Abstract
The turnover of phospholipids plays an essential role in membrane lipid homeostasis by impacting both lipid head group and acyl chain composition. This review focusses on the degradation and acyl chain remodeling of the major phospholipid classes present in the ER membrane of the reference eukaryote Saccharomyces cerevisiae, i.e. phosphatidylcholine (PC), phosphatidylinositol (PI) and phosphatidylethanolamine (PE). Phospholipid turnover reactions are introduced, and the occurrence and important functions of phospholipid remodeling in higher eukaryotes are briefly summarized. After presenting an inventory of established mechanisms of phospholipid acyl chain exchange, current knowledge of phospholipid degradation and remodeling by phospholipases and acyltransferases localized to the yeast ER is summarized. PC is subject to the PC deacylation-reacylation remodeling pathway (PC-DRP) involving a phospholipase B, the recently identified glycerophosphocholine acyltransferase Gpc1p, and the broad specificity acyltransferase Ale1p. PI is post-synthetically enriched in C18:0 acyl chains by remodeling reactions involving Cst26p. PE may undergo turnover by the phospholipid: diacylglycerol acyltransferase Lro1p as first step in acyl chain remodeling. Clues as to the functions of phospholipid acyl chain remodeling are discussed.
Keywords: Acyl chain exchange, Membrane phospholipids, Acyltransferase, Membrane lipid homeostasis, Phospholipid degradation
1. Introduction
The ER is a highly dynamic organelle that is involved in many aspects of the biogenesis of cellular membranes, including the production and distribution of most cellular lipids. Most membrane lipids (or their lipid precursors) are synthesized de novo by enzymes localized in the ER membrane, and can be transported to other cellular membrane compartments by vesicular trafficking and lipid transfer at membrane contact sites. For proper function of these and a multitude of other processes it is imperative that the ER membrane is flexible while maintaining barrier function. Membrane flexibility is a reflection of physical parameters such as membrane fluidity and intrinsic curvature that, to a large extent, are determined by membrane lipid composition. The lipid matrix of the ER membrane is mainly composed of diacyl glycerophospholipids enriched in unsaturated acyl chains, with up to 10 mol% sterol [1].
Membrane fluidity depends on the ratio of saturated-to-unsaturated acyl chains and on the membrane’s content of phosphatidylethanolamine (PE) and sterol [2], whereas membrane intrinsic curvature is determined by the proportion of lipids with non-bilayer propensity, diunsaturated PE molecular species first and foremost [3]. To maintain membrane physical parameters in an optimal range, glycerophospholipid head group and acyl chain composition need to be regulated in concert, i.e. at the phospholipid molecular species level. The ER membrane harbors sensors that monitor membrane physical properties and signal the transcriptional machineries governing fatty acid desaturation and de novo phospholipid and sterol biosynthesis to make adjustments as necessary (reviewed by [4,5]). In addition to transcriptional regulation of membrane lipid composition, lipid biosynthetic enzymes are regulated by posttranslational modification such as phosphorylation (see e.g. [6]), and acetylation [7].
The synthesis of membrane lipid components, as well as the regulation of those processes, have been extensively studied in yeast [8,9] and eukaryotes in general [10]. Far fewer studies have addressed the role of phospholipid turnover, let alone the regulation of those events, in membrane lipid homeostasis. Several processes contribute to the turnover of the main membrane phospholipids of the ER, i.e. phosphatidylserine (PS), phosphatidylinositol (PI), PE, and phosphatidylcholine (PC) [11]. First of all, PS and PE serve as biosynthetic precursors of PE and PC, respectively, making those turnover/conversion reactions the most important quantitatively. Superimposed on the lipid headgroup conversions, the molecular species selectivity (i.e. selectivity at the level of the specific acyl chains esterified at the sn-1 and sn-2 positions of the glycerol backbone) of the decarboxylation of PS to PE [12] and of the triple methylation of PE to PC [13] impacts the membrane’s biophysical properties. Since the conversion of PS to PE is for the most part localized outside the ER [14,15], interorganellar lipid transport out of and back into the ER and the local supply of substrate species also play a role.
PE and PI are subject to turnover in several additional biosynthetic reactions. PE serves as precursor in an essential step of the synthesis of the glycosylphosphatidylinositol (GPI) anchor that attaches a subset of plasma membrane proteins to the membrane [16,17], and it becomes covalently bound to Atg8 in the assembly of autophagosomes [18]. Whether these conversions are PE molecular species-selective and affect the molecular species composition of the PE pool is unknown.
PI is the biosynthetic precursor in the synthesis of phosphoinositides, GPI anchors and yeast sphingolipids. The phosphoinositides, phosphatidylinositol phosphate (PIP) and phosphatidylinositol bisphosphate (PIP2) show roughly similar fatty acyl chain compositions as PI in several mammalian cell lines and tissues [19] as well as in yeast [20]. These studies suggest a single rapidly interconverting metabolic pool, which could be interpreted to argue against pronounced molecular species selectivity by the kinases and phosphatases involved. In support of molecular species-selective substrate use, inactivation of the PI-kinases Vps34 and Pik1 or the phosphoinositide phosphatase Sac1 in yeast mutants was found to affect the molecular species profile of PIP [21]. However, the molecular species profile of the bulk membrane lipid PI was largely unaffected in these mutants, underscoring the low abundance of phosphoinositides.
The PI-derived moiety of GPI-anchors is subject to acyl chain remodeling during the assembly of GPI-anchored proteins in the ER and Golgi [17], but no data exists to suggest that GPI anchor synthesis is substrate selective at the level of PI molecular species. In the synthesis of yeast sphingolipids, PI serves as head group donor [22], generating diacylglycerol. To the best of our knowledge no PI molecular species preference has been reported for the Golgi-localized inositol phosphorylceramide synthase, Aur1p. Similarly, no preference for certain PC molecular species has been reported in the synthesis of sphingomyelin in mammalian cells [23].
Finally, bulk membrane phospholipids are subject to turnover by phospholipases and transacylases as part of lipid acyl chain remodeling or degradation. This review will focus on the role of these reactions in membrane lipid homeostasis in the reference eukaryote Saccharomyces cerevisiae, with emphasis on the acyl chain remodeling reactions that the bulk membrane phospholipids PC, PI and PE undergo in the ER membrane. First, we will put acyl chain remodeling (also referred to as acyl chain editing or retailoring) in a broader perspective and briefly summarize its discovery and occurrence in eukaryotes.
2. Occurrence and functions of phospholipid acyl chain remodeling in eukaryotes
Phospholipid acyl chain exchange was first reported in a pioneering study by W.E. Lands [24]. He discovered that lysophosphatidylcholine (lysoPC) formed in rat liver microsomes by the action of a phospholipase A2 (PLA2), is reacylated by an acyltransferase dependent on the presence of acyl-CoA. These sequential reactions are known as the Lands’ cycle, which was proposed to adapt the molecular species profile of PC to obtain the desired thickness and fluidity of cellular membranes [25]. The substrate specificity of the acyltransferases involved leads to the enrichment of saturated acyl chains at the sn-1 and unsaturated acyl chains at the sn-2 position of the glycerol backbone [26].
Mammalian cells host a multitude of intracellular phospholipases A and acyltransferases, often with tissue-specific distributions, that catalyze phospholipid acyl chain exchange [27–29]. Several of these enzymes play key roles in essential processes, such as the generation of lysophospholipid and eicosanoid messengers in signal transduction. Polyunsaturated fatty acyl chains (PUFAs) including arachidonic acid (C20:4), mainly enter the pool of membrane phospholipids by acyl chain remodeling via the Lands’ cycle (reviewed by [30]). Mobilization of PUFAs from the phospholipid pool by highly regulated phospholipases presents an essential step in inflammatory responses of the immune system, in which arachidonic acid is converted to eicosanoids (reviewed by [31]). Mobilization is followed by regeneration of the phospholipid.
