Abstract
Yeast (CUP1) and mammalian (HMT-1A) metallothioneins (MTs) have been efficiently expressed in Escherichia coli as fusions to the outer membrane protein LamB. A 65-amino-acid sequence from the CUP1 protein of Saccharomyces cerevisiae (yeast [Y] MT) was genetically inserted in permissive site 153 of the LamB sequence, which faces the outer medium. A second LamB fusion at position 153 was created with 66 amino acids recruited from the form of human (H) MT that is predominant in the adipose tissue, HMT-1A. Both LamB153-YMT and LamB153-HMT hybrids were produced in vivo as full-length proteins, without any indication of instability or proteolytic degradation. Each of the two fusion proteins was functional as the port of entry of lambda phage variants, suggesting maintenance of the overall topology of the wild-type LamB. Expression of the hybrid proteins in vivo multiplied the natural ability of E. coli cells to bind Cd2+ 15- to 20-fold, in good correlation with the number of metal-binding centers contributed by the MT moiety of the fusions.
Widespread pollution by heavy metals has important consequences for human health and environmental quality (32). Higher organisms respond systematically to the presence of heavy metals with the production of metallothioneins (MTs). This name was first used in 1957 for a Cd2+-binding protein from mammalian kidneys (27), and it is currently applied to a number of low-molecular-weight Cys-rich proteins that bind metal ions (e.g., Zn2+, Cd2+, Cu2+, Hg2+, and Ag+) and sequester them in a biologically inactive form (8, 17). MTs are widely distributed among living organisms, and they are fairly well conserved in mammals, plants, and fungi (8, 20). Based on structure, MTs have been subdivided into two classes. Class I includes those polypeptides related to mammalian species (25), while those that are more divergent but are still able to chelate metal ions efficiently are considered class II. Mammalian MTs are usually composed of 61 amino acids (molecular mass, 6 to 7 kDa) and lack aromatic amino acids and histidines. Two distinct domains of these proteins coordinate 7 divalent or 12 monovalent metal ions with 20 Cys residues. These are present along the sequence in the form of Cys-X-Cys or Cys-Cys motifs (X is any other amino acid residue), which are characteristic and invariant for this class of proteins. Class II MTs originate from nonanimal sources, such as yeasts (e.g., Saccharomyces cerevisiae, Candida glabrata, and Candida albicans [28]), algae (36), or cyanobacteria (e.g., Synechococcus sp. [33]). A well-known member of class II is the S. cerevisiae MT responsible for copper tolerance, called CUP1. This product is synthesized as a polypeptide of 61 amino acids, but its leading 8 residues are posttranslationally cleaved off, resulting in a 53-residue polypeptide. This protein contains 12 cysteine residues organized in Cys-X-Cys, Cys-Cys, and Cys-X-X-Cys motifs which originate eight binding sites for monovalent and four binding sites for divalent metal ions (44).
Earlier attempts to produce MTs in bacterial cells (i.e., Escherichia coli) as a way to increase their metal-binding ability were successful in some cases (2, 22, 31, 37). However, expression of such Cys-rich proteins is not devoid of problems because of the predicted interference with the redox pathways in the cytosol (1, 30, 34, 35, 45). Intracellular expression of MTs has been difficult to detect, perhaps because they become quickly degraded by host proteases (14, 39) unless they are associated with stabilizing moeities (13). More practically, intracellular expression of MTs may prevent the recycling of the biomass by desorption of the accumulated metal (16).
In this study, we examine the performance of the outer membrane protein of E. coli designated LamB as a carrier in vivo for expression of eukaryotic MTs and the resulting increase in the metalloadsorption by the bacterial cells. LamB is the port of entry for maltose and maltodextrins through the outer membrane as well as the receptor for phage lambda (19). The active LamB includes three monomers, each containing 18-stranded antiparallel β sheets arranged as a barrel and connected to each other by rather long loops and turns (19). The protein segments spanning amino acid positions 153 to 154 and 183 to 184 (located on the outer surface and in the cell periplasm, respectively [Fig. 1]) tolerate insertions of heterologous peptides of various sizes without disrupting the overall structure of the protein or its ability to assemble in functional trimers (5, 9, 12, 18). These two sites were used to construct and express LamB-MT hybrids in various configurations. Our results show that expression in vivo of LamB derivatives in which the yeast (YMT) or the human MT (HMT) 1A sequences were genetically grafted to an external permissive site multiplies the natural ability of E. coli cells to accumulate metal ions such as Cd2+ 15- to 20-fold.
