Abstract
Mitochondrial homeostasis is tightly regulated by two major processes: mitochondrial biogenesis and mitochondrial degradation by autophagy (mitophagy). Research in mitochondrial biogenesis in skeletal muscle in response to endurance exercise training has been well established, while the mechanisms regulating mitophagy and the interplay between mitochondrial biogenesis and degradation following endurance exercise training are not yet well defined. The purpose of this study was to examine the effects of a short-term inhibition of autophagy in response to acute endurance exercise on skeletal muscle mitochondrial biogenesis and dynamics in an exercise-trained condition. Male wild-type C57BL/6 mice performed five daily bouts of 1-h swimming per week for 8 weeks. In order to measure autophagy flux in mouse skeletal muscle, mice were treated with or without 2 days of 0.4 mg/kg/day intraperitoneal colchicine (blocking the degradation of autophagosomes) following swimming exercise training. The autophagic flux assay demonstrated that swimming training resulted in an increase in the autophagic flux (~100 % increase in LC3-II) in mouse skeletal muscle. Mitochondrial fusion proteins, Opa1 and MFN2, were significantly elevated, and mitochondrial fission protein, Drp1, was also increased in trained mouse skeletal muscle, suggesting that endurance exercise training promotes both mitochondrial fusion and fission processes. A mitochondrial receptor, Bnip3, was further increased in exercised muscle when treated with colchicine while Pink/Parkin protein levels were unchanged. The endurance exercise training induced increases in mitochondrial biogenesis marker proteins, SDH, COX IV, and a mitochondrial biogenesis promoting factor, PGC-1α but this effect was abolished in colchicine-treated mouse skeletal muscle. This suggests that autophagy plays an important role in mitochondrial biogenesis and this coordination between these opposing processes is involved in the cellular adaptation to endurance exercise training.
Keywords: Autophagy, Mitochondrial biogenesis, Mitophagy, Exercise, Skeletal muscle
Introduction
Macroautophagy (autophagy) is a bulk degradation process that mediates the clearance of long-lived proteins and organelles. This process requires the following steps: (1) phagophore initiation and elongation, (2) autophagosome formation, (3) autophagosome fusion with acidic lysosomes, and (4) lysosomal degradation [1]. In skeletal muscle, autophagy is activated by stimuli such as nutrient deprivation, drugs (e.g., rapamycin) and exercise [2]. Autophagy can also selectively target protein aggregates and organelles including mitochondria. Mitochondria are very dynamic organelles that are continuously remodeled through regulated turnover [3]. Mitochondrial turnover occurs by three mechanisms; (1) mitochondrial biogenesis, (2) mitochondrial fusion and fission, (3) degradation by autophagy, which is called mitophagy [4].
Endurance exercise has long been recognized to increase mitochondrial content assessed by the expression of mitochondrial markers (e.g., cytochrome c oxidase-IV, COX IV) and mitochondrial function measured by mitochondrial enzyme activities (e.g., citrate synthase, CS) and whole-body peak oxygen consumption [5]. Mitochondrial biogenesis induced by exercise training can be promoted by the action of the transcriptional co-activator, PGC-1α. Activated PGC-1α controls the expression of genes encoding proteins involved in mitochondrial biogenesis, oxidative phosphorylation, and other features of oxidative muscle fibers [6].
Mitochondria undergo successive rounds of fusion and fission with a dynamic exchange of components to segregate functional and damaged elements [4]. Mitochondrial fission is regulated by two proteins: fission protein 1 (Fis1) and dynamin-related protein 1 (Drp1). In contrast, mitochondrial fusion requires two outer membrane proteins, mitofusin 1 and 2 (MFN1, MFN2) and optic atrophy 1 (Opa1) [7]. Despite insufficient research, exercise training appears to promote both mitochondrial fusion and fission processes in skeletal muscle [8, 9]. However, it has been reported that excessive mitochondrial fission was linked to mitochondrial degradation [10].