In recent years, acyltransferases catalyzing acyl chain remodeling have been implicated in processes where maintenance of the proper physical state of the (ER) membrane appears crucial. For example, genetic studies in mice have shown that the ER-localized lysophosphatidylcholine acyltransferase LPCAT3 with a preference for PUFA-CoA is required for VLDL assembly in hepatocytes, and for the regulation of cholesterol biosynthesis and, as a consequence, intestinal stem cell proliferation (reviewed in [32]). The lysophosphatidylcholine acyltransferase LPCAT1 expressed in lung alveolar type II cells, is an important contributor to the synthesis of dipalmitoyl-PC (PC 16:0/16:0), an essential component of pulmonary lung surfactant [33]. Although their names suggest otherwise, most acyltransferases are promiscuous with regard to the lysophospholipid acceptor [30].
In plant cells phospholipid remodeling is crucial for the production of polyunsaturated fatty acids. The chloroplast produces saturated and monounsaturated fatty acids that are exported to the cytosol, and after activation to acyl-CoA, channeled into glycerophospholipids and triacylglycerol [34,35]. A major proportion of C18:1-CoA is incorporated at the sn-2 position of lysoPC by acyl-CoA:lysophosphatidylcholine acyltransferase (LPCAT) in the ER. PC then serves as substrate for further desaturation of the acyl chain at the sn-2 position by the desaturases FAD2 and FAD3, yielding linoleic (18:2cis9,12) and α-linolenic (18:3cis9,12,15) acids, which are released for use in de novo lipid synthesis [36]. The latter step, regenerating lysoPC, is mediated by phospholipase A activity (Lands’ cycle), phospholipid:diacylglycerol acyltransferase (PDAT) or by the reversed LPCAT reaction, yielding free fatty acid, triacylglycerol or acyl-CoA, respectively [35].
Phosphatidylcholine deacylation and reacylation has been proposed to facilitate lipid transport at membrane contact sites between ER and the chloroplast outer envelope membrane [37]. Transfer of more polar lysoPC and acyl-CoA between membranes would be energetically more favorable than of intact PC.
In the model eukaryote Saccharomyces cerevisiae, remodeling of PC in microsomes was discovered by Yamada and colleagues [38]. Phospholipid acyl chain exchange was demonstrated in vivo in yeast by the fast, on a time scale of minutes, incorporation of radiolabeled oleic and palmitic acid supplied in the culture medium into the major membrane phospholipids [39]. While studies in yeast have often paved the way for understanding mechanisms and functions of the homologous processes in so-called higher eukaryotes, this does not (yet) apply to lipid acyl chain remodeling, with the notable exception of Taz1p (tafazzin)-mediated remodeling of cardiolipin in mitochondria [40].
3. Definition and mechanisms of phospholipid (acyl chain) remodeling
We define phospholipid acyl chain remodeling, or phospholipid remodeling in short, as comprising reactions in which one or both phospholipid acyl chains are exchanged by deacylation and subsequent reacylation, with a lysophospholipid or glycerophosphodiester as intermediate. This implies that the phospholipid headgroup stays attached to the glycerol backbone in acyl chain remodeling reactions, i.e. the glycerophosphodiester moiety remains intact. Thus, the recycling of choline (or ethanolamine) derived from PC (or PE) degradation back into PC (or PE) via the CDP-choline (or CDP-ethanolamine) route is considered de novo phospholipid biosynthesis rather than lipid remodeling. Moreover, phosphatidic acid (PA) can only be remodeled with lysoPA as intermediate since reacylation of its glycerophosphomonoester moiety, glycerol-3-phosphate, is indistinguishable from de novo synthesis.
In addition to the acyl-CoA dependent Lands’ cycle introduced above, several other mechanisms of acyl chain remodeling have been described (Fig. 1; reviewed in [28]). These include CoA-dependent transacylation reactions in which an acyl chain is transferred from a diacyl phospholipid to a lysophospholipid in the presence of CoA. This transacylation consists of the reverse (i.e. ATP-independent acyl-CoA synthesis) and forward reactions of acyl-CoA-dependent acyltransferases, with acyl-CoA serving as short-lived intermediate. In CoA-independent transacylation an acyl chain is directly transferred from a diacyl- or lysophospholipid donor to a lysophospholipid without requirement for any cofactor. The lecithin:cholesterol acyltransferase (LCAT) transfers the sn-2 acyl chain from PC to cholesterol generating lysoPC and cholesteryl ester in mammalian systems. Likewise, a phospholipid:diacylglycerol acyltransferase (PDAT) transfers an acyl chain from a phospholipid to diacylglycerol (DAG) generating an sn-2 lysophospholipid and triacylglycerol in plants and yeast [41]. Recently, the exchange of both phospholipid acyl chains with a glycerophosphodiester as intermediate was reported for PC. This reaction involves acyl-CoA:glycerophosphocholine acyltransferase (GPCAT) activity and was first demonstrated in plants and yeast [42,43].
Fig. 1.
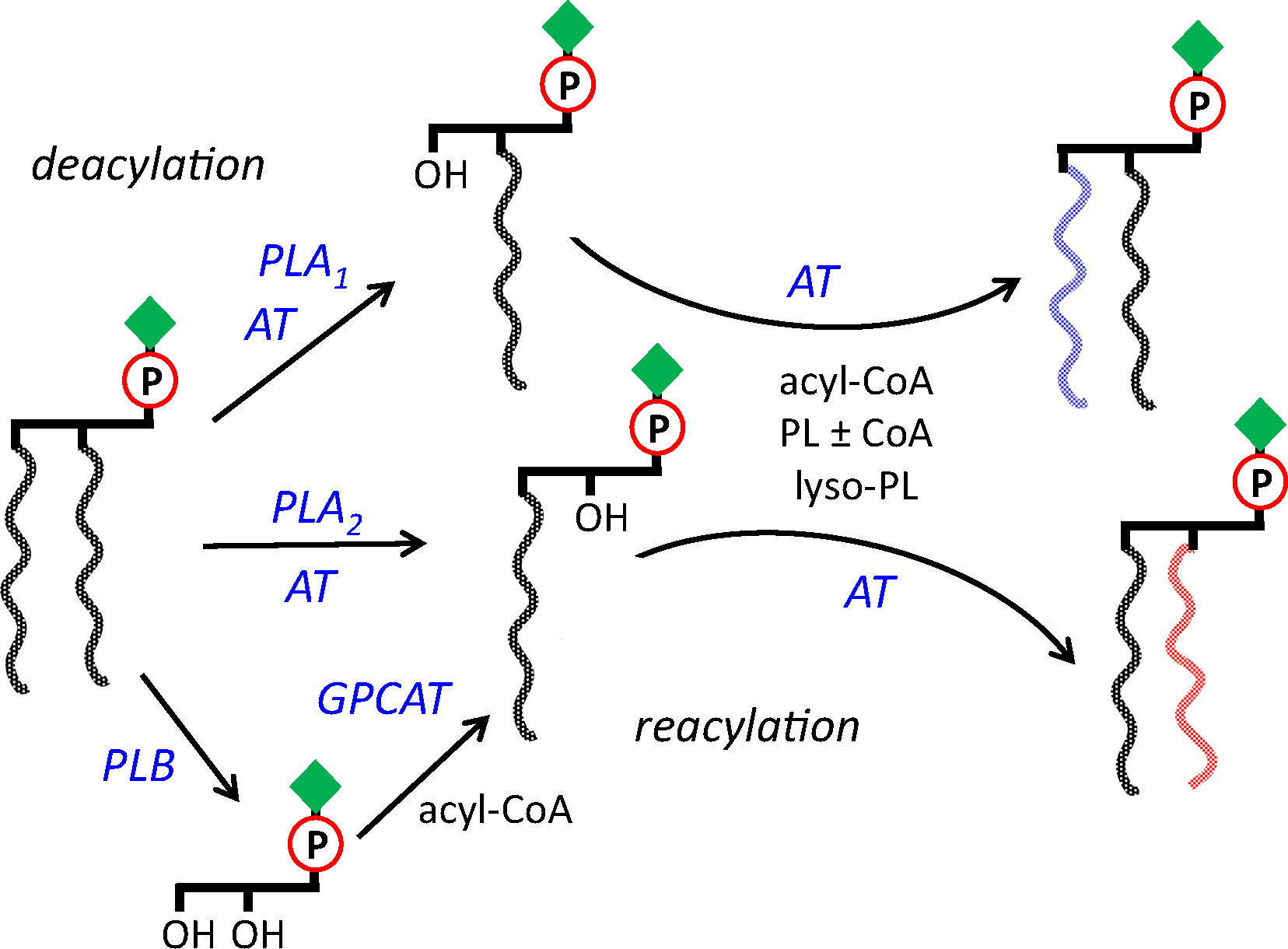
Phospholipid acyl chain remodeling reactions in eukaryotes, as found for phosphatidylcholine (PC). PC is first deacylated by a phospholipase A1 (PLA1), A2 (PLA2), or B (PLB), yielding sn-1 lyso-PC, sn-2 lyso-PC or glycerophosphocholine (GPC), respectively. Acyltransferases (AT) may generate lyso-PC in transacylation reactions. Subsequently GPC is reacylated to sn-2 lyso-PC by glycerophosphocholine acyltransferase (GPCAT), and lyso-PC’s are reacylated by AT that use acyl-CoA’s, phospholipids (in CoA-dependent or independent reactions) or lyso-phospholipids as acyl donor.