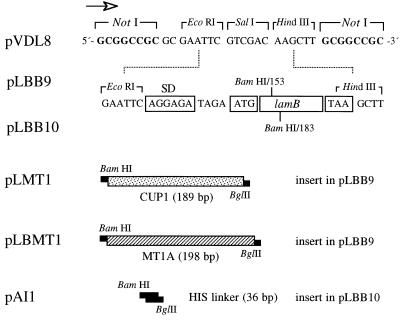
FIG. 1.
Organization of lamB-MT and lamB-HIS hybrid genes. The figure shows relevant sequences of the constructs used to insert metal-chelating sequences within LamB and their expression as fusions in the outer membrane of E. coli. pLBB9 and pLBB10 are derivatives of low-copy-number vector pVDL8 which harbor a 1.4-kb EcoRI-HindIII restriction fragment spanning the lamB sequence with BamHI sites engineered at structural codons 153 and 183, respectively. The orientation of the lac promoter present in the vector is marked with an arrow. The ribosomal binding site (SD), the first structural codon (ATG), and the stop codon TAA are indicated at the extremes of the structural sequence. The inserts born by the different plasmids listed to the left are indicated (not to scale) as follows: CUP1 of S. cerevisiae, encoding YMT; MT1A, encoding HMT-1A; and the HIS linker (41), encoding an artificial six-His sequence.
MATERIALS AND METHODS
Strains, plasmids, and general procedures.
E. coli TG1 (supE hsd his Δlac-proAB F′[traD36 proAB+ lacIq lacZΔM15]) and E. coli CC118 F′SURE were used to host recombinant plasmids. E. coli C600 (40) was used as the reference λ phage-sensitive strain. The lamB-minus E. coli K-12 strain pop6510 (thr leu tonB thi lacY1 recA dex5 metA supE) was used as the recipient of all plasmids bearing LamB variants (11, 12). The vector for expression of protein fusions to the position 153 or 154 of the amino acid sequence of LamB, called pLBB9, has been described before (9). pLBB9 is a derivative of the pSC101-based Cmr vector (15) bearing a lac promoter in front of a lamB sequence variant (11, 12) with a BamHI site overlapping codon 153 of the gene. The equivalent vector for expression of fusions to position 183 of LamB (which faces the periplasm) is called pLBB10 (Fig. 1).
Low-phosphate MJS medium contained 12.5 mM HEPES (pH 7.1), 50 mM NaCl, 20 mM NH4Cl, 1 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 0.05 mM MnCl2, 0.8% (wt/vol) Casamino Acids, 0.4% (vol/vol) glycerol, and 0.005% (wt/vol) thiamine. Both MJS medium and complete Luria-Bertani medium (29) were supplemented, when required, with 30 μg of chloramphenicol and 100 μM isopropyl thiogalactopyranoside (IPTG). Recombinant DNA techniques were carried out according to standard protocols (38). Insertion and orientation of the HIS linkers, YMT, and HMT-1A within the lamB sequence (Fig. 2) were verified by subjecting the clones under examination to a PCR under the conditions specified in reference 9.
FIG. 2.
