Abstract
Most uveal melanoma cases harbor activating mutations in either GNAQ or GNA11. Despite activation of the mitogen-activated protein kinase (MAPK) signaling pathway downstream of Gαq/11, there are no effective targeted kinase therapies for metastatic uveal melanoma. The human genome encodes numerous understudied kinases, also called the “dark kinome”. Identifying additional kinases regulated by Gαq/11 may uncover novel therapeutic targets for uveal melanoma. In this study, we treated GNAQ-mutant uveal melanoma cell lines with a Gαq/11 inhibitor, YM-254890, and conducted a kinase signaling proteomic screen using multiplexed-kinase inhibitors followed by mass spectrometry. We observed downregulated expression and/or activity of 22 kinases. A custom siRNA screen targeting these kinases demonstrated that knockdown of microtubule affinity regulating kinase 3 (MARK3) and serine/threonine kinase 10 (STK10) significantly reduced uveal melanoma cell growth and decreased expression of cell cycle proteins. Additionally, knockdown of MARK3 but not STK10 decreased ERK1/2 phosphorylation. Analysis of RNA-sequencing and proteomic data showed that Gαq signaling regulates STK10 expression and MARK3 activity. Our findings suggest an involvement of STK10 and MARK3 in the Gαq/11 oncogenic pathway and prompt further investigation into the specific roles and targeting potential of these kinases in uveal melanoma.
Keywords: cancer, microtubule affinity-regulating kinase 3, melanoma, g protein, mitogen-activated protein kinase (MAPK), guanine nucleotide-binding protein G(q) subunit alpha, proteomics, serine/threonine-protein kinase 10
The human genome encodes over 500 kinases that act by phosphorylating target proteins to activate or deactivate downstream signaling (1). Kinases regulate various cellular functions, including cell cycle progression, cytoskeletal structure, and cellular differentiation. Numerous cancer types are driven by dysfunctional kinase activity, often due to genetic mutations, as seen in cutaneous melanoma (BRAF) or lymphoma (BCR-ABL). Mutations that cause constitutive kinase activation can result in uncontrolled cell cycle progression and tumor growth. Beyond specific mutations, upregulated expression or activation of kinases may be a biomarker for aggressive cancer and result in aberrant downstream signaling. Although targeted therapies have been developed against well-studied kinases, many other kinases are understudied and may still be relevant to cancer progression (2, 3).
Uveal melanoma is the most common primary ocular malignancy in adults. Approximately 50% of patients experience metastasis, most commonly to the liver, and the 1-year survival rate of metastatic uveal melanoma patients is 15% (4, 5). Currently, there are only two FDA-approved therapies for metastatic uveal melanoma, Tebentafusp and Hepzato kit. Tebentafusp is an immune-based therapy limited to a subset of patients, and the Hepzato kit is a liver-directed administration of a chemotherapeutic agent, melphalan (6). Uveal melanoma accounts for 5% of all melanomas and is genetically distinct from cutaneous melanoma. It is characterized by a low mutational burden and harbors mutually exclusive somatic activating mutations in GNAQ or GNA11, encoding the heterotrimeric G protein guanine nucleotide-binding protein subunit αq or α11 (Gαq/11) (7). These mutations initiate and promote uveal melanoma growth by activating the phospholipase C (PLC)/protein kinase C (PKC)/MAPK and the Trio-Rho/Rac/yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) pathways (8, 9). Targeted kinase inhibitors have effectively treated cutaneous melanoma but show limited efficacy in uveal melanoma (10). For example, selumetinib, a MEK inhibitor used to treat cutaneous melanoma, did not significantly improve progression-free survival (PFS) in phase III clinical trials in uveal melanoma (11, 12). Additional inhibitors targeting PKC, MEK, and FAK (downstream of Rac and Rho) are being developed and tested in various combinations (13). Furthermore, direct targeting of mutant Gαq/11 is clinically challenging because of non-specific inhibition of wild-type Gαq/11 and systemic side effects (14). Due to concerns regarding the toxicity of and intrinsic resistance to existing targeted therapies, there is an urgent unmet clinical need to characterize downstream effectors of Gαq/11 to identify new therapeutic strategies for uveal melanoma.
Previous attempts to study the kinome landscape of uveal melanoma have focused on transcriptomic data (15, 16). The downside to this approach is that it does not consider protein expression or activity. In contrast, a functional proteomic technique using multiplexed inhibitor beads and mass spectrometry (MIB-MS) measures both kinase activity and expression levels (2, 3). This method is an unbiased, quantitative approach to detecting understudied kinases that may be relevant to tumor growth and cell survival. In this study, we employed MIB-MS kinome profiling to study changes in kinase expression and/or activation status after treatment with YM-254890, a Gαq/11 inhibitor. MIB-MS identified several understudied kinases that were downregulated after Gαq/11 treatment. A secondary targeted siRNA screen was conducted and demonstrated a dependency of uveal melanoma cell growth on microtubule affinity regulating kinase 3 (MARK3) and serine/threonine kinase 10 (STK10). Overall, this study highlights the importance of unexplored kinases in uveal melanoma.
Results
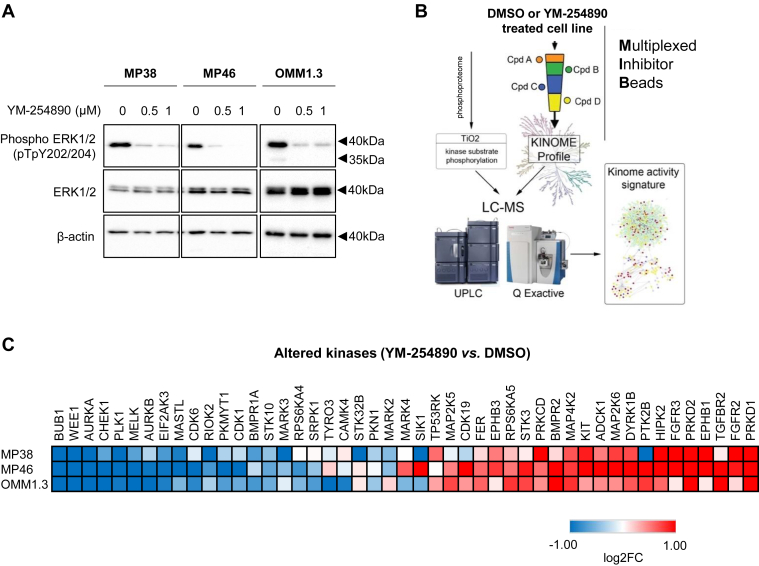
To identify kinases regulated by Gαq/11, we treated GNAQ-mutant MP38, MP46, and OMM1.3 uveal melanoma cells with the Gαq/11 inhibitor, YM-254890, for 24 h. Decreased ERK1/2 phosphorylation at the Thr202/Tyr204 sites following YM-254890 treatment was confirmed in all three cell lines (8, 17) (Fig. 1A). Lysates from DMSO (vehicle control) or YM-254890-treated cells were purified through MIB columns composed of competitive pan-kinase inhibitors, pulling down active and highly expressed kinases that were then identified by MS (3) (Fig. 1B). Approximately 250 kinases were captured with this technique in the three uveal melanoma cell lines. Comparing YM-254890-treated samples to DMSO control showed significant downregulation of cell cycle kinases such as Aurora A and WEE1 and cell division kinases such as PLK1. Additionally, YM-254890 led to an upregulation of PRKD1 and PRKD2, and receptor tyrosine kinases, including KIT, FGFR2, and FGFR3 (Figs. 1C and S1, A and B).