The enzymes involved in phospholipid remodeling at the ER mostly contain conserved motifs that predict their generalized function. Most acyltransferases and transacylases catalyzing reacylation belong to the acylglycerophosphate acyltransferase (AGPAT) family or to the membrane bound O-acyltransferase (MBOAT) family. The characteristic AGPAT domain contains four conserved motifs [44], and is widespread, but not found in viruses or archaea. This family comprises taffazin as well as 1-acyl-sn-glycerol-3-phosphate acyltransferases [28,45,46], including mammalian LPCAT1 mentioned above. The AGPAT family is a member of the larger Acyltransferase Clan (CL0228) which includes several families of acyltransferases, including DAGAT and LipA acyltransferases [45,46].
Members of the MBOAT superfamily (Clan CL0517) are found in all kingdoms of life and are characterized by the ability to transfer organic acids onto hydroxyl groups of membrane-embedded targets [47]. Many, but not all MBOAT-containing enzymes, utilize acyl-CoA as the acyl donor and targets include lipids, amino acids residues within proteins, and other hydroxyl group-containing compounds [47,48]. All confirmed MBOAT proteins (including mammalian LPCAT3 mentioned above) contain a conserved histidine residue that appears important for catalysis [47,49]. A recent paper presents the crystal structure of DltB, an MBOAT that catalyzes the alanylation of cell-wall teichoic in Gram-positive bacteria [50], providing the most detailed structural information on this class of enzymes to date. The structure reveals an extra-cellular funnel extending into the membrane with the conserved histidine located at the bottom. A narrow tunnel connects the histidine to the cytoplasm suggesting a mechanism of cross-membrane catalysis.
4. ER-based phospholipid turnover by acyl chain remodeling and degradation
Here we will review current knowledge of degradation and acyl chain remodeling of PC, PE and PI, the bulk phospholipid constituents found in the yeast ER membrane [11], and the enzymes involved. Since PC is at the end of the phospholipid biosynthetic pathways, and has a relatively long half-life compared to PE and PI, which also serve as biosynthetic intermediates to other lipids [51], PC turnover has been most extensively characterized.
4.1. PC
PC turnover, acyl chain remodeling, and biosynthesis are intertwined (Fig. 2). At any point in time, the PC molecular species profile observed in cellular membranes reflects both the molecular specificity of the biosynthetic routes and post-synthetic acyl chain exchange [52–54]. Beginning with the basics, the fatty acids esterified to the sn-1 and sn-2 positions of the glycerol backbone in PC are mainly C16 and C18 with one or no double bonds, although shorter chain fatty acids occur in lesser amounts. As a result, four major PC species occur: PC 32:1 (monounsaturated) consisting of C16:0 and C16:1, PC 32:2 (diunsaturated) consisting of C16:1 and C16:1, PC 34:2 (diunsaturated) consisting of C16:1 and C18:1, PC 34:1 (monounsaturated) consisting of either C16:0 and C18:1 (this being the predominant acyl chain combination) or C16:1 and C18:0 [53,55]. In general, the unsaturated species predominate at the sn-2 position [39,56]. PC biosynthesis occurs through the PE methylation and the CDP-choline pathways. The methylation pathway produces primarily diunsaturated species and the CDP-choline pathway a more heterogeneous set of PC molecules [52] that appears to reflect the available pool of diacylglycerol species [57]. In choline-free media, the CDP-choline pathway does not contribute to net synthesis, but rather utilizes choline released through PC turnover events for re-synthesis [58].
Fig. 2.
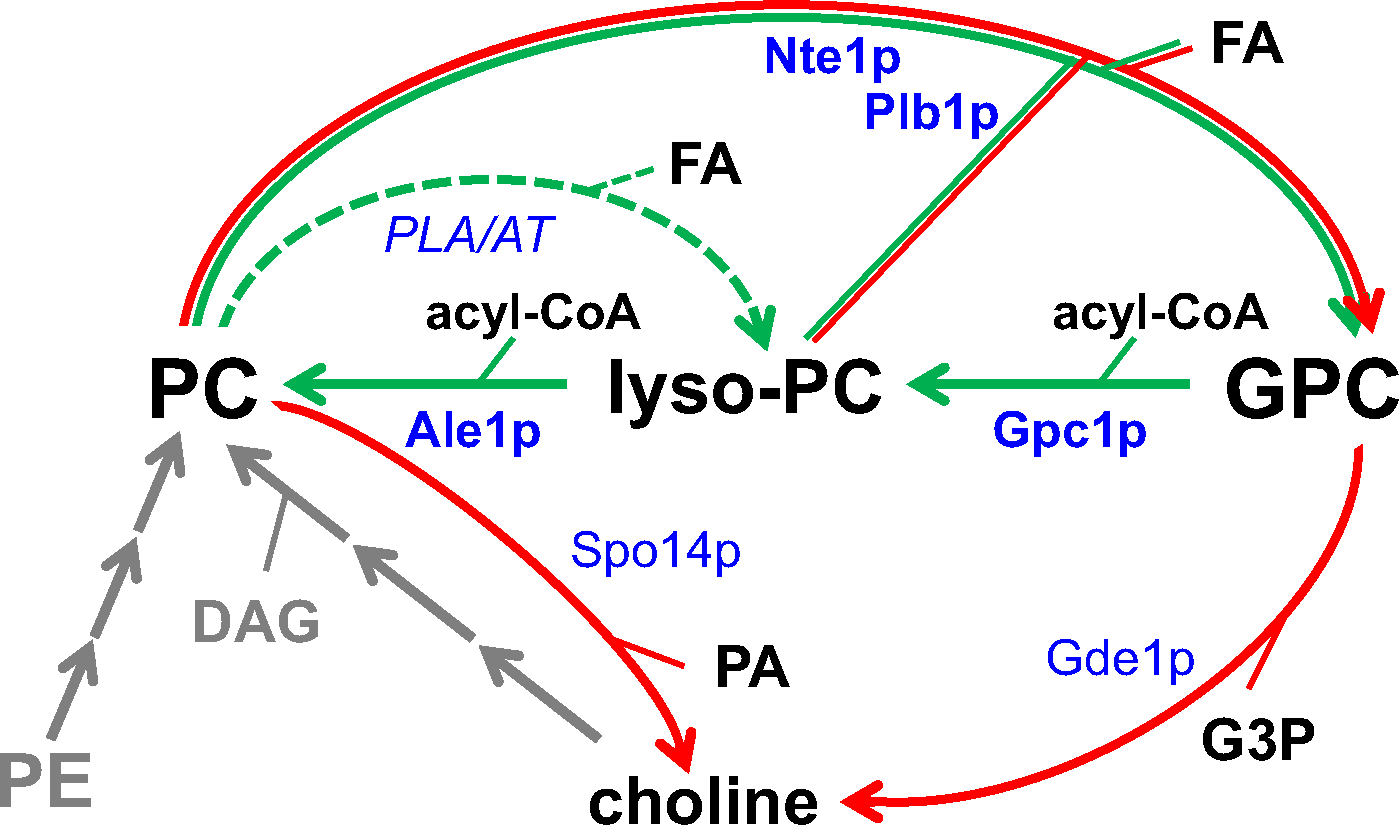
Phosphatidylcholine (PC) turnover in the yeast ER. Turnover reactions degrading PC are indicated by red arrows, acyl chain exchange reactions (PC-DRP) by green arrows, and the enzymes involved in blue, with those localized to the ER in bold. The putative conversion of PC to lyso-PC by a phospholipase A (PLA) or transacylase (AT) referred to by the dashed green arrow has not been demonstrated in vivo. The methylation of phosphatidylethanolamine (PE) and the CDP-choline pathway both synthesizing PC de novo are indicated in grey. DAG, diacylglycerol; FA, fatty acid; G3P, glycerol-3-phosphate; GPC, glycerophosphocholine.