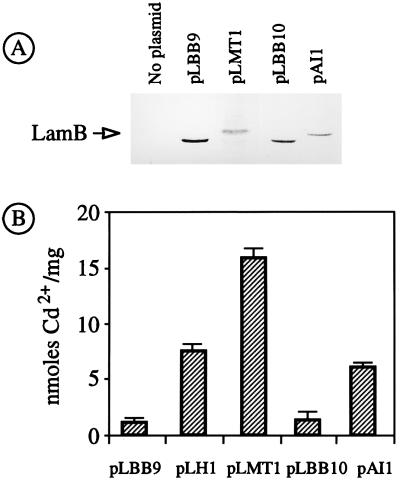
(A) Expression of LamB hybrid proteins in E. coli pop6510. Approximately 108 E. coli pop6510 cells transformed with each of the plasmids indicated were run in a denaturing polyacrylamide gel, blotted on a membrane and probed with a preadsorbed polyclonal anti-LamB serum, and further developed with protein A conjugated to horseradish peroxidase. (B) Accumulation of Cd2+ by cells expressing hybrid LamB proteins. The same transformants as for panel A were grown in a medium with 20 μM Cd2+ and examined for their metal content, with the results shown in the plot. The data shown are the mean values (+ standard deviation) of three separate experiments. pLH1 is a previously reported control plasmid (41) in which a six-His peptide is inserted in position 153 of the LamB sequence.
Construction and expression of hybrid LamB proteins.
The source of the YMT gene sequence was plasmid pUC7-YMTL, which contains the CUP1 coding sequence of S. cerevisiae (2). Two PCR primers, 5′-AAAGGATCCGTTCAGCGAATTAATTAACTTCCAA-3′ and 5′-GTTAGATCTGTTTTCCCAGAGCAGCATGACTTCTT-3′, were designed to amplify the entire gene sequence as a BamHI/BglII DNA fragment. This fragment was inserted into the BamHI sites of vectors pLBB9 and pLBB10, resulting in plasmids pLMT1 and pLMT2. The source of the edited HMT-1A gene sequence was phagemid clone HAWAU68 (ATCC 100746), which spans the α and β domains of the protein, originated in a cDNA isolated from adipose tissue. Similarly to the case of the yeast counterpart, PCR primers 5′-AAGGATCCCGAAATGGACCCCAACTGC-3′ and 5′-CATAGATCTGAGGCACAGCAGCTGCACTTC-3′ were designed to amplify the HMT-1A sequence from HAWAU68. These primers introduced BamHI and BglII sites for the purposes of allowing insertion of the sequence into the unique BamHI site of pLBB9 and originating the corresponding fusion at position 153 of the protein sequence. The resulting plasmid was named pLBMT1. Plasmid pLH1, encoding a LamB variant with a poly-His insert in position 153, has been described before (41). The equivalent plasmid expressing the same His cluster fused to position 183 of LamB was constructed by inserting a synthetic HIS linker encoding the sequence Asp-Pro-Ser-Gly-His-His-His-His-His-His-Ser-Gly in the BamHI site of vector pLBB10. The resulting plasmid was called pAI1.
Sensitivity of E. coli to λ phage variants.
Lambda phages λh+ (wild type), λh0, (single mutant), and λhh* (double mutant) have been described elsewhere (6, 10). High-titer phage stocks were prepared by infection and lysis of the permissive strain E. coli C600 as reported before (40). Approximately 100 μl of each of these lysates (titer, ∼1010 CFU/ml) was spread in a vertical line across the surface of Luria-Bertani agar plates (supplemented with chloramphenicol and 100 μM IPTG) and allowed to dry. E. coli pop6510 transformants bearing each of the plasmids encoding LamB variants were then spread perpendicular to and across the phage line in a single swath. Overnight incubation of the plates at 37°C revealed the sensitivity or resistance of each transformant to the corresponding phage lysate.
Protein techniques.
Whole-cell extracts were examined by denaturing 12% polyacrylamide gel electrophoresis (26). When required, proteins were transferred onto Immobilon membranes (Millipore) by electroblotting (43), treated with 2% skim milk in phosphate-buffered saline (PBS; 10 mM sodium phosphate [pH 7.4], 150 mM NaCl, 3 mM KCl) for 30 min, and then washed three times for 10 min each time with the same buffer. Anti-LamB polyclonal rabbit serum (a kind gift of M. Hofnung) was preadsorbed with a cell extract of E. coli pop6510 and added at a 1:1,000 dilution to the blotted and blocked membranes. Following 1 h of incubation with the serum, the blots were washed three times for 10 min each time with PBS buffer and then incubated with 0.5 mg of protein A-peroxidase conjugate (Sigma)/ml. This was followed by another wash with PBS for 15 min and rinsing with distilled water. The position of the LamB protein and its derivatives was revealed with 0.02% diaminobenzidine tetrahydrochloride (Sigma) and 0.03% oxygen peroxide. Alternatively, gels were electroblotted on nitrocellulose membranes (Bio-Rad) which were blocked with 10% skim milk in TBS (20 mM Tris-Cl, 250 mM NaCl, 3 mM KCl). In this case, anti-LamB serum was applied at a 1:5,000 dilution in TBST (0.1% Tween 20 in TBS) with 2% skim milk for 2 h. Then the blots were washed with TBST, incubated with swine anti-rabbit antibody conjugated with alkaline phosphatase (Bio-Rad), and finally washed again with TBST. The LamB and LamB-MT1 were visualized with 5-bromo-4-chloro-3-indolylphosphate as a substrate along with nitroblue tetrazolium.