Figure 1.
Profiling the uveal melanoma kinome using quantitative MIB-MS after YM-254890 treatment shows downregulation of cell cycle, signal transduction, and cell division kinases.A, GNAQ-mutant uveal melanoma cell lines MP38, MP46, and OMM1.3 were treated for 24 h with 0.5 μM or 1.0 μM YM-254890. Cell lysates were analyzed by Western blotting for phospho-ERK1/2 and total ERK1/2. β-actin was used as loading control. B, schematic representation of kinome profiling using MIB-MS. Lysates from GNAQ-mutant uveal melanoma cells treated with either DMSO (vehicle control) or 1 μM YM-254890 were run through MIB columns followed by MS (n = 3). C, heatmap of altered kinases after 24 h of 1 μM YM-254890 treatment in uveal melanoma cell lines. Statistical significance was determined by the Student’s t test p-value < 0.05.
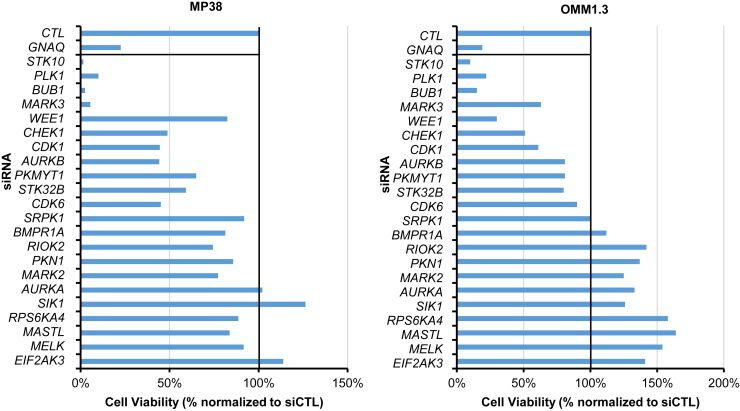
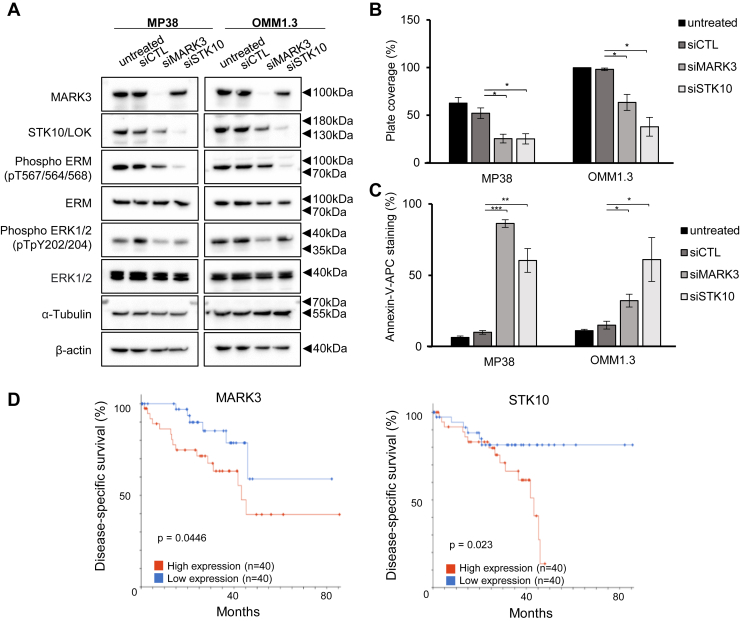
Next, we investigated kinases that were significantly downregulated in activity or expression after YM-254890 treatment in at least one cell line. We identified 22 downregulated kinases, some previously unexplored in uveal melanoma (Fig. 1C). We tested whether these 22 kinases were required for uveal melanoma cell growth using siRNA. Knockdown of GNAQ was used as a positive control (18, 19). Knockdown of STK10, PLK1, MARK3, BUB1, and WEE1 led to the greatest inhibition of viability in all three cell lines, although these effects were modest in MP46 cells (Figs. 2 and S2). Among these kinases, MARK3 and STK10 are unexplored in uveal melanoma. The knockdown efficiency of MARK3 and STK10 was confirmed by Western blotting (Fig. 3A). We also blotted for phosphorylation of putative downstream targets of MARK3 and STK10, including ERK1/2 and the ezrin-radixin-moesin (ERM) protein complex (20). As expected, the knockdown of STK10 reduced ERM complex phosphorylation, and interestingly, the knockdown of MARK3 reduced ERK1/2 phosphorylation and STK10 protein levels (Figs. 3A and S3). We validated the growth inhibitory effects of siRNA-mediated knockdown of STK10 and MARK3 by measuring cell confluency using an IncuCyte live cell imager. MARK3 or STK10 knockdown elicited significant growth inhibition in MP38 and OMM1.3 cells (39%–65% inhibition) (Fig. 3B). MARK3 or STK10 knockdown also increased cell death, as measured by the percentage of cells positive for annexin-V staining (Figs. 3C and S3B). MARK3 knockdown elicited a more pronounced effect on cell growth in MP38 cells, while OMM1.3 cells were more sensitive to STK10 knockdown. Knockdown of MARK3 or STK10 did not result in growth inhibition of BRAF mutant cutaneous melanoma cells, 1205Lu, suggesting that the effect of MARK3 or STK10 knockdown is Gαq/11-specific (Fig. S4, A and B). To investigate the clinical relevance of these kinases, we analyzed the disease-specific survival of uveal melanoma patients based on the expression of MARK3 and STK10 in TCGA. High expression of MARK3 or STK10 was associated with poor disease-specific survival amongst uveal melanoma patients (Fig. 3D). These data suggest that MARK3 and STK10 may be critical kinases downstream of Gαq/11 that are important in uveal melanoma cell growth.
Figure 2.
Loss-of-function screen identifies MARK3 and STK10 as potential therapeutic targets. Histogram of cell viability measured by CellTiter-Glo post-siRNA knockdown of 22 candidate kinases in MP38 (left panel) and OMM1.3 (right panel) cells. Targets are ordered based on average cell viability in all three cell lines.
Figure 3.
MARK3 and STK10 knockdown effects on target proteins and cell growth.A, representative Western blots of MARK3, STK10, and respective downstream targets in knockdown cells. B, effects of siRNA-mediated knockdown of MARK3 or STK10 on MP38 and OMM1.3 cell growth were analyzed using the IncuCyte Live Cell Analysis Imaging System by measuring % plate coverage (n = 3). ∗p < 0.05 as determined by t test, and error bars are ± SEM. C, annexin-V staining (%) following MARK3 or STK10 knockdown in MP38 or OMM1.3 (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by t test, and error bars are ± SEM. D, analysis of TCGA data for uveal melanoma patient disease-specific survival according to MARK3 and STK10 expression, above or below median RNA expression. Log-rank test was used to determine the significance of disease-specific survival.