Phospholipase D (encoded by SPO14/PLD1) produces choline and PA, the choline being converted to choline-phosphate by Cki1p during the first step of the CDP-choline pathway [59]. Other than cardiolipin-specific Cld1p [60] and Ddl1p, a PLA1 with a broad substrate specificity [61,62], both of which are localized to mitochondria, no dedicated phospholipases A have been identified in yeast. The ER-localized PDAT Lro1p and the mitochondrial CoA-independent transacylase Taz1p were shown to convert PC into sn-1 lysoPC in vitro [41,63,64]. However, whether these reactions also occur in vivo remains to be established.
PC turnover via the activity of phospholipases of the B type, primarily Plb1p and Nte1p, results in the formation of free fatty acids and glycerophosphocholine (GPC, Fig. 2) [65,66]. Plb1p is a protein of 73 kDa consisting of 664 amino acids [65]. Although first thought to be plasma membrane-associated and secreted, it was later shown to be primarily ER-associated, as well as secreted from the cell [67]. Plb1p contains no predicted transmembrane domains, suggesting that it enters the ER lumen and associates transiently with the membrane to perform its enzymatic activities. Plb1p contains the Pfam [45,46] defined PLA2_B domain (PF01735). This family includes lysophospholipase/phospholipase B (EC:3.1.1.5) [65] and cytosolic phospholipase A2 [68] activities (EC:3.1.4). The phospholipase Nte1p consists of 1679 amino acids with a predicted MW of 187 kDa [66]. It derives its name from its similarity to the human neuropathy target esterase gene [66,69]. Nte1p is ER-associated and has four predicted transmembrane domains. It contains cyclic nucleotide-binding domains (PF00027) as well as a patatin-like phospholipase domain (PF01734). The patatin domain is found widely across eukaryotic and prokaryotic organisms and contains the consensus serine lipase motif, Gly-X-Ser-X-Gly [70,71]. Radiolabeling showed that Nte1p deacylates PC derived from CDP-choline rather than PC synthesized by the methylation of PE [66], in agreement with the observation that the supply of choline induces deacylation of PC [72]. An explanation for these intriguing findings is lacking, and requires future experiments that distinguish between the stimulatory effects of choline on PC synthesis on the one hand and PC degradation on the other. To date there have been no indications that the phospholipases that turn over PC exhibit molecular species selectivity.
Some of the GPC produced is released extracellularly [72,73] and can be transported back into the cell via the Git1p permease under low phosphate conditions [74]. Git1p, which transports both GPI (Section 4.2) and GPC, provides a mechanism for adding exogenous radiolabeled GPC in order to track its metabolism [74]. Through a series of such experiments, GPC has been shown to have two potential metabolic fates. The first metabolic fate does not impact remodeling as defined here, but does impact synthesis. The glycerophosphodiesterase encoded by Gde1p hydrolyzes GPC to produce glycerol-3-phosphate and choline [74,75]. The choline released through Gde1p activity can be incorporated into the CDP-choline pathway for PC biosynthesis (Fig. 2).
The second metabolic fate for GPC, its acylation to lysoPC, directly impacts PC remodeling. This atypical acyltransferase activity, one acting on GPC instead of glycerol-3-phosphate, was first detected in yeast [43] and plant [42] extracts. Identification of GPC1, the GPC acyltransferase (GPCAT)-encoding gene, took longer, as it required a biochemical screen of a portion of the yeast knockout collection due to the lack of similarity of Gpc1p to other known acyltransferases [76]. Unlike other enzymes involved in lipid remodeling in yeast, Gpc1p has no domains that would have predicted its role as an acyltransferase [54,76]. Gpc1p consists of 432 amino acid residues and has a MW of 53 kDa. The protein has 8 predicted transmembrane segments and a DUF2838 domain (PF10998). DUF2838 is one of several domains of unknown function (DUFs) collected in the Pfam database. The results of high throughput analysis suggest that Gpc1p localizes to the ER [77]. Gpc1p homologs are widely distributed in eukaryotes, including fungi, plants, animals, algae and protists [76]. Notably, Gpc1p homologs are not found in prokaryotes nor in the animal subclades of chordates and arthropods. Gpc1p homologs cloned from Arabidopsus thaliana, Ricinus communis (castor bean) and Brassica napus (oilseed rape) were shown to also have GPCAT activity when expressed in yeast [76], suggesting that other homologs will also have this function.
Once Gpc1p has formed lysoPC, the lysophospholipid acyltransferase, Ale1p, is primarily responsible for the second acylation step to produce PC, although other acyltransferases may be involved [54,76]. Ale1p is an acyl-CoA-dependent acyltransferase that consists of 619 amino acids (MW 72 kDa) and belongs to the MBOAT family (PF03062). Ale1p preferentially esterifies unsaturated acyl chains at the sn-2 position of a variety of lysophospholipids in vitro [78–82]. However, deletion of ALE1 hardly affects the molecular species profiles of the major phospholipid classes [78]. In support of a role for Ale1p in PC remodeling, ale1Δ cells contain more lysoPC than wild type [80], and accumulate lysoPC when supplied with GPC [54]. Ale1p is ER-localized and enriched in mitochondria-associated membranes (MAM) [81]. It contains multiple membrane spanning regions and its conserved His residue was localized to the ER lumen like that of other yeast MBOAT enzymes [49]. If the conserved His residue is part of the active site, this would imply that the acylation of lysoPC occurs in the ER lumen. We speculate that the cytosol-facing tunnel found in the crystal structure of DltB and most likely conserved in other MBOAT proteins [50], allows access of cytosolic acyl-CoA to the enzyme’s active site at the bottom of the luminal funnel.
The identification of Gpc1p, coupled with in vitro and in vivo experiments in various mutant backgrounds, defined a novel route for PC biosynthesis. Perhaps more importantly, it provided the last player in a post-synthetic PC deacylation/reacylation remodeling pathway (PC-DRP) by which both acyl chains can be exchanged [54,76]. Indeed, prior to the identification of Gpc1p, an enzyme with such an activity was predicted through experiments in which post synthetic acyl chain remodeling of PC was detected [52,83]. In particular, pulse-chase studies in a pct1Δ mutant in which the CDP-choline route is inactivated, using deuterium labeled (methyl-D3)-methionine followed by ESI-MS/MS analysis, detected a post-synthetic increase in monounsaturated PC species (PC 32:1, PC 34:1) at the expense of diunsaturated PC species (PC 32:2, PC 34:2) that stems from the preferential incorporation of C16:0 at the sn-1 position [52,56]. Gpc1p activity appears to be primarily responsible for this remodeling at the sn-1 position to more saturated species based on experiments with strains lacking and overexpressing the gene [54]. Accordingly, in vitro assays show that microsomal Gpc1p has a preference for C16:0-CoA as acyl donor [76]. Similar assays revealed that Gpc1p acylates GPC at about tenfold reduced rate when lysoPC is supplied as acyl donor instead of acyl-CoA [76].