Measurement of Cd2+ adsorption to and desorption from the biomass.
Bioaccumulation of Cd2+ was measured in cells growing at 37°C in MJS medium with chloramphenicol. The cells were induced with 100 μM IPTG when cultures reached an absorbance of 0.4, and then 20 μM Cd2+ was added in order to allow expression of the LamB-MT hybrids in the presence of the cation (—SH groups not bound by metal ions quickly become oxidized). The cultures were grown for another 4 h. Prior to determination of metal content, the bacterial cells were pelleted, washed twice with 0.85% NaCl in 5 mM HEPES (pH 7.1), and treated overnight with 70% nitric acid (37). Cd2+ concentrations were measured directly from the soluble fraction resulting from this acid treatment by atomic absorption with a spectrophotometer (Hitachi Z-8200 or Varian Spectra A300). Alternatively, cells collected after being washed were incubated on ice for 15 min with an excess volume of 5 mM EDTA (pH 8.0) to remove the surface-bound metal. The supernatant resulting from this treatment was then subjected to atomic absorption analysis as described above. The same procedure was used to determine bioaccumulation of Cu2+ and Zn2+.
RESULTS AND DISCUSSION
Structural tolerance of LamB to yeast MT and His fusions.
To address the issue of whether LamB could act as a molecular anchor for the expression of eukaryotic MTs in E. coli, we started by inserting the YMT sequence at two different sites in LamB, i.e., positions 153 and 183. To this end, the same PCR fragment spanning the YMT sequence was inserted into the BamHI sites of equivalent vectors pLBB9 (around 154) and pLBB10 (around 183), generating plasmids pLMT1 (lamB154-YMT+) and pLMT2 (lamB183-YMT+).
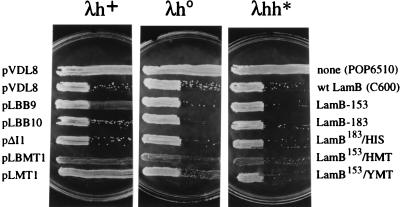
E. coli pop6510(pLMT1) displayed normal growth in both liquid and solid media compared to the same strain carrying pLBB9 vector without an insert. The growth rate (approximately 2 h−1 in MJS medium during exponential growth) was not significantly altered when expression of LamB154-YMT was induced with IPTG. Immunoblotting of crude bacterial extracts with a polyclonal anti-LamB serum demonstrated expression of the full-length hybrid protein (Fig. 2A). As expected, the hybrid protein displayed an increase in its apparent molecular weight in comparison to that of the wild-type protein. The same experiment (Fig. 2A) indicated that lamB154-YMT is expressed to a level in the range of that of LamB devoid of inserts. Furthermore, Western blot assays of cell fractions from E. coli pop6510(pLMT1) indicated that the LamB variant encoded by the plasmid is entirely located at the outer membranes of the cells (data not shown). In order to determine whether the hybrid was not only expressed and secreted but also assembled in the outer membrane of E. coli with a topology not unlike that of wild-type LamB, we examined the sensitivity of E. coli pop6510(pLMT1) to lambda phages λh+ (wild type) and its variants λh0 and λhh* (Fig. 3). In spite of the permissiveness of these variants to some changes on the LamB surface, infection in all cases requires the assembly of a defined LamB trimer (6, 10). Therefore, sensitivity of the LamB hybrids to one or more of these phages is evidence of the correct folding and domain positioning of the fusion protein at the outer membrane. Figure 3 shows that E. coli pop6510(pLMT1) was resistant to wild-type phage but sensitive to the λhh* variant. Taken together, these results indicated that LamB154-YMT was expressed in E. coli to roughly the same extent and with roughly the same properties as the wild-type protein. On the contrary, when pLMT2 plasmid carrying lamB183-YMT was introduced into E. coli pop6510, no expression of the corresponding hybrid could be detected by any of these procedures. When the MT insert at position 183 of LamB was replaced by a shorter poly-His peptide, the resulting LamB183-HIS protein (encoded by pAI1) was detectable in Western blots (Fig. 2), could be located in the outer membrane, and endowed E. coli pop6510 with sensitivity to all phage variants (Fig. 3). These data suggested that it was the size of the YMT insertion and not its metal-binding properties that hindered expression of lamB183-YMT.