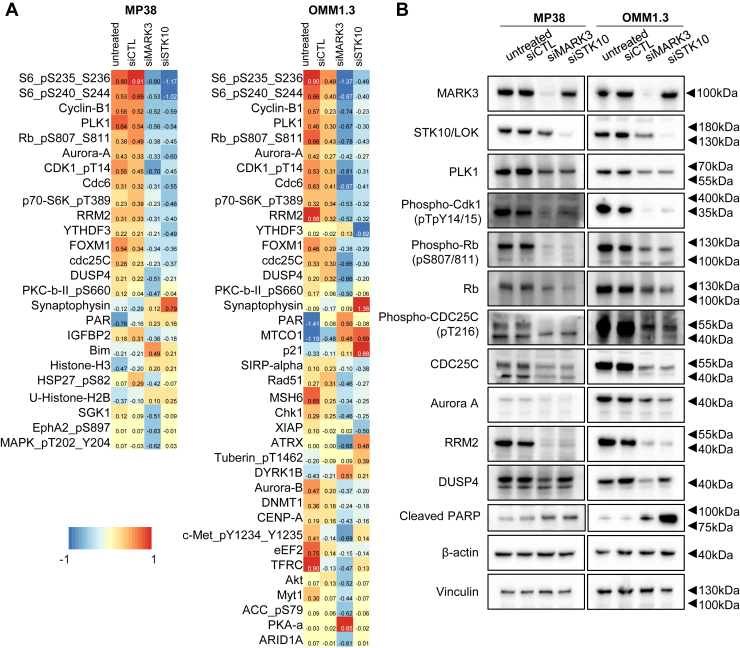
To understand signaling and protein expression changes associated with MARK3 or STK10 knockdown, we performed a reverse-phase protein array (RPPA). In both MP38 and OMM1.3 cell lines, we observed a reduction of ribosomal protein phospho-S6 (S235/236), master cell cycle regulators Aurora kinase A, FOXM1, and phospho-Rb (S807/811), G2/M associated proteins PLK1, cyclin B, phospho-CDK1 (T14/Y15), and phospho-CDC25C (T216), and S phase proteins CDC6 and RRM2 with MARK3 or STK10 knockdown (Fig. 4A). Knockdown of STK10 and MARK3 significantly induced cell cycle arrest at G0 phase in both MP38 and OMM1.3 cells (Fig. S4C). Consistent with Figure 1C, Aurora A and PLK1 were also downregulated with YM-254890 treatment. Of note, synaptophysin, a neuronal protein, was the only protein detected on RPPA as upregulated after STK10 knockdown (Fig. 4A). Reduced DUSP4 expression was observed with the knockdown of MARK3 but not STK10. We validated the RPPA results by Western blot and showed decreased expression of DUSP4 with knockdown of MARK3 only and decreased expression of phospho-Rb (S807/811), PLK1, Aurora A, RRM2, phospho-CDK1 (T14/Y15), and phospho-CDC25C (T216) with knockdown of either MARK3 or STK10 (Fig. 4B). Consistent with the increase in the % of annexin-V-positive cells, cleaved PARP, a marker of apoptosis, was elevated following the knockdown of MARK3 and STK10 (Fig. 4B). Overall, these data demonstrate that knockdown of MARK3 or STK10 downregulates proteins involved in various stages of cell cycle progression.
Figure 4.
MARK3 and STK10 knockdown effects on cell cycle proteins.A, heatmap of median-centered, log2-transformed group average expression RPPA data for differentially expressed proteins (p < 0.05, log2fc > 0.58496) when comparing siRNA-mediated knockdown of MARK3 or STK10 cells to untreated or siCTL. Each lysate was collected in quadruplets (n = 4). B, representative Western blots validating the effects of MARK3 or STK10 knockdown on cell cycle and cell growth proteins.
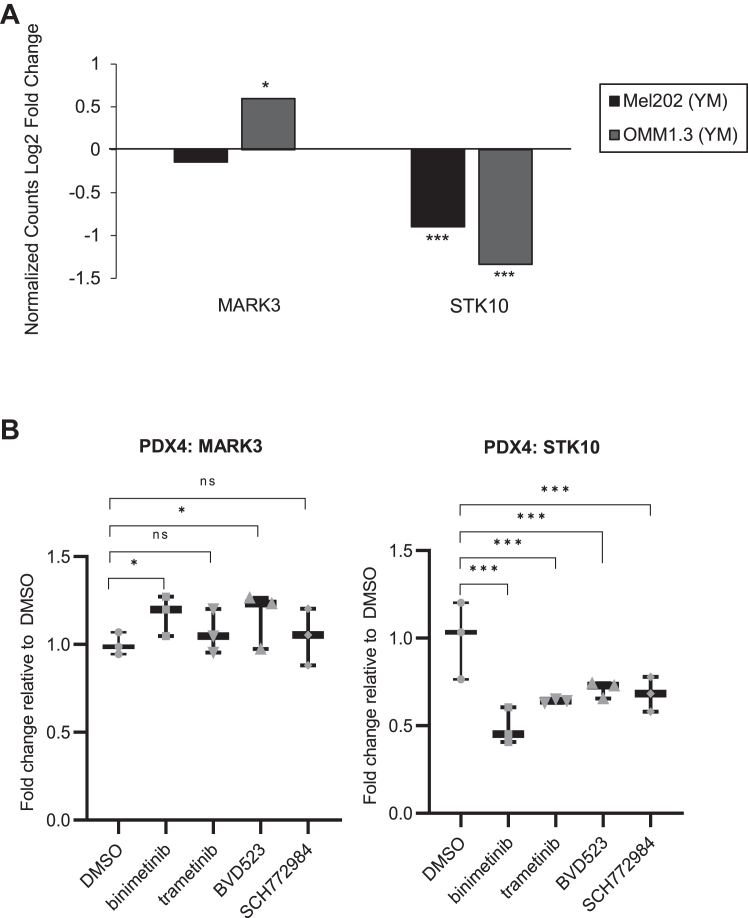
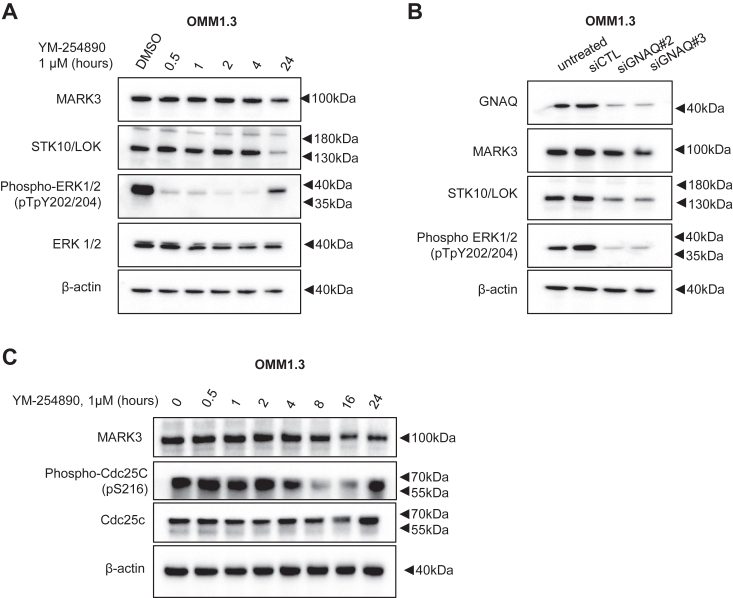
Our kinome profiling results suggest that Gαq/11 regulates MARK3 and STK10 expression or activity. To investigate the exact mechanism of regulation, we examined RNA-seq data from a publicly available dataset of two uveal melanoma cell lines, Mel202 and OMM1.3, treated with YM-254890 (17). Differential gene expression analysis showed that STK10 transcript levels were reduced after 24 h of YM-254890 treatment, while MARK3 levels were either unchanged or slightly increased (Fig. 5A). Treatment with either MEK inhibitor or ERK1/2 inhibitor also reduced STK10 transcript levels without affecting MARK3 transcript levels (Figs. 5B and S5). To determine if MARK3 regulation by Gαq/11 occurs at the protein level, we conducted a YM-254890 treatment time-course experiment over 24 h. Western blot analysis showed that STK10, but not MARK3, protein expression was significantly reduced at 24 h (Figs. 6A and S6, A and B). Additionally, the knockdown of Gαq reduced STK10 protein levels (Figs. 6B and S6C). We identified whether MEK or ERK1/2 inhibition induces similar effects on STK10 and MARK3 as with YM-254890 treatment. Trametinib (MEK inhibitor) and SCH772984 similarly decreased the phosphorylation of ERK1/2 after 30 min of treatment and markedly downregulated STK10 after 16 to 24 h of treatment (Fig. S7). Overall, we determined that STK10 expression was regulated by Gαq and MAPK signaling. To determine YM-254890-driven changes in MARK3 activity, we analyzed the phosphorylation of CDC25C, a known MARK3 substrate, after YM-254890 treatment. Consistent with MARK3 knockdown, phosphorylation of endogenous CDC25C was reduced after YM-254890 treatment (Fig. 6C). Thus, transcriptomic and proteomic data indicate that STK10 expression is regulated by Gαq/11 activation of the MAPK pathway, while MARK3 activity is likely regulated further downstream of mutant Gαq/11.
Figure 5.
Gαq/11 and MAPK pathway regulates STK10 mRNA expression.A, publicly available RNA-seq data was analyzed for MARK3 and STK10 expression following 24-h YM-254890 treatment in Mel202 and OMM1.3 cells. Expression data is shown as a log2 fold change of normalized counts compared to DMSO. B, RNA-seq of PDX4 cells treated with MEK (5 μM binimetinib or 50 nM trametinib) or ERK1/2 (2 μM BVD523 or 500 nM SCH772984) inhibitors for 24 h. Data are shown as fold change relative to DMSO (n = 3). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Figure 6.
