Abstract
The U.S. Environmental Protection Agency Endocrine Disruptor Screening Program and the Organization for Economic Cooperation and Development (OECD) have used the human adrenocarcinoma (H295R) cell-based assay to predict chemical perturbation of androgen and estrogen production. Recently, a high-throughput H295R (HT-H295R) assay was developed as part of the ToxCast program that includes measurement of 11 hormones, including progestagens, corticosteroids, androgens, and estrogens. To date, 2012 chemicals have been screened at 1 concentration; of these, 656 chemicals have been screened in concentration-response. The objectives of this work were to: (1) develop an integrated analysis of chemical-mediated effects on steroidogenesis in the HT-H295R assay and (2) evaluate whether the HT-H295R assay predicts estrogen and androgen production specifically via comparison with the OECD-validated H295R assay. To support application of HT-H295R assay data to weight-of-evidence and prioritization tasks, a single numeric value based on Mahalanobis distances was computed for 654 chemicals to indicate the magnitude of effects on the synthesis of 11 hormones. The maximum mean Mahalanobis distance (maxmMd) values were high for strong modulators (prochloraz, mifepristone) and lower for moderate modulators (atrazine, molinate). Twenty-five of 28 reference chemicals used for OECD validation were screened in the HT-H295R assay, and produced qualitatively similar results, with accuracies of 0.90/0.75 and 0.81/0.91 for increased/decreased testosterone and estradiol production, respectively. The HT-H295R assay provides robust information regarding estrogen and androgen production, as well as additional hormones. The maxmMd from this integrated analysis may provide a data-driven approach to prioritizing lists of chemicals for putative effects on steroidogenesis.
Keywords: ToxCast, H295R, steroidogenesis, high-throughput screening, Mahalanobis distance
Endocrine disruption is a toxicity of both physiological and regulatory importance; as steroid hormones regulate reproduction, development, and other biological processes, it is a priority to identify chemicals that may interact with production of these hormones. The U.S. Environmental Protection Agency (EPA) Endocrine Disruptor Screening Program (EDSP) currently employs a 2-tiered system for screening chemicals for endocrine-disrupting potential, with Tier 1 including in vitro and short-term in vivo assays to characterize this potential activity. However, the time and resources needed to conduct Tier 1 screening has limited the number of chemicals evaluated, and as such, the EPA has pursued high-throughput methodologies to more rapidly prioritize and identify chemicals with potential endocrine activity via programs including ToxCast and Tox21 (Browne et al., 2015; Dix et al., 2007; EPA, 2011; Judson et al., 2010; Judson et al., 2015; Kavlock et al., 2012; Kleinstreuer et al., 2017; Tice et al., 2013). Currently, 1 low-throughput assay used in the EDSP Tier 1 is a steroidogenesis assay employing the human adrenocarcinoma cell line (H295R). Steroidogenesis is the complex process in which cholesterol is converted into bioactive steroid hormones with important physiological functions including sexual differentiation and development, metabolism, physiological homeostasis, and reproduction. With 4 major classes of steroid hormones (progestagens, corticosteroids, androgens, and estrogens), synthesized in steroidogenic in vivo, disruption of steroidogenic enzymes can result in the development of a wide range of disorders such as congenital adrenal hyperplasia, virilization, sterility, salt retention, and hypertension (as reviewed by Miller and Auchus [2011]). The H295R assay is an in vitro method for detecting chemical disruption of the catalytic events of steroidogenesis, and has been used predominantly to predict chemical perturbation of 17b-estradiol (E2) and testosterone (T) synthesis (OECD, 2011). The H295R cell line demonstrates the biological characteristics of zonally undifferentiated human fetal adrenal cells, but produces steroid hormones found in adult adrenal cortex, ovaries, and testes (Figure 1) (Gazdar et al., 1990; Gracia et al., 2006). H295R cells have been used to evaluate effects of xenobiotics on hormone production, as well as steroidogenic enzyme activity and expression (Hilscherova et al., 2004; Maglich et al., 2014; Sanderson et al., 2000). Recently, a high-throughput adaptation of the H295R assay was developed as part of the ToxCast program (Karmaus et al., 2016) that might expedite the use of this assay as a screening tool. The current work provides a new analysis of the concentration-response data for the ToxCast high-throughput (HT) H295R assay to enable a fit-for-purpose evaluation of this assay as an alternative for the low-throughput H295R assay. Then, a novel, integrated analysis of chemical-mediated effects on steroidogenesis in the high-throughput H295R assay was developed to yield a metric that may be useful in future prioritization tasks.
Figure 1.
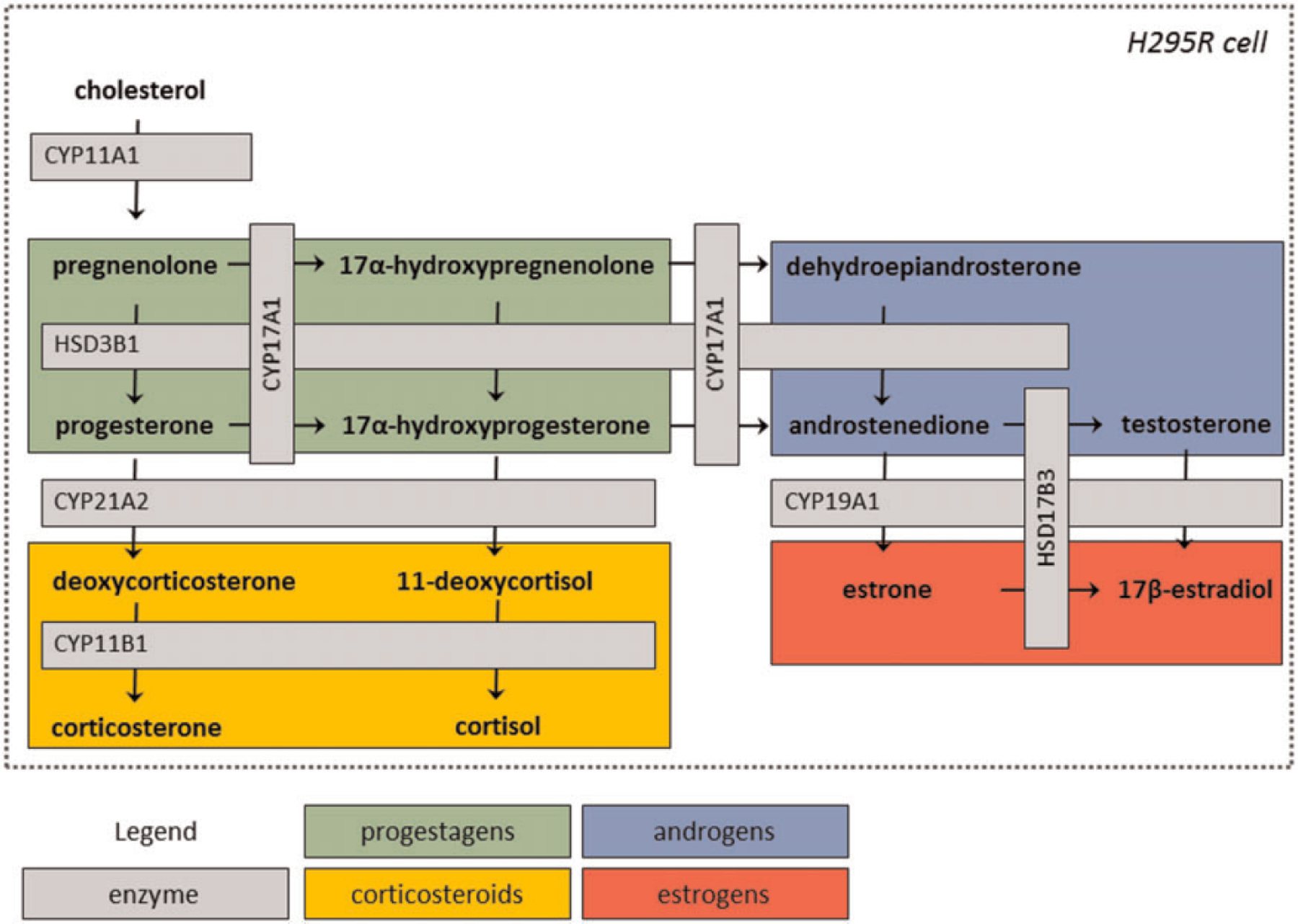
Representation of the steroid biosynthesis pathway expressed in H295R cells.
The utility of the H295R assay in screening for putative endocrine disruptors has been recognized internationally; both the Organization for Economic Cooperation and Development (OECD) and the U.S. Environmental Protection Agency (EPA) Endocrine Disruptor Screening Program (EDSP) have developed test guidelines for utilizing H295R cell-based steroidogenesis assays to detect potential chemical perturbation of E2 and T production (EPA, 2009; Hecker et al., 2011; OECD, 2011). Conduct of the assay by the OECD Test Guideline (TG) 456 or the EPA test guideline (OCSPP 890.1550) involves measurement of only E2 and T in the cell culture medium from exposed H295R cells as indicators of steroidogenesis disruption (EPA, 2009; OECD, 2011). Briefly, when performed to guideline specifications, H295R cells are acclimated in 24-well plates for 24 h, exposed for 48 h to test chemical in triplicate, and then medium is removed for steroid hormone measurement by ELISA or analytical detection. The cells are then used to assess cell viability. This assay procedure was previously adapted (Karmaus et al., 2016) for high-throughput (HT) application in the US EPA ToxCast program, with primary modifications including: the use of a single concentration pre-screen to determine chemicals most likely to perturb steroidogenesis in multi-concentration screening; the uniform use of a 48 h pre-stimulation period with forskolin; measurement of 13 steroid hormones using high-performance liquid chromatography followed by tandem mass spectrometry (HPLC-MS/MS); and the use of a 96-well format. These modifications were all intended to increase screening efficiency and fill data gaps related to in vitro steroidogenesis for large numbers of chemicals. This HT-H295R screening effort demonstrated that the assay performed reproducibly and robustly with positive controls, forskolin and prochloraz, and prototypical modulators including conazole fungicides (Karmaus et al., 2016). Statistical analysis using the Z’-factor, an indicator of assay robustness, produced values of 0.5–1, indicating an assay readout with sufficiently large signal-to-background difference and low inter-sample variability to distinguish positive and negative test chemicals from noise (Zhang et al., 1999). In a previously published analysis, 10 of the 13 measured steroid hormones in the HT-H295R assay were reported and demonstrated a median Z’ ≥ 0.5 under stimulation with forskolin and inhibition with prochloraz (Karmaus et al., 2016). Strictly standardized median difference (SSMD), a measure of effect size, was calculated to demonstrate overall assay quality and directionality. Forskolin generally increased hormone quantities with good dynamic range (SSMD values ≥7), whereas prochloraz generally inhibited hormone production with good dynamic range (SSMD values ≤–7) (Karmaus et al., 2016). These assay quality metrics suggest that the HT-H295R screening assay may be useful not only for prediction of estrogen and androgen synthesis disruption, but also in understanding broader effects on the steroid biosynthesis pathway as part of a weight-of-evidence approach (Ankley and Jensen, 2014; EPA, 2015a; Juberg et al., 2013) for predicting the endocrine bioactivity of chemicals.
Evident from the network of enzymes expressed and steroid hormones produced in the H295R cell (Figure 1), many different mechanisms of disruption may be captured in measurement of hormones from the H295R model system. For instance, prochloraz, an imidazole fungicide, is used as a reference chemical in the H295R assay that inhibits the hydroxylase activity of cytochrome P450 (CYP) 17A1, and it clearly decreases androgen production in the H295R cell line (Blystone et al., 2007). Similar observations have been reported previously for other triazole fungicides (Goetz et al., 2009). Vinclozolin mediates a complex interaction that increases E2 and decreases T in H295R cells, along with reported effects on progestagen synthesis (Hecker et al., 2006; Sanderson et al., 2002; Villeneuve et al., 2007). Forskolin, used often as a control in the H295R assay to stimulate steroidogenesis, increases hormone production by activating cyclic adenosine monophosphate-dependent signaling (Hilscherova et al., 2004). Thus, the H295R assay represents an important screening tool for identifying chemicals that may act through diverse mechanisms to affect production of T, E2, other steroid hormones in vitro.
H295R cells have been used previously in non-guideline applications to examine the effect of chemicals on hormones beyond E2 and T, as measurement of other steroid hormones produced in H295R cells may provide additional evidence for disruption of estrogen or androgen synthesis, mechanisms of steroidogenesis disruption, and/or information about effects on other specific steroid hormone classes, namely the corticosteroids and progestagens (Asser et al., 2014; Nielsen et al., 2012; Tinfo et al., 2011; Zhang et al., 2011). H295R cells also produce androstenedione and estrone as precursors of E2 and T, respectively, and thus information on these hormones may support hypotheses about enzymatic involvement, eg, upregulation of aromatase should increase both estrone and E2 production, as observed with atrazine (Tinfo et al., 2011). However, the same report also found that atrazine weakly induced progesterone production in H295R cells (Tinfo et al., 2011), suggesting that atrazine likely has other effects on the steroidogenesis pathway not mediated by aromatase and not captured by measurement of E2 and T alone, as supported by other work (Karmaus & Zacharewski, 2015; Kucka et al., 2012). Bisphenol A (BPA) was reported to decrease T and androstenedione production, increase estrone and E2 production, and produced various effects on progestagens and corticosteroids, suggesting that BPA may inhibit or otherwise down-regulate CYP17A1 in addition to having other pathway effects (Zhang et al., 2011). The pharmacologic agent metyrapone is known to reversibly inhibit CYP11B1, blocking cortisol synthesis in H295R cells (Breen et al., 2010). Using dexamethasone and mifepristone to stimulate or inhibit glucocorticoid receptor signaling in H295R cells demonstrated that the glucocorticoid receptor exerts control at least in part on adrenal hormone synthesis in H295R cells (Asser et al., 2014), which may impact the production of other steroid hormones in this interrelated system (Karmaus et al., 2016; Nielsen et al., 2012). Nielsen et al. (2012) measured 7 hormones including pregnenolone, progesterone, dehydroepiandrosterone (DHEA), androstenedione, T, estrone, and E2 to develop putative mechanisms of pathway disruption for genistein, prochloraz, and ketoconazole, and demonstrated that all 3 chemicals appeared to affect >1 enzyme in the system. Mechanistic computational models of the metabolic network represented by the steroidogenesis pathway present in H295R cells provide quantitative evidence for the interdependence of progestagen, corticosteroid, androgen, and estrogen production downstream of cholesterol importation, in part due to competitive substrate inhibition of enzymes in the pathway by the hormones produced (Breen et al., 2010; Saito et al., 2016). For example, upregulated cholesterol importation may drive more progestagen production and reduced corticosteroid and sex hormone production in H295R cells, and the relative expression or activity of CYP17A1 and 3β-hydroxysteroid dehydrogenase may determine the emphasis of steroidogenic output in H295R between corticosteroids and androgens (Saito et al., 2016). Understanding of the biological mechanisms that affect steroid hormone production in H295R cells continues to grow, eg, the exact nature of the interdependence of enzyme activities and the effect of various nuclear receptor-mediated activities on the steroid biosynthesis pathway. A K-means clustering of the pattern of steroid hormone responses in the HT-H295R assay suggests that it may be possible to hypothesize additional potential modes of action of screened chemicals by examining the putative mechanisms of the chemicals that have similar bioactivity profiles (Karmaus et al., 2016). These examples highlight not only the possible advantage of gaining mechanistic insight into the action of chemicals within the steroidogenesis pathway, but also the utility of understanding the magnitude of effect across the pathway as a whole. One of the aims of the present work was to integrate these hormone responses into a metric that could be used to distinguish chemicals of highest priority for further evaluation, including possible confirmatory screening in orthogonal assays or assays of greater biological complexity.
Previously, pathway-based models of estrogen and androgen receptor activity have been developed using multiple in vitro assays that targeted different key events involved in steroid hormone receptor activation, from binding to activation of receptor-mediated transcription (Judson et al., 2015; Kleinstreuer et al., 2017). These models were then used to rank chemicals for further evaluation or screening using the model score in the context of exposure (EPA, 2014). In contrast to these ER and AR pathway models, the data needed to computationally model the complex biological mechanisms that may affect production of steroid hormones in the HT-H295R assay specifically are still under development. Thus, to enable rapid, data-driven prioritization of chemical lists for further screening, a statistical method was developed in the present work to enable simultaneous consideration of all of the available steroid hormone endpoints from concentration-response data obtained from the HT-H295R assay.
To enable prioritization beyond only E2 and T production, a novel statistical approach was developed that utilized the effects on 11 of the hormones measured (OH-pregnenolone, progesterone, OH-progesterone, 11-deoxycortisol, deoxycorticosterone (DOC), cortisol, corticosterone, androstenedione, T, estrone, and E2) for each concentration of a chemical tested. A statistical measure based on Mahalanobis distance (De Maesschalck et al., 2000; SAS, 2012), an extension to Euclidean distance that enables consideration of the correlation of the measurement error, was calculated to characterize the magnitude of the concentration-dependent effects on the steroidogenesis pathway. Using this approach, 11 available hormone responses were considered simultaneously in the computation of a single unitless value per chemical to indicate the magnitude of effect on the steroid biosynthesis pathway in H295R cells. Analysis of this Mahalanobis distance-based metric demonstrated that increased potency generally correlated with increased maximum mean Mahalanobis distance, suggesting that this metric may be useful in understanding potency and efficacy. This analysis was attempted as part of a fit-for-purpose effort to derive a single value that might be useful in prioritization of chemicals screened in the HT-H295R assay for further evaluation of their potential endocrine bioactivity.
Additionally, to bolster confidence in the utility of HT-H295R assay as well as the Mahalanobis distance-based approach, a comparison of the HT-H295R assay data with existing OECD reference chemical information was performed. A qualitative comparison was made first between the results of the interlaboratory validation report for the OECD TG 456 and the HT-H295R data for prediction of effects on E2 and T production for 25 reference chemicals. To enable this comparison, all of the HT-H295R steroid hormone data, including E2 and T, were analyzed per a similar methodology to the one outlined in the OECD interlaboratory validation study for OECD TG 456 (Hecker et al., 2011), rather than using the ToxCast data pipeline as used previously for a subset of these data (Karmaus et al., 2016). This comparison enables an evaluation of the hypothesis that the HT-H295R assay may function as a possible alternative with the potential to decrease the resources needed to obtain screening level information about chemical effects on in vitro production of E2 and T specifically. The integrative statistical ranking metric developed herein was then added to the comparison in order to demonstrate the added quantitative value of this metric beyond simple consideration of the number of steroid hormone analytes perturbed.
In total, the research outlined in this study demonstrates the value of the HT-H295R assay as an alternative to the OECD TG 456, and further creates a summary value that can be used to prioritize chemicals for further consideration of potential steroidogenesis pathway disruption.
