. 3, lane 2). Transcription at this second lacR promoter is initiated 22 bp upstream of the lacR start codon (Fig. 1). Therefore, lacR is transcribed from two promoters which differ in strength and inducibility. Transcription initiated at the major promoter, P1, occurs independently from lactose in the growth medium, whereas initiation at the minor promoter, P2, relies on lactose for induction. Accordingly, primer extension experiments in the lacR mutant, TX258, showed that the major transcript is present, whereas the minor one could not be detected (data not shown). Therefore, transcription starting from P2 requires a functional LacR activator.
FIG. 3.
Primer extension analysis of lacR transcription in the S. xylosus wild type, C2a. RNA was isolated from wild-type cells grown in B medium without additional sugar (lane 1), with lactose (lane 2), or with lactose and glucose (lane 3). Fifteen micrograms of RNA and a 32P-labeled lacR-specific primer were used in the primer extension reactions. One-fourth of the reaction mixtures were separated on 5% polyacrylamide–urea gels next to a sequencing reaction mixture obtained with the same primer. The autoradiograph and the sequence interpretation around the +1 sites are shown.
Glucose repression of lacR expression would be a conceivable mechanism to prevent induction of the lacPH operon. Therefore, lacR transcription in the presence of glucose was studied. As shown in Fig. 3, lane 3, transcription at P1 is hardly affected by glucose. Initiation at this promoter is constitutive with respect to the carbon source in the medium. On the other hand, the minor transcript from P2 is subject to glucose repression. No P2-specific reverse transcript was obtained with RNA from cells grown in the presence of lactose and glucose (Fig. 3, lane 3).
DISCUSSION
The lactose utilization system of S. xylosus is comprised of a lactose permease of the GPH family of sugar transporters (37), a β-galactosidase, which belongs to family 2 of glycosyl hydrolases (21), and a regulator of the AraC/XylS group of proteins (12). The system clearly differs from that found in Staphylococcus aureus, where lactose uptake is mediated by a phosphoenolpyruvate-dependent phosphotransferase system and internalized lactose-phosphate is cleaved by a phospho-β-galactosidase (4, 5, 35). In S. xylosus, β-galactosidase produces glucose and galactose from incoming lactose. Therefore, utilization of lactose depends on the phosphorylation of glucose and galactose by respective kinases. A gene encoding glucose kinase has been characterized in S. xylosus (51), and a genomic fragment that complemented an E. coli galactokinase mutant has been identified but not further characterized (8). It appears that both kinases needed for lactose utilization are present in S. xylosus.
The lacPH genes of S. xylosus are positively controlled by LacR, which constitutes, to our knowledge, the first example of a member of the AraC/XylS family regulating lac genes. In Staphylococcus aureus, Lactococcus lactis, and Streptococcus mutans, these genes are negatively controlled by repressors with similarity to DeoR of E. coli (9, 36, 38, 48, 49), whereas lac regulation in Lactobacillus casei is achieved by antitermination (1, 14, 40). Two other sugar catabolic operons in gram-positive bacteria appear to be controlled by AraC/XylS-type proteins, the multiple-sugar metabolism (msm) gene cluster in Streptococcus mutans (30, 39) and the raffinose utilization genes in Pediococcus pentosaceus (L32093).
The putative binding site for LacR in the lacPH promoter region could not be identified by sequence inspection. No sequences resembling operators for other AraC/XylS members could be detected (29, 34, 47, 53). In addition, no extended direct or inverted repeats are present in this area.
Apart from lactose-specific, LacR-mediated control, lacPH transcription is subject to CcpA-dependent carbon catabolite repression. The cre-like palindrome located from +7 to +20 with respect to the lacPH promoter is most likely the target for CcpA.
In the ccpA mutant strain, glucose still reduces β-galactosidase activity about fourfold (Table 3). Expression of lacH under control of a constitutive promoter showed that the activity of the enzyme is not affected by the carbon source in the medium (data not shown). Likewise, the β-galactosidase assay was not sensitive to the addition of glucose (data not shown). Therefore, the observed reduction of β-galactosidase activity is due to diminished lacPH expression.
It is well documented that glucose in the growth medium can reduce internal inducer concentrations by processes termed inducer exclusion and expulsion (41). Therefore, this regulatory mode should affect the activity of LacR in S. xylosus, resulting in less efficient initiation of transcription at the lacPH promoter. The comparable intensities of the reverse transcripts in the ccpA mutant strain (Fig. 2B, lanes 2 and 3) strongly indicate that inducer exclusion or expulsion plays a minor role, if any, in the CcpA-independent regulation of lacPH expression.
Additional evidence for lacPH expression control, which is not operating at the initiation of transcription, is provided by the β-galactosidase assays and lacPH primer extension analysis with the wild type. The β-galactosidase activity in glucose-repressed cells was lower than that in uninduced cells (Table 3). The opposite was true for intensities of the respective reverse transcripts, however (Fig. 2A).