The overexpression of the GPAT SCT1 increases cellular C16:0 content, and strongly enhances the incorporation of C16:0 into PC by acyl chain remodeling in pct1 cells, thus providing a more sensitive read-out of PC remodeling by ESI-MS/MS [53]. Under these conditions, deletion of the PLB1 gene was found to reduce the rate of PC remodeling and the cellular lysoPC content [56], implicating Plb1p in PC-DRP.
4.2. PI
PI synthase encoded by the PIS1 gene uses CDP-DAG and inositol to synthesize PI [84]. Compared to the other phospholipid classes PI is strongly enriched in saturated acyl chains, which are present at 50% of total [55,85], some 10% being stearic acid (C18:0). A major fate of PI is its deacylation by Plb3p to produce free fatty acids and intracellular and extracellular GPI [51,73,86]. Under conditions of low phosphate availability, extracellular GPI is taken up by the Git1p transporter [87]. There is no evidence that GPI can be reacylated or contributes to PI remodeling. Whether the deacylation of PI is molecular species-selective is not known.
The enrichment of C18:0 results from acyl chain remodeling by the acyl-CoA dependent acyltransferase Cst26p/Psi1p [88]. Cst26p/Psi1p consists of 397 amino acids, has a MW of 46 kDa, and is localized to the ER [8]. It has 3 predicted transmembrane domains and contains the acyltransferase domain (PF01553). Deletion of CST26 causes the virtual disappearance of stearic acid from the sn-1 position of PI, and accordingly, microsomes isolated from the mutant lack acyltransferase activity for sn-2-acyl-1-lysoPI [88]. Interestingly, the deletion mutant has an overall reduced cellular C18:0 content, indicating the loss of a metabolic sink for C18:0 chains [89]. The loss in C18:0 is compensated for by a rise in the proportion of C16:0 that appears to preferentially end up in PC [88]. The phospholipase or transacylase generating 2-acyl-lysoPI remains elusive.
Deletion of CST26 was recently reported to exhibit strong negative genetic interactions with deletion of ELO2 and ELO3, genes that encode the acyl-CoA elongases responsible for the synthesis of C26:0-CoA required for sphingolipid synthesis [90]. It turned out that Cst26p is important for synthesizing a minor molecular PI species with C26:0 attached at the sn-1 position [91]. In elo2Δ or elo3Δ, Cst26p most likely generates PIs with a very long saturated acyl chain at sn-1 that compensate for the lack of mature sphingolipids [90].
The majority of PI and derived phosphoinositides in mammalian cells consists of the 1-stearoyl-2 arachidonoyl molecular species [92]. The post-synthetic incorporation of C18:0 at the sn-1 position of PI by Psi1p homologs (AGPAT8/ALCAT1) has been conserved in higher eukaryotes including C. elegans and mice [93,94]. An acyltransferase belonging to the MBOAT family, LPIAT1 (MBOAT7) incorporates C20:4 at the sn-2 position of PI via the Lands’ cycle [95].
4.3. PE
Data on the degradation and remodeling of PE in yeast is scarce. A Ca2+-dependent PE-hydrolyzing phospholipase D activity was reported that has not yet been assigned to an ORF [96,97]. Turnover of PE to glycerophosphoethanolamine (GPE) has been reported in a cho2opi3 strain that accumulates PE due to inactivation of the methylation of PE to PC [72]. Plb1p that hydrolyzes PE in vivo [65], and/or the broad specificity phospholipase Plb2p [86] may be involved. Ale1p is the major acyltransferase acylating lysoPE [81], and accordingly, ale1Δ cells contain more lysoPE than wild type [80].
Remodeling of exogenously supplied short-chain PE 10:0/10:0 by exchange of both acyl chains was demonstrated in a psd1Δ psd2Δ strain with impaired PE synthesis [98]. Whereas Plb1p, Plb2p, Plb3p, Nte1p, Ddl1 and the potential phospholipase Spo1p were not required for the remodeling, Ale1p and, to a less extent, Slc1p, the major lysoPA acyltransferase [78], were involved in the incorporation of C16:1 and C18:1 acyl chains at the sn-2 position.
The involvement of additional enzymes in PE turnover and remodeling may be inferred from data obtained in vitro. Microsomal Gpc1p acylates GPE [76], presumably at the sn-1 position. The dual function enzyme Tgl3p with both triacylglycerol lipase and acyltransferase activity, acylates sn-2 lysoPE in vitro [99]. The PDAT Lro1p uses PE and PC as acyl chain donor in vitro [41,63], with a preference for PE [100]. Based on the observations that both the synthesis of triacylglycerol and the activity of Lro1p are decreased in mutants impaired in PE biosynthesis via the Kennedy pathway, Lro1p was proposed to preferably deacylate PE in vivo [101].
Lro1 consists of 661 amino acids with a MW of 75 kDa, and contains an LCAT (lecithin: cholesterol acyltransferase) domain (PF02450). This family also encompasses enzymes with phospholipid:diacylglycerol acyltransferase (PDAT) activity (EC 2.3.1.158) that, like Lro1, transfer acyl groups from the sn-2 position of a phospholipid to diacylglycerol, thus forming an sn-1 lysophospholipid. The LCAT family is part of the Alpha/Beta (AB) hydrolase clan. The AB hydrolase fold, is found in a wide range of enzymes [102]. Lro1p has been localized to the ER membrane, contains a single transmembrane domain, and has a large luminal domain containing the active site [103].
In mammals, LPEAT1, an MBOAT, and LPEAT2, an AGPAT, acylate lysoPE using unsaturated acyl-CoA as preferred acyl donor [28]. Both have been implicated in the synthesis of cone-shaped PE, required for organogenesis [30] and osteoclast fusion [104], respectively.
5. Functions of phospholipid acyl chain remodeling in yeast
In the absence of any strong phenotypes associated with the inactivation of enzymes implicated in phospholipid acyl chain remodeling in the ER, we can only speculate about its biological functions. The most obvious function for phospholipid acyl chain remodeling is the fine-tuning of membrane lipid composition at the molecular species level to ensure optimal membrane physical properties and to sustain specific lipid functions. The latter may be exemplified by the changes in the cellular distribution of PI(4)P and PI(4,5)P2 impacting cell polarity that were observed in a cst26 deletion mutant [20]. Inactivation of Cst26p not only depletes PI of C18:0 acyl chains, but PIP and PIP2 as well. Furthermore, acyl chain remodeling may serve to remove acyl chains that have been damaged, e.g. by oxidation.
Phospholipid acyl chain remodeling of PC by PC-DRP has been proposed to buffer acyl chain composition [56], based on the increased channeling of C16:0 into PE-derived PC by, presumably, Gpc1p-mediated acyl chain remodeling under conditions of Sct1p overexpression. Studies in which free fatty acids were supplied in the culture medium provided further clues as to the buffering function of acyl chain remodeling. Yeast dga1Δ lro1Δ mutants lack the ability to store excess acyl chains in triacylglycerol, and as a consequence exhibit sensitivity to unsaturated but not to saturated fatty acids supplied in the culture medium [105,106]. Exogenous C16:0 was found to be efficiently incorporated in the phospholipid fraction, PC and PS in particular, in the absence of TAG synthesis [106]. Accordingly, if dga1Δ lro1Δ cells are disturbed in PC synthesis by deletion of either of the phospholipid methyltransferase-encoding genes CHO2 or OPI3 they become sensitive to C16:0 [106].
Interestingly, PC deacylation via Plb1p is regulated in coordination with sphingolipid biosynthesis. Abrogation of sphingolipid biosynthesis via deletion of the Ypk1 kinase or treatment with myriocin results in increased Plb1p expression and increased PC turnover. Moreover, deletion of PLB1 exacerbates the phenotype of a ypk1Δ mutant, while overexpression of PLB1 (but not of NTE1) rescues cells defective in Ypk1p and Ypk2p, suggesting that increased PC turnover (or remodeling) mitigates the consequences of defects in sphingolipid biosynthesis [67].