MATERIALS AND METHODS
Chemical Library
Previously, data were collected using the HT-H295R assay for 1998 unique test chemicals at a single high concentration, with 514 of these chemicals screened in multi-concentration response (EPA, 2015b; Karmaus et al., 2016). Including this current study, 2012 unique test chemicals have been screened at a single high concentration (100 μM, solubility- and viability-permitting), with 656 chemicals assayed in concentration-response ranging from 0.041 nM to 100 lM. One chemical, triadimenol, was assayed in concentration-response with 2 different concentration ranges, and as such is given 2 unique chemical identifiers in any analyses (for a total of 657 chemicals, but 656 unique CAS numbers). The chemicals were selected from the ToxCast phase I, II, III, and endocrine 1000 (E1K) libraries, which were compiled based on commercial availability and solubility in dimethyl sulfoxide (DMSO) among other considerations to capture a broad chemical space (Richard et al., 2016). Phase I contained a high percentage of pesticide active ingredients and chemicals for which additional in vivo data were available; phases II and III broadened the chemical landscape and included a greater diversity of chemical use types (Richard et al., 2016). The E1K chemical library, a set of roughly 800 chemicals enriched for endocrine-active chemicals, was also included. Information on the complete ToxCast chemical library is publicly available for download (http://www.epa.gov/ncct/dsstox/ or https://comptox.epa.gov/dashboard/chemical_lists). A top nominal stock concentration of 100 μM in DMSO was attempted, solubility-permitting, for the entire library.
The majority (approximately 80%) of the 656 chemicals advanced for concentration-response screening in the HT-H295R assay demonstrated changes of 1.5-fold or greater relative to control in single concentration screening for ≥3 steroid hormone analytes from the pathway at the maximum tested concentration that maintained ≥70% cell viability. Most of the chemicals advanced to concentration-response screening (approximately 60%) affected ≥4 steroid hormones in single concentration screening (Karmaus et al., 2016), with some exceptions to include additional positive and negative chemicals for reference and specific chemical classes of interest. The justification for this pre-selection screening workflow was 3-fold: (1) on a hypothetical basis, modulation of even 1 enzyme in the pathway would theoretically perturb the concentrations of at least 4 steroid hormones; (2) empirically, the recall sensitivity or percentage of positive responses that repeated between single concentration and concentration-response screening was high (86%) when a cumulative total of ≥4 hormones were affected in single concentration screening (Karmaus et al., 2016); and (3) identification of chemicals with the greatest perturbation of the interrelated steroidogenic pathway responses represented a sensible approach to reducing the resources needed to screen a chemical set in concentration-response.
HT-H295R Assay and Quantification of Steroid Hormones
The HT-H295R assay (Karmaus et al., 2016) is comprised of 4 main experimental components: (1) H295R cell culture and treatment; (2) cell viability assay using the MTT (3-[4,5-dimethylthiazol-2-y]2,5-diphenyltetrazoliumbromide) tetrazolium reduction assay; (3) quantification of steroid hormones in the media from exposed H295R cells; and (4) statistical analysis of steroid hormone concentrations. The HT-H295R assay was conducted in accordance with the OECD TG 456 (OECD 2011), with modification to increase the throughput of the assay. Key aspects of the assay design, conduct, and analysis by the OECD Test No. 456 and the HT-H295R assay are summarized and compared in Table 1. The following steroid hormone abbreviations are used throughout the manuscript: OHPREG, 17α-hydroxypregnenolone; PROG, progesterone; OHPROG, 17α-hydroxyprogesterone; DOC, deoxycorticosterone; CORTIC, corticosterone; 11DCORT, 11-deoxycortisol; CORTISOL or CORT, cortisol; ANDR, androstenedione; TESTO or T, testosterone; ESTRONE or E1, estrone; ESTRADIOL or E2, 17β-estradiol.
Table 1.
H295R Steroidogenesis Assay Methodology Comparison
| Design Phase | Aspect | OECD TG 456 | HT-H295R |
|---|---|---|---|
| Cell culture | Plate format | 24-well, but OECD TG specifies other plate formats can be used (eg, 48-well) | 96-well |
| Experimental timeline | 24 h acclimatization of cells, followed by 48 h chemical exposure, terminated at sample collection. | Overnight acclimatization of cells, followed by 48 h pre-stimulation with forskolin, followed by 48 h chemical exposure, terminated at sample collection. | |
| Cell passage | 5–10 | 5–10 | |
| Target cell confluency | 50%–60% | 50%–60% | |
| Replicates | Triplicate technical replicates | Duplicate technical replicates | |
| Triplicate biological replicates | Most of the library had 1 biological replicate; approximately 16% was screened with 2–3 biological replicates. | ||
| Viability testing | Viability measures | Live/Dead® or MTT assay | MTT assay |
| Cell viability threshold | ≥80% | ≥70% | |
| Hormone detection | Baseline stimulation | Nonea | Cells are pre-stimulated for 48 h in medium containing 10 μM forskolin. |
| Minimum basal production | 500 pg/ml or ≥ 5-fold method detection limit (MDL) for T and 40 pg/ml or ≥ 2.5-fold MDL for E2. | Following forskolin stimulation, DMSO-exposed H295R demonstrated 2.19 ± 0.32 ng/ml and 1.57 ± 0.36 ng/ml for T and E2, respectively. This is ≥ 5-fold the LLOQ for both hormones.b | |
| Accuracy | Within 30% of nominal concentrations. | 98.1%–101.7% recovery for 13 hormones (Karmaus et al. 2016).b | |
| Precision | Variation between replicate samples should be ≤ 25%. | Percent relative standard deviation for controls ranging from 3.3 to 10.0% during assay optimization for the 13 hormones measured (Karmaus et al. 2016).b | |
| Steroid hormone data analysis | Analysis | Normally distributed data: an analysis of variance (ANOVA) with differences from vehicle control evaluated using a Dunnett’s test. Non-normally distributed data: Kruskal–Wallis test followed by a Mann–Whitney U test. | Initial data analysis used the ToxCast data pipeline (tcpl) (Filer et al., 2017) to enable standardization of the data with other HT data and a first look at the data. Data analysis for comparison to the OECD reference chemicals involved use of an ANOVA with differences from vehicle control evaluated by Dunnett’s test (new in this work). |
| Criteria for positive | Two consecutive concentrations and/or maximum non-cytotoxic concentration significantly different from control. | See Karmaus et al. (2016) for a description of the tcpl analysis employed for a first analysis. For the ANOVA approach presented here: 2 consecutive concentrations and/or maximum non-cytotoxic concentration significantly different from control. |
A summary of the OECD TG 456 requirements versus the performance of the HT-H295R assay (as currently implemented).
22-R-Hydroxycholesterol has been suggested as a medium supplement (20–40 μM) to increase basal E2 production as needed, but it is not part of the standard protocol. Further, the OECD validation report (2008) noted that, “during the qualifying experiments it was only expected that the laboratory showed conformance with the performance criteria for E2 induction after exposure to the stimulator forskolin.”
Note these are reported performance results and not criteria for acceptance of the HT-H295R assay data.
Cell Culture and Treatment
The cell culture, treatment, and assay conditions of the HT-H295R assay have been described previously in detail (Karmaus et al., 2016). All cell culture and treatments were conducted by Cyprotex US, LLC (formerly CeeTox, Inc.) (Kalamazoo, MI). Briefly, H295R cells (ATCC CRL-2128) were expanded for 5 passages and frozen in batches in liquid nitrogen. Prior to experimentation, batches of H295R cells were thawed and passed at least 4 times, taking care that the maximum passage number used for experimentation was 10. Cells were maintained in a 1:1 mixture of Dulbecco’s Modified Eagle’s Medium with Ham’s F-12 Nutrient mixture (DMEM/F12) supplemented with 5 ml/l ITS+ Premix (BD Bioscience) and 12.5 ml/l Nu-Serum (BD Bioscience). Cells seeded at 50%–60% confluency into 96-well plates were acclimated overnight. Culture medium was then replaced with 175 μl of medium containing 10 μM forskolin to stimulate steroidogenesis for 48 h. The forskolin stimulus medium was replaced with medium supplemented with test chemical or controls (forskolin, prochloraz, or digitonin) added to a final concentration of 0.1% DMSO. On each 96-well plate, duplicate treatment wells were included for all chemical treatments as well as controls (10 μM forskolin and 3 μM prochloraz), in addition to 2 or 4 DMSO solvent control wells and 4 or 6 cell viability control wells (250 μM digitonin). The test chemicals were assayed on 8 different dates, and each experimental date is used to indicate block throughout the study in order to account for observed block effects. Most test chemicals were assayed in 1 plate-block combination with technical duplicates only; approximately 16% of the screened library (107 of 656 unique chemicals screened in concentration-response) were assayed on >1 plate-block combination. Following 48 h of test chemical exposure, medium was removed, split into 2 vials of approximately 75 μl media each, and stored at −80 °C prior to steroid hormone quantification.
Cell Viability Assay
Cell viability was evaluated by MTT cytotoxicity assay after chemical treatment in all studies, and was previously described in Karmaus et al. (2016). Briefly, after chemical exposure and removal of media, 100 μl of 0.5 mg/ml 3-[4,5-dimethylthiazol-2-y]2,5-diphenyltetrazoliumbromide (MTT) solution was added to the cells remaining in the 96-well treatment plates. Following a 3 h incubation at 37 °C and 5% CO2 to allow formazan-MTT crystal formation, the MTT solution was removed and blue formazan salt crystals were solubilized using 100 μL anhydrous isopropanol with shaking for 20 min. Absorbance at 570 and 650 nm was measured using a BioTek Synergy H4 plate reader. Background correction of absorbance units was used to determine percent change relative to controls. All plates contained multiple control wells including 10 μM forskolin (n = 4; control for stimulation of steroidogenesis), 3 μM prochloraz (n = 4; control for the inhibition of steroidogenesis) and digitonin (n = 4–6; control for cell death).
For the first 1998 chemicals screened, cytotoxicity was used to establish a maximum tolerated concentration (MTC) per chemical sample with a target cell viability ≥70%, as reported previously (Karmaus et al., 2016). ToxCast chemicals were evaluated at a maximum nominal concentration of 100 μM, where possible. MTT cytotoxicity evaluation was also conducted for the duplicates of all concentrations for chemicals tested in the concentration-response studies (CR; 6-point CR established by 3-fold serial dilutions from the MTC).
For the 85 additional chemicals with multi-concentration data reported herein for the first time, the MTT assay was also run for all concentrations attempted in the HT-H295R assay, but the MTC was not used to limit the concentration-response curve. If a stock concentration of 100 mM was achieved, then each chemical was tested at 100, 33.33, 11.11, 3.70, 1.23, and 0.41 μM in the MTT assay for these 85 chemicals. Otherwise, the same dilution series was performed using the highest possible stock concentration of test chemical. The purpose of this change in the experimental workflow was to enable full concentration-response curves for the steroid hormone analysis to be visualized without implementing the MTC logic, which may have limited the ability to observe effects on steroid biosynthesis in cases when the difference between a cytotoxic concentration and a viable, efficacious concentration may have been small.
Steroid Hormone Quantification
Frozen medium samples from treated HT-H295R assays were shipped on dry ice to OpAns, LLC (Durham, NC) for extraction and quantification of steroid hormones. As described previously (Karmaus et al., 2016), samples were thawed to room temperature prior to liquid-liquid extraction. Steroid hormones were extracted from media samples using methyl tert-butyl ether (MTBE). An extra derivatization with dansyl chloride was included for estrogen (estrone and E2) detection only. Steroid hormones were separated and quantified using HPLC-MS/MS. Specifically, reverse phase C18 gradient elution with electrospray positive ionization was used followed by MS/MS detection. All data were acquired using MassHunter Workstation Acquisition version B03.01 (Agilent Technologies, Inc.), and processed using MassHunter Quantitative Analysis for QQQ.
The lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ) were reported previously (Karmaus et al., 2016) using a 7-point standard curve. The precision and accuracy of the extraction and quantification methods are briefly reviewed in Table 1; the recovery for all 13 hormones ranged from 98.1% to 101.7% recovery and the percent relative standard deviation (%RSD) of the spiked standards and percent spiked standard recovered ranged from 3.3 to 10.0%, as reported by Karmaus and colleagues (2016). During the sample analysis process, samples were flagged as “not-detected” or “not-quantifiable” when the sample was available, but the steroid hormone analyte was below the LLOQ; in such cases, a surrogate value of the LLOQ divided by the square root of 2 was substituted for analyses herein (CDC, 2009; Hornung and Reed, 1990). Any sample measurement flagged as “not reportable” was set to “NA” for any subsequent analysis. A comparison of the method detection limits (ng/ml) for OECD TG 456 and the HT-H295R assay (Karmaus et al., 2016) is provided in Table 2.
Table 2.
Comparison of Method Detection Limit (from OECD TG 456) and Reported LLOQs for HT-H295R
| Hormone Family | Steroid Hormone | OECD TG 456 |
HT-H295R Assay |
|
|---|---|---|---|---|
| Method detection limit (ng/mL) | Lower limit of quantitation (LLOQ) (ng/mL) | Upper limit of quantitation (ULOQ) (ng/mL) | ||
| Androgen | Testosterone (T) | 0.1 | 0.1 | 20 |
| Dehydroepiandrosteronea | NA | 3 | 600 | |
| Androstenedione | NA | 1 | 200 | |
| Estrogen | Estradiol (E2) | 0.01 | 0.03 | 6 |
| Estrone | NA | 0.03 | 6 | |
| Progestagen | 17α-hydroxyprogesterone | NA | 0.2 | 40 |
| 17α-hydroxypregnenolone | 5 | 1000 | ||
| Progesterone | 0.2 | 40 | ||
| Pregnenolonea | 2 | 400 | ||
| Corticosteroid | 11-Deoxycortisol | 5 | 1000 | |
| Deoxycorticosterone | 0.5 | 100 | ||
| Cortisol | 0.5 | 100 | ||
| Corticosterone | 0.5 | 100 | ||
The gray cells highlight the comparison of testosterone (T) and estradiol (E2), as these are the only 2 hormones with minimum method detection limits (MDL) in the performance criteria for the test guidelines. The LLOQs for all of the hormones measured in Karmaus et al. (2016) are listed.
Dehydroepiandrosterone and pregnenolone were excluded from further analysis in the work herein as 69.5% and 53.1% of the measured values for these 2 steroid hormones, respectively, were below the LLOQ.
Data Analysis
All data were analyzed in R (R 3.3.2; R Foundation for Statistical Computing). The R scripts are available at: ftp://newftp.epa.gov/COMPTOX/NCCT_Publication_Data/Haggard/2017_Prediction_of_H295R_steroidogenesis_Pathway_Perturbation/
Cell Viability Assay Data Processing
Initially, and as described previously (Karmaus et al., 2016), the MTT assay was used to establish a MTC per chemical sample for the first 571 chemicals that were assayed in concentration-response by evaluation of each chemical sample at a target top concentration of 100 μM, solubility-permitting, and then seeking to find a concentration that would maintain cell viability of ≥70%. Chemicals that yielded H295R cell viability of 20%–70% were diluted 10-fold, while those with <20% viability were diluted 100-fold and re-evaluated. Dilutions were made until ≥70% viability was achieved for chemicals to establish the MTC. The MTT method differed from Karmaus et al. (2016) for the additional 85 chemicals (Supplementary File 1 unique plate IDs for 04112017) reported for the first time in this manuscript in that no MTC was determined. MTT assay data were collected for all 6 concentrations tested, with a target top concentration of 100 μM and decreasing half-log increments (33.33, 11.11, 3.70, 1.23, and 0.41), with adjustments made based on chemical solubility.
The concentration-response MTT data for all 656 chemicals screened were processed using the ToxCast data pipeline (tcpl) (Filer et al., 2017) for comparison with the HT-H295R steroid hormone data. The data were analyzed as percent control, where the baseline value was defined as the plate-wise baseline of the DMSO control wells, as shown in Equation 1.
| (1) |
Consistent with previous estimations of the variability around the baseline response for this assay, a 70% cutoff criterion (Karmaus et al., 2016) was established for the purpose of filtering steroid hormone data. This cutoff criterion (allowing up to 30% cell viability loss) corresponded to approximately 4.4-times the baseline median absolute deviation (6.81). Cell viability filtering was performed by matching the MTT percent control response to the steroid hormone data; if the cell viability decreased by >30%, the steroid hormone data for that concentration of a chemical was excluded from any further analysis.
The normalized data by concentration and the resultant plots of these data for 655 of 656 chemical samples are available in Supplementary Files 2 and 1, respectively. Two chemicals, colchicine (CASRN 64-86-8) and digoxigenin (CASRN 1672-46-4), were included as viability controls, and as expected resulted in substantial loss of cell viability, leaving only 1 concentration with viable cells. As such, steroid hormone data were not analyzed for these 2 chemicals. A third chemical, quizalofop-ethyl (CASRN 76578-14-8) had data quality flags in the source files from the assay vendor that suggested these data should not be used; these data were excluded from any further analysis of steroid hormones or cytotoxicity. This reduced the set of chemicals with concentration-response hormone data available from 656 to 654 unique chemicals, corresponding to 766 chemical samples. Of these 766 chemicals samples, when a 70% cell viability filter was applied, 715, 36, 6, 5, and 4 chemicals retained 6, 5, 4, 3, and 2 concentrations for analysis of the concentration-responses for steroid hormones (see Supplementary File 3 for the master steroid hormone data table).
Analysis of Variance for Significance of Effects on Steroid Hormone Profiles
When concentration-response data were available, the vendor-provided source files with raw steroid hormone data (quantified as ng/ml) were converted to micromolar (μM) units and each steroid hormone assay component was analyzed, per the analysis methodology in the OECD TG 456, by an analysis of variance (ANOVA) followed by a post hoc Dunnett’s test with alpha set to 0.05 (a complete table of these values is available as Supplementary File 4). The DMSO control data originating from the same plate the chemical was tested on were used as the sample for comparison. In most cases, a minimum of 2 technical replicate samples within 1 plate were available for each chemical-concentration-hormone test. In some cases, a chemical may have appeared in multiple blocks of the study (ie, assayed on >1 date); in this case, the data for each block were analyzed separately due to the presence of block effects.
Per the OECD TG 456 (OECD, 2011) and the interlaboratory validation report (Hecker et al., 2011), for a positive result, 2 consecutive concentrations (not necessarily including the top concentration) had to produce results significantly different from control for a steroid hormone analyte (only 8% of positive responses in the HT-H295R assay did not include a significant maximum concentration). A positive result was also counted if the significant effect occurred only at the maximum concentration tested that still maintained ≥70% cell viability. A minimum efficacy threshold of a 1.5-fold change from DMSO control was applied for context as suggested by the OECD interlaboratory analysis, as some results were deemed statistically significant by ANOVA but were still <1.5-fold different from DMSO control.