Interestingly, this alternative level of glucose control seems to depend on a functional glucose kinase. In the glucose kinase mutant strain, initiation of transcription at the lacPH promoter is not altered but β-galactosidase activity in the presence of glucose is higher than in the wild type (Table 3). Premature termination of transcription, mRNA stability, or even posttranscriptional events could perhaps be affected by the presence of glucose and a functional glucose kinase. Further detailed analyses are needed to elucidate the role of glucose kinase in regulation. It appears that the lac genes constitute a good model system to study CcpA-dependent as well as CcpA-independent glucose repression in S. xylosus.
ACKNOWLEDGMENTS
We thank F. Götz, in whose laboratory the work was carried out, for his continuous interest and support, P. L. Huynh for expert technical assistance, and E. Knorpp for photographic work.
The work was supported in part by the European Community Biotech Programme (BIO2-CT92-0137) and by the Deutsche Forschungsgemeinschaft (BR 947/3-1).
REFERENCES
- 1.Alpert C-A, Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the BglG family of transcriptional antiterminators. J Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson T J, MacInnes J I. Expression and phylogenetic relationships of a novel lacZ homologue from Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 1997;152:117–123. doi: 10.1111/j.1574-6968.1997.tb10417.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Breidt F, Jr, Hengstenberg W, Finkeldei U, Stewart G C. Identification of the genes for the lactose-specific components of the phosphotransferase system in the lac operon of Staphylococcus aureus. J Biol Chem. 1987;262:16444–16449. [PubMed] [Google Scholar]
- 5.Breidt F, Jr, Stewart G C. Nucleotide and deduced amino acid sequences of the Staphylococcus aureus phospho-β-galactosidase gene. Appl Environ Microbiol. 1987;53:969–973. doi: 10.1128/aem.53.5.969-973.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brückner R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene. 1992;122:187–192. doi: 10.1016/0378-1119(92)90048-t. [DOI] [PubMed] [Google Scholar]
- 7.Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 8.Brückner R, Wagner E, Götz F. Characterization of a sucrase gene from Staphylococcus xylosus. J Bacteriol. 1993;175:851–857. doi: 10.1128/jb.175.3.851-857.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Vos W M, Vaughan E E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 10.Egeter O, Brückner R. Characterization of a genetic locus essential for maltose-maltotriose utilization in Staphylococcus xylosus. J Bacteriol. 1995;177:2408–2415. doi: 10.1128/jb.177.9.2408-2415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egeter O, Brückner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 12.Gallegos M-T, Michán C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gering M, Brückner R. Transcriptional regulation of the sucrase gene of Staphylococcus xylosus by the repressor ScrR. J Bacteriol. 1996;178:462–469. doi: 10.1128/jb.178.2.462-469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosalbes M J, Monedero V, Alpert C-A, Pérez-Martínez G. Establishing a model to study the regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol Lett. 1997;148:83–89. doi: 10.1111/j.1574-6968.1997.tb10271.x. [DOI] [PubMed] [Google Scholar]
- 15.Götz F. Applied genetics in the Gram-positive bacterium Staphylococcus carnosus. Food Biotechnol. 1990;4:505–513. [Google Scholar]
- 16.Götz F, Zabielski J, Philipson L, Lindberg M. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid. 1983;9:126–137. doi: 10.1016/0147-619x(83)90015-x. [DOI] [PubMed] [Google Scholar]
- 17.Hama H, Wilson T H. Primary structure and characteristics of the melibiose carrier of Klebsiella pneumoniae. J Biol Chem. 1992;267:18371–18376. [PubMed] [Google Scholar]
- 18.Hammes W P. Bacterial starter cultures in food production. Food Biotechnol. 1990;4:383–397. [Google Scholar]
- 19.Henikoff S, Haughn G W, Calvo J M, Wallace J C. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 21.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann K, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 23.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 24.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 25.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylS on the Pseudomonas putida TOL plasmid and identification of the protein product. Gene. 1986;44:235–242. doi: 10.1016/0378-1119(86)90187-3. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson R H, Zhang X-J, DuBose R F, Matthews B W. Three-dimensional structure of β-galactosidase from E. coli. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 27.Kalnins A, Otto K, Rüther U, Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Lu Y, Flaherty C, Hendrickson W. AraC protein contacts asymmetric sites in the Escherichia coli araFGH promoter. J Biol Chem. 1992;267:24848–24857. [PubMed] [Google Scholar]