Mitophagy is a highly selective process that can promote the elimination of dysfunctional or unnecessary mitochondria [11]. Mitophagy requires two processes; (1) recognizing damaged mitochondria, and (2) inducing autophagy [12]. Selective mitophagy is recognized by PINK1/Parkin signaling system or Bnip3 (Bcl-2 and 19 kDa interacting protein-3) and NIX (Nip3-likeprotein X). Mitophagy by PINK1/Parkin recognizes damaged mitochondria through poly-ubiquitination of mitochondrial proteins. In contrast, Bnip3 and NIX are mitochondrial proteins and function as mitophagy receptors recruit autophagosomes for clearance [13].
Several recent studies have implicated that autophagy/mitophagy plays an important role in skeletal muscle adaptation to endurance exercise and interacts with mitochondrial biogenesis, for example, mice with deficient/defective autophagy (e.g., Becn1 +/−, Atg6 +/−, and Atg7 −/−) have been shown to result in an attenuation of exercise-induced metabolic benefits, decreased in mitochondrial function and exercise performance [14–17]. Additionally, autophagy has also been shown to be associated with fiber type shifting from type IIX to IIA after exercise training [18] and prevention of mitochondrial damage during exercise, especially eccentric contraction [16]. PGC-1α involved in mitochondrial biogenesis appears to play a role in the regulation of exercise-induced autophagy. Mice with muscle-specific overexpression of PGC-1α have elevated basal autophagy and mitophagy protein expression in plantaris muscle [15] while PGC-1α knockout mice have been demonstrated attenuated exercise-induced autophagy/mitophagy signaling and flux [17]. Nonetheless, the mechanisms regulating autophagy/mitophagy and the crosstalk between mitochondrial biogenesis and degradation following endurance exercise training are not yet well defined. Therefore, it seems important to study further the intimate relationship between mitochondrial synthesis and degradation processes in response to exercise training. The purpose of this study was to examine the effects of basal autophagy/mitophagy flux on skeletal muscle mitochondrial biogenesis and dynamics in response to endurance exercise in an exercise-trained condition. In this current study, by using the autophagy flux strategy which is an in vivo autophagy/mitophagy marker turnover assay with or without colchicine (blocking autophagy) administration described previously [19], we have evaluated whether 8 weeks of swimming exercise training enhances basal autophagy simultaneously with changes in mitochondrial biogenesis and dynamics in mouse skeletal muscle. We have also examined whether a short-term inhibition of autophagy/mitophagy flux influences skeletal muscle mitochondrial biogenesis and dynamics in response to endurance exercise in an exercise-trained condition.
Materials and methods
Materials and antibodies
Reagents for SDS-PAGE were from Bio-Rad Laboratories. Reagents for ECL were obtained from Thermo Fisher Scientific (32106). Colchicine (C9754), anti-LC3B (L7543) and anti-actin (A2066) polyclonal antibodies were purchased from Sigma-Aldrich (www.sigmaaldrich.com). Anti-Bnip3 (13795), anti-Beclin-1 (3738), anti-pyruvate dehydrogenase (2784), anti-succinate dehydrogenase (SDH, 5839) and anti-COX IV (4844) antibodies were from Cell Signaling Technology (www.cellsignal.com). Anti-LAMP1 (MABC39) antibodies were from Millipore (www.emdmillipore.com). Anti-FIS1 (10956-1-AP), p62/SQSTM1 (18420-1-AP), Cytochrome C (10993-1-AP) polyclonal antibodies were from Proteintech (www.ptglab.com). Anti-PINK1 (ab75487), anti-Parkin (ab15954), anti-δ-ALA synthase (ab84962) antibodies were from Abcam (www.abcam.com). Anti-Atg7 (BS6046) polyclonal antibodies were purchased from Bioworld Technology (www.bioworld.com). Mitochondrial Isolation Kit (K288-50) and Citrate Synthase Activity Colorimetric Assay Kit (K318-100) were from BioVision, Inc. (www.biovision.com).
Animals
Ten-week-old male wild-type C57BL/6 mice were obtained from Samtaco BioKorea (Korea) and used in this study. The animals were housed four per cage in a temperature (~22 °C) and light-controlled environment with a 12:12-h light–dark cycle and provided with food (a normal chow diet) and water ad libitum. After 1-week acclimation period, the animals were divided into two groups: sedentary control and swimming training. All protocols for animals use and euthanasia were approved by the University of Suwon Animal Research Ethics Board.