Computation of the Mean Mahalanobis Distance to Derive a Maximum Mean Mahalanobis Distance by Chemical
A statistical approach based on the Mahalanobis distance (Mahalanobis, 1936) was employed to characterize the magnitude of change for 11 steroid hormones produced by H295R cells; this analysis was intended to yield a value that could be used to prioritize chemicals for evaluation of potential endocrine activity using the HT-H295R assay data. The purpose in employing this analysis methodology was to account for the correlation in the residuals of these steroid hormone measures when considering a chemical response for multiple hormones simultaneously. As the 11 steroid hormones were measured from the same experimental well, and the synthesis of these steroid hormones is interdependent, a statistical method that could account for the interrelatedness of these measurements, in the absence of complete enzyme kinetic information in the HT-H295R model, was developed and described here. A mean Mahalanobis distance () was calculated to summarize the 11 steroid hormone responses measured following exposure to each chemical concentration screened in the assay. Then, the maximum mean Mahalanobis distance (maxmMd) was selected from the set of values generated for a chemical. The maxmMd then serves as a single numeric value to characterize the magnitude of effect on synthesis of 11 steroid hormones for a given chemical screened in the HT-H295R assay. Below, the computation of the and maxmMd are described, followed by a detailed description of the computation of the covariance matrix used to compute values.
Calculation of the Mahalanobis distance metrics.
A Mahalanobis distance is a generalization of Euclidian distance that adjusts for the variance and covariance among the hormone measures at each concentration screened (De Maesschalck et al., 2000; SAS, 2012). Although 13 hormones were measured in the HT-H295R assay, measurements of 2 of these hormones frequently indicated a value below the LLOQ; pregnenolone and DHEA were often missing (53.1% and 69.5% of all measurements) and have been excluded from this approach, leaving 11 hormone measures for analysis. Thus, a Mahalanobis distance-based approach was used to indicate the effect of each test chemical concentration in 11-dimensional space.
To calculate the Mahalanobis distance, the response at each concentration of a test chemical was considered as a point in an 11-dimensional space; each axis corresponds to the natural logarithm of the measured concentration of 1 of the hormones included in this analysis, respectively. In brief, the degree to which variation among replicates is correlated across hormones was estimated, and a covariance matrix that characterizes both the noise variance and correlation among hormone levels across replicates, after taking chemical and concentration into account, was constructed. Conceptually, this is equivalent to rotating and scaling the hormone concentrations to a set of new variables that are uncorrelated with each other and have the same standard deviation, followed by computation of the Euclidean distance in this new space (Supplementary File 5).
Due to the need to compare distances based on different numbers of hormone analyte data for a given test chemical (eg, due to missing data), a mean Mahalanobis distance () statistic was computed, ie, the Mahalanobis distance divided by the square root of the number of hormones used to compute it. The for a given test compound between the hormone concentration at the th concentration relative to that at the DMSO vehicle control concentration was computed as shown in Equation 2.
| (2) |
For this analysis, is the vector of natural log-transformed steroid hormone concentrations at the concentration, is the vector of natural log-transformed steroid hormone concentrations for the DMSO control, is the number of hormones with measurements for this chemical, and is the estimate of the covariance matrix.
The maximum (maxmMd) is the maximum of the set of values computed for all concentrations of a test chemical.
Multivariate linear modeling and computation of the covariance matrix for the .
The steroid hormone responses measured in the HT-H295R assay represent a multivariate response, and as such, a variance-covariance matrix was computed to account for the variation and covariation of the multiple steroid hormone measurements. An estimate of the covariance matrix that characterizes both the noise variance and correlation among measured steroid hormone concentrations across replicates, after taking chemical and concentration into account, was needed to compute the as indicated above. Due to the presence of block effects between chemicals assayed on different days, separate covariance matrices were computed for each assay date, resulting in a total of 8 individual covariance matrices. The covariance matrix used in the computation was constructed per the following procedure:
A multivariate linear model of the unique set of chemicals within each block was fit using the natural log-transformed hormone concentrations from the HT-H295R assay. The model includes terms for plate-specific values for all DMSO controls, and a separate mean for each test chemical concentration across all the measured steroid hormone analytes. All these entities were replicated on the same plate. Outlier detection was performed by fitting all data to the multivariate linear model and identifying where the standard deviation of the residuals for a chemical-concentration technical replicate pair was >1 for any steroid hormone analyte measured (indicating an approximately 2.7 fold-change difference in steroid hormone concentration between technical replicates). This resulted in the removal of 18 of 4655 unique chemical-concentration replicate pairs. The matrix of residuals from the fits of the filtered data across all the plates within each block were used to estimate a variance and covariance matrix.
To retain estimates for the largest possible number of chemicals and to keep the estimation process simple, if data for a particular hormone were missing for a chemical within a block, the hormone measure was dropped from that block prior to linear model fitting. This affected only 1 of the 8 blocks, which contained some missing data for estrone and E2, representing 81 unique test chemicals. In this case, the computed covariance matrix for this block included only 9 of the 11 steroid hormone analytes.
The full pooled 11 × 11 covariance matrix (omitting DHEA and pregnenolone) used for the calculation was estimated as the unweighted average of the 8 block-specific covariance matrices.
The resulting pooled covariance matrix was positive-definite (a requirement for a proper covariance matrix).
Critical value for positive steroidogenesis pathway results using the .
A critical value to assess significance was derived to distinguish values that are greater than what would likely result from sampling noise. The critical value accounts for the multiple comparisons arising from comparing each concentration group to the control. The critical value reflects the similarity between and the Hotelling statistic used to compare 2 groups with multiple measurements (Mardia et al., 1979). Hotelling’s is analogous to the usual t- or F-statistics used for comparisons of single characteristics in that evaluates the difference between 2 groups (ie, the response of 1 concentration compared to that of its plate DMSO control) relative to the variability among measurements within the groups. Instead of simply computing the variance within the groups, as would be performed for a univariate response, a variance-covariance matrix was computed and accounts for the variation and covariation of the multiple steroid hormone measurements in the HT-H295R assay (described in the next section). For this analysis, all the test chemical concentrations and control groups were used to determine this within-group variability. This yields an estimate of the within-group variance-covariance matrix which is more precise than the one that would be used for . With the variance-covariance matrix known, we employed the method developed by Nakamura and Imada (2005) to adjust for multiple comparisons for multivariate tests. This is analogous to adjusting for multiple comparisons for univariate tests such as Dunnett’s procedure. Nakamura and Imada’s method requires equal sample sizes across comparison groups, so a critical value for the set of values for a test chemical was derived by assigning the sample size for a concentration group as the largest of the sample sizes across hormones evaluated in that group, and the sample size for all the comparisons for a given test chemical as the median sample size across concentration groups. The critical value was derived for a nominal Type I error of 0.01. Because of the sample size decision just described, and the fact that the covariance matrix is estimated, even though the sample used was large, this approach should only approximate the actual Type I error. The resulting critical value for the varied across the set of chemicals as the critical value is related to the number of hormones with data for each chemical. The critical values ranged from 1.15 to 1.81, with a median of 1.64 and a mean of 1.58, for all of the chemicals with available data for computation.
Any observed value for a chemical exceeding the critical value was considered a positive for potential steroidogenesis pathway disruption. The maxmMd was adjusted for the critical value (maxmMd — critical value = adjusted maxmMd) to more clearly flag maxmMd values of interest; this difference should be >0 for a positive pathway result.
prediction for the maxmMd.
Calculated values for each test chemical were fit to a 4-parameter logistic function as shown in Equation 3.
| (3) |
In Equation 3, a is the background level which was set equal to 1, is the ratio of the maximum and minimum asymptotes, is the Hill’s slope of the curve, is the concentration of test chemical , and is a function of the inflection point, , as described in Equation 4 below.
| (4) |
The parameters , , and were optimized using the Nelder-Mead method. The benchmark dose () was then calculated as the concentration of test chemical where the fitted value equals the calculated critical value, as shown below in Equation 5.
| (5) |
In Equation 5, , , and are the parameters as Equation 3, and is the critical value for test chemical .
Determination of the 95% confidence interval of the maxmMd.
Natural log-transformed maxmMd values for the 107 replicated test chemicals were fit to a linear model to determine the standard deviation for residuals around the chemical-specific means across replicates. This residual standard deviation, 0.33, was then used to approximate a 95% prediction interval for the chemical-specific maxmMd, ie, the exponential raised to 2times 0.33, yielding a value of 1.93. The highest critical value for this dataset was 1.81; multiplying this critical value by 2 times the residual deviation (1.93) yields a maxmMd value of 3.5.
Comparison Methodology for HT-H295R to OECD Reference Chemicals
Chemicals With Comparable Data for Comparison
Ten of the 12 core reference chemicals included in the OECD H295R interlaboratory validation study (Hecker et al., 2011) have been screened using the HT-H295R assay, including: aminoglutehimide, atrazine, benomyl, butylparaben, ethylene dimethanesulfonate, forskolin, letrozole, molinate, nonoxynol-9 (Polyoxyethylene(10) nonylphenyl ether), and prochloraz (Table 3). Trilostane and a protein hormone, human chorionic gonadotropin, have not been screened in the HT-H295R assay. In addition to the 12 core chemicals for reference, 16 chemicals were included as “supplemental” verification for the interlaboratory validation, with testing in only 2 laboratories in the OECD interlaboratory validation instead of 5 laboratories (Hecker et al., 2011). These data have a greater amount of uncertainty than the core reference chemicals due to disagreements reported between the 2 testing laboratories. Fifteen of these 16 chemicals have HT-H295R data for comparison (Table 3).
Table 3.
DSSTox Reference Information for the Chemicals Used for Comparison of OECD and HT-H295R Approaches
| DTXSID | Preferred Name | HT-H295R Data | CASRN | INCHI KEY | Average MW |
|---|---|---|---|---|---|
| Core reference chemicals | |||||
| DTXSID8022589 | Aminoglutethimide | Y | 125-84-8 | ROBVIMPUHSLWNV-UHFFFAOYSA-N | 232.28 |
| DTXSID9020112 | Atrazine | Y | 1912-24-9 | MXWJVTOOROXGIU-UHFFFAOYSA-N | 215.69 |
| DTXSID5023900 | Benomyl | Y | 17804-35-2 | RIOXQFHNBCKOKP-UHFFFAOYSA-N | 290.32 |
| DTXSID3020209 | Butylparaben | Y | 94-26-8 | QFOHBWFCKVYLES-UHFFFAOYSA-N | 194.23 |
| DTXSID40196931 | Ethylene dimethanesulfonate | Y | 4672-49-5 | QSQFARNGNIZGAW-UHFFFAOYSA-N | 218.24 |
| DTXSID8040484 | Forskolin | Y | 66575-29-9 | OHCQJHSOBUTRHG-KGGHGJDLSA-N | 410.51 |
| DTXSID4023202 | Letrozole | Y | 112809-51-5 | HPJKCIUCZWXJDR-UHFFFAOYSA-N | 285.31 |
| DTXSID6024206 | Molinate | Y | 2212-67-1 | DEDOPGXGGQYYMW-UHFFFAOYSA-N | 187.30 |
| DTXSID2036588 | Nonoxynol | Y | 26027-38-3 | NA | NA |
| DTXSID4024270 | Prochloraz | Y | 67747-09-5 | TVLSRXXIMLFWEO-UHFFFAOYSA-N | 376.66 |
| DTXSID9023706 | Trilostane | N | 13647-35-3 | KVJXBPDAXMEYOA-CXANFOAXSA-N | 329.44 |
| DTXSID4036770 | Human chorionic gonadotropin | N | NA | NA | NA |
| Supplemental reference chemicals | |||||
| DTXSID0020523 | 2,4-Dinitrophenol | Y | 51-28-5 | UFBJCMHMOXMLKC-UHFFFAOYSA-N | 184.11 |
| DTXSID7020182 | Bisphenol A | Y | 80-05-7 | IISBACLAFKSPIT-UHFFFAOYSA-N | 228.29 |
| DTXSID2022880 | Danazol | Y | 17230-88-5 | POZRVZJJTULAOH-LHZXLZLDSA-N | 337.46 |
| DTXSID5020607 | Di(2-ethylhexyl) phthalate | Y | 117-81-7 | BJQHLKABXJIVAM-UHFFFAOYSA-N | 390.56 |
| DTXSID7020479 | Dimethoate | Y | 60-51-5 | MCWXGJITAZMZEV-UHFFFAOYSA-N | 229.25 |
| DTXSID2032390 | Fenarimol | Y | 60168-88-9 | NHOWDZOIZKMVAI-UHFFFAOYSA-N | 331.20 |
| DTXSID3020625 | Finasteride | Y | 98319-26-7 | DBEPLOCGEIEOCV-WSBQPABSSA-N | 372.55 |
| DTXSID7032004 | Flutamide | Y | 13311-84-7 | MKXKFYHWDHIYRV-UHFFFAOYSA-N | 276.21 |
| DTXSID5022308 | Genistein | Y | 446-72-0 | TZBJGXHYKVUXJN-UHFFFAOYSA-N | 270.24 |
| DTXSID1024122 | Glyphosate | N | 1071-83-6 | XDDAORKBJWWYJS-UHFFFAOYSA-N | 169.07 |
| DTXSID7029879 | Ketoconazole | Y | 65277-42-1 | XMAYWYJOQHXEEK-OZXSUGGESA-N | 531.43 |
| DTXSID5023322 | Mifepristone | Y | 84371-65-3 | VKHAHZOOUSRJNA-GCNJZUOMSA-N | 429.60 |
| DTXSID1021166 | Piperonyl butoxide | Y | 51-03-6 | FIPWRIJSWJWJAI-UHFFFAOYSA-N | 338.44 |
| DTXSID6022341 | Prometon | Y | 1610-18-0 | ISEUFVQQFVOBCY-UHFFFAOYSA-N | 225.30 |
| DTXSID6034186 | Spironolactone | Y | 52-01-7 | LXMSZDCAJNLERA-ZHYRCANASA-N | 416.58 |
| DTXSID4021391 | Tricresyl phosphate | Y | 1330-78-5 | NA | NA |
One of the 10 core reference chemicals with data for comparison, “nonoxynol-9,” presented some uncertainties with respect to the nature and concentration of the substance tested in the OECD interlaboratory validation. The CAS number provided for nonoxynol-9 in the OECD interlaboratory report is 26027-38-3. According to the “definitive” CAS registry listing in SciFinder, this CAS number corresponds to an oligomer, sometimes also named Polyoxyethylene(10)nonylphenyl ether, which is a mixture of repeating ethoxy (oxy-1,2-ethanediyl) groups, (C2H4O) × C15H24O, of undefined composition. Therefore, without additional substance details, this CAS number cannot be definitively mapped to a specific structure. The molecular weight (MW) selected for use by EPA’s contractor in the management of ToxCast chemical samples, solely for the purposes of computing a concentration, was 264.4 g/mol. This MW corresponds to a SMILES (CCCCCCCCCC1 = CC = C(C = C1)OCCO) and structure described by CAS number 104-35-8 for a specific chemical, 2-(4-Nonylphenoxy) ethanol, that has been used as an approximate “representative” structure for nonoxynol-9. Further chemical information on the nonoxynol-9 used in the OECD interlaboratory validation was not in the report (Hecker et al., 2008) or peer-reviewed publication (Hecker et al., 2011). Therefore, it is unclear if the substance, and the nominal concentration tested, are comparable between the OECD interlaboratory validation study and the HT-H295R screening. This uncertainty is further supported by discrepancies between the OECD interlaboratory validation report and the HT-H295R screening for cytotoxicity. Though there was variability among labs, in the OECD interlaboratory validation, cell viability appeared to range from 80% to 100% at 1 μM, and from 25% to 100% at 10 μM (interpolated from graphs; Hecker, 2008). Due to cytotoxicity, the MTC for nonoxynol-9 in the HT-H295R assay was 0.4 μM. It is unknown if these differences in cytotoxicity are due to variability in testing between the assay systems, or due to differences in the composition and/or computed concentration of the substance.
Interpretation of the OECD Interlaboratory Validation Results
E2 and T were measured as biomarkers of estrogen and androgen biosynthesis, respectively. These data were analyzed per OECD TG 456 (Hecker et al., 2011; OECD, 2011). This analysis of the HT-H295R data was completely independent of the tcpl-based analysis of these data. For normally distributed data, an analysis of variance (ANOVA) was performed and differences from vehicle control were evaluated using a Dunnett’s test. For data that were not normally distributed, as evaluated by standard probability plots or Shapiro-Wilk’s test, a Kruskal-Wallis test followed by a Mann-Whitney U test was employed (see Hecker et al., 2011 for details). These data are summarized in Hecker et al. (2011) as part of the OECD interlaboratory validation study, and were extracted for this comparison. A lowest effect concentration (LOEC) was reported for each laboratory. However, there was an error in the published work, and these LOECs from Tables 3 and 4 of Hecker et al. (2011) were really in μM units (not mg/ml as reported, as detailed in a recent erratum (Hecker et al., 2017). If no LOEC was reported, the LOEC was assigned a value of “not detected” (ND). E2 and were annotated as being increased (up) or decreased (dn). For the core chemicals, in the event that the results of ≥2 of the 5 laboratories qualitatively disagreed, an effect on E2 or was considered equivocal. For the 16 supplemental chemicals, a response was considered equivocal if the anticipated response failed to match qualitatively between the 2 laboratories.
Table 4.
Positive ANOVA (plus post hoc Dunnett’s test) Results by Steroid Hormone Analyte
| # | Steroid Hormone Analyte | Abbreviation | # Positive Chemical Samples | % of Tested Library |
|---|---|---|---|---|
| 1 | OH-Pregnenolone | OHPREG | 387 | 59.2 |
| 2 | Progesterone | PROG | 509 | 77.8 |
| 3 | OH-Progesterone | OHPROG | 562 | 85.9 |
| 4 | DOC | DOC | 511 | 78.1 |
| 5 | Corticosterone | CORTIC | 386 | 59.0 |
| 6 | 11-deoxycortisol | 11DCORT | 504 | 77.1 |
| 7 | Cortisol | CORTISOL or CORT | 376 | 57.5 |
| 8 | Androstenedione | ANDR | 438 | 67.0 |
| 9 | Testosterone | TESTO or T | 397 | 60.7 |
| 10 | Estrone | ESTRONE or E1 | 425 | 65.0 |
| 11 | Estradiol | ESTRADIOL or E2 | 408 | 62.4 |
Of 654 chemical samples, positive hit rates for the 11 hormones used in this analysis ranged from 57.5% to 85.9%.
Interpretation of the HT-H295R Results
E2 (assay component CEETOX_H295R_ESTRADIOL) and T (assay component CEETOX_H295R_TESTO) were used as biomarkers of estrogen and androgen biosynthesis, respectively. The data used for this comparison were analyzed by ANOVA as described above. Per the procedure in Hecker et al. (2011), chemicals were indicated as positives, but were flagged accordingly, if they fell into any of the following categories: (1) effects were seen at only the maximum concentration; (2) effects were observed for a minimum of 2 consecutive concentrations, but with the highest concentration corresponding to a loss in cell viability; (3) effects were seen at 2 consecutive concentrations, but no effect was seen at the highest concentration tested; or (4) positive effects were seen, but they were within 1.5-fold of control.