FIG. 3.
Sensitivity of E. coli pop6510 expressing LamB hybrids to λ phage variants. The lamB mutant strain E. coli pop6510 was transformed with each of the plasmids listed to the left and subjected to a sensitivity assay with phages λh+ (wild type), λh0 (single mutant), and λhh* (double mutant), as described in Materials and Methods. E. coli C600 was used as the reference phage-sensitive strain. The proteins expressed by the transformants in each case are indicated to the right.
Expression of a HMT fusion to LamB.
In view of the fact that only site 153 of LamB appeared to be adequate for production in vivo of YMT fusions, we also employed it to insert the HMT-1A gene (17, 25). This MT species was chosen on the basis of its superior metal-binding capacity compared to that of its yeast counterpart, owing to the presence of twice as many metal-binding centers in its structure (three sites at its α domain and four sites at its β domain). The insertion of the entire DNA segment spanning 198 bp of the HMT-1A sequence within LamB was predicted to result in a hybrid protein with an extra 66 amino acids anchored at position 153 and facing the external medium. To verify these predictions, E. coli pop6510(pLBMT1) cells expressing LamB153-HMT were passed through the same battery of assays as the other hybrids described above to determine the expression, location, and correct assembly of the protein. The results shown in Fig. 3 and 4 indicated that like LamB153-YMT, the LamB hybrid with the human gene product, was present in a stable fashion in the outer membrane fractions of the bacteria. Interestingly, E. coli pop6510(pLBMT1) was fully resistant to λh+ and partially resistant to λh0 but sensitive to λhh* (Fig. 3), a behavior somewhat different from that of cells expressing the yeast gene.
FIG. 4.
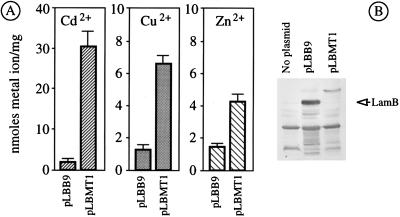
(A) Bioaccumulation of divalent ions by lamB153-HMT+ strain E. coli pop6510(pLBMT1). E. coli pop6510 cells transformed with either pLBMT1 (lamB153-HMT+) or the control plasmid pLBB9 (devoid of inserts in lamB), as indicated, were grown in MJS medium and induced with IPTG, and 30 μM of either Cd2Cl, Cu2Cl, or Zn2Cl was added. Their metal content after expression of the LamB hybrids is shown. The bars represent the mean values of three separate measurements. Note the different scales, depending on the metal. (B) Expression of LamB153-HMT in E. coli pop6510(pLBMT1). Approximately 2 × 108 E. coli pop6510 cells transformed with the plasmids indicated were run in a polyacrylamide gel electrophoresis system, blotted and probed with a crude (not preadsorbed) polyclonal anti-LamB serum, and further developed with swine anti-rabbit antibody conjugated with alkaline phosphatase.
Metal-binding properties of MT fusions to LamB in vivo.