Gαq/11 regulates STK10 expression and MARK3 activity.A, representative Western blots of MARK3 and STK10 after OMM1.3 cells were treated with 1 μM of YM-254890 over a 24-h time course (n = 3). B, representative Western blots showing the effect of siGNAQ on MARK3 and STK10 expression. C, Western blot of lysates from OMM1.3 cells treated with YM-254890 for 0 to 24 h were analyzed for phosphorylation of the endogenous MARK3 substrate, CDC25C; WCL, whole cell lysate.
Discussion
Aberrant kinase signaling contributes to cancer initiation and progression. In this study, we inhibited constitutively active mutant Gαq/11 in uveal melanoma cells and performed a novel MIB-MS technique followed by a loss-of-function screen to identify kinases relevant to uveal melanoma cell growth that are potential drug targets. Since direct targeting of Gαq/11 is clinically difficult due to toxicity, we focused on kinases downregulated following YM-254890 treatment and determined if they could be targeted to reduce uveal melanoma growth. Our loss-of-function screen identified MARK3 and STK10 as potential therapeutic targets that had not been previously investigated in uveal melanoma. Bioinformatic patient data analysis underscored the importance of these kinases, as increased expression was correlated with worse prognosis in uveal melanoma. Functionally, MARK3 and STK10 knockdown reduced cell growth, induced cell death, and reduced activation of downstream cell cycle proteins. Knockdown of MARK3, but not STK10, reduced ERK1/2 phosphorylation and DUSP4 expression, suggesting that MARK3 is active within the MAPK pathway and may function as a tumor promoter in uveal melanoma. Knockdown of MARK3 also reduced STK10 expression, suggesting MARK3 may regulate STK10 via activation of the MAPK pathway.
MARK3, also known as CDC25C-associated kinase 1 (C-TAK1), was first identified as a kinase that phosphorylates CDC25C, targeting it for proteasomal degradation (21). Our RPPA results suggest that cell cycle alterations are prevalent after the knockdown of MARK3. There was a decrease in phosphorylated CDC25C, which was also observed with Gαq/11 inhibition, suggesting an effect mediated by reduced MARK3 activity. MARK3 has been identified as a negative regulator of the MAPK pathway through decreasing KSR1 activation of RAF (22). However, our results show that MARK3 promotes MAPK signaling, as knockdown of MARK3 reduced cell growth and decreased ERK1/2 activity. There is no consensus on whether MARK3 behaves as a tumor suppressor or promoter. In hepatocellular carcinoma and glioma, increased MARK3 activity promotes tumor growth (23, 24). In a kinome-wide knockout screen in a leukemic cell line, MARK3 was shown to be critical in developing resistance to DNA damage-inducing chemotherapies, suggesting a pro-tumorigenic effect (25). In contrast, in a kinome-wide siRNA screen in HEK293T cells, MARK3 was identified downstream of tumor suppressor LKB1 and negatively regulates YAP signaling in high-grade serous ovarian cancer (26, 27). These findings suggest that in some contexts, MARK3 inhibits YAP/TAZ-TEAD signaling and suppresses cancer growth. Further experiments are necessary to elucidate the pro-tumorigenic signaling axis driven by MARK3 in uveal melanoma.
STK10, also known as lymphocyte-oriented kinase (LOK), was first characterized as a serine/threonine kinase highly expressed in proliferating cells, lymphocytes, and numerous cancer tissues that plays a role in cell cycle progression (28, 29). STK10 knockout models in lymphocytes and prostate cancer have shown that STK10 is necessary for cell migration (20, 30). Additionally, our data corroborate previous findings showing that STK10 phosphorylates the ERM complex and does not activate the MAPK pathway (Fig. 2B) (20, 29). Once phosphorylated and active, ERM proteins connect the cytoskeleton to the plasma membrane and associate with receptor tyrosine kinases (RTKs) essential to tumor progression, such as EGFR or c-Met (31). Rather than activating the MAPK pathway, our results suggest that the MAPK pathway regulates STK10. Further cell cycle analysis assays may be necessary to determine the exact function of STK10 in uveal melanoma.
There are no FDA-approved targeted kinase therapies for metastatic uveal melanoma, despite constitutive activation of Gαq/11 and downstream MAPK and YAP/TAZ-TEAD signaling. However, kinase inhibitors are still the standard of care in many cancer types and continue to be developed as first- and second-line therapies. BRAF inhibitors and MEK inhibitors revolutionized the treatment of cutaneous melanoma by reducing tumor burden and improving survival in BRAF mutant melanoma patients (10). Thus, novel targetable dependencies in uveal melanoma need to be identified. Results from our study can be used to inform future drug design and combination therapies for uveal melanoma.