Calculation of Confusion Matrices
Confusion matrices were constructed for E2 and for increased and decreased production, using the OECD interlaboratory validation results (Hecker et al., 2011; Tables 3 and 4) as the source of “true” positives and negatives. The HT-H295R assay data, analyzed by an ANOVA and post hoc Dunnett’s procedure, along with the OECD logic used for positive responses (Hecker et al., 2008; Hecker et al., 2011), were used for comparison. Equivocal data from the OECD interlaboratory validation results for the specific effect type were excluded from the calculation of sensitivity, specificity, and accuracy; increased and decreased T and increased and decreased E2 sets excluded 4, 1, 4, and 2 equivocal results, respectively, yielding 21, 24, 21, and 23 chemicals total in the analysis of these effect types. A set of revised confusion matrices and associated sensitivity, specificity, and accuracy values were also generated following removal of nonoxynol-9 (due to uncertainties in the substance evaluated for the OECD interlaboratory validation) from all effect types and letrozole from decreased T (due to effects on T in the OECD interlaboratory validation occurring at concentrations that greatly exceeded the MTC in the HT-H295R assay), leaving 20, 22, 20, and 22 chemicals for increased and decreased T and E2, respectively. The sensitivity or true positive rate was calculated per Equation 6, below.
| (6) |
The specificity or true negative rate was calculated per Equation 7, below.
| (7) |
And finally, the accuracy was calculated per Equation 8, below.
| (8) |
RESULTS
The results of this study include the analyzed hormone concentration-response outputs using significance testing by ANOVA and post hoc Dunnett’s results for chemicals assayed in HT-H295R assay, a comparison of the results for chemicals included in the OECD interlaboratory validation and HT-H295R assay, and the pathway-based results from computation of the maximum mean Mahalanobis distance (maxmMd) for each concentration of each chemical. Additonal data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.385j7
Analysis of HT-H295R Data by ANOVA and Post Hoc Dunnett’s Test
An ANOVA and post hoc Dunnett’s test was computed for raw hormone quantification data (converted to μM units) collected for 766 chemicals samples, composed of 654 unique chemicals with concentration-response data. The complete results of this analysis are provided in Supplementary File 4 as a table of the p-values from the ANOVA procedure. Supplementary File 6 contains a summary of the significant effects of a chemical sample for each hormone, denoted as a 0 for no effect or a 1 for a significant effect. Supplementary File 7 contains binary strings that represent the significant effects (p ≤ .05) by concentration for each chemical-steroid hormone analyte pair. These binary strings were used to determine when significant effects were observed for a given hormone, ie, when 2 consecutive concentrations demonstrated significant effects, or if a significant effect was demonstrated at the top concentration only, a chemical was labeled as a “positive” response for a particular steroid hormone analyte. The complete graphical results are presented in Supplementary File 8, with concentrations that demonstrated a significant effect of treatment colored red, and dotted horizontal lines demarcating the 1.5-fold control boundaries.
The number of positive chemicals, and the positive percentage of the library tested in concentration response, are summarized in Table 4. The relatively high rate of hits for the chemical library (with positives on all steroid hormones exceeding 50% of the screened chemical library) screened in concentration-response was expected, as chemicals screened in concentration-response were selected predominantly from positive responses in single concentration screening (with positive responses for ≥3 steroid hormones for approximately 80% of the chemicals screened in concentration-response). All of the p values by steroid hormone analyte for each comparison of concentrations for a chemical, and binary assessment of the positive/negative behavior of each chemical for each steroid hormone analyte, are presented in Supplementary Files 4 and 6. An example of the ANOVA results for the prototypical pathway inhibitor, prochloraz, are presented in Figure 2. The high positive rate (Table 4) was further explored via determination of the correlation of residuals between steroid hormone analytes, discussed in subsequent explanation of the Mahalanobis distance results.
Figure 2.
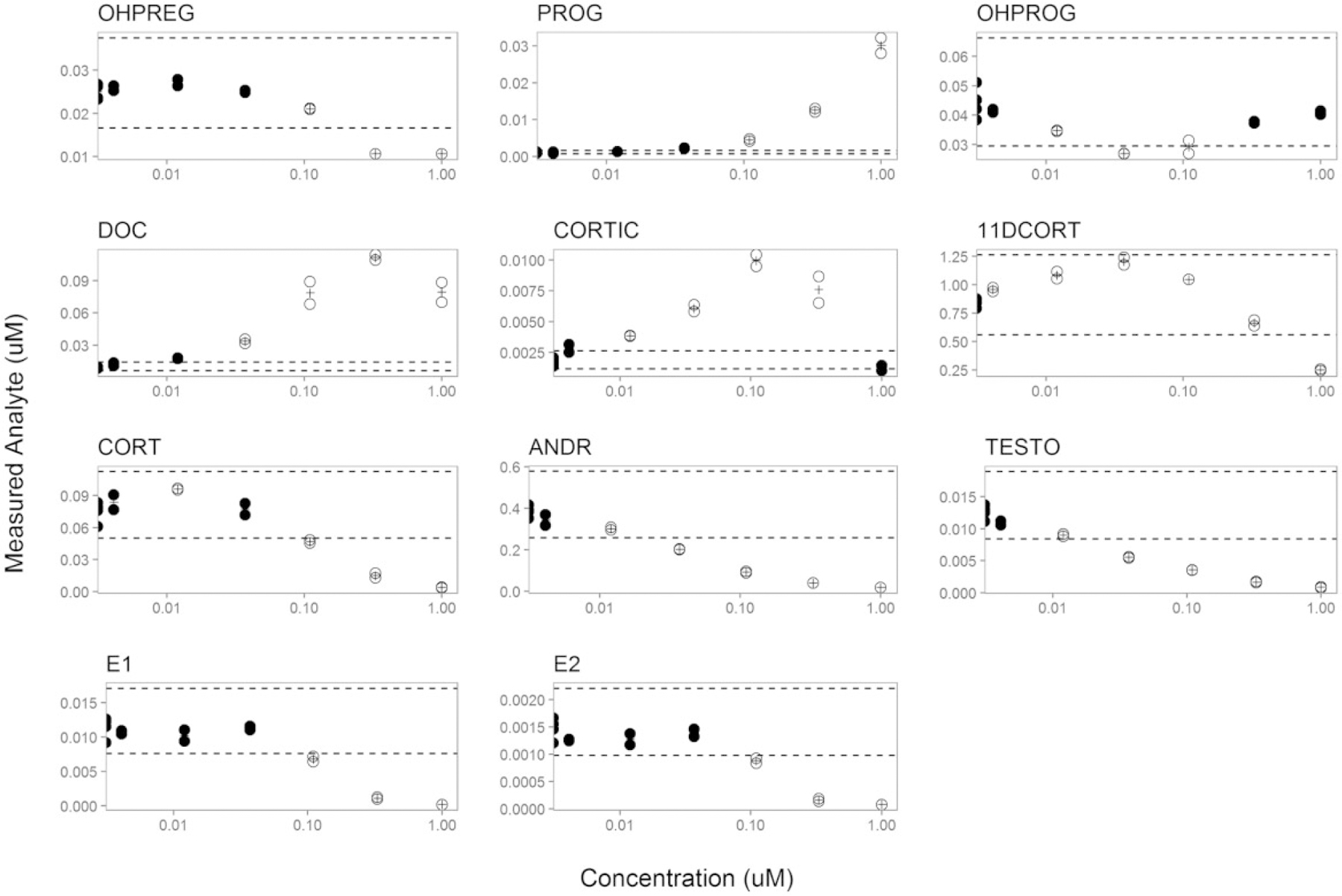
Example visualization of the ANOVA results for prochloraz. Replicates and the mean response values are denoted as filled circles and plus signs, respectively. Open circles indicate data points that were significantly different from control (p < .05). Dashed horizontal lines indicate 61.5-fold versus DMSO control to give additional context for low magnitude, but positive, responses. Data are plotted as concentration (μM) of prochloraz versus the measured steroid hormone analyte concentration (μM).
The results of the ANOVA analysis for all steroid hormone analyte data were also considered in terms of how each chemical may have affected different hormone classes across the steroid biosynthetic pathway, ie, progestagen, corticosteroid, androgen, or estrogen production. Considering these steroid hormone classes (highlighted in Figure 1), the results for the 654 chemicals evaluated are represented in a Venn diagram (Figure 3) to illustrate the number of chemicals that affected each hormone class or combination of classes. Of the 654 chemicals with concentration-response data amenable to ANOVA, 25 chemicals failed to produce a positive result on any hormone; the remaining 629 chemicals produced a positive result on at least 1 hormone class. Three hundred seven chemicals, or 47% of chemicals tested in concentration-response, demonstrated positive results for at least 1 hormone from each of the 4 classes. This finding is not unexpected, as chemicals evaluated in concentration-response were largely pre-selected for demonstrated effects in single concentration screening for 3 to 4 hormone analytes. Interestingly, few chemicals affected only estrogens (estrone and E2; 8 chemicals) or androgens (androstenedione and T; 1 chemical), or both (1 chemical), even though 4 hormone analytes comprise these 2 classes combined. Due to the relatively high percentage of the screened chemicals that affected androgens or estrogens in addition to corticosteroid and/or progestagens, it appears that integration of data for the corticosteroid and progestagen hormone measurements with the data for estrogen and androgen hormone measurements may provide important information on the magnitude of overall steroid biosynthetic pathway perturbation. Sixty-seven chemicals, or approximately 10% of the chemicals screened in concentration-response, affected progestagens only (13), corticosteroids only (10), or progestagens and corticosteroids only (44). Thus, consideration of corticosteroid and progestagen hormone levels in the HT-H295R assay may identify chemicals that perturb portions of the steroid biosynthesis pathway expressed in H295R cells that are overlooked in the H295R assay when only E2 and are reported.
Figure 3.
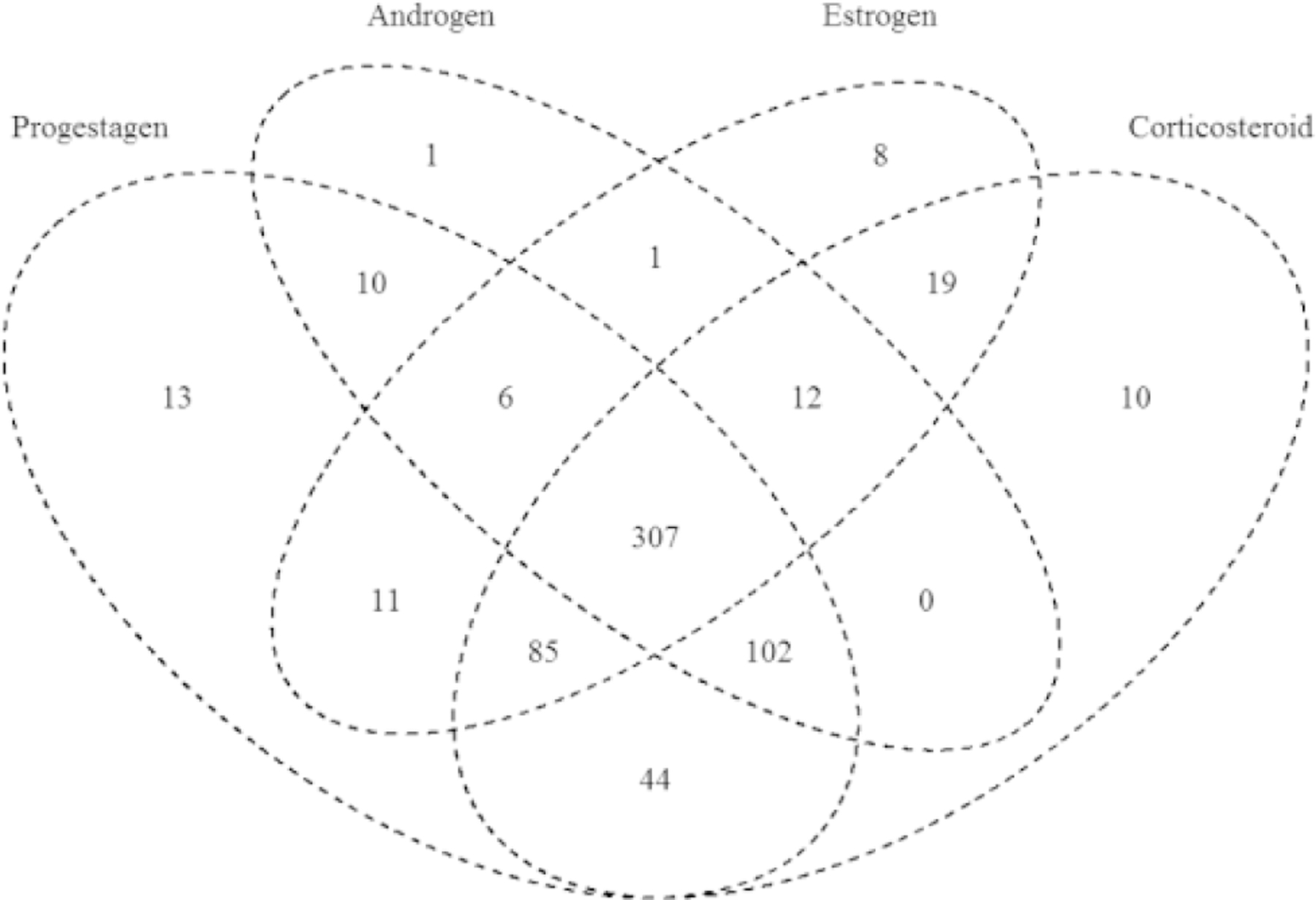
Venn diagram of ANOVA results for effects on steroid hormone synthesis, grouped by steroid class. The number of chemicals with positive results for progestagens (OH-pregnenolone, progesterone, OH-progesterone), corticosteroids (DOC, Corticosterone, 11-deoxycortisol, Cortisol), androgens (androstenedione, T), and estrogens (estrone, E2) are shown. A total of 629 chemical samples are represented in the Venn diagram (25 chemicals tested in concentration-response with data available for analysis failed to produce positive ANOVA results for any hormone class).
Pathway-Based Results Using the Mahalanobis Distance Approach
The Mahalanobis distance adjusts the distances, or effect sizes, for the variance and covariance among the hormone measures at each concentration, thereby accounting for knowledge of the interrelatedness of the steroid hormone measurements (Supplementary File 5). To support selection of the Mahalanobis distance as a basis for the new statistical approach, the correlation matrix corresponding to the covariance matrix used in calculation of the for the steroid hormone analytes was examined. As anticipated from knowledge of the steroidogenesis pathway in H295R cells (Figure 1), the covariance of the residuals for several steroid hormone analytes in the HT-H295R assay were highly correlated with one another (Figure 4). For example, the residuals for estrone and E2 were highly correlated (Pearson’s R = .75), as were androstenedione and T (R = .66). Residuals for cortisol and 11-deoxycortisol were also highly correlated (R = .69). In contrast, the residuals for both progesterone and DOC had very weak correlations, in some cases negative correlations, with residuals for all of the steroid hormones measured. This correlation matrix demonstrated high correlation of the residuals of many of the steroid hormone measures, which suggests that the Mahalanobis distance is one appropriate analysis metric for interpretation of these data.
Figure 4.
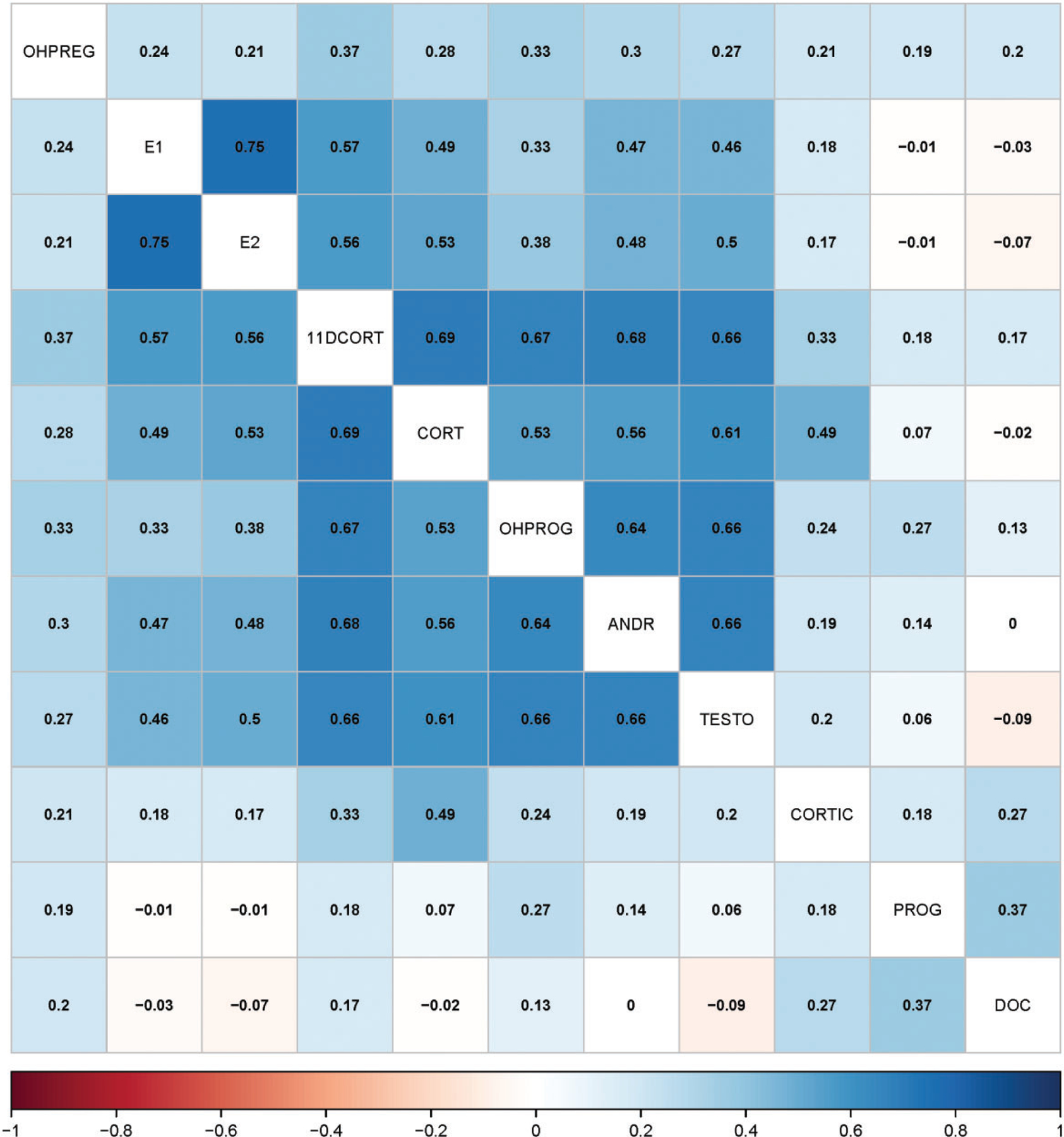
Hierarchically clustered heatmap summarizing correlation of the covariance of steroid hormone analytes. The correlation coefficients for each steroid hormone pair are provided.