Training program
The mice were accustomed to swimming for 15 min/day for 2 days. An endurance exercise training program was conducted 5 days/week, for 8 weeks. The program consisted of 1 h of swimming. Swimming was performed in the fed state in a plastic tank (45 × 60 × 40 cm) filled with ~30 cm in depth water, maintained at a temperature of 35–36 °C. Sedentary control animals were handled and immersed for a few minutes in the warm water as the trained mice in order to submit them similar stress. In order to measure autophagic flux in mouse skeletal muscle, mice were treated with saline or the microtubule depolarizing agent, colchicine (0.4 mg/kg/day, intraperitoneally) which blocks autophagosome degradation for 2 days prior to sacrifice [19]. In the last day of swimming training following the last exercise bout, sedentary control mice (n = 16) were divided into two groups: sedentary plus saline (Sed+Sal, n = 8) and sedentary plus colchicine (Sed+Col, n = 8). Trained mice (n = 16) were also grouped into two: exercise plus saline (Exe+Sal, n = 8) and exercise plus colchicine (Exe+Col, n = 8). To avoid the effect of acute exercise, triceps muscles were harvested 48 h later following the last bout of swimming exercise.
Immunoblot analysis
Homogenates for immunoblot analysis were made by grinding muscle tissues with ice-cold RIPA lysis buffer (150 mM NaCl; 10 mM Tris–HCl, pH 7.2; 0.1 % Triton X-100; 1 % sodium deoxycholate; 5 mM EDTA) containing protease inhibitor cocktail (Sigma-Aldrich) with grinding tubes resting in ice-water baths. Homogenates were centrifuged at 12,000g for 10 min. The protein concentration of the supernatant was quantitated using a BCA protein assay kit (Thermo Fisher Scientific). Aliquots of homogenate were further solubilized in Laemmli sample buffer and boiled for 5 min, and 20–80 μg of protein was subjected to SDS-PAGE (10–12 % resolving gel). Proteins were transferred to nitrocellulose membranes (Trans-Blot; 0.2-μm nitrocellulose; Bio-Rad Laboratories). Membranes were blocked in a solution of Tris-buffered saline containing 5 % nonfat dry milk. Membranes were incubated with a 1:2000 dilution of LC3B antibody in 1 % milk and TBS-T, a 1:2500 dilution of anti-p62 antibody, a 1:5000 dilution of anti-actin antibody, or a 1:1000 dilution of other antibodies. After incubation with the appropriate secondary antibody (horseradish peroxidase–conjugated IgG) (Enzo Life Sciences), bands were visualized by ECL solution. Immunoblots were scanned with a film scanner (Scanjet G4010; HP), and images were collected in Photoshop CS3. Densitometry was measured with ImageJ (NIH).
RT-PCR
Total RNA from triceps muscle isolated from mice was extracted using E.Z.N.A™ DNA/RNA Kit (Omega Bio-tek) and purified following the manufacturer’s instructions. Complementary DNA was generated with a PrimeScript Reverse Transcriptase (TAKARA) and was analyzed by quantitative real-time RT-PCR using AmpiGene aPCR Green Mix (Enzo Life Sciences) and a Lightcycler480 (Roche Applied Science). Expression levels, calculated as copy number in each sample, were normalized to the expression level of GAPDH. Sequences of primers were used as follows: p62, (f) 5′-CCAGAGGGTCCACTGTGACT-3′ and (r) 5′-CTGACTCCCCTTGACTCTGG-3′; Bnip3, (f) 5′-GTCTCATCTGCTGGCCATTG-3′ and (r) 5′-AACACCCAAGGACCATGCTA-3′; PGC-1α, (f) 5′-AAGTGTGGAACTCTCTGGAACTG-3′ and (r) 5′-GGGTTATCTTGGCTTTATG-3′; Parkin, (f) 5′-TGGAAAGCTCCGAGTTCAGT-3′ and (r) 5′-CCTTGTCTGAGGTTGGGTGT-3′; PINK1, (f) 5′-CCCACACCCTAACATCATCC-3′ and (r) 5′-ACTGGGAGTCTGCTCCTCAA -3′; GAPDH, (f) 5′-GGCATTGTGAAGGGCTCAT-3′ and (r) 5′-GACACATTGGGGTAGGAACAC-3′.