Our study demonstrates the validity and power of kinome profiling by MIB-MS. Previous molecular and therapeutic screens in uveal melanoma have focused mainly on transcriptomic data or well-studied therapeutic targets with known cancer drugs (16, 32). Limitations of these approaches are that they fail to consider the functional proteome, do not assess beyond mRNA or protein expression, and overlook understudied kinases, which could be crucial nodes regulating cancer growth. It avoids an inherent bias in screening for a limited number of proteins and kinases for example kinases involved in specific pathways such as mTOR, MAPK, or cell cycle signaling (33). Consistent with previous reports, our kinome profiling data show downregulation of the cell cycle and mitotic spindle kinases after Gαq/11 inhibition (33). Conversely, upregulated kinases included RTKs such as c-Kit (KIT) and fibroblast growth factor receptors 2 and 3 (FGFR2/3), which have been studied in the context of uveal melanoma (Fig. 1C) (34, 35). The expression or activity of these RTKs may be upregulated following inhibition of Gαq/11 in a compensatory manner, but further studies are required to see if these can be targeted in combination with Gαq/11 inhibition. Additionally, protein kinase D1 and 2 (PRKD1/2), which have not been previously studied in uveal melanoma, were upregulated following Gαq/11 inhibition. However, preliminary experiments knocking down PRKD1/2/3 in combination with YM-254890 treatment did not increase sensitivity to the drug (data not shown).
In conclusion, kinome profiling using MIB-MS combined with loss-of-function studies led to the identification of MARK3 and STK10 as potential therapeutic targets in uveal melanoma regulated by Gαq/11. The strength of our research lies in the functional proteomics of the MIB-MS kinome profiling method. This technique allows for the identification of both active and highly expressed kinases (2). Additional studies, such as inducible knockdown of MARK3 or STK10 in vivo, are needed to confirm the validity of targeting these kinases in uveal melanoma patients. Furthermore, small molecular inhibitors with increased specificity for MARK3 or STK10 are being developed (23, 36). Characterization of these inhibitors in vitro and in vivo is necessary to determine the therapeutic potential of targeting these kinases in uveal melanoma.
Experimental procedures
Analysis of The Cancer Genome Atlas (TCGA) data
Patient survival evaluation was performed for The Cancer Genome Atlas (TCGA) uveal melanoma cohort using cBioPortal (37). Survival outcome data originated from the TCGA Pan-Cancer Clinical Data Resource (38). Patients were stratified into high- and low-expression groups based on median RNA expression. The Log-rank test was used to determine statistical significance for disease-specific survival.
Annexin/PI staining
Cells were trypsinized, washed with PBS, and resuspended in 100 μl binding buffer. Cells were stained with 5 μl annexin V-APC (BD Biosciences #550475) for 15 min at room temperature. Cells were washed with binding buffer, resuspended in 1 ml binding buffer, and stained with 2 μl of 1 mg/ml propidium iodide (PI; MP Biomedicals #195458). Staining was analyzed by flow cytometry on a BD FACSCelesta flow cytometer (BD Biosciences). Data were analyzed by FlowJo software (BD Biosciences). Experiments were performed in triplicate, and statistical analysis was performed using a two-tailed t test assuming equal variance with error bars representing SEM.
Cell cycle analysis
Cells were trypsinized, washed with PBS, and fixed in 4% paraformaldehyde/PBS for 15 min. Cells were then washed and incubated with 100 μg/ml RNAse A (Thermo Fisher Scientific) and 40 μg/ml propidium iodide. After 30 min of incubation at room temperature, propidium iodide staining was determined by flow cytometry.
Cell lines
Details on the acquirement and maintenance of MP46, MP38, PDX4, UM004, and OMM1.3 cell lines have been reported (39, 40, 41). Briefly, MP46 and MP38 cells were provided by Dr Sergio Roman (Institute of Curie, France) and cultured in RPMI supplemented with 20% FBS. PDX4 and UM004 were generated by Dr Takami Sato (Thomas Jefferson University) and cultured in MEM supplemented with 10% heat-inactivated FBS and 2 mM L-glutamine. OMM1.3 cells were acquired from Dr Bruce Ksander (Harvard Medical School) and maintained in RPMI supplemented with 10% FBS. 1205Lu cells were cultured in MCDB 153 medium containing 20% Leibovitz-L15 medium, 2% FBS, 0.2% sodium bicarbonate, and 5 μg/ml insulin. All cell lines were routinely tested for mycoplasma and authenticated by STR analysis.
Cell viability assay
Cells were trypsinized and seeded at 5 to 10 × 103 cells/well in a 96-well plate. Cells were transfected with pooled siRNAs targeting the 22 downregulated kinases identified via MIB-MS profiling. After 6 days, cell viability was measured using the CellTiter-Glo 2.0 Cell Viability Assay (Promega) according to the manufacturer’s instructions. Percent cell viability was normalized to cells treated with pooled non-targeting control siRNAs. siRNA targeting GNAQ was included as a positive control.
Immunoprecipitation and kinase activity assay
All steps were performed at 4 °C unless otherwise noted. Cells were plated at 2 × 106 cells per 100 mm plate, treated with YM-254890 or DMSO for 24 h, washed once in PBS, and lysed in RIPA Lysis Buffer System (Santa Cruz). After a 30-min incubation, lysates were centrifuged at 10,000g for 10 min, and the supernatant was incubated with MARK3 antibody (#9311; Cell Signaling Technology) overnight at 4 °C. 20 microliters of Protein A/G PLUS-Agarose was added and incubated at 4° C for 3 h. IPs were collected by centrifugation at 3000 rpm for 5 min, washed 2× with PBS and 2× with kinase buffer (25 mM Tris (pH 7.5), 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2). The pellet was suspended in 40 μl kinase buffer supplemented with 200 μM ATP (Cell Signaling Technology) and recombinant CDC25C (Sigma-Aldrich) and incubated for 30 min at 30 °C. The reaction was stopped by boiling in 2X SDS sample buffer.
IncuCyte live cell growth assay
Cells were trypsinized and seeded at 2 to 4 × 105 cells/well onto a 6-well plate. Photomicrographs were taken every 2 h by an IncuCyte Live Cell Analysis Imaging System using a 10× or 20× objective (Essen Biosciences). Plate confluence was measured using IncuCyte software. Experiments were performed in triplicate, and statistical analysis was performed using a two-tailed t test assuming equal variance with error bars representing SEM.
Inhibitors
YM-254890 from Cayman Chemical Company (Ann Arbor, MI) and binimetinib (MEK162), trametinib (GSK1120212), ulixertinib (BVD-523), and SCH772984 from Selleck Chemicals LLC were dissolved in DMSO and used at the indicated concentrations.