The results from measurement of 11 steroid hormone analytes were used in the derivation of the mean Mahalanobis distance () at each concentration for chemicals screened in concentration-response. Radar plots were used to visualize the response for a single chemical for these 11 assayed hormones, with examples for atrazine, benfluralin, and mifepristone illustrated in Figure 5 (radar plots for all tested chemicals available in Supplementary File 9). Next to the example radar plots in Figure 5, the plot of the estimated by concentration is shown, with a horizontal red dashed line to indicate the critical value. If a exceeds the critical value, it is considered a positive result for this pathway approach. The maxmMd is the maximum of the set of values produced for all concentrations of a tested chemical. Atrazine moderately affected a number of hormones, including estrogens, progestagens, corticosteroids, and androgens, yielding a moderate adjusted maxmMd of 3.14. Benfluralin provides an example of a chemical with a negative pathway result, with no significant concentration-response for the values, as the maxmMd failed to exceed the critical value (adjusted maxmMd of −0.14). In contrast to the moderate effects of atrazine on multiple steroid hormones, mifepristone strongly modulated progestagens with significant effects on progesterone and OH-progesterone and moderate but non-significant trends on corticosteroids and androgens, resulting in a relatively high adjusted maxmMd of 33. The steroid hormone response data, annotated by the ANOVA results, and plots of the for all tested chemicals are available as Supplementary File 9.
Figure 5.
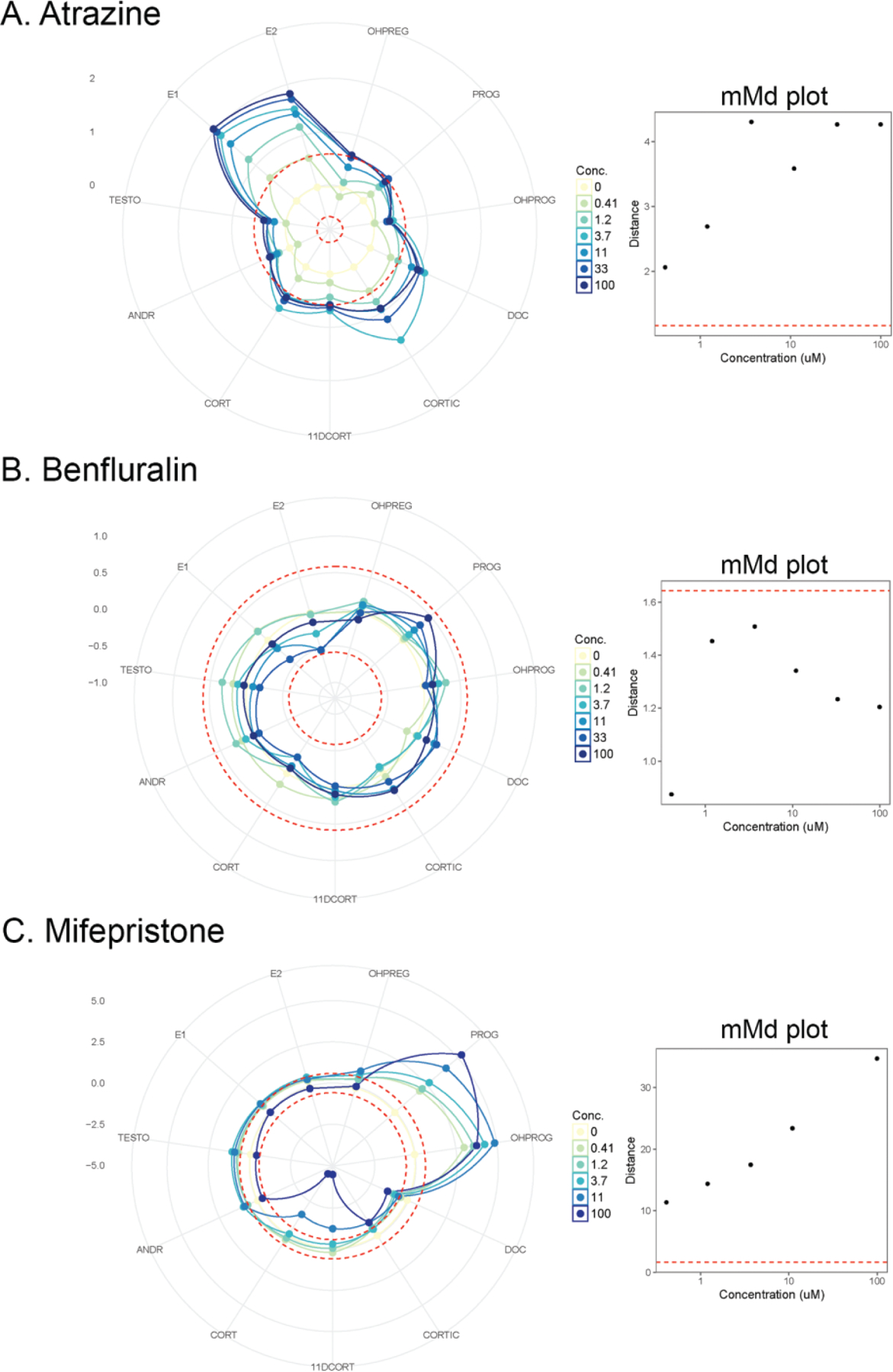
Example radar plots of the 11-dimensional dataset used to derive a mean Mahalanobis distance () for each concentration assayed. The 11 steroid hormone analytes are represented as the “spokes” of the radar plot, and each concentration of the chemical is annotated by a different color. The dotted, concentric circles denote ±1.5-fold control as threshold to contextualize the responses, as the y-axes vary by chemical to allow for visualization of the relative magnitude of effects. The numbers on the left of each radar plot denotes the fold change values of the major gridlines of the plots. Next to each radar plot is a plot of by concentration, with the critical value for the annotated using a horizontal dashed red line. A, atrazine (CASRN 1912-24-9); B, benfluralin (CASRN 1861-40-1); C, mifepristone (CASRN 84371-65-3). Radar plots and plots are supplied for all chemicals in Supplemental File 10.
To provide context for the relative maxmMd responses, the distribution of the maxmMd values for the 766 chemical samples with concentration-response data that cleared the cell viability filter is illustrated in Supplementary File 10. These maxmMd values are adjusted for the critical value (maxmMd — critical value = adjusted maxmMd), such that a positive maxmMd should be >0. The range of adjusted-maxmMd values for this dataset is −0.64 to 51.8. The median of the distribution was 3.52, is annotated by a vertical dashed red line. The mean of the distribution was 5.92. The distribution would likely be more informative if the chemical set had not been pre-selected predominantly from single concentration screening for positivesAll of the maxmMd values, the critical values, and the adjusted maxmMd values are provided by chemical sample in Supplementary File 11.
Comparison and Evaluation of the ANOVA and maxmMd Results
Comparison of the HT-H295R data with the OECD interlaboratory validation results.
Utilizing an ANOVA procedure and a post hoc Dunnett’s test enabled a comparison of the HT-H295R screening data with the summary results available from the OECD interlaboratory validation (Hecker et al., 2011). A detailed comparison of the effects on estrogen synthesis and androgen synthesis is illustrated in Supplementary File 13 Tables A and B, respectively, and summarized by confusion matrices and a table of sensitivity and specificity values by effect type in Figure 6. For the confusion matrix, a chemical was excluded from the sensitivity and specificity calculations if the OECD interlaboratory validation results for E2 or T in a particular direction were equivocal. OECD interlaboratory results for a chemical were considered equivocal if there was significant disagreement among labs, as specified here: (1) 2 or more laboratories failed to detect a LOEC for a “core” reference chemical tested in all 5 laboratories; or (2) if only 1 of 2 laboratories reported a LOEC for the “supplemental” reference chemicals that were tested in only 2 labs. A revised confusion matrix along with sensitivity, specificity, and accuracy values were also generated based on exclusion of 1 chemical, nonoxynol-9, from all effect types, and letrozole from decreased T (Figure 6).
Figure 6.
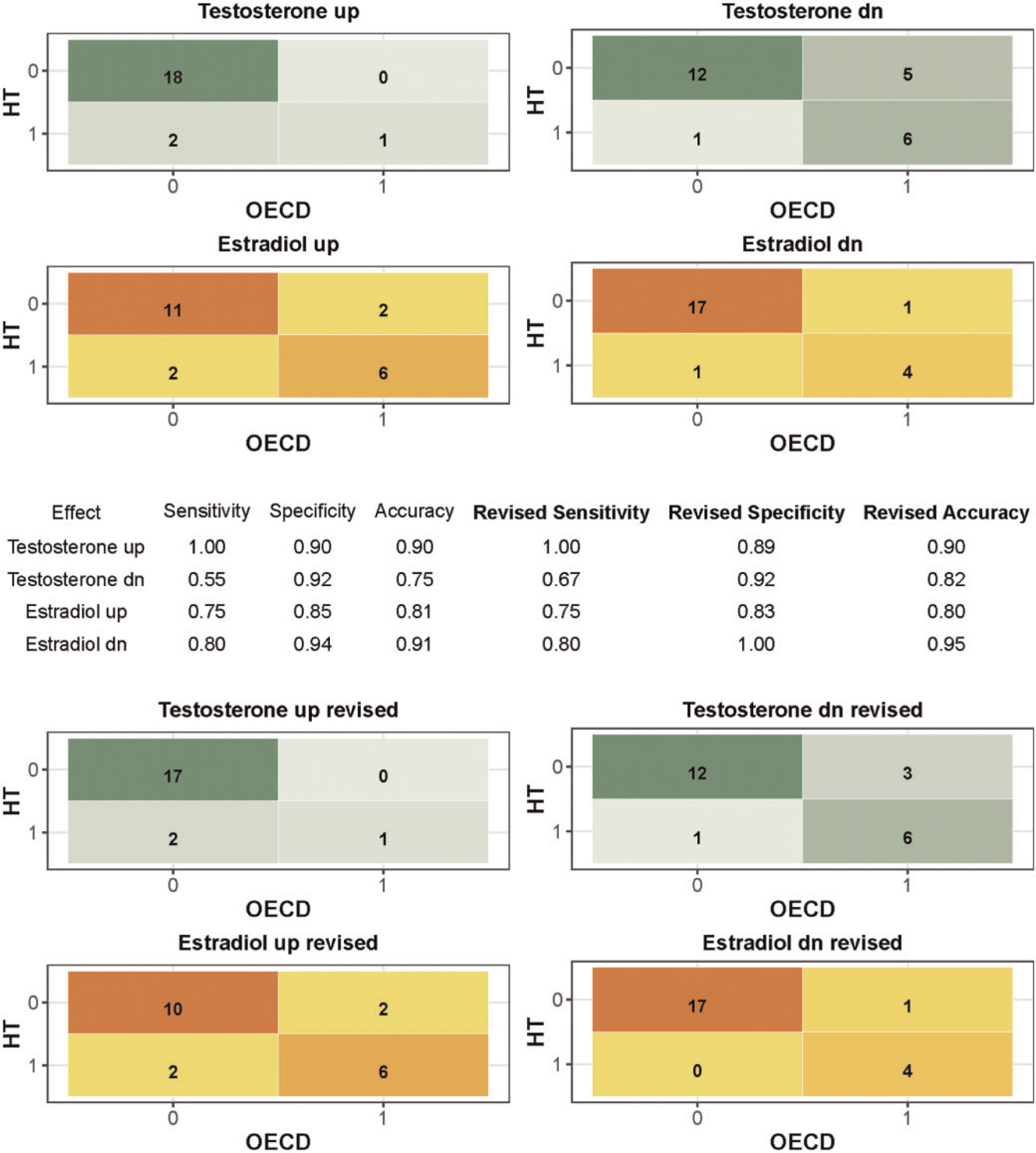
Confusion matrices for effects on T and E2. The OECD interlaboratory validation study results (Hecker et al., 2011) were interpreted as true outcomes, and the HT-H295R results analyzed by ANOVA with a post hoc Dunnett’s test were interpreted as predicted outcomes. Four effect types were considered: increased (up) and decreased (dn) testosterone (T) and estradiol (E2). The number of chemicals included for each effect type varied because chemicals with equivocal results for the effect type (4 for T up, 1 for T down, 4 for E2 up, 2 for E2 down) were removed. Revised confusion matrices present the comparison without nonoxynol-9 and omitting letrozole from testosterone dn.
Confusion matrices summarizing the comparison of OECD interlaboratory validation results and the HT-H295R screening data analyzed by ANOVA, excluding the OECD interlaboratory equivocal results by effect type, demonstrated sensitivities of 0.75 and 0.80, specificities of 0.85 and 0.94, and accuracies of 0.81 and 0.91 for increased and decreased estradiol, respectively (Figure 6). For T synthesis, sensitivities of 1 and 0.55, specificities of 0.90 and 0.92, and accuracies of 0.90 and 0.75 were observed for increased and decreased T, respectively. Revision of the confusion matrices to exclude nonoxynol-9 and letrozole (from decreased T only) increased the sensitivity for decreased T to 0.67. It should be noted that the reference chemical sets were not balanced, with strong weighting toward true negatives and limited true positives. True positives ranged from only >5% to approximately 29% of the result sets used for the confusion matrices. Further, inclusion of the supplemental reference chemicals, tested in only 2 laboratories for the OECD interlaboratory validation, was complicated by additional equivocal findings due to discordance between labs.
Qualitative comparison of the effects of the OECD reference chemicals on E2 synthesis in both the OECD interlaboratory validation and HT-H295R assay demonstrated good concordance (Supplementary File 14). For increased E2 for the core reference chemicals, 1 chemical had equivocal findings (butylparaben), and of the remaining 9 chemicals, 8 chemicals agreed (aminoglutehimide, atrazine, benomyl, forskolin, letrozole, molinate, nonoxynol-9, and prochloraz). For decreased E2 for the core reference chemicals, there were no equivocal findings, and 8 of the 10 chemicals agreed (atrazine, benomyl, butylparaben, ethylene dimethanesulfonate, forskolin, letrozole, molinate, and prochloraz). Five of the 15 supplemental reference chemicals with data for comparison produced equivocal results for effects on E2 synthesis in the OECD interlaboratory validation: 3 chemicals, dimethoate, flutamide, and tricresyl phosphate demonstrated equivocal findings for increased estradiol, and 2 chemicals, fenarimol and finasteride, demonstrated equivocal findings for decreased estradiol. For these 5 chemicals, the “true” result is uncertain. Three of the 15 chemicals produced equivocal results for increased E2, leaving 12 chemicals for comparison; of these 12, 9 chemicals agreed for increased E2 (bisphenol A, danazol, di(2-ethylhexyl)phthalate, 2,4-dinitrophenol, fenarimol, finasteride, ketoconazole, prometon, spironolactone). Two chemicals (fenarimol and finasteride) were equivocal for decreased E2, leaving 13 chemicals for comparison; of these 13 chemicals, all agreed for decreased E2 (bisphenol A, danazol, di(2-ethylhexyl)phthalate, dimethoate 2,4-dinitrophenol, flutamide, genistein, ketoconazole, mifepristone, piperonyl butoxide, prometon, spironolactone, and tricresyl phosphate).
Qualitative comparison of the effects of the OECD reference chemicals on T synthesis was similarly concordant. For increased T for the core reference chemicals, 2 chemicals had equivocal findings (atrazine and butylparaben), and of the remaining 8 chemicals, 6 chemicals agreed (aminoglutehimide, forskolin, letrozole, molinate, nonoxynol-9, prochloraz). For decreased T for the core reference chemicals, there were no equivocal findings, and 8 of the 10 chemicals agreed (aminoglutehimide, atrazine, benomyl, butylparaben, ethylene dimethanesulfonate, forskolin, molinate, and prochloraz). However, if nonoxynol-9 is excluded based on uncertainty regarding the chemical identity, and letrozole is excluded as the MTC in the HT-H295R assay (14 μM) is less than the LOECs reported by the OECD interlaboratory study (100 μM), then 8 of 8 core reference chemicals agree for decreased T. Two of the 15 supplemental chemicals produced equivocal results for increased T, leaving 13 chemicals for comparison; all of which agreed for increased T (bisphenol A, danazol, di(2-ethylhexyl)phthalate, dimethoate, 2,4-dinitrophenol, fenarimol, finasteride, flutamide, genistein, ketoconazole, piperonyl butoxide, prometon, and spironolactone). One of the 15 supplemental chemicals produced equivocal results for decreased T, leaving 14 chemicals for comparison; of these 14, 10 chemicals agreed for decreased T (bisphenol A, di(2-ethylhexyl)phthalate, dimethoate, flutamide, genistein, ketoconazole, mifepristone, prometon, spironolactone, and tricresyl phosphate).
Equivocal findings and discordances included the following chemicals:
Aminoglutehimide. Aminoglutehimide was likely a borderline positive for decreased E2 in the OECD interlaboratory validation; 3 of the 5 labs reported a LOEC at the greatest non-cytotoxic concentration (100 μM) with no concentration-response, and 1 lab reported a LOEC that was annotated as not significantly different from control (p value of .051). Aminoglutehimide was negative for E2 effects in the HT-H295R assay, but it did significantly decrease several hormones (11-deoxycortisol, DOC, progesterone, OH-progesterone, androstenedione, and testosterone) and increase progesterone at 100 μM. These responses produced a weak pathway positive, with a low but significant adjusted maxmMd (1.56), and so would not constitute a false negative for effects on steroid biosynthesis when using all of the available screening data.
Atrazine. Two of 5 laboratories failed to detect a LOEC for atrazine-induced increases in T. The HT-H295R assay was a positive, but the effects did not exceed 1.5-fold control. The pathway analysis produced a significant adjusted maxmMd (3.14) as atrazine moderately, but significantly, affected 10 of the 11 hormones in the pathway.
Benomyl. Benomyl was negative in the OECD interlaboratory validation for effects on T, but produced a borderline positive in the HT-H295R assay for increased T (ie, effects were not concentration-dependent and failed to exceed the threshold of 1.5-fold control). The adjusted maxmMd was positive but small (0.16).
Butylparaben. Butylparaben was negative for effects on E2 in the HT-H295R assay, but produced equivocal results for increased E2 in the OECD interlaboratory validation, as 3 of 5 labs failed to detect a LOEC. Three of 5 laboratories in the OECD validation failed to detect a LOEC for butylparaben-induced increases in , and the HT-H295R results were negative; however, butylparaben was a pathway positive (adjusted maxmMd = 4.64), as it significantly affected 2 progestagen hormones (progesterone and OH-progesterone) in the pathway.
Danazol. Danazol decreased in the HT-H295R assay but was negative in the OECD interlaboratory validation; danazol in this comparison is classed as a false positive, but appeared to affect several hormones across the pathway in the HT-H295R in a concentration-consistent manner (adjusted maxmMd = 15.3–21.5).
2,4-dinitrophenol. 2,4-dinitrophenol decreased in the OECD interlaboratory validation with LOECs for the 2 laboratories that ranged 5 orders of magnitude on a log10 scale (0.0001–100 μM). 2,4-dinitrophenol was negative in the HT-H295R assay, but was screened only at the MTC (10 μM); no concentration-response data were available for pathway-based analysis and so a maxmMd value was not computed.
Ethylene dimethanesulfonate (EDS). EDS was negative in the OECD interlaboratory validation for effects on E2 synthesis, but was a conditional positive in the HT-H295R assay for increased E2; though multiple concentrations were positive, the effects were not concentration-responsive and were not significant at the maximum concentration; further these effects did not exceed 1.5-fold of the control. As such, this positive result for EDS in the HT-H295R assay was a borderline positive. EDS was also negative in the OECD interlaboratory validation for effects on T, but produced a conditional or borderline positive in the HT-H295R assay for increased (ie, effects were not concentration-responsive and failed to exceed the threshold of 1.5-fold control). Supportive of these borderline findings for E2 and is the negative result for the pathway-based approach due to a maxmMd that failed to exceed the critical value (adjusted maxmMd of −0.433).