Studies on the physiological importance of Gpc1p are just beginning, but it is, like other genes involved in lipid metabolism in yeast, upregulated by inositol limitation [8]. In addition, gpc1Δ mutants growing in inositol-free medium display a defect in stationary phase viability [54]. Thus, PC remodeling to more saturated molecular species may be important for maintaining long-term survival under certain culture conditions.
6. Conclusion and future directions
Acyl chain remodeling of the bulk membrane phospholipids PC and PI has been unequivocally demonstrated in yeast, and several of the enzymes involved have been identified. As detailed in Section 5, we are beginning to catch the first glimpses of possible functions of acyl chain remodeling. Many questions remain: e.g., what is the added value of PC-DRP given that choline-recycling via the CDP-choline pathway also increases the saturated acyl chain content of PC [52]? How are PC-DRP and recycling of choline via the CDP-choline route regulated? The membrane topology of Plb1p, Ale1p and Lro1p raises the question of whether acyl chain remodeling is restricted to the luminal leaflet of the ER membrane, and prompts elucidation of the active site and membrane topology of Gpc1p and Cst26p. Which additional enzymes are involved?
With the availability of dynamic lipidomics approaches and the ease of obtaining yeast mutants, there are no technical obstacles to increase our insight into yeast acyl chain remodeling and its role in membrane lipid homeostasis. We expect it to soon reach and surpass knowledge about acyl chain remodeling in higher eukaryotes.
Supplementary Material
Acknowledgments
We thank the (former) members of our research groups for useful discussions and their input in the research described here. This work was supported by the Division of Chemical Sciences in the Netherlands with financial aid from The Netherlands Organization for Scientific Research (NWO) and by National Institutes of Health Grant R15 GM104876 (to J. P.-V.).
Footnotes
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- [1].van Meer G, de Kroon AI, Lipid map of the mammalian cell, J. Cell Sci. 124 (2011) 5–8. [DOI] [PubMed] [Google Scholar]
- [2].Dawaliby R, Trubbia C, Delporte C, Noyon C, Ruysschaert JM, Van Antwerpen P, Govaerts C, Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells, J. Biol. Chem. 291 (2016) 3658–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Renne MF, de Kroon AIPM, The role of phospholipid molecular species in determining the physical properties of yeast membranes, FEBS Lett. 592 (2018) 1330–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jacquemyn J, Cascalho A, Goodchild RE, The ins and outs of endoplasmic reticulum-controlled lipid biosynthesis, EMBO Rep. 18 (2017) 1905–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Covino R, Hummer G, Ernst R, Integrated functions of membrane property sensors and a hidden side of the unfolded protein response, Mol. Cell 71 (2018) 458–467. [DOI] [PubMed] [Google Scholar]
- [6].Carman GM, Han GS, Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae, Annu. Rev. Biochem. 80 (2011) 859–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li TY, Song L, Sun Y, Li J, Yi C, Lam SM, Xu D, Zhou L, Li X, Yang Y, Zhang CS, Xie C, Huang X, Shui G, Lin SY, Reue K, Lin SC, Tip60-mediated lipin 1 acetylation and ER translocation determine triacylglycerol synthesis rate, Nat. Commun. 9 (2018) (1916–018-04363-w). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Henry SA, Kohlwein SD, Carman GM, Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae, Genetics 190 (2012) 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de Kroon AI, Rijken PJ, De Smet CH, Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective, Prog. Lipid Res. 52 (2013) 374–394. [DOI] [PubMed] [Google Scholar]
- [10].Harayama T, Riezman H, Understanding the diversity of membrane lipid composition, Nat. Rev. Mol. Cell Biol. 19 (2018) 281–296. [DOI] [PubMed] [Google Scholar]
- [11].Tuller G, Nemec T, Hrastnik C, Daum G, Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild-type strain grown on different carbon sources, Yeast 15 (1999) 1555–1564. [DOI] [PubMed] [Google Scholar]
- [12].Somerharju P, Is spontaneous translocation of polar lipids between cellular organelles negligible? Lipid Insights 8 (2016) 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Boumann HA, Chin PT, Heck AJ, De Kruijff B, De Kroon AI, The yeast phospholipid N-methyltransferases catalyzing the synthesis of phosphatidylcholine preferentially convert di-C16:1 substrates both in vivo and in vitro, J. Biol. Chem. 279 (2004) 40314–40319. [DOI] [PubMed] [Google Scholar]
- [14].Voelker DR, Bridging gaps in phospholipid transport, Trends Biochem. Sci. 30 (2005) 396–404. [DOI] [PubMed] [Google Scholar]
- [15].Friedman JR, Kannan M, Toulmay A, Jan CH, Weissman JS, Prinz WA, Nunnari J, Lipid homeostasis is maintained by dual targeting of the mitochondrial PE biosynthesis enzyme to the ER, Dev. Cell 44 (2018) 261–270.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wilson-Zbinden C, dos Santos AX, Stoffel-Studer I, van der Vaart A, Hofmann K, Reggiori F, Riezman H, Kraft C, Peter M, Autophagy competes for a common phosphatidylethanolamine pool with major cellular PE-consuming pathways in Saccharomyces cerevisiae, Genetics 199 (2015) 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kinoshita T, Fujita M, Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling, J. Lipid Res. 57 (2016) 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ohsumi Y, Molecular dissection of autophagy: two ubiquitin-like systems, Nat. Rev. Mol. Cell Biol. 2 (2001) 211–216. [DOI] [PubMed] [Google Scholar]
- [19].Traynor-Kaplan A, Kruse M, Dickson EJ, Dai G, Vivas O, Yu H, Whittington D, Hille B, Fatty-acyl chain profiles of cellular phosphoinositides, Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862 (2017) 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Doignon F, Laquel P, Testet E, Tuphile K, Fouillen L, Bessoule JJ, Requirement of phosphoinositides containing stearic acid to control cell polarity, Mol. Cell. Biol. 36 (2015) 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, De Camilli P, Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry, Nat. Biotechnol. 21 (2003) 813–817. [DOI] [PubMed] [Google Scholar]
- [22].Dickson RC, Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast, J. Lipid Res. 49 (2008) 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taniguchi M, Okazaki T, The role of sphingomyelin and sphingomyelin synthases in cell death, proliferation and migration-from cell and animal models to human disorders, Biochim. Biophys. Acta 1841 (2014) 692–703. [DOI] [PubMed] [Google Scholar]
- [24].Lands WE, Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin, J. Biol. Chem. 235 (1960) 2233–2237. [PubMed] [Google Scholar]
- [25].Lands WE, Lipid Metabolism, Annu. Rev. Biochem. 34 (1965) 313–346. [DOI] [PubMed] [Google Scholar]
- [26].Lands WE, Hart P, Metabolism of glycerolipids. VI. Specificities of acyl coenzyme A: phospholipid acyltransferases, J. Biol. Chem. 240 (1965) 1905–1911. [PubMed] [Google Scholar]
- [27].Shindou H, Shimizu T, Acyl-CoA:lysophospholipid acyltransferases, J. Biol. Chem. 284 (2009) 1–5. [DOI] [PubMed] [Google Scholar]
- [28].Yamashita A, Hayashi Y, Nemoto-Sasaki Y, Ito M, Oka S, Tanikawa T, Waku K, Sugiura T, Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms, Prog. Lipid Res. 53 (2014) 18–81. [DOI] [PubMed] [Google Scholar]
- [29].Hermansson M, Hokynar K, Somerharju P, Mechanisms of glycerophospholipid homeostasis in mammalian cells, Prog. Lipid Res. 50 (2011) 240–257. [DOI] [PubMed] [Google Scholar]
- [30].Hishikawa D, Hashidate T, Shimizu T, Shindou H, Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells, J. Lipid Res. 55 (2014) 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Astudillo AM, Balboa MA, Balsinde J, Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization, Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864 (2018) 772–783. [DOI] [PubMed] [Google Scholar]