MIB-MS profiling and data analysis of YM-254890-treated cells
Cells were treated with 1 μM of YM-254890 for 24 h and lysed on ice in buffer containing 50 mM HEPES (pH 7.5), 0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 mM sodium fluoride, 2.5 mM sodium orthovanadate, 1X protease inhibitor cocktail (Roche), and 1% each of phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich). Lysates were sonicated and particulate was removed by centrifugation at 21,000g for 15 min at 4 °C and filtration through 0.45 μm syringe filters. Protein concentrations were determined by standard bicinchoninic acid assay (BCA) (Thermo Fisher Scientific). Lysates were processed and analyzed by MIB-MS as described previously (3). Kinases were isolated by flowing lysates over kinase inhibitor-conjugated Sepharose beads (purvalanol B, VI16832, PP58, and CTx-0294885) in 10 ml gravity-flow columns. Eluted kinases were reduced by incubation with 5 mM DTT at 65 °C for 25 min following alkylation with 20 mM iodoacetamide at room temperature for 30 min in the dark. Proteins were digested with sequencing-grade modified trypsin (Promega) overnight at 37 °C. C18-purified peptides were dried in a speed vac, and subsequent LC-MS/MS analysis was performed. Proteolytic peptides were resuspended in 0.1% formic acid and separated with a Thermo Scientific RSLCnano Ultimate 3000 LC on a Thermo Scientific Easy-Spray C18 PepMap 75 μm × 50 cm C18 2 μm column. A 240 min gradient of 4 to 25% acetonitrile with 0.1% formic acid was run at 300 nl/min at 50 °C. Eluted peptides were analyzed by a Thermo Scientific Q Exactive Plus mass spectrometer utilizing a top 15 methodology in which the 15 most intense peptide precursor ions were subjected to fragmentation. The AGC for MS1 was set to 3 × 106 with a max injection time of 120 milliseconds, the AGC for MS2 ions was set to 1 × 105 with a max injection time of 150 milliseconds, and the dynamic exclusion was set to 90 s. Raw data analysis of LFQ experiments was performed using MaxQuant software 1.6.0.1 and searched using Andromeda 1.5.6.0 against the Swiss-Prot human protein database (downloaded on April 24, 2019, 20,402 entries). The search was set up for full tryptic peptides with a maximum of two missed cleavage sites. All settings were defaulted and searched using acetylation of protein N-terminus and oxidized methionine as variable modifications. Carbamidomethylation of cysteine was set as a fixed modification. The precursor mass tolerance threshold was set at 10 ppm and the maximum fragment mass error was 0.02 Da. LFQ quantitation was performed using MaxQuant with the following parameters: LFQ minimum ratio count: Global parameters for protein quantitation were as follows: label minimum ratio count: 1, peptides used for quantitation: unique, only use modified proteins selected and with normalized average ratio estimation selected. Match between runs was employed for LFQ quantitation, and the significance threshold of the ion score was calculated based on a false discovery rate of <1%.
Measurement of MIB-enriched kinase abundance in cell lines was performed by LFQ using MaxQuant software version 1.6.1.0. MaxQuant normalized LFQ values were filtered for human protein kinases in Excel and then imported into Perseus software (1.6.2.3) for quantitation. LFQ values were filtered in the following manner: kinases identified by site only were removed, and reverse or potential contaminants were removed. Kinase LFQ intensity values were then log2 transformed. No imputation of missing values was performed. Log2 LFQ intensity values were subjected to a Student's t test comparing treatment versus DMSO with the parameters S0 = 2, side both. All relevant proteomics files are available through the PRIDE partner repository (http://www.ebi.ac.uk) with the data set identifier PXD.
Reverse phase protein array analysis
Cells were plated in 6-well dishes at 2 to 4 × 105 cells per well, treated with siRNA for 72 h, washed twice in ice-cold PBS, and then lysed in 50 μl reverse phase protein array (RPPA) lysis buffer [1% Triton X-100, 50 mM HEPES (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM NaPPI, 1 mM Na3VO4, 10% glycerol, protease and phosphatase inhibitors (Boehringer/Roche) for 20 min with occasional shaking on ice. Lysates were centrifuged for 10 min at 14,000 rpm, and the supernatant was collected. Protein concentration was determined by Bradford assay. Lysates were analyzed at the MD Anderson Functional Proteomics Core Facility, where antibodies are extensively validated before being included in the panel. Serial dilutions of samples were arrayed on nitrocellulose-coated slides and run against 456 validated antibodies. A DAB (3, 3′-diaminobenzidin) colorimetric reaction for a tyramide-based signal amplification approach was used to produce stained slides. The slides were scanned on a Huron TissueScope scanner, and spot densities were determined using Array-Pro Analyzer. Relative protein levels were quantified using SuperCurve fitting and normalized for protein loading. Reverse-phase protein array (RPPA) data were used to determine proteins/phospho-proteins that were significantly different between siMARK3 or siSTK10 and untreated or control groups for MP38 and OMM1.3 cell line samples. Comparisons were performed in matlab(R) using the two-sample t test method with 1000 permutations and assumed unequal variance. The Benjamini-Hochberg False Discovery Rate (BHFDR) method was used to determine statistical significance. Data analyses were performed in R (v3.5.1 http://www.R-project.org/). The RPPA results were validated by Western blotting for key targets.
RNA sequencing (RNA-seq) sample processing
Bulk sequencing: total RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit and normalized to 5 ng/μl. Following plating, 2 μl of ERCC controls (using a 1:1000 dilution) were spiked into each sample. 200 ng aliquots of each sample were transferred into library preparation which uses an automated variant of the Illumina TruSeq Stranded mRNA Sample Preparation Kit. This method preserves the strand orientation of the RNA transcript, uses oligo dT beads to select mRNA from the total RNA sample, and is followed by heat fragmentation and cDNA synthesis from the RNA template. The resultant 400 base pairs (bp) cDNA were then processed for dual-indexed library preparation: ‘A’ base addition, adapter ligation using P7 adapters, and PCR enrichment using P5 adapters. After enrichment, the libraries were quantified using Quant-iT PicoGreen (1:200 dilution). After normalizing samples to 5 ng/μl, the set was pooled and quantified using the KAPA Library Quantification Kit for Illumina Sequencing Platforms. The entire process was performed in a 96-well format with pipetting performed by Agilent Bravo or Hamilton Starlet. For Illumina sequencing, pooled libraries were normalized to 2 nM and denatured using 0.1 M NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using the NovaSeq 6000. Each run was a 101 bp paired-end with an eight-base index barcode read. Data were analyzed using the Broad Picard Pipeline, which includes de-multiplexing and data aggregation. RNA-seq data were aligned to the human reference genome (GRCh38) using Star aligner (42) and GENCODE (43) annotations. RSEM (44) was used to quantify gene and transcript-level expression. Gene differential analysis was performed by comparing treated and DMSO samples using DESeq2 (45). Data analyses were performed in R (v3.5.1 http://www.R-project.org/). RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) database with accession code GSE228090.
siRNA transfections
A total of 5 to 10 × 103 cells/well were plated on a 96-well plate with the indicated siRNAs at a final concentration of 25 nM using Lipofectamine RNAi-MAX (Invitrogen). For Western blotting, 2 to 4 × 103 cells/well were plated on a 6-well plate, transfected with siRNA at the same final concentration, and allowed to grow for 72 h before cell lysis. The siGENOME targeting human GNAQ (D-008562-02) and non-targeting control (D-001210-02-20) from Horizon Discovery (Lafayette, CO, USA) were used. Additional pooled ON-TARGET plus siRNAs targeting humans are found in Table S1.