Finasteride. Finasteride decreased in the OECD interlaboratory validation; though it failed to significantly perturb in the HT-H295R assay (only 1 concentration, 10 μM, was significant), it significantly affected production of OH-pregnenolone, progesterone, OH-progesterone, DOC, 11-deoxycortisol, and androstenedione, yielding a pathway positive (adjusted maxmMd of 12.3).
Genistein. Genistein increased E2 in the OECD interlaboratory validation, but failed to increase E2 in the HT-H295R assay. Genistein did produce a strong pathway positive, based on significant effects on OH-pregnenolone, progesterone, OH-progesterone, DOC, 11-deoxycortisol, cortisol, androstenedione, and T, with a significant, high adjusted maxmMd (31.8). One concentration, 11.11 μM, appeared to significantly increase estrone and estradiol, but did not meet the minimum criteria for a positive result (2 consecutive concentrations with significant results or the highest non-cytotoxic concentration with significant results). Genistein was a strong positive using a pathway approach.
Letrozole. Letrozole was reported to decrease , but all 5 laboratories in the OECD interlaboratory validation reported a LOEC at the maximum tested concentration only (100 μM), which exceeded the MTC used in the HT-H295R to maintain cell viability (14 μM). Based on differences in the concentration range tested, letrozole was excluded from the confusion matrix for decreased T. Letrozole was maintained in the confusion matrices for the other effect types that would not have been affected by inability to screen up to 100 μM. Letrozole, a pharmacologic CYP19A1 inhibitor, inhibited estrone and E2 production at submicromolar concentrations such that these hormones dropped below the LLOQ in addition to moderate effects on several other hormones in the pathway (adjusted maxmMd = 12.4).
Mifepristone. Mifepristone increased E2 in the OECD interlaboratory validation, but failed to increase E2 in the HT-H295R assay. Mifepristone produced significant effects on 2 hormones (progesterone, OH-progesterone), with trends toward decreased DOC, corticosterone, 11-deoxycortisol, cortisol. The responses across the pathway produced a high adjusted maxmMd (33.1).
Nonoxynol-9. Nonoxynol-9 was negative for effects on E2 synthesis in the OECD validation, but positive in the HT-H295R assay for decreased E2; it is unclear if this was a false positive in the HT-H295R assay or not due to uncertainties associated with the identity of the substance tested in the OECD validation (see Materials and Methods for detailed discussion). The magnitude of the effect on E2 synthesis was low. Nonoxynol-9 decreased T in the OECD validation, but was negative in HT-H295R; however, the LOEC reported by 4 of the 5 labs in the OECD validation (10 μM) exceeded the maximum tested concentration in HT-H295R (0.4 μM) (1 lab failed to detect a LOEC, and all reported LOECs reflected a single significant concentration). Nonoxynol-9 was just barely positive in the pathway-based approach (adjusted maxmMd of 0.078). Uncertainties regarding the chemical substance, and the disparity in the tested concentration range due to cytotoxicity concerns, supported revision of the confusion matrix to exclude nonoxynol-9.
Piperonyl butoxide. Piperonyl butoxide was negative in the OECD interlaboratory validation but produced a conditional positive for increased E2 in the HT-H295R assay, with multiple concentrations significantly different from control that failed to exceed 1.5-fold of the control. Piperonyl butoxide minimally decreased T synthesis in the OECD interlaboratory validation at 10 μM. Piperonyl butoxide failed to affect T synthesis in the HT-H295R assay, but did demonstrate minor effects on a number of hormones in the pathway, often without a monotonic concentration-response, yielding a weak pathway positive and adjusted maxmMd of 2.30.
Combined comparison of E2 and T results and maxmMd for OECD reference chemicals.
A summary comparison of the OECD interlaboratory and HT-H295R results for E2 and for each reference chemical is provided in Figure 7 along with a positive or negative designation for the pathway-based maxmMd analysis. In Figure 7, the reference chemicals are rank-ordered by log10-maxmMd. The maxmMd value appears to separate known strong steroidogenesis disruptors largely comprised of pharmacological modulators of hormone biosynthesis (eg, mifepristone, prochloraz, ketoconazole, danazol, letrozole) from moderate disruptors (eg, atrazine, molinate, di(2-ethylhexyl-phthalate) and from non-active chemicals (eg, EDS). However, effects of these reference chemicals on progestagen and glucorticoid biosynthesis is unknown in some cases. Known activities of these reference chemicals, approximations of the magnitude of perturbation, the classes of steroid biosynthesis perturbed in the HT-H295R assay, the number of steroid hormones perturbed in the HT-H295R assay, and the maxmMd values are briefly summarized in Supplementary File 13.
Figure 7.

Geometric tiling to compare the OECD validation and HT-H295R results. For each chemical in the core and supplemental OECD chemical reference sets, a binary comparison of the OECD interlaboratory validation result (OECD_) and the HT-H295R results (HT_) is presented. Positive E2 responses are blocked as yellow, positive T responses are blocked as green, equivocal responses in the OECD interlaboratory validation are blocked as gray, and negatives are blocked as white. Blue blocks denote positive pathway responses (defined as the maxmMd exceeding the critical value for a chemical), and the annotation bar ranks all of the chemicals in the set by their log10 maxmMd from high (red) to low (yellow), white blocks indicating negative pathway results. “OECD Summary” is a text annotation to indicate whether an effect (up or dn) was observed for E2 or T in the OECD interlaboratory validation.
Consideration of the maxmMd as a ranking metric.
A data-driven approach to understanding the added value of the maxmMd metric involved comparison of the number of steroid hormones significantly affected by a chemical using the ANOVA-based logic with the maxmMd value for that chemical. A boxplot of the maxmMd values, binned by the steroid hormone hit-count, is presented in Figure 8. The primary purpose of this visualization is to demonstrate that the sum of steroid hormone hits does not necessarily relay the magnitude of the effect of a test chemical on the set of 11 steroid hormones, whereas the maxmMd value allows for quantitative distinction of chemicals that affect similar numbers of hormones but with varying efficacy. The median of the maxmMd values generally increased as the steroid hormone hit-count increased; however, the maxmMd values enabled distinction of chemicals with the same steroid hormone hit-count, in some cases by >1 order of magnitude on a log10 scale. For example, both tricresyl phosphate and letrozole significantly perturbed synthesis of 7 steroid hormones in the set based on the ANOVA logic employed, but their adjusted maxmMd values were 0.94 and 12.4, respectively. Mifepristone significantly affected only 2 steroid hormones, but with great magnitude, such that it had a high maxmMd. BPA was replicated on 3 plates in 2 different screening blocks, and across these 3 replicates, perturbed 5–7 hormones based on minor effects for a few steroid hormones near the threshold for positive activity; however, the maxmMd values were relatively stable (adjusted maxmMd values for BPA ranged from 4.21 to 5.22). Open symbols in Figure 8 indicate chemicals with maxmMd values that failed to exceed the critical value, ie, pathway-based negatives; these negatives are distributed across steroid hit count bins of zero to 6, indicating that though effects of low magnitude may produce positive results in the ANOVA-based logic, the maxmMd provides a more quantitatively robust indicator of pathway perturbation than the sum of steroid hormone hit calls. Of the OECD reference chemicals, EDS yielded a negative adjusted maxmMd value, with a corresponding steroid hormone hit-count of 6. Benomyl, nonoxynol-9, and tricresyl phosphate produced weak pathway positives with adjusted maxmMd values of 0.16, 0.078, and 0.94 that corresponded to steroid hormone hit-counts of 4, 4, and 7. Conversely, small trends in the data for multiple steroid hormones that are not significant may result in a positive maxmMd; in the case of dimethoate, the adjusted maxmMd is 0.12, just above zero, indicating a very low pathway response that corresponds to no significant steroid hormone perturbations by the ANOVA-based logic. Thus, the maxmMd value appears to provide added value above steroid hormone hit-count alone for description of the magnitude of steroid biosynthesis pathway effects.
Figure 8.
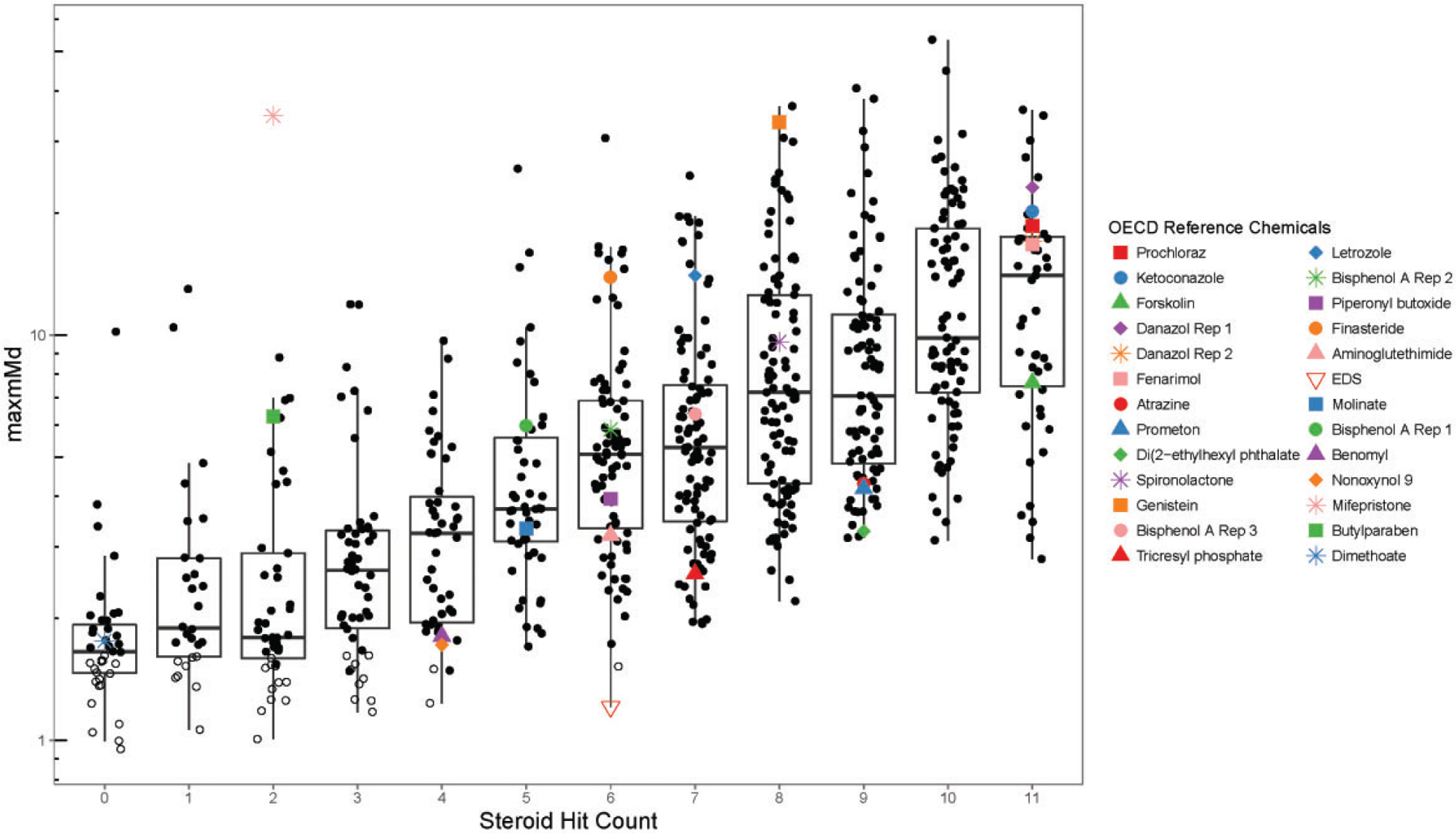
Boxplot of adjusted maxmMd values versus sum of steroid hormone positive responses. The maxmMd values for all 654 chemicals were binned by steroid hit count (ranging from 0 to 11 steroid hormones, as analyzed by the ANOVA-based logic employed herein), with the y-axis is log10-scaled. OECD reference chemicals are annotated within the plot. Closed symbols for all chemicals, including OECD reference chemicals, indicate positive maxmMd values that exceeded the critical value; open symbols for all chemicals, including OECD reference chemicals, indicate negative maxmMd values.
In Figure 9, an estimate of potency is related to the maxmMd using non-parametric local regression, or loess. The (μM) is the concentration at which the 4-parameter logistic fit of the mMd data intersects with the critical value for a given chemical, giving the potency at the threshold for a positive maxmMd. An inset table summarizes the dataset considering the maxmMd value, ie, negative (maxmMd < critical value) and positive (maxmMd > critical value), and whether or not a non-parametric trend, using Spearman’s correlation, was observed in the versus concentration data. Most of the negative maxmMd values failed to demonstrate a trend (48 of 51 chemical samples), whereas low to moderate positive maxmMd values often failed to demonstrate a trend (308 chemical samples) and moderate to high maxmMd values more often demonstrated a trend (407 chemical samples). A loess smooth regression line demonstrates a general relationship between and maxmMd: decreased , ie, greater potency, corresponds to higher maxmMd value, with the strongest relationship evident for the very weak or negative chemicals (top portion of the loess curve) and the strongest maxmMd positive values (bottom portion of the loess curve) that also demonstrated a trend in the mMd. The reproducibility of the maxmMd metric was also evaluated as part of considering its strengths and weaknesses. As suggested by the appearance of BPA on 3 plates across 2 separate screening blocks, a subset of the chemicals were replicated in screening the ToxCast Phase I, II, and E1K chemicals libraries, which allowed the reproducibility of the maxmMd to be examined. A total of 107 chemicals were screened in >1 experimental blocks (all other chemicals appeared in technical duplicate in 1 screening block only). A plot of the maxmMd values for all 107 replicated chemicals and the associated standard deviations is shown in Figure 10. The approximated standard deviation appeared nearly constant across the distribution of maxmMd values, so as the maxmMd increases, the likelihood that replicate block measures will produce a positive pathway finding (maxmMd > critical value) also increases. The standard deviation for residuals around the chemical-specific means of natural log-transformed maxmMd values across replicate blocks was 0.33. This value was used to approximate a 95% prediction interval for the chemical-specific maxmMd yielding a value of 1.93. Given that the highest critical value for this dataset was 1.81, maxmMd values of 3.5 or greater would be more likely to reproduce a positive pathway finding in additional block replicates. This is evident from the observation that chemicals failing to replicate positive pathway effects, as measured by a maxmMd exceeding the critical value for all replicates, were concentrated at lower maxmMd values (negative maxmMd values represented as open circles in Figure 10). As an example, the 95% confidence interval for bisphenol A, with a median maxmMd of 5.98 across replicates, would be 3.10–11.5. One chemical, 1,2,4-butanetriol, stands out due to the large difference between replicate blocks. 1,2,4-butanetriol was missing data for most of the steroid hormones in the block replicate that produced a larger pathway positive. The median maximum difference between maxmMd values across blocks was approximately 1.47 units on the arithmetic scale, demonstrating fairly good agreement between block-replicates of maxmMd, which ranged from 0.996 to 34.7 for this 107 chemical subset. Considering the maxmMd metric as a binary determinant of pathway positive or negative results, 94 of the 107 chemicals (87.9%) replicated a positive (maxmMd > critical value) or negative (maxmMd < critical value) pathway response across blocks. In contrast, the average recall for the 11 steroid hormone hit-calls across replicate blocks using the ANOVA-based logic was approximately 65% (analysis not shown).
Figure 9.
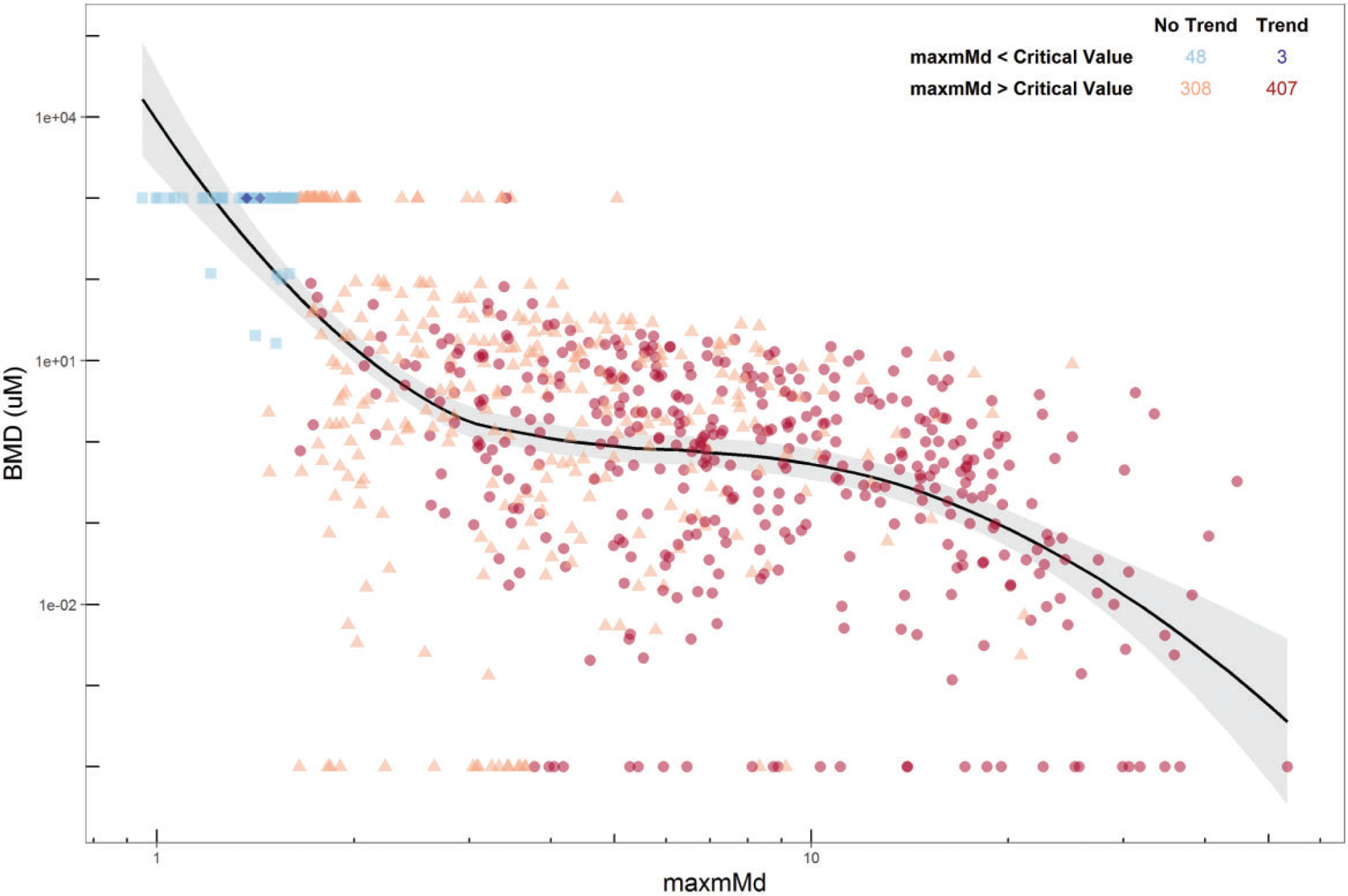
Estimate of potency versus the maxmMd. An estimate of potency, or benchmark dose () is compared to the maxmMd value. The (μM) is the concentration at which the Hill fit of the data intersects with the critical value for a given chemical. A loess smooth regression line demonstrates a general relationship between and maxmMd. A table summarizing the effects of a Spearman-based trend analysis versus negative and positive maxmMd values is provided in the upper right. Light blue squares = negative maxmMd response and no trend (48 chemical samples); dark blue diamonds = negative maxmMd response and trend (3 chemical samples); orange triangles = positive maxmMd value and no trend (308 chemical samples); red circles = positive maxmMd value and trend (407 chemical samples).