In order to test the ability of bacterial cells expressing various LamB hybrids with metal-binding peptides to increase bioaccumulation in vivo of Cd2+, we monitored binding of the metal to E. coli pop6510 cells transformed independently with pLMT1 (lamB153-YMT+), pLBMT1 (lamB153-HMT+), and pAI1 (lamB183-HIS+) through atomic absorption spectrometry. As a control, we employed cells expressing LamB devoid of inserts (i.e., bearing pLBB9 or pLBB10) as well as E. coli pop6510 cells transformed with pLH1, a plasmid which expresses the metal-binding LamB variant LamB153-HIS (41). For these experiments, we grew the cells in the low-phosphate MJS medium (to avoid chelation of the metal ions) supplemented with a subinhibitory concentration of Cd2+ (20 to 30 μM). As shown in Fig. 2 and 4, cells expressing LamB hybrids displayed metal-binding capacities superior to those of the controls, albeit to different extents.
Strains bearing plasmids pLH1 (lamB153-HIS+) and pAI1 (lamB183-HIS+), which express the same metal-binding peptide at different cell compartments, increased their accumulation of Cd2+ by ca. fivefold. E. coli pop6510(pLMT1), which expresses the hybrid with the YMT, LamB153-YMT, more than doubled this amount (Fig. 2) and increased metalloadsorption to >15 nmol of Cd2+/mg (dry weight) of cells. Finally, cells expressing the human hybrid LamB153-HMT (Fig. 4) further increased metalloadsorption by ca. 30 nmol of Cd2+/mg (dry weight) of cells. These results are consistent with the increased number of metal-binding centers present in each of the hybrid proteins.
The best strain in terms of metal accumulation, E. coli pop6510(pLBMT1), was subjected to additional tests to gain some insight into the physiology of the adsorption. Figure 4 shows the data on accumulation of Zn2+ and Cu2+ by this strain under the same conditions employed with Cd2+. Although there was an evident increase in metal ion binding, the absolute figures were below those observed with Cd2+. In no case was the level of tolerance of E. coli pop6510(pLBMT1) to any of the metals assayed increased by the expression of the hybrid (not shown), thus reinforcing the notion that heavy metal resistance and heavy metal adsorption are independent phenomena (24). Finally, E. coli pop6510(pLBMT1) cells preloaded with 31.9 ± 6.4 nmol of Cd2+/mg were subjected to a desorption assay with EDTA. Under the conditions specified in Materials and Methods, which did not affect cell viability, only 40% of the metal could be released from the bacteria. This result suggested that only about half of the bound cations were available on the cell surface, whereas the rest were occluded in other cell compartments. This is consistent with the realization that the total increase in bioaccumulation of Cd2+ by cells expressing LamB153-HMT exceeds by at least 1 order of magnitude the theoretical increase of metal-chelating centers contributed by the MT moiety of the fusion. This can only be explained if, besides direct binding of cations, the hybrid protein helps to increase the local metal concentration around the cells and thus facilitates interactions of the ions with other cell structures (3). The fate of the metal ions bound to LamB hybrids deserves further study (41).
Conclusion.
In this work, we have constructed and characterized LamB fusions in which the complete MT sequences are anchored by their N termini and C termini to the permissive site 153 of the protein. Such a double anchor appears to result in increased stability and maintenance of the topology of the hybrid and the properties of the two separate proteins. LamB-MT fusions increase by more than 1 order of magnitude the natural ability of E. coli cells to bind Cd2+, a trait that can be unequivocally traced to the expression and surface presentation of the metal-binding polypeptide. On this basis, it seems that the LamB protein is a versatile vector to expose not only peptides but even heterologous proteins of considerable size in an active form on the surfaces of different bacteria, such as E. coli, Salmonella typhimurium (42), and even nonenteric bacteria such as Pseudomonas (9). Expression of LamB-MT hybrids in environmentally robust strains of Ralstonia eutropha (formerly Alcaligenes eutrophus) and Pseudomonas putida (4) is under way in view of the potential for increasing the bioadsorption of cations from sites polluted with heavy metals (7, 16, 21, 23).
ACKNOWLEDGMENTS
We are indebted to M. Hofnung (Institut Pasteur, Paris, France) for the gift of various strains and anti-LamB serum. We also thank T. Sevilla and J. Rodríguez (F. Ciencia, U.A.M., Madrid, Spain) for atomic absorption measurements.