Western blot analysis
Protein lysates were boiled in Laemmli sample buffer, separated by SDS-PAGE, and transferred to PVDF membranes. The following primary antibodies were used: α-tubulin (#2144), Aurora A (#3092), cleaved PARP (#9541), cyclin B1 (#4135), DUSP4 (#5149S), Ezrin/Radixin/Moesin (ERM) (#3142), FOXM1 (#5436), MARK3 (#9311), phospho-CDC25C (Ser216) (#9528S), phospho-Ezrin (Thr567)/Radixin (Thr564)/Moesin (Thr558) (#3726), phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (#9101), phospho-Rb (Ser807/811) (#9308S), PLK1 (#4513S), Rb (#9309S), and RRM2 (#65939S) from Cell Signaling Technology (Danvers, MA, US); phospho-CDK1 (Thr14/Tyr15) (#44686G) from Invitrogen; CDC25C (#sc-327), ERK1 (#sc-93), Gαq (GNAQ) (#sc-393), LOK/STK10 (#398083), and vinculin (#sc-73614) from Santa Cruz; and β-actin (#A2066) from Sigma-Aldrich. Immunoreactivity was detected using HRP-conjugated secondary antibodies (CalBioTech) and chemiluminescence substrate (Thermo Scientific) on a Versadoc Imaging System (Bio-Rad).
Statistical analysis
Data were analyzed using the two-sided Student’s t test with Microsoft Excel software (∗p value < 0.05; ∗∗p value < 0.01, and ∗∗∗p value < 0.001).
Data availability
The data generated in this study are within the article or in Supporting Information files. The publicly available data analyzed in this study were obtained from GEO at GSE152705. All relevant proteomics files are available through the PRIDE partner repository (http://www.ebi.ac.uk) with the data set identifier PXD. New RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) database with accession code GSE228090.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: A. E. A. has an ownership interest in patent number 9880150 and a pending patent, PCT/US22/76492. No potential conflicts of interest were disclosed by the other authors. M. A. D. has been a consultant to Roche/Genentech, Array, Pfizer, Novartis, BMS, GSK, Sanofi-Aventis, Vaccinex, Apexigen, Eisai, Iovance, and ABM Therapeutics, and he has been the PI of research grants to MD Anderson by Roche/Genentech, GSK, Sanofi-Aventis, Merck, Myriad, Oncothyreon, ABM Therapeutics, and LEAD Pharma. J. S. G. has been a consultant for Domain Pharmaceuticals, Pangea Therapeutics, and io9, and is the founder of Kadima Pharmaceuticals, all unrelated to the current study. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Ms Signe Caksa and Drs Claudia Capparelli, Nicole Wilski, and Scott Varney for valuable feedback during the preparation of this article.
Author contributions
U. B., J. S. D., and A. E. A. conceptualization; U. B., A. M. K., and T. J. P. formal analysis; U. B., A. M. K., I. V. T., T. J. P., J. I. H., A. H., K. L., N. F. P., and A. J. investigation; U. B., A. M. K., I. V. T., T. J. P., J. I. H., A. H., K. L., N. F. P., and A. J. data curation; V. C., M. A. D., J. S. G., J. L. B., J. S. D., and A. E. A. resources; U. B. writing original draft; A. M. K., J. I. H., V. C., J. S. G., J. S. D., and A. E. A. writing review and editing; J. S. D. and A. E. A. funding acquisition.
Funding and additional information
This work was supported by the Melanoma Research Foundation Medical Student Award 2020 to U. B. and a U.S. Department of Defense (DoD) grant to A. E. A. and J. S. D. It was also supported by the National Institutes of Health (NIH)/National Cancer Institute (NCI) R01 grant, CA253977 to A. E. A. The Sidney Kimmel Cancer Center Flow Cytometry and Translational Pathology core facilities are supported by NIH/NCI (P30 CA056036). The RPPA studies were performed at the Functional Proteomics Core Facility at The University of Texas, MD Anderson Cancer Center, which is supported by an NCI Cancer Center Support Grant (P30 CA16672). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Donita C. Brady
Supporting information
References
- 1.Essegian D., Khurana R., Stathias V., Schürer S.C. The clinical kinase index: a method to prioritize understudied kinases as drug targets for the treatment of cancer. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan J.S., Whittle M.C., Nakamura K., Abell A.N., Midland A.A., Zawistowski J., et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurimchak A.M., Herrera-Montávez C., Brown J., Johnson K.J., Sodi V., Srivastava N., et al. Functional proteomics interrogation of the kinome identifies MRCKA as a therapeutic target in high-grade serous ovarian carcinoma. Sci. Signal. 2020;13:1–18. doi: 10.1126/scisignal.aax8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakalian S., Marshall J.-C., Logan P., Faingold D., Maloney S., Di Cesare S., et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin. Cancer Res. 2008;14:951–956. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- 5.Carvajal R.D., Schwartz G.K., Tezel T., Marr B., Francis J.H., Nathan P.D. Metastatic disease from uveal melanoma: treatment options and future prospects. Br. J. Ophthalmol. 2017;101:38–44. doi: 10.1136/bjophthalmol-2016-309034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathan P., Hassel J.C., Rutkowski P., Baurain J.-F., Butler M.O., Schlaak M., et al. Overall Survival benefit with Tebentafusp in metastatic uveal melanoma. N. Engl. J. Med. 2021;385:1196–1206. doi: 10.1056/NEJMoa2103485. [DOI] [PubMed] [Google Scholar]
- 7.Chua V., Lapadula D., Randolph C., Benovic J.L., Wedegaertner P.B., Aplin A.E. Dysregulated GPCR signaling and therapeutic options in uveal melanoma. Mol. Cancer Res. 2017;15:501–506. doi: 10.1158/1541-7786.MCR-17-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Weng L., Bastian B.C., Chen X. Functional characterization of uveal melanoma oncogenes. Oncogene. 2021;40:806–820. doi: 10.1038/s41388-020-01569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng X., Degese M.S., Iglesias-Bartolome R., Vaque J.P., Molinolo A.A., Rodrigues M., et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty K.T., Infante J.R., Daud A., Gonzalez R., Kefford R.F., Sosman J., et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvajal R.D., Sosman J.A., Quevedo J.F., Milhem M.M., Joshua A.M., Kudchadkar R.R., et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA. 2014;311:2397–2405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvajal R.D., Piperno-Neumann S., Kapiteijn E., Chapman P.B., Frank S., Joshua A.M., et al. Selumetinib in combination with dacarbazine in patients with metastatic uveal melanoma: a phase III, multicenter, randomized trial (SUMIT) J. Clin. Oncol. 2018;36:1232–1239. doi: 10.1200/JCO.2017.74.1090. [DOI] [PubMed] [Google Scholar]