Figure 10.
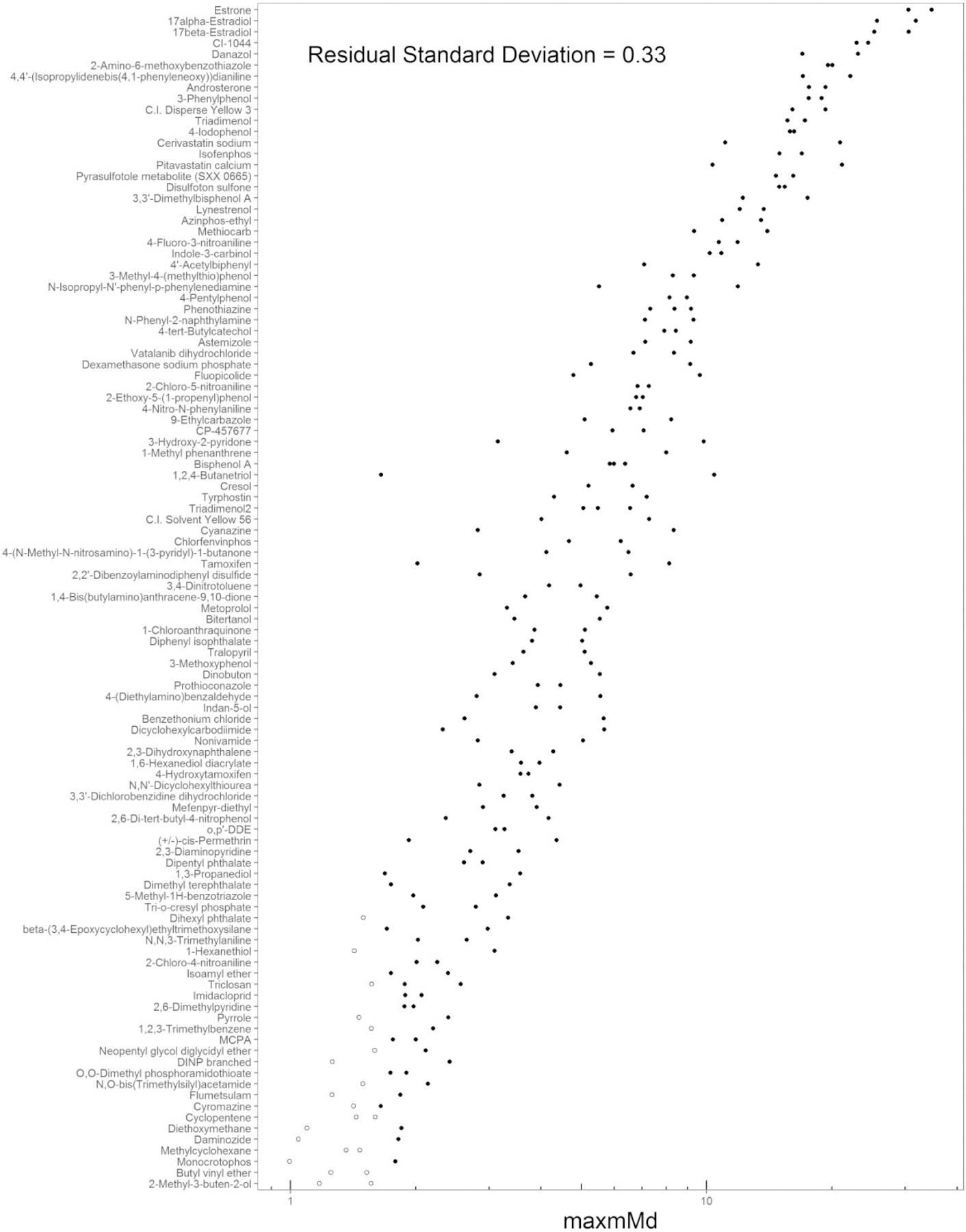
Reproducibility of the maxmMd values. A 107 chemical subset was screened in multiple experimental block replicates, enabling an evaluation of reproducibility. The residual standard deviation of the natural log-transformed maxmMd values was determined and annotated in the plot (0.33). Open symbols indicate negative maxmMd values (failed to exceed the critical value).
DISCUSSION
The current work demonstrates the utility of the HT-H295R screening assay as an alternative for the OECD-validated, low throughput H295R assay (OECD TG 456). The ANOVA analysis and logic used herein for the HT-H295R dataset to determine effects on the steroid biosynthesis pathway enabled a direct comparison of the OECD interlaboratory validation data and the HT-H295R data. This detailed, performance-based comparison highlights good concordance of results, with accuracies that range 0.75–0.91 for effects on E2 and . Understanding that E2 and provide limited perspective on the impact of chemicals on the steroidogenesis pathway present in H295R cells, this work also presents a novel evaluation of hormone data from more of the steroid biosynthesis pathway. To integrate 11 steroid hormone analytes for pathway-level analysis using the HT-H295R assay data, a mean Mahalanobis distance () was computed for each chemical concentration screened. The provided a set of unitless values from which the maximum mean Mahalanobis distance (maxmMd) could be calculated across the concentration range screened. We suggest that this maxmMd may be useful for prioritizing chemicals by the relative magnitude of their overall impact on the steroid biosynthesis pathway. Thus, this work, through demonstration of the HT-H295R as an alternative and a novel data analysis approach, advances efforts to rapidly identify and prioritize large numbers of chemicals as potential steroidogenesis disruptors for further evaluation or confirmatory screening.
Evaluation of the concordance of the OECD reference chemical effects on E2 and synthesis in the OECD interlaboratory validation exercise and the HT-H295R screening campaign demonstrated similarity in the findings, despite some differences in experimental assay design. In addition, it also underscored some of the thematic challenges of comparing alternative screening approaches to traditional methods. The OECD reference chemical set was heavily weighted with “true” negatives for E2 or T, yielding relatively high specificity values (0.85–0.94). However, only 1 of the 25 chemicals with data from the OECD interlaboratory validation that were screened in the HT-H295R assay (EDS and benomyl) were negative in the OECD interlaboratory validation for effects on both E2 and synthesis (Supplementary File 12). Despite a small number of “true” positives, the sensitivity values (0.55–1.0) demonstrated that the HT-H295R assay was capable of detecting these chemical effects on E2 and alone. The sensitivity without adjustment for decreased T was 0.55, but increased to 0.67 if nonoxynol-9 and letrozole were omitted (for reasons of chemical uncertainty and a LOEC > the MTC, respectively).
Using a pathway approach, rather than solely measures of E2 and , appeared to increase screening sensitivity and identified chemicals as pathway positives that were potential HT-H295R false negatives for effects on E2 (aminoglutehimide failed to decrease E2; mifepristone and genistein failed to increase E2), and T (2,4-dintrophenol, finasteride, and piperonyl butoxide failed to decrease T). One hypothesis for the false negative findings for mifepristone and genistein and increased E2 is that the HT-H295R system may be slightly less sensitive to E2 increases due to pre-stimulation with forskolin. However, a critical strength of collecting data for multiple steroid hormones in the pathway and combining these data into a single metric, the maxmMd, is that weak effects on multiple hormones, or strong effects on 1 or 2 hormones, can contribute to a pathway-based positive. Indeed, all of the aforementioned potential false negatives were pathway positives using this approach. In addition to the need for a higher number of curated reference chemicals with data from multiple studies on which to base evaluations, it would be helpful to have reference chemicals to better evaluate the steroidogenesis pathway as a whole, including known negatives for the entire pathway, and chemicals with effects on corticosteroid and progestagen synthesis. The small number of “true” negatives for both E2 and in the OECD reference chemical set, and a lack of information regarding “true” pathway negatives, limits determination of the negative predictive value of the maxmMd approach. Another challenge in comparing these datasets includes the variability in the reference data set; data insufficient for comparison due to laboratory disagreements, and reported potency and efficacy values that were highly variable, are difficult to evaluate for validation purposes. However, ranking screened chemicals by the magnitude of perturbation induced across the steroid biosynthesis pathway appears to represent an effective and efficient means of understanding the priority of particular chemicals within a list, above and beyond tabulation of the number of steroid hormones perturbed (Figure 8). As suggested in Figure 8, though the maxmMd generally increased with increasing number of steroid hormones affected, as measured by the ANOVA-based logic, the maxmMd metric appeared to provide the ability to distinguish chemicals with the same steroid hormone hit count, but different magnitude of effects. For example, finasteride and EDS both had a hit-count of 6, but had maxmMd values of 13.9 and 1.2, respectively. Finasteride had significant effects on OHPREG, PROG, OHPROG, DOC, 11DCORT, and ANDR which were >1.5-fold from the DMSO controls (Supplementary File 8). In contrast, EDS had significant effects on OHPROG, 11DCORT, ANDR, T, E1, and E2; however, all of these significant effects were within 1.5-fold of the DMSO controls (Supplementary File 8). Therefore, the difference in magnitude of effect on the overall steroidogenesis pathway between these 2 chemicals, and equally the priority of these chemicals for further study, is captured by the maxmMd metric employed here, but not by the ANOVA-based logic. Further, as detailed in Supplementary File 13, the maxmMd appeared to distinguish strong modulators of steroidogenesis (eg, mifepristone, genistein, prochloraz, ketoconazole, danazol, letrozole, with adjusted maxmMds ranging from 33.1 to 12.4) from moderate modulators of steroidogenesis (eg, BPA, butylparaben, atrazine, prometon, with adjusted maxmMd values ranging from 5.22 to 3.10) and minor or borderline modulators (eg, piperonyl butoxide, molinate, benomyl, and nonoxynol with maxmMd values ranging from 2.30 to 0.078) or negative chemicals showing no effect on steroidogenesis (eg, EDS, flutamide, 2,4-dinitrophenol, with adjusted maxmMd values of ≤0 or NA). As with nearly any alternative approach, additional reference chemicals with full steroid biosynthesis pathway information would enable additional consideration of the quantitative and qualitative value of using the maxmMd approach.
Additional key questions in evaluating maxmMd as a potential prioritization metric are: (1) whether this metric relates to both efficacy and potency, and (2) if the maxmMd is reproducible across experimental blocks. An estimate of potency, the concentration corresponding to the mMd at the critical value, is related to the maxmMd using a smoothed loess fit in Figure 9. With some exceptions, increased potency (a smaller value in Figure 9) appeared to correspond to increased maxmMd; the strongest relationship between potency and maxmMd was apparent for the weak or negative responses and for the highest maxmMd responses. The relationship between potency and maxmMd reflects the monotonicity of the values for a chemical versus concentration; a chemical with higher potency in the HT-H295R assay is likely to have increasing magnitude of response with increasing concentration screened. The maxmMd response appeared reproducible for approximately 88% of the 107 chemicals that were screened in multiple experimental blocks, with failure to replicate largely attributable to responses near the critical value (Figure 10). Calculation of the set of values reduced an 11-dimensional question to a single dimension, and selection of the maxmMd appeared to provide a reproducible approximation of efficacy and potency in a single metric, emphasizing the advantages of the analysis described herein.
In addition to the ToxCast HT-H295R screening implementation, other research efforts have measured multiple steroid hormones in the pathway expressed in H295R cells (Abdel-Khalik et al., 2013; Hansen et al., 2017; Maglich et al., 2014; Nielsen et al., 2012; Rijk et al., 2012; Tonoli et al., 2015; Zhang et al., 2011), but to date the ToxCast screening implementation remains the largest publicly reported screening effort in terms of number of chemicals and concentrations evaluated for effects on steroid hormones. The number of recent reports that measure multiple hormones in the steroidogenesis pathway support the concept that the synthesis of steroids other than E2 and contribute important insight into chemically induced steroidogenesis disruption. Existing computational models for chemical modulation of interdependent hormone profiles in H295R cells have employed a systems biology approach, incorporating biological and kinetic information to quantitatively estimate the anticipated levels of multiple steroid hormones in the pathway following chemical exposure (Breen et al., 2010; Saito et al., 2016). The necessary time course information used to inform such a pathway-based model has not been generated for the HT-H295R assay. Thus, the current work uses an empirical approach to statistically integrate screening data for 11 steroid hormones and compute a mean Mahalanobis distance (or ) for each chemical concentration screened. Using values statistically accounts for the correlation of the residuals of the steroid hormone measures rather than using information about the enzyme reaction kinetics to describe their interrelatedness. The concentration-response behavior of the values can also be condensed to a single value, the maximum mean Mahalanobis distance or maxmMd, which may be useful in prioritizing chemicals to more accurately reflect their effects on the broader steroidogenesis pathway. Future work would be needed to develop time-course information for control chemicals in the HT-H295R assay to inform model parameters to describe the kinetics accurately for a systems biology approach. Further, the potential interaction of biological mechanisms beyond cholesterol transport and enzymatic steroid synthesis reactions, eg, the contributions of steroid hormone nuclear receptors expressed in H295R cells such as the glucocorticoid and androgen receptors (Asser et al., 2014; Hecker et al., 2006; Robitaille et al., 2015; Yanes and Romero, 2009) is the subject of ongoing research, and suggests additional mechanisms that could be included in a model.
As demonstrated in the current study, the maxmMd may be a useful ranking metric, but areas of uncertainty in applying these pathway data to prioritization should be noted. Clearly more work is needed to understand how to translate in vitro steroidogenesis findings to prediction of in vivo effects. Three adverse outcome pathways (AOPs) in development within the AOP Wiki (aopwiki.org/aops), AOPs 7, 25, and 153, and a review of available literature, support a relationship between aromatase (CYP19A1) inhibition and impaired fertility and reproduction in female mammals and fish (Breen et al., 2013; Villeneuve et al., 2007; Villeneuve et al., 2013). A quantitative AOP relates aromatase inhibition to reduced population fecundity in fathead minnow (Conolly et al., 2017). Aromatase inhibitors are also known to alter spermatogenesis in male rats (Gerardin and Pereira, 2002; Pouliot et al., 2013) and impact neuroendocrine function in many species (Charlier et al., 2013; Cornil et al., 2013; Vierk et al., 2014). Inhibition of CYP17A1 by chemicals like prochloraz causes reduction in serum dihydrotestosterone concentrations (Robitaille et al., 2015). However, in previous validation efforts for guideline-based H295R assays, in vitro results have not always predicted the effect or correct direction of effect for serum E2 and/or T findings (Hecker et al., 2011; LeBaron et al., 2014; Paul Friedman et al., 2016). It could be that contributions of absorption, distribution, metabolism, and excretion must also be accounted for in a computational model that could incorporate HT-H295R assay data, in addition to other potentially relevant assay data for predicting in vivo steroidogenesis, such as aromatase and other enzymes necessary for steroidogenesis (eg, conversion of T to 5α-dihydrotestosterone by steroid-5α-reductase), indicators of cholesterol transport, and markers of mitochondrial toxicity.
Another area of uncertainty in understanding how HT-H295R assay data might be translated involves interpretation of changes in progestagen and corticosteroid hormones. Disruption of the enzymes responsible for corticosteroid synthesis in the adrenal cortex result in the development of congenital adrenal hyperplasia (CAH), which can present with different phenotypes specific to the steroidogenic enzyme impacted, such as lipoid CAH (mutations in steroid acute regulatory protein, StaR, or CYP11A1), salt-wasting CAH (mutations in CYP21A2), virilizing CAH (mutations in CYP21A1 or CYP11B1), and other forms (as reviewed by Miller and Auchus, 2011). Altered synthesis of progestagen and corticosteroid hormones may inform hypothetical mechanisms of action, particularly for potential inhibition of enzymes that act early in the steroidogenesis pathway in H295R cells, eg, StaR, CYP11A1, and 3β-hydroxysteroid dehydrogenases. Further, given that H295R cells present a dynamic system, one might hypothesize that modulation of progestagens in particular would eventually propagate to changes in downstream corticosteroid, estrogen, and androgen production in vitro given enough exposure and time (Saito et al., 2016). In vitro studies have suggested that H295R cells are useful for identifying chemicals that may perturb only progestagens and/or corticosteroids and modulate hypothalamic-pituitary-adrenal axis function in vivo, leading to pathologies associated with hyper- or hypofunction of the adrenal (Oskarsson et al., 2016; Strajhar et al., 2017). However, the database of animal toxicology information to connect these in vitro findings and in vivo measures is lacking.; it is unclear if changes in progestagen and corticosteroid synthesis in the H295R model are only helpful in identifying putative mechanisms of steroidogenesis disruption, or if effects on these steroid classes would correspond to changes in serum steroid hormone concentrations or adrenal pathology, such as CAH. If it was important to focus a particular prioritization task for chemicals that may affect estrogen and androgen synthesis specifically, chemicals with effects on estrogens and/or androgens could be ranked using the maxmMd approach, ie, separating chemicals that affected progestagens and/or corticosteroids only (ie, an absence of effects on any other hormones in the pathway) into a list for future consideration. However, based on the current chemical set screened in HT-H295R in concentration-response, it is unlikely that many chemicals affect only 1 steroid hormone class in the H295R assay. From the 654 chemicals with data for the maxmMd analysis, 596 chemicals had positive maxmMd responses, and of these, 10 chemicals affected only estrogen and/or androgen synthesis, and 67 chemicals affected only progestagen and/or corticosteroid synthesis. One conclusion from the Venn diagram presented in Figure 3 is that though it is possible to identify chemicals that only perturb estrogen and/or androgen synthesis, most of the chemicals in the screened set affected other steroid hormones as well, and using these data to evaluate the magnitude of overall pathway effect appears useful. Interestingly, examples of the chemicals that affected only synthesis of progestagens and/or corticosteroids include butylparaben, in line with an independent report of its activity in the H295R model (Taxvig et al., 2008), and prednisone, which has known clinical interactions with the mineralocorticoid receptor as a glucocorticoid prodrug (Ferraldeschi et al., 2013). Thus, excluding chemicals that only affect progestagen and/or corticosteroid synthesis from prioritization tasks may exclude chemicals with activities of potential interest, and using the maxmMd approach for the whole pathway would be more inclusive. The ratio of observed positives in this screened chemical set by steroid hormone class might shift if a naı¨ve screening approach was taken without pre-selecting positive chemicals based on single concentration screening. However, this high rate of pathway positives in this pre-selected set demonstrates the original success of the HT-H295R ToxCast screening workflow in terms of identifying chemicals that may disrupt steroidogenesis by performing single concentration screening followed by concentration-response screening.