Quantification of mitochondrial DNA (mtDNA)
Total DNA containing nuclear and mtDNA was extracted using E.Z.N.A™ DNA/RNA Kit (Omega Bio-tek). Real-time PCR was performed as described above to quantify mtDNA (16S rRNA). The copy number of mtDNA was normalized with nuclear DNA (hexokinase 2). Sequences of primers were used as follows: 16S rRNA: (f) 5′- CCGCAAGGGAAAGATGAAAGAC-3′; (r) 5′-TCGTTTGGTTTCGGGGTTTC-3′ and hexokinase 2, intron 9: (f) 5′-GCCAGCCTCTCCTGATTTTAGTGT-3′; (r) 5′-GGGAACACAAAAGACCTCTTCTGG-3′.
Mitochondrial isolation and citrate synthase activity assay
Mitochondria were isolated using the Mitochondria Isolation Kit (BioVision, Milpitas, CA, USA). Approximately 200 mg of the tissue in 1 ml of ice-cold Mitochondrial Isolation Buffer was used to mince the tissue on ice into small pieces using scissors. The minced tissue was centrifuged in a tabletop centrifuge at 10,000 × g for 2 min. The buffer was discarded and replaced with 1 ml of fresh ice-cold Mitochondrial Isolation Buffer. The tissue in Mitochondria Isolation Buffer was ground using a precooled glass homogenizer. The homogenate was transferred to a tube and centrifuged at 600 × g for 10 min at 4 °C. The resulting supernatant was collected in a separate tube and centrifuged at 7000 × g for 10 min at 4 °C. The supernatant (cytosolic fraction) was collected and stored at −80 °C while the mitochondria pellet was washed with Mitochondria Isolation Buffer and resuspended in 0.1 ml of Storage Buffer and stored at −80 °C until analyses were performed. Mitochondria were kept in ice bath throughout the isolation process. CS activity from isolated mitochondria was determined using a citrate assay kit (BioVision, Milpitas, CA, USA) according to the protocol provided by the manufacturer. Isolated mitochondria samples (10 μl) and 2 mM GSH Standard (0, 4, 8, 12, 16, 20 μl) was added into a 96-well plate and the volume was adjusted to 50 μl with CS Assay Buffer in the kit. The principle of the assay was to initiate the reaction of acetyl-CoA with Oxaloacetate and link the release of free CoA-SH to a colorimetric reagent, which generates the colored product. The rate change in color was monitored at wavelength of 412 nm at 60-s intervals for a period of 30 min by using an Epoch Microplate Spectrophotometer (Bio-Tek). All measurements were performed in duplicate, in the same setting at 20–22 °C. Two time points (T 1 and T 2) were selected in the linear range to calculate the enzyme activity of the samples. The solubilized protein extracts of the homogenates were quantified in triplicate by using a BCA protein assay kit (Thermo Fisher Scientific) and bovine serum albumin standards. The CS activity was then normalized to the total protein content and was reported in as micromoles per gram protein per minute (μmol/g/min).
Statistical analysis
Data are presented as means ± SE and were evaluated by a univariate analysis of variance (ANOVA) followed by Fisher’s LSD post hoc comparisons at p < 0.05.
Results
Body weight did not differ among the groups before the beginning of the exercise protocol. At the end of training, body weight gain was lower in the swimming exercise group (10.2 ± 0.5 %) compared to the sedentary control group (16.5 ± 0.6 %) (p < 0.05) (Table 1). In contrast, the food intake of the swimming exercise group was greater compared to that of the sedentary control group. Table 2 shows that the swimming exercise mice, despite consumption of significantly more food compared to that of the sedentary mice, underwent a smaller weight increase (Table 2). There were no changes in body weight and food consumption after 2 days of colchicine treatment. We measured the concentrations of mitochondrial marker enzymes, to see whether the swimming exercise training induced an increase in mitochondrial biogenesis in triceps muscle. As shown in Fig. 1, 8 weeks of exercise training induced significant increases in δ-ALA synthase (delta-aminolevulinic synthase) (~88 %) and cytochrome C protein (~53 %) concentrations (p < 0.05).
Table 1.