- 13.Caksa S., Baqai U., Aplin A.E. The future of targeted kinase inhibitors in melanoma. Pharmacol. Ther. 2022;239 doi: 10.1016/j.pharmthera.2022.108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlegel J.G., Tahoun M., Seidinger A., Voss J.H., Kuschak M., Kehraus S., et al. Macrocyclic Gq protein inhibitors FR900359 and/or YM-254890-fit for translation? ACS Pharmacol. Transl. Sci. 2021;4:888–897. doi: 10.1021/acsptsci.1c00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y.J., Park S.J., Maeng K.J., Lee S.C., Lee C.S. Multi-platform omics analysis for identification of molecular characteristics and therapeutic targets of uveal melanoma. Sci. Rep. 2019;9:1–8. doi: 10.1038/s41598-019-55513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey F.P., Clarke K., Kalirai H., Kenyani J., Shahidipour H., Falciani F., et al. Kinome-wide transcriptional profiling of uveal melanoma reveals new vulnerabilities to targeted therapeutics. Pigment Cell Melanoma Res. 2018;31:253–266. doi: 10.1111/pcmr.12650. [DOI] [PubMed] [Google Scholar]
- 17.Hitchman T.D., Bayshtok G., Ceraudo E., Moore A.R., Lee C., Jia R., et al. Combined inhibition of Gαq and MEK enhances therapeutic efficacy in uveal melanoma. Clin. Cancer Res. 2021;27:1476–1490. doi: 10.1158/1078-0432.CCR-20-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O’Brien J.M., et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrosini G., Musi E., Ho A.L., de Stanchina E., Schwartz G.K. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol. Cancer Ther. 2013;12:768–776. doi: 10.1158/1535-7163.MCT-12-1020. [DOI] [PubMed] [Google Scholar]
- 20.Belkina N.V., Liu Y., Hao J.J., Karasuyama H., Shaw S. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4707–4712. doi: 10.1073/pnas.0805963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogg S., Gabrielli B., Piwnica-Worms H. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J. Biol. Chem. 1994;269:30461–30469. [PubMed] [Google Scholar]
- 22.Müller J., Ory S., Copeland T., Piwnica-Worms H., Morrison D.K. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell. 2001;8:983–993. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 23.Li F., Liu Z., Sun H., Li C., Wang W., Ye L., et al. PCC0208017, a novel small-molecule inhibitor of MARK3/MARK4, suppresses glioma progression in vitro and in vivo. Acta Pharm. Sin. B. 2020;10:289–300. doi: 10.1016/j.apsb.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato T., Satoh S., Okabe H., Kitahara O., Ono K., Kihara C., et al. Isolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis. Neoplasia. 2001;3:4–9. doi: 10.1038/sj.neo.7900132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owusu M., Bannauer P., Ferreira da Silva J., Mourikis T.P., Jones A., Májek P., et al. Mapping the human kinome in response to DNA damage. Cell Rep. 2019;26:555–563.e6. doi: 10.1016/j.celrep.2018.12.087. [DOI] [PubMed] [Google Scholar]
- 26.Mohseni M., Sun J., Lau A., Curtis S., Goldsmith J., Fox V.L., et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat. Cell Biol. 2014;16:108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machino H., Kaneko S., Komatsu M., Ikawa N., Asada K., Nakato R., et al. The metabolic stress-activated checkpoint LKB1-MARK3 axis acts as a tumor suppressor in high-grade serous ovarian carcinoma. Commun. Biol. 2022;5:1–15. doi: 10.1038/s42003-021-02992-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter S.A., Cutler R.E., Martinez R., Gishizky M., Hill R.J. Stk10, a new member of the polo-like kinase kinase family highly expressed in hematopoietic tissue. J. Biol. Chem. 2003;278:18221–18228. doi: 10.1074/jbc.M212556200. [DOI] [PubMed] [Google Scholar]
- 29.Kuramochi S., Moriguchi T., Kuida K., Endo J., Semba K., Nishida E., et al. LOK is a novel mouse STE20-like protein kinase that is expressed predominantly in lymphocytes. J. Biol. Chem. 1997;272:22679–22684. doi: 10.1074/jbc.272.36.22679. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L., Lu S.-Y., Guo R., Ma J.-X., Tang L.-Y., Wang J.-J., et al. STK10 knockout inhibits cell migration and promotes cell proliferation via modulating the activity of ERM and p38 MAPK in prostate cancer cells. Exp. Ther. Med. 2021;22:1–8. doi: 10.3892/etm.2021.10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clucas J., Valderrama F. ERM proteins in cancer progression. J. Cell Sci. 2014;127:267–275. doi: 10.1242/jcs.133108. [DOI] [PubMed] [Google Scholar]
- 32.Feng X., Arang N., Rigiracciolo D.C., Lee J.S., Yeerna H., Wang Z., et al. A platform of synthetic lethal gene interaction networks reveals that the GNAQ uveal melanoma oncogene controls the Hippo pathway through FAK. Cancer Cell. 2019;35:457–472.e5. doi: 10.1016/j.ccell.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lapadula D., Farias E., Randolph C.E., Purwin T.J., McGrath D., Charpentier T.H., et al. Effects of oncogenic Gαq and Gα11 inhibition by FR900359 in uveal melanoma. Mol. Cancer Res. 2019;17:963–973. doi: 10.1158/1541-7786.MCR-18-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calipel A., Landreville S., De La Fouchardière A., Mascarelli F., Rivoire M., Penel N., et al. Mechanisms of resistance to imatinib mesylate in KIT-positive metastatic uveal melanoma. Clin. Exp. Metastasis. 2014;31:553–564. doi: 10.1007/s10585-014-9649-2. [DOI] [PubMed] [Google Scholar]
- 35.Chua V., Orloff M., Teh J.L., Sugase T., Liao C., Purwin T.J., et al. Stromal fibroblast growth factor 2 reduces the efficacy of bromodomain inhibitors in uveal melanoma. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201809081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serafim R.A.M., Sorrell F.J., Berger B.T., Collins R.J., Vasconcelos S.N.S., Massirer K.B., et al. Discovery of a potent dual SLK/STK10 inhibitor based on a maleimide scaffold. J. Med. Chem. 2021;64:13259–13278. doi: 10.1021/acs.jmedchem.0c01579. [DOI] [PubMed] [Google Scholar]
- 37.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:1–20. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han A., Purwin T.J., Bechtel N., Liao C., Chua V., Seifert E., et al. BAP1 mutant uveal melanoma is stratified by metabolic phenotypes with distinct vulnerability to metabolic inhibitors. Oncogene. 2021;40:618–632. doi: 10.1038/s41388-020-01554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paradis J.S., Acosta M., Saddawi-Konefka R., Kishore A., Gomes F., Arang N., et al. Synthetic lethal screens reveal cotargeting FAK and MEK as a multimodal precision therapy for GNAQ-driven uveal melanoma. Clin. Cancer Res. 2021;27:3190–3200. doi: 10.1158/1078-0432.CCR-20-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teh J.L.F., Purwin T.J., Han A., Chua V., Patel P., Baqai U., et al. Metabolic adaptations to MEK and CDK4/6 cotargeting in uveal melanoma. Mol. Cancer Ther. 2020;19:1719–1726. doi: 10.1158/1535-7163.MCT-19-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are within the article or in Supporting Information files. The publicly available data analyzed in this study were obtained from GEO at GSE152705. All relevant proteomics files are available through the PRIDE partner repository (http://www.ebi.ac.uk) with the data set identifier PXD. New RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) database with accession code GSE228090.