The work described herein demonstrates the performance of the HT-H295R assay as an alternative to the OECD TG 456 H295R assay, and proposes use of a novel statistical approach to integrate the information from 11 steroid hormones in the pathway to yield a relative rank of steroidogenesis perturbation. The approach based on Mahalanobis distances accounts for the correlation of the residuals of the hormone measures. A clear advantage of the mean Mahalanobis distance approach is that the concentration at which effects across the pathway begin to occur can be identified. The pathway analysis approach reduced an 11-dimensional analysis to a single dimension and appears to increase the sensitivity of detecting chemicals that are known to perturb the steroidogenesis pathway expressed in H295R cells. The prioritization metric derived, the maxmMd, demonstrated increased reproducibility on a per chemical basis when compared to responses using the ANOVA results. Further, the maxmMd appears to provide a single metric that relates to efficacy and potency, allowing for discrimination of strong positives from weak or negative reference chemicals. As the data-base of reference chemicals for perturbation of in vitro steroidogenesis grows, further characterization of the strengths and weaknesses of this approach will develop. Potential use of the maxmMd in prioritization tasks represents a data-driven option for evaluating lists of chemicals for putative effects on steroidogenesis.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge Ann Richard, Indira Thillainadarajah, and Antony Williams for assistance with chemical curation and annotation of nonoxynol-9; Nathaniel Rush for assistance with the MTT data extraction and analysis; and, Briana Franz of Cyprotex for assistance in interpreting assay protocols. They would also like to thank Russell Thomas, Kevin Crofton, Keith Houck, Christopher Grulke, and Nicole Kleinstreuer for insightful comments on previous versions of this manuscript.
FUNDING
D.E.H. was supported by appointment to the Research Participation Program of the U.S. Environmental Protection Agency, Office of Research and Development, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. EPA.
Footnotes
Disclaimer: The United States Environmental Protection Agency (U.S. EPA) through its Office of Research and Development has subjected this article to Agency administrative review and approved it for publication. Mention of trade names or commercial products does not constitute endorsement for use. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the US EPA.
SUPPLEMENTARY DATA
Supplemental Files available at DRYAD DOI: https://doi.org/10.5061/dryad.385j7.
REFERENCES
- Abdel-Khalik J, Bjorklund E, and Hansen M (2013). Development of a solid phase extraction method for the simultaneous determination of steroid hormones in H295R cell line using liquid chromatography-tandem mass spectrometry. J. Chromatog.r B Analyt. Technol. Biomed. Life Sci 935, 61–69. [DOI] [PubMed] [Google Scholar]
- Ankley GT, and Jensen KM (2014). A novel framework for interpretation of data from the fish short-term reproduction assay (FSTRA) for the detection of endocrine-disrupting chemicals. Environ. Toxicol. Chem 33, 2529–2540. [DOI] [PubMed] [Google Scholar]
- Asser L, Hescot S, Viengchareun S, Delemer B, Trabado S, and Lombes M (2014). Autocrine positive regulatory feedback of glucocorticoid secretion: Glucocorticoid receptor directly impacts H295R human adrenocortical cell function. Mol. Cell. Endocrinol 395, 1–9. [DOI] [PubMed] [Google Scholar]
- Blystone CR, Lambright CS, Howdeshell KL, Furr J, Sternberg RM, Butterworth BC, Durhan EJ, Makynen EA, Ankley GT, Wilson VS, et al. (2007). Sensitivity of fetal rat testicular steroidogenesis to maternal prochloraz exposure and the underlying mechanism of inhibition. Toxicol. Sci 97, 512–519. [DOI] [PubMed] [Google Scholar]
- Breen M, Villeneuve DL, Ankley GT, Bencic DC, Breen MS, Watanabe KH, Lloyd AL, and Conolly RB (2013). Developing predictive approaches to characterize adaptive responses of the reproductive endocrine axis to aromatase inhibition: II. Computational modeling. Toxicol. Sci 133, 234–247. [DOI] [PubMed] [Google Scholar]
- Breen MS, Breen M, Terasaki N, Yamazaki M, and Conolly RB (2010). Computational model of steroidogenesis in human H295R cells to predict biochemical response to endocrine-active chemicals: Model development for metyrapone. Environ. Health Perspect 118, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, and Thomas RS (2015). Screening chemicals for estrogen receptor bioactivity using a computational model. Environ. Sci. Technol 49, 8804–8814. [DOI] [PubMed] [Google Scholar]
- CDC (2009). Fourth National Report on Human Exposure to Environmental Chemicals Available at: https://www.cdc.gov/exposurereport/pdf/FourthReport.pdf. Accessed August 11, 2017.
- Charlier TD, Cornil CA, and Balthazart J (2013). Rapid modulation of aromatase activity in the vertebrate brain. J. Exp. Neurosci 7, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conolly RB, Ankley GT, Cheng W, Mayo ML, Miller DH, Perkins EJ, Villeneuve DL, and Watanabe KH (2017). Quantitative adverse outcome pathways and their application to predictive toxicology. Environ. Sci. Technol 51, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornil CA, Seredynski AL, de Bournonville C, Dickens MJ, Charlier TD, Ball GF, and Balthazart J (2013). Rapid control of reproductive behaviour by locally synthesised oestrogens: Focus on aromatase. J. Neuroendocrinol 25, 1070–1078. [DOI] [PubMed] [Google Scholar]
- De Maesschalck R, Jouan-Rimbaud D, and Massart DL (2000). The Mahalanobis distance. Chemometr. Intell. Lab 50, 1–18. [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, and Kavlock RJ (2007). The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci 95, 5–12. [DOI] [PubMed] [Google Scholar]
- EPA (2011). Endocrine Disruptor Screening Program for the 21st Century: EDSP21 Work Plan; The Incorporation of In Silico Models and In Vitro High Throughput Assays in the Endocrine Disruptor Screening Program (EDSP) for Prioritization and Screening https://www.epa.gov/sites/production/files/2015-07/documents/edsp21_work_plan_summary_overview_final.pdf. Retrieved June 23, 2017.
- EPA (2009). OPPTS 890.1550: Steroidogenesis (Human Cell Line H295R). Office of Prevention, and Toxic Substances Docket # EPA-HQ-OPPT-2009-05676-0011.pdf, Endocrine Disruptor Screening Program Test Guidelines. [Google Scholar]
- EPA (2014). Integrated Bioactivity and Exposure Ranking: A Computational Approach for the Prioritization and Screening of Chemicals in the Endocrine Disruptor Screening Program Available at: https://www.regulations.gov/document?D=EPA-HQ-OPP-2014-0614-0003; last accessed August 11, 2017.
- EPA (2015a). Endocrine Disruptor Screening Program Tier 1 Screening Determinations and Associated Data Evaluation Records Available at: https://www.epa.gov/endocrine-disruption/en-docrine-disruptor-screening-program-tier-1-screening-determinations-and. Accessed October 18, 2017.
- EPA (2015b). ToxCast and Tox21 Data from invitrodb_v2 Available at: http://www2.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data. Accessed October 28, 2015.
- Ferraldeschi R, Sharifi N, Auchus RJ, and Attard G (2013). Molecular pathways: Inhibiting steroid biosynthesis in prostate cancer. Clin. Cancer Res 19, 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer DL, Kothiya P, Setzer RW, Judson RS, and Martin MT (2017). tcpl: The ToxCast pipeline for high-throughput screening data. Bioinformatics 33(4), 618–620. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Oie HK, Shackleton CH, Chen TR, Triche TJ, Myers CE, Chrousos GP, Brennan MF, Stein CA, and La Rocca RV (1990). Establishment and characterization of a human adrenocortical carcinoma cell line that expresses multiple pathways of steroid biosynthesis. Cancer Res 50, 5488–5496. [PubMed] [Google Scholar]
- Gerardin DC, and Pereira OC (2002). Reproductive changes in male rats treated perinatally with an aromatase inhibitor. Pharmacol. Biochem. Behav 71, 301–305. [DOI] [PubMed] [Google Scholar]
- Goetz AK, Rockett JC, Ren H, Thillainadarajah I, and Dix DJ (2009). Inhibition of rat and human steroidogenesis by triazole antifungals. Syst. Biol. Reprod. Med 55, 214–226. [DOI] [PubMed] [Google Scholar]
- Gracia T, Hilscherova K, Jones PD, Newsted JL, Zhang X, Hecker M, Higley EB, Sanderson JT, Yu RM, Wu RS, et al. (2006). The H295R system for evaluation of endocrine-disrupting effects. Ecotoxicol. Environ. Saf 65, 293–305. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Larsen LW, Sorensen AM, Halling-Sorensen B, and Styrishave B (2017). The six most widely used selective serotonin reuptake inhibitors decrease androgens and increase estrogens in the H295R cell line. Toxicol. in Vitro 41, 1–11. [DOI] [PubMed] [Google Scholar]
- Hecker M, et al. (2017). Erratum to: The OECD validation program of the H295R steroidogenesis assay: Phase 3. Final inter-laboratory validation study. Environ Sci Pollut Res Int [DOI] [PubMed]
- Hecker M, Giesy J, and Timm G (2008). Multi-Laboratory Validation of the H295R Steroidogenesis Assay to Identify Modulators of Testosterone and Estradiol Production, 89 pp. Report prepared for the U.S. Environmental Protection Agency. [Google Scholar]
- Hecker M, Hollert H, Cooper R, Vinggaard AM, Akahori Y, Murphy M, Nellemann C, Higley E, Newsted J, Laskey J, et al. (2011). The OECD validation program of the H295R steroidogenesis assay: Phase 3. Final inter-laboratory validation study. Environ. Sci. Pollut. Res. Int 18, 503–515. [DOI] [PubMed] [Google Scholar]
- Hecker M, Newsted JL, Murphy MB, Higley EB, Jones PD, Wu R, and Giesy JP (2006). Human adrenocarcinoma (H295R) cells for rapid in vitro determination of effects on steroidogenesis: Hormone production. Toxicol. Appl. Pharmacol 217, 114–124. [DOI] [PubMed] [Google Scholar]
- Hilscherova K, Jones PD, Gracia T, Newsted JL, Zhang X, Sanderson JT, Yu RM, Wu RS, and Giesy JP (2004). Assessment of the effects of chemicals on the expression of ten steroidogenic genes in the H295R cell line using real-time PCR. Toxicol. Sci 81, 78–89. [DOI] [PubMed] [Google Scholar]
- Hornung RW, and Reed LD (1990). Estimation of average concentration in the presence of nondetectable values. Appl. Occup. Environ. Hyg 5, 46–51. [Google Scholar]
- Juberg DR, Gehen SC, Coady KK, LeBaron MJ, Kramer VJ, Lu H, and Marty MS (2013). Chlorpyrifos: Weight of evidence evaluation of potential interaction with the estrogen, androgen, or thyroid pathways. Regul. Toxicol. Pharmacol 66, 249–263. [DOI] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, et al. (2010). In vitro screening of environmental chemicals for targeted testing prioritization: The ToxCast project. Environ. Health Perspect 118, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia MH, Huang RL, Rotroff DM, Filer DL, et al. (2015). Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmaus AL, and Zacharewski TR (2015). Atrazine-Mediated Disruption of Steroidogenesis in BLTK1 Murine Leydig Cells. Toxicol. Sci 148(2), 544–554. [DOI] [PubMed] [Google Scholar]
- Karmaus AL, Toole CM, Filer DL, Lewis KC, and Martin MT (2016). High-throughput screening of chemical effects on steroidogenesis using h295r human adrenocortical carcinoma cells. Toxicol. Sci 150, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, et al. (2012). Update on EPA’s ToxCast program: Providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Ceger P, Watt ED, Martin M, Houck K, Browne P, Thomas RS, Casey WM, Dix DJ, Allen D, et al. (2017). Development and validation of a computational model for androgen receptor activity. Chem. Res. Toxicol 30, 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucka M, et al. (2012). “Atrazine acts as an endocrine disrupter by inhibiting cAMP-specific phosphodiesterase-4.” Toxicol. Appl. Pharmacol 265(1), 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBaron MJ, Coady KK, O’Connor JC, Nabb DL, Markell LK, Snajdr S, and Sue Marty M (2014). Key learnings from performance of the U.S. EPA Endocrine Disruptor Screening Program (EDSP) Tier 1 in vitro assays. Birth Defects Res. B Dev. Reprod. Toxicol 101, 23–42. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Kuhn M, Chapin RE, and Pletcher MT (2014). More than just hormones: H295R cells as predictors of reproductive toxicity. Reprod. Toxicol 45, 77–86. [DOI] [PubMed] [Google Scholar]
- Mahalanobis PC (1936). On the generalized distance in statistics. Proc. Natl. Inst. Sci. India 2, 49–55. [Google Scholar]
- Mardia KV, Kent JT, Bibby JM (1979). Multivariate analysis New York, NY: Academic Press. [Google Scholar]
- Miller WL, and Auchus RJ (2011). The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev 32, 81–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, and Imada T (2005). Multiple comparison procedure of Dunnetts type for multivariate normal means. JJSCS 18, 21–32. [Google Scholar]
- Nielsen FK, Hansen CH, Fey JA, Hansen M, Jacobsen NW, Halling-Sorensen B, Bjorklund E, and Styrishave B (2012). H295R cells as a model for steroidogenic disruption: A broader perspective using simultaneous chemical analysis of 7 key steroid hormones. Toxicol. in Vitro 26, 343–350. [DOI] [PubMed] [Google Scholar]
- OECD (2011). Test No. 456: H295R Steroidogenesis Assay OECD Publishing, Paris. [Google Scholar]
- Oskarsson A, Ulleras E, and Ohlsson Andersson A (2016). acetaminophen increases aldosterone secretion while suppressing cortisol and androgens: A possible link to increased risk of hypertension. Am. J. Hypertens 29, 1158–1164. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K, Papineni S, Marty MS, Yi KD, Goetz AK, Rasoulpour RJ, Kwiatkowski P, Wolf DC, Blacker AM, and Peffer RC (2016). A predictive data-driven framework for endocrine prioritization: A triazole fungicide case study. Crit. Rev. Toxicol 46, 785–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot L, Schneider M, DeCristofaro M, Samadfam R, Smith SY, and Beckman DA (2013). Assessment of a nonsteroidal aromatase inhibitor, letrozole, in juvenile rats. Birth Defects Res. B Dev. Reprod. Toxicol 98, 374–390. [DOI] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, et al. (2016). ToxCast chemical landscape: Paving the road to 21st century toxicology. Chem. Res. Toxicol 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Rijk JC, Peijnenburg AA, Blokland MH, Lommen A, Hoogenboom RL, and Bovee TF (2012). Screening for modulatory effects on steroidogenesis using the human H295R adrenocortical cell line: A metabolomics approach. Chem. Res. Toxicol 25, 1720–1731. [DOI] [PubMed] [Google Scholar]
- Robitaille CN, Rivest P, and Sanderson JT (2015). Antiandrogenic mechanisms of pesticides in human LNCaP prostate and H295R adrenocortical carcinoma cells. Toxicol. Sci 143, 126–135. [DOI] [PubMed] [Google Scholar]
- Saito R, Terasaki N, Yamazaki M, Masutomi N, Tsutsui N, and Okamoto M (2016). Estimation of the mechanism of adrenal action of endocrine-disrupting compounds using a computational model of adrenal steroidogenesis in NCI-H295R cells. J. Toxicol doi: 10.1155/2016/4041827. [DOI] [PMC free article] [PubMed]
- Sanderson JT, Boerma J, Lansbergen GW, and van den Berg M (2002). Induction and inhibition of aromatase (CYP19) activity by various classes of pesticides in H295R human adrenocortical carcinoma cells. Toxicol. Appl. Pharmacol 182, 44–54. [DOI] [PubMed] [Google Scholar]
- Sanderson JT, Seinen W, Giesy JP, and van den Berg M (2000). 2-Chloro-s-triazine herbicides induce aromatase (CYP19) activity in H295R human adrenocortical carcinoma cells: A novel mechanism for estrogenicity? Toxicol. Sci 54, 121–127. [DOI] [PubMed] [Google Scholar]
- SAS (2012). What Is Mahalanobis Distance? http://blogs.sas.com/content/iml/2012/02/15/what-is-mahalanobis-distance.html. Accessed June 23, 2017.
- Strajhar P, Tonoli D, Jeanneret F, Imhof RM, Malagnino V, Patt M, Kratschmar DV, Boccard J, Rudaz S, and Odermatt A (2017). Steroid profiling in H295R cells to identify chemicals potentially disrupting the production of adrenal steroids. Toxicology 381, 51–63. [DOI] [PubMed] [Google Scholar]
- Taxvig C, Vinggaard AM, Hass U, Axelstad M, Boberg J, Hansen PR, Frederiksen H, and Nellemann C (2008). Do parabens have the ability to interfere with steroidogenesis?. Toxicol. Sci 106, 206–213. [DOI] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, and Bucher JR (2013). Improving the human hazard characterization of chemicals: A Tox21 update. Environ. Health Perspect 121, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinfo NS, Hotchkiss MG, Buckalew AR, Zorrilla LM, Cooper RL, and Laws SC (2011). Understanding the effects of atrazine on steroidogenesis in rat granulosa and H295R adrenal cortical carcinoma cells. Reprod. Toxicol 31, 184–193. [DOI] [PubMed] [Google Scholar]
- Tonoli D, Furstenberger C, Boccard J, Hochstrasser D, Jeanneret F, Odermatt A, and Rudaz S (2015). Steroidomic footprinting based on ultra-high performance liquid chromatography coupled with qualitative and quantitative high-resolution mass spectrometry for the evaluation of endocrine disrupting chemicals in H295R cells. Chem. Res. Toxicol 28, 955–966. [DOI] [PubMed] [Google Scholar]
- Vierk R, Brandt N, and Rune GM (2014). Hippocampal estradiol synthesis and its significance for hippocampal synaptic stability in male and female animals. Neuroscience 274, 24–32. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Ankley GT, Makynen EA, Blake LS, Greene KJ, Higley EB, Newsted JL, Giesy JP, and Hecker M (2007). Comparison of fathead minnow ovary explant and H295R cell-based steroidogenesis assays for identifying endocrine-active chemicals. Ecotoxicol. Environ. Saf 68, 20–32. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Breen M, Bencic DC, Cavallin JE, Jensen KM, Makynen EA, Thomas LM, Wehmas LC, Conolly RB, and Ankley GT (2013). Developing predictive approaches to characterize adaptive responses of the reproductive endocrine axis to aromatase inhibition: I. Data generation in a small fish model. Toxicol. Sci 133, 225–233. [DOI] [PubMed] [Google Scholar]
- Yanes LL, and Romero DG (2009). Dihydrotestosterone stimulates aldosterone secretion by H295R human adrenocortical cells. Mol. Cell. Endocrinol 303, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TDY, and Oldenburg KR (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chang H, Wiseman S, He Y, Higley E, Jones P, Wong CK, Al-Khedhairy A, Giesy JP, and Hecker M (2011). Bisphenol A disrupts steroidogenesis in human H295R cells. Toxicol. Sci 121, 320–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