- [32].Wang B, Tontonoz P, Phospholipid remodeling in physiology and disease, Annu. Rev. Physiol. 81 (2019) 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goss V, Hunt AN, Postle AD, Regulation of lung surfactant phospholipid synthesis and metabolism, Biochim. Biophys. Acta 1831 (2013) 448–458. [DOI] [PubMed] [Google Scholar]
- [34].Ohlrogge J, Browse J, Lipid biosynthesis, Plant Cell 7 (1995) 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bates PD, Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis, Biochim. Biophys. Acta 1861 (2016) 1214–1225. [DOI] [PubMed] [Google Scholar]
- [36].Dyer JM, Stymne S, Green AG, Carlsson AS, High-value oils from plants, Plant J. 54 (2008) 640–655. [DOI] [PubMed] [Google Scholar]
- [37].Block MA, Jouhet J, Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites, Curr. Opin. Cell Biol. 35 (2015) 21–29. [DOI] [PubMed] [Google Scholar]
- [38].Yamada K, Okuyama H, Endo Y, Ikezawa H, Acyltransferase systems involved in phospholipid metabolism in Saccharomyces cerevisiae, Arch. Biochem. Biophys. 183 (1977) 281–289. [DOI] [PubMed] [Google Scholar]
- [39].Wagner S, Paltauf F, Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions, Yeast 10 (1994) 1429–1437. [DOI] [PubMed] [Google Scholar]
- [40].Schlame M, Greenberg ML, Biosynthesis, remodeling and turnover of mitochondrial cardiolipin, Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862 (2017) 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S, Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants, Proc. Natl. Acad. Sci. U. S. A. 97 (2000) 6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lager I, Glab B, Eriksson L, Chen G, Banas A, Stymne S, Novel reactions in acyl editing of phosphatidylcholine by lysophosphatidylcholine transacylase (LPCT) and acyl-CoA:glycerophosphocholine acyltransferase (GPCAT) activities in microsomal preparations of plant tissues, Planta 241 (2015) 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stalberg K, Neal AC, Ronne H, Stahl U, Identification of a novel GPCAT activity and a new pathway for phosphatidylcholine biosynthesis in S. cerevisiae, J. Lipid Res. 49 (2008) 1794–1806. [DOI] [PubMed] [Google Scholar]
- [44].Lewin TM, Wang P, Coleman RA, Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction, Biochemistry 38 (1999) 5764–5771. [DOI] [PubMed] [Google Scholar]
- [45].Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M, Pfam: the protein families database, Nucleic Acids Res. 42 (2014) D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A, The Pfam protein families database: towards a more sustainable future, Nucleic Acids Res. 44 (2016) D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hofmann K, A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling, Trends Biochem. Sci. 25 (2000) 111–112. [DOI] [PubMed] [Google Scholar]
- [48].Stahl U, Stalberg K, Stymne S, Ronne H, A family of eukaryotic lysophospholipid acyltransferases with broad specificity, FEBS Lett. 582 (2008) 305–309. [DOI] [PubMed] [Google Scholar]
- [49].Pagac M, de la Mora HV, Duperrex C, Roubaty C, Vionnet C, Conzelmann A, Topology of 1-acyl-sn-glycerol-3-phosphate acyltransferases SLC1 and ALE1 and related membrane-bound O-acyltransferases (MBOATs) of Saccharomyces cerevisiae, J. Biol. Chem. 286 (2011) 36438–36447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W, Crystal structure of a membrane-bound O-acyltransferase, Nature 562 (2018) 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA, Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation, J. Biol. Chem. 272 (1997) 20873–20883. [DOI] [PubMed] [Google Scholar]
- [52].Boumann HA, Damen MJ, Versluis C, Heck AJ, de Kruijff B, de Kroon AI, The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling, Biochemistry 42 (2003) 3054–3059. [DOI] [PubMed] [Google Scholar]
- [53].Renne MF, Bao X, De Smet CH, de Kroon AI, Lipid Acyl Chain Remodel. Yeast 8 (2016) 33–40 Lipid Insights. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Anaokar S, Kodali R, Jonik B, Renne MF, Brouwers JFHM, Lager I, de Kroon AIPM, Patton-Vogt J, The glycerophosphocholine acyltransferase Gpc1 is part of a phosphatidylcholine (PC)-remodeling pathway that alters PC species in yeast, J. Biol. Chem. 294 (2019) 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A, Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry, Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].De Smet CH, Cox R, Brouwers JF, de Kroon AI, Yeast cells accumulate excess endogenous palmitate in phosphatidylcholine by acyl chain remodeling involving the phospholipase B Plb1p, Biochim. Biophys. Acta 1831 (2013) 1167–1176. [DOI] [PubMed] [Google Scholar]
- [57].Boumann HA, Gubbens J, Koorengevel MC, Oh CS, Martin CE, Heck AJ, Patton-Vogt J, Henry SA, de Kruijff B, de Kroon AI, Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote, Mol. Biol. Cell 17 (2006) 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].McMaster CR, Bell RM, Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases, J. Biol. Chem. 269 (1994) 28010–28016. [PubMed] [Google Scholar]
- [59].Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA, A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast, J. Biol. Chem. 273 (1998) 16635–16638. [DOI] [PubMed] [Google Scholar]
- [60].Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R, Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast, J. Biol. Chem. 284 (2009) 11572–11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yadav PK, Rajasekharan R, Misregulation of a DDHD domain-containing lipase causes mitochondrial dysfunction in yeast, J. Biol. Chem. 291 (2016) 18562–18581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Urafuji K, Arioka M, Yor022c protein is a phospholipase A1 that localizes to the mitochondrial matrix, Biochem. Biophys. Res. Commun. 480 (2016) 302–308. [DOI] [PubMed] [Google Scholar]
- [63].Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL, A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast, J. Biol. Chem. 275 (2000) 15609–15612. [DOI] [PubMed] [Google Scholar]
- [64].Abe M, Hasegawa Y, Oku M, Sawada Y, Tanaka E, Sakai Y, Miyoshi H, Mechanism for remodeling of the acyl chain composition of cardiolipin catalyzed by Saccharomyces cerevisiae tafazzin, J. Biol. Chem. 291 (2016) 15491–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lee KS, Patton JL, Fido M, Hines LK, Kohlwein SD, Paltauf F, Henry SA, Levin DE, The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity, J. Biol. Chem. 269 (1994) 19725–19730. [PubMed] [Google Scholar]
- [66].Zaccheo O, Dinsdale D, Meacock PA, Glynn P, Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells, J. Biol. Chem. 279 (2004) 24024–24033. [DOI] [PubMed] [Google Scholar]
- [67].Surlow BA, Cooley BM, Needham PG, Brodsky JL, Patton-Vogt J, Loss of Ypk1, the yeast homolog to the human serum- and glucocorticoid-induced protein kinase, accelerates phospholipase B1-mediated phosphatidylcholine deacylation, J. Biol. Chem. 289 (2014) 31591–31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nalefski EA, Sultzman LA, Martin DM, Kriz RW, Towler PS, Knopf JL, Clark JD, Delineation of two functionally distinct domains of cytosolic phospholipase A2, a regulatory Ca(2+)-dependent lipid-binding domain and a Ca (2+)-independent catalytic domain, J. Biol. Chem. 269 (1994) 18239–18249. [PubMed] [Google Scholar]
- [69].Fernandez-Murray JP, McMaster CR, Phosphatidylcholine synthesis and its catabolism by yeast neuropathy target esterase 1, Biochim. Biophys. Acta 1771 (2007) 331–336. [DOI] [PubMed] [Google Scholar]
- [70].Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ, Characterization of the human patatin-like phospholipase family, J. Lipid Res. 47 (2006) 1940–1949. [DOI] [PubMed] [Google Scholar]
- [71].Kienesberger PC, Oberer M, Lass A, Zechner R, Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions, J. Lipid Res. 50 (2009) S63–S68 Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dowd SR, Bier ME, Patton-Vogt JL, Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway, J. Biol. Chem. 276 (2001) 3756–3763. [DOI] [PubMed] [Google Scholar]