- 30.McLaughlin R E, Ferretti J J. The multiple-sugar metabolism (msm) gene cluster of Streptococcus mutans is transcribed as a single operon. FEMS Microbiol Lett. 1996;140:261–264. doi: 10.1016/0378-1097(96)00191-7. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima K, Awakihara S, Kuroda M, Ishikawa T, Tsuda M, Tsuchiya T. Cloning and sequencing of the melB gene encoding the melibiose permease of Salmonella typhimurium LT2. Mol Gen Genet. 1992;234:74–80. doi: 10.1007/BF00272347. [DOI] [PubMed] [Google Scholar]
- 32.Morse M L, Alire M L. An agar medium indicating acid production. J Bacteriol. 1958;76:270–271. doi: 10.1128/jb.76.3.270-271.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller-Hill B. The lac operon: a short history of a genetic paradigm. Berlin, Germany: Walter de Gruyter; 1996. [Google Scholar]
- 34.Niland P, Hühne R, Müller-Hill B. How AraC interacts specifically with its target DNAs. J Mol Biol. 1996;264:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- 35.Oskouian B, Rosey E L, Breidt F, Jr, Stewart G C. The lactose operon of Staphylococcus aureus. In: Novick R P, Skurray R A, editors. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 99–112. [Google Scholar]
- 36.Oskouian B, Stewart G C. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J Bacteriol. 1990;172:3804–3812. doi: 10.1128/jb.172.7.3804-3812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poolman B, Knol J, van der Does C, Henderson P J F, Liang W-J, Leblanc G, Pourcher T, Mus-Veteau I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol Microbiol. 1996;19:911–922. doi: 10.1046/j.1365-2958.1996.397949.x. [DOI] [PubMed] [Google Scholar]
- 38.Rosey E L, Stewart G C. Nucleotide and deduced amino acid sequences of the lacR, lacABCD, and lacFE genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon of Streptococcus mutans. J Bacteriol. 1992;174:6159–6170. doi: 10.1128/jb.174.19.6159-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell R R B, Aduse-Opoku J, Sutcliffe I C, Tao L, Ferretti J J. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem. 1992;267:4631–4637. [PubMed] [Google Scholar]
- 40.Rutberg B. Antitermination of transcription of catabolic operons. Mol Microbiol. 1997;23:413–421. doi: 10.1046/j.1365-2958.1997.d01-1867.x. [DOI] [PubMed] [Google Scholar]
- 41.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1300–1309. [Google Scholar]
- 44.Schleifer K H, Kloos W E. Isolation and characterization of staphylococci from human skin. I. Amended descriptions of Staphylococcus epidermidis and Staphylococcus saprophyticus and descriptions of three new species: Staphylococcus cohnii, Staphylococcus haemolyticus and Staphylococcus xylosus. Int J Syst Bacteriol. 1975;25:50–61. [Google Scholar]
- 45.Sizemore C, Wieland B, Götz F, Hillen W. Regulation of Staphylococcus xylosus xylose utilization genes at the molecular level. J Bacteriol. 1992;174:3042–3048. doi: 10.1128/jb.174.9.3042-3048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinmetz M. Carbohydrate catabolism: pathways, enzymes, genetic regulation, and evolution. In: Hoch J A, Losick R, Sonenshein A L, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 157–170. [Google Scholar]
- 47.Tobin J F, Schleif R F. Purification and properties of RhaR, the positive regulator of the l-rhamnose operons of Escherichia coli. J Mol Biol. 1990;211:75–89. doi: 10.1016/0022-2836(90)90012-B. [DOI] [PubMed] [Google Scholar]
- 48.Valentin-Hansen P, Hojrup P, Short S. The primary structure of the DeoR repressor from Escherichia coli K-12. Nucleic Acids Res. 1985;13:5927–5936. doi: 10.1093/nar/13.16.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 50.Wagner E, Götz F, Brückner R. Cloning and characterization of the scrA gene encoding the sucrose-specific enzyme II of the phosphotransferase system from Staphylococcus xylosus. Mol Gen Genet. 1993;241:33–41. doi: 10.1007/BF00280198. [DOI] [PubMed] [Google Scholar]
- 51.Wagner E, Marcandier S, Egeter O, Deutscher J, Götz F, Brückner R. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J Bacteriol. 1995;177:6144–6152. doi: 10.1128/jb.177.21.6144-6152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace R G, Lee N, Fowler A V. The araC gene of Escherichia coli: transcriptional and translational start-points and complete nucleotide sequence. Gene. 1980;12:179–190. doi: 10.1016/0378-1119(80)90100-6. [DOI] [PubMed] [Google Scholar]
- 53.Webster C, Gardner L, Busby S. The Escherichia coli melR gene encodes a DNA-binding protein with affinity for specific sequences located in the melibiose-operon regulatory region. Gene. 1989;83:207–213. doi: 10.1016/0378-1119(89)90106-6. [DOI] [PubMed] [Google Scholar]
- 54.Yazyu H, Shiota-Niiya S, Shimamoto T, Kanazawa H, Futai M, Tsuchiya T. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J Biol Chem. 1984;259:4320–4326. [PubMed] [Google Scholar]
- 55.Youngman P, Poth H, Green B, York K, Olmedo G, Smith K. Methods for genetic manipulation, cloning, and functional analysis of sporulation genes in Bacillus subtilis. In: Smith I, Slepecky R A, Setlow P, editors. Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. Washington, D.C: American Society for Microbiology; 1989. pp. 65–87. [Google Scholar]