This work was funded by grants 937062IL (ALAMED) and ENV4-CT95-0141 (Environment) from the EC, grant 980157114 from the ICT, and grant BIO95-788 from the CICYT.
REFERENCES
- 1.Bardwell J C A. Building bridges: disulphide bond formation in the cell. Mol Microbiol. 1994;14:199–205. doi: 10.1111/j.1365-2958.1994.tb01281.x. [DOI] [PubMed] [Google Scholar]
- 2.Berka T, Shatzman A, Zimmermen J, Strickler J, Rosenberg M. Efficient expression of the yeast metallothionein gene in Escherichia coli. J Bacteriol. 1988;170:21–26. doi: 10.1128/jb.170.1.21-26.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beveridge T J. Role of cellular design in bacterial metal accumulation and mineralization. Annu Rev Microbiol. 1989;43:147–171. doi: 10.1146/annurev.mi.43.100189.001051. [DOI] [PubMed] [Google Scholar]
- 4.Bordons A, Jofre J. Extracellular adsorption of nickel by a strain of Pseudomonas sp. Enzyme Microb Technol. 1987;9:709–713. [Google Scholar]
- 5.Boulain J C, Charbit A, Hofnung M. Mutagenesis by random linker insertion into lamB gene of E. coli K-12. Mol Gen Genet. 1986;205:339–348. doi: 10.1007/BF00430448. [DOI] [PubMed] [Google Scholar]
- 6.Braun-Breton C, Hofnung M. In vivo and in vitro functional alterations of the bacteriophage lambda receptor in lamB missense mutants of Escherichia coli K-12. J Bacteriol. 1981;148:845–852. doi: 10.1128/jb.148.3.845-852.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S. Metal-recognition by repeating polypeptides. Nat Biotechnol. 1997;15:269–272. doi: 10.1038/nbt0397-269. [DOI] [PubMed] [Google Scholar]
- 8.Butt T R, Ecker D J. Yeast metallothionein and applications in biotechnology. Microbiol Rev. 1987;51:351–364. doi: 10.1128/mr.51.3.351-364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cebolla A, Guzmán C, de Lorenzo V. Nondisruptive detection of activity of catabolic promoters of Pseudomonas putida with an antigenic surface reporter system. Appl Environ Microbiol. 1996;62:214–220. doi: 10.1128/aem.62.1.214-220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charbit A, Hofnung M. Isolation of different bacteriophages using the LamB protein for adsorption on Escherichia coli K-12. J Virol. 1985;53:667–671. doi: 10.1128/jvi.53.2.667-671.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charbit A, Boulain J C, Ryter A, Hofnung M. Probing the topology of a bacterial membrane protein by genetic insertion of a foreign epitope: expression at the cell surface. EMBO J. 1986;5:3029–3037. doi: 10.1002/j.1460-2075.1986.tb04602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charbit A, Ronco J, Michel V, Werts C, Hofnung M. Permissive sites and topology of an outer membrane protein with a reporter epitope. J Bacteriol. 1991;173:262–275. doi: 10.1128/jb.173.1.262-275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Wilson D B. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2+-contaminated environments. Appl Environ Microbiol. 1997;63:2442–2445. doi: 10.1128/aem.63.6.2442-2445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derman A I, Prinz W A, Belin D, Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 15.Fernández S, de Lorenzo V, Pérez-Martín J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 16.Gadd G M, White C. Microbial treatment of metal pollution: a working biotechnology? Trends Biotechnol. 1993;11:353–359. doi: 10.1016/0167-7799(93)90158-6. [DOI] [PubMed] [Google Scholar]
- 17.Hamer D H. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- 18.Hofnung M. Expression of foreign polypeptides at the Escherichia coli cell surface. Methods Cell Biol. 1991;4:77–105. doi: 10.1016/s0091-679x(08)61677-5. [DOI] [PubMed] [Google Scholar]
- 19.Hofnung M. An intelligent channel (and more) Science. 1995;267:473–474. doi: 10.1126/science.7824946. [DOI] [PubMed] [Google Scholar]
- 20.Huckle J W, Morby A, Turner J, Robinson N. Isolation of prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol Microbiol. 1993;7:177–187. doi: 10.1111/j.1365-2958.1993.tb01109.x. [DOI] [PubMed] [Google Scholar]