- [73].Angus WW, Lester RL, Turnover of inositol and phosphorus containing lipids in Saccharomyces cerevisiae; extracellular accumulation of glycerophosphorylinositol derived from phosphatidylinositol, Arch. Biochem. Biophys. 151 (1972) 483–495. [DOI] [PubMed] [Google Scholar]
- [74].Fisher E, Almaguer C, Holic R, Griac P, Patton-Vogt J, Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae, J. Biol. Chem. 280 (2005) 36110–36117. [DOI] [PubMed] [Google Scholar]
- [75].Fernandez-Murray JP, McMaster CR, Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway, J. Biol. Chem. 280 (2005) 38290–38296. [DOI] [PubMed] [Google Scholar]
- [76].Glab B, Beganovic M, Anaokar S, Hao MS, Rasmusson AG, Patton-Vogt J, Banas A, Stymne S, Lager I, Cloning of glycerophosphocholine acyltransferase (GPCAT) from fungi and plants: a novel enzyme in phosphatidylcholine synthesis, J. Biol. Chem. 291 (2016) 25066–25076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK, Global analysis of protein localization in budding yeast, Nature 425 (2003) 686–691. [DOI] [PubMed] [Google Scholar]
- [78].Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A, SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast, J. Biol. Chem. 282 (2007) 30845–30855. [DOI] [PubMed] [Google Scholar]
- [79].Chen Q, Kazachkov M, Zheng Z, Zou J, The yeast acylglycerol acyltransferase LCA1 is a key component of Lands cycle for phosphatidylcholine turnover, FEBS Lett. 581 (2007) 5511–5516. [DOI] [PubMed] [Google Scholar]
- [80].Tamaki H, Shimada A, Ito Y, Ohya M, Takase J, Miyashita M, Miyagawa H, Nozaki H, Nakayama R, Kumagai H, LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae, J. Biol. Chem. 282 (2007) 34288–34298. [DOI] [PubMed] [Google Scholar]
- [81].Riekhof WR, Wu J, Jones JL, Voelker DR, Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae, J. Biol. Chem. 282 (2007) 28344–28352. [DOI] [PubMed] [Google Scholar]
- [82].Riekhof WR, Wu J, Gijon MA, Zarini S, Murphy RC, Voelker DR, Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation, J. Biol. Chem. 282 (2007) 36853–36861. [DOI] [PubMed] [Google Scholar]
- [83].Tanaka K, Fukuda R, Ono Y, Eguchi H, Nagasawa S, Nakatani Y, Watanabe H, Nakanishi H, Taguchi R, Ohta A, Incorporation and remodeling of extracellular phosphatidylcholine with short acyl residues in Saccharomyces cerevisiae, Biochim. Biophys. Acta 1781 (2008) 391–399. [DOI] [PubMed] [Google Scholar]
- [84].Gardocki ME, Jani N, Lopes JM, Phosphatidylinositol biosynthesis: biochemistry and regulation, Biochim. Biophys. Acta 1735 (2005) 89–100. [DOI] [PubMed] [Google Scholar]
- [85].Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, Paltauf F, Wieland FT, Kohlwein SD, Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane, J. Cell Biol. 146 (1999) 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Merkel O, Oskolkova OV, Raab F, El-Toukhy R, Paltauf F, Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae, Biochem. J. 387 (2005) 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Almaguer C, Mantella D, Perez E, Patton-Vogt J, Inositol and phosphate regulate GIT1 transcription and glycerophosphoinositol incorporation in Saccharomyces cerevisiae, Eukaryot. Cell 2 (2003) 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Le Guedard M, Bessoule JJ, Boyer V, Ayciriex S, Velours G, Kulik W, Ejsing CS, Shevchenko A, Coulon D, Lessire R, Testet E, PSI1 is responsible for the stearic acid enrichment that is characteristic of phosphatidylinositol in yeast, FEBS J. 276 (2009) 6412–6424. [DOI] [PubMed] [Google Scholar]
- [89].De Smet CH, Vittone E, Scherer M, Houweling M, Liebisch G, Brouwers JF, de Kroon AI, The yeast acyltransferase Sct1p regulates fatty acid desaturation by competing with the desaturase Ole1p, Mol. Biol. Cell 23 (2012) 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Vazquez HM, Vionnet C, Roubaty C, Mallela SK, Schneiter R, Conzelmann A, Chemogenetic E-MAP in Saccharomyces cerevisiae for identification of membrane transporters operating lipid flip flop, PLoS Genet. 12 (2016) e1006160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Schneiter R, Brugger B, Amann CM, Prestwich GD, Epand RF, Zellnig G, Wieland FT, Epand RM, Identification and biophysical characterization of a very-long-chain-fatty-acid-substituted phosphatidylinositol in yeast subcellular membranes, Biochem. J. 381 (2004) 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Balla T, Phosphoinositides: tiny lipids with giant impact on cell regulation, Physiol. Rev. 93 (2013) 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Imae R, Inoue T, Kimura M, Kanamori T, Tomioka NH, Kage-Nakadai E, Mitani S, Arai H, Intracellular phospholipase A1 and acyltransferase, which are involved in Caenorhabditis elegans stem cell divisions, determine the sn-1 fatty acyl chain of phosphatidylinositol, Mol. Biol. Cell 21 (2010) 3114–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Imae R, Inoue T, Nakasaki Y, Uchida Y, Ohba Y, Kono N, Nakanishi H, Sasaki T, Mitani S, Arai H, LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice, J. Lipid Res. 53 (2012) 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lee HC, Inoue T, Sasaki J, Kubo T, Matsuda S, Nakasaki Y, Hattori M, Tanaka F, Udagawa O, Kono N, Itoh T, Ogiso H, Taguchi R, Arita M, Sasaki T, Arai H, LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice, Mol. Biol. Cell 23 (2012) 4689–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Waksman M, Tang X, Eli Y, Gerst JE, Liscovitch M, Identification of a novel (Ca2+)-dependent, phosphatidylethanolamine-hydrolyzing phospholipase D in yeast bearing a disruption in PLD1, J. Biol. Chem. 272 (1997) 36–39. [DOI] [PubMed] [Google Scholar]
- [97].Mayr JA, Kohlwein SD, Paltauf F, Identification of a novel, Ca(2+)-dependent phospholipase D with preference for phosphatidylserine and phosphatidylethanolamine in Saccharomyces cerevisiae, FEBS Lett. 393 (1996) 236–240. [DOI] [PubMed] [Google Scholar]
- [98].Deng L, Fukuda R, Kakihara T, Narita K, Ohta A, Incorporation and remodeling of phosphatidylethanolamine containing short acyl residues in yeast, Biochim. Biophys. Acta 1801 (2010) 635–645. [DOI] [PubMed] [Google Scholar]
- [99].Rajakumari S, Daum G, Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions, Mol. Biol. Cell 21 (2010) 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ghosal A, Banas A, Stahl U, Dahlqvist A, Lindqvist Y, Stymne S, Saccharomyces cerevisiae phospholipid:diacylglycerol acyl transferase (PDAT) devoid of its membrane anchor region is a soluble and active enzyme retaining its substrate specificities, Biochim. Biophys. Acta 1771 (2007) 1457–1463. [DOI] [PubMed] [Google Scholar]
- [101].Horvath SE, Wagner A, Steyrer E, Daum G, Metabolic link between phosphatidylethanolamine and triacylglycerol metabolism in the yeast Saccharomyces cerevisiae, Biochim. Biophys. Acta 1811 (2011) 1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, The alpha/beta hydrolase fold, Protein Eng. 5 (1992) 197–211. [DOI] [PubMed] [Google Scholar]
- [103].Choudhary V, Jacquier N, Schneiter R, The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmatic reticulum: implications for the biogenesis of lipid droplets, Commun. Integr. Biol. 4 (2011) 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Irie A, Yamamoto K, Miki Y, Murakami M, Phosphatidylethanolamine dynamics are required for osteoclast fusion, Sci. Rep. 7 (2017) 46715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD, Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast, J. Biol. Chem. 284 (2009) 30981–30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Garbarino J, Padamsee M, Wilcox L, Oelkers PM, D’Ambrosio D, Ruggles KV, Ramsey N, Jabado O, Turkish A, Sturley SL, Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death, J. Biol. Chem. 285 (2009) 30994–31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