- 21.Hutchins S R, Davidson M, Brierley J, Brierly C. Microorganisms in reclamation of metals. Annu Rev Microbiol. 1986;40:311–336. doi: 10.1146/annurev.mi.40.100186.001523. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs F A, Romeyer F M, Beaucheim M, Brousseau R. Human metallothionein-II is synthesized as a stable membrane-localized fusion protein in Escherichia coli. Gene. 1989;83:95–103. doi: 10.1016/0378-1119(89)90407-1. [DOI] [PubMed] [Google Scholar]
- 23.Jeong B C, Hawes C, Bonthrone K M, Macaskie L E. Localization of enzymatically enhanced heavy metal accumulation by Citrobacter sp. and metal accumulation in vitro by liposomes containing entrapped enzyme. Microbiology. 1997;143:2497–2507. doi: 10.1099/00221287-143-7-2497. [DOI] [PubMed] [Google Scholar]
- 24.Ji G, Silver S. Bacterial resistance mechanisms for heavy metals of environmental concern. J Ind Microbiol. 1995;14:61–75. doi: 10.1007/BF01569887. [DOI] [PubMed] [Google Scholar]
- 25.Kagi J H R. Overview of metallothionein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Margoshes M, Vallee B. A cadmium protein from equine kidney cortex. J Am Chem Soc. 1957;79:4813–4814. [Google Scholar]
- 28.Mehra R, Winge D. Metal ion resistance in fungi: molecular mechanisms and their regulated expression. J Cell Biochem. 1991;45:30–40. doi: 10.1002/jcb.240450109. [DOI] [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 30.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murooka Y, Nagaoka T. Expression of cloned monkey metallothionein in Escherichia coli. Appl Environ Microbiol. 1987;53:204–207. doi: 10.1128/aem.53.1.204-207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nriagu J O, Pacyna J M. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature. 1989;333:134–139. doi: 10.1038/333134a0. [DOI] [PubMed] [Google Scholar]
- 33.Olafson R W, McCubbin W, Kay C. Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from Synechococcus sp. cyanobacterium. Biochem J. 1988;251:691–699. doi: 10.1042/bj2510691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pugsley A P. Translocation of folded protein across the outer membrane in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina S, Missiakas D. Making and breaking disulphide bonds. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- 36.Reed R H, Gadd G. Metal tolerance in eukaryotic and prokaryotic algae. In: Shaw A J, editor. Heavy metal tolerance in plants: evolutionary aspects. Boca Raton, Fla: CRC Press; 1991. pp. 105–118. [Google Scholar]
- 37.Romeyer F M, Jacobs F A, Masson L, Hana Z, Brousseau R. Bioaccumulation of heavy metals in Escherichia coli expressing an inducible synthetic human metallothionein gene. J Biotechnol. 1988;8:207–220. [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Silhavy T J, Benson S A, Emr S D. Mechanisms of protein localization. Microbiol Rev. 1983;47:313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silhavy T J, Berman M, Enquist L. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 41.Sousa C, Cebolla A, de Lorenzo V. Enhanced metallo-adsorption of bacterial cells displaying poly-His peptides. Nat Biotechnol. 1996;14:1017–1020. doi: 10.1038/nbt0896-1017. [DOI] [PubMed] [Google Scholar]
- 42.Su G-F, Brahmbhatt H N, Wehland J, Rohde M, Timmis K N. Construction of stable LamB-Shiga toxin B subunit hybrids: analysis of expression in Salmonella typhimurium aroA strains and stimulation of B subunit-specific mucosal and serum antibody responses. Infect Immun. 1992;60:3345–3359. doi: 10.1128/iai.60.8.3345-3359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winge D R, Nielson K B, Gray W, Hamer D. Yeast metallothionein: sequence and metal-binding properties. J Biol Chem. 1985;260:14464–14470. [PubMed] [Google Scholar]
- 45.Yasukama T, Kanei-Ishii C, Maekama T, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J Biol Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]