Body weight was measured approximately weekly for 8 weeks following the start of the swimming exercise protocol
| Pre (g) | Post (g) | Increase (%) | |
|---|---|---|---|
| Sedentary | 25.4 ± 0.3 | 29.6 ± 0.4 | 16.5 ± 0.3 |
| Exercise | 2505 ± 0.4 | 28.1 ± 0.2 | 10.2 ± 0.6* |
Changes in body weights are shown using two time points, pre- and post-training
Values represent the mean ± SE; * p < 0.05 vs. sedentary controls
Table 2.
Food intake is reported as average grams of chow food per mouse each day
| Sedentary | Exercise | |
|---|---|---|
| g/mouse | 4.12 ± 0.1 | 4.88 ± 0.3* |
A significant difference was assessed by unpaired t test
Values represent the mean ± SE; * p < 0.05 vs. sedentary controls
Fig. 1.
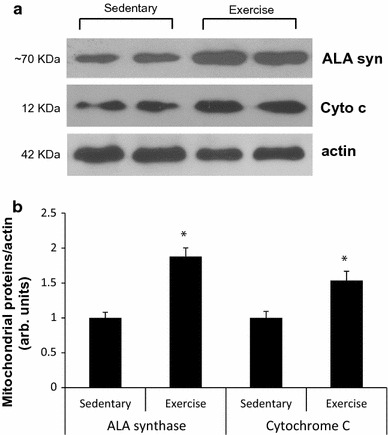
Eight-week swimming exercise training increases expression of mitochondrial enzymes in triceps muscle. Representative blots are shown at the top of the figure (a). Average ALA synthase and cytochrome C protein values in muscles of control and exercised mice (b). Each bar represents the mean ± SE for muscles from eight mice. *p < 0.05 vs. sedentary controls
Next, to determine whether the swimming training provides an adequate stimulus for inducing an increase in autophagy, we measured the levels of autophagosome marker proteins, LC3 (microtubule-associated protein 1 light chain 3) and p62 (SQSTM1, sequestosome) by using autophagic flux assay in mouse triceps muscle. Conversion of LC3 I into LC3 II and the degradation of LC3 II have been frequently evaluated by immunoblot. However, static levels of LC3 II protein may render possible misunderstandings since LC3 II levels can increase, decrease, or remain unchanged in the setting of autophagic induction [19]. To avoid this, we treated 2-month-old male mice with intraperitoneal (i.p.) saline or 0.4 mg/kg/day colchicine for 2 days (on the last day and one day after exercise training) prior to sacrifice. Colchicine administration for 2 days was able to effectively block autophagosome degradation in vivo [19], and this strategy allowed us to accurately quantitate autophagic flux which denotes the rates of initiation and resolution of autophagic events [14, 20] in an in vivo system. As shown in Fig. 2, colchicine treatment increased LC3-II proteins levels significantly in two groups, both the Sed+Col and the Exe+Col groups compared with the Sed+Sal group in skeletal muscle of mice. The protein levels of LC3-II were significantly higher (~100 %) in the Exe+Col group than the Sed + Col group (p < 0.05) (Fig. 2a, b). Similarly, swimming exercise training led to a further increase in the ratio of LC3-II/LC3-I (the conversion of the nonlipidated form of LC3 I into the autophagosome membrane-bound form of LC3 II) than colchicine alone (the Sed+Col group) (p < 0.05) (Fig. 2a, d) but LC3-I was not significantly affected by exercise training (Fig. 2a, c). These results suggest that 8 weeks of the exercise training increases basal autophagic flux in mouse skeletal muscle. It is noted that LC3 II protein levels were in the Exe+Sal group were almost undetectable, which is believed to be the activation of autophagy and it demonstrates the importance of the autophagic flux assay in vivo system. It is notable that the mRNA levels of p62 were not significantly changed following exercise training (Fig. 2g) whereas an 8 week swimming training led to ~75 % reduction in p62 protein levels (p < 0.05) (Fig. 2a, e). The lysosomal marker, LAMP1 (Lysosomal-associated membrane protein 1) protein levels were increased by ~95 % following 8 weeks of swimming training (Fig. 2a, f). Exercise training also resulted in increased protein expression of Beclin-1/Atg6 and Atg7, which are involved in the early phases of autophagosome formation (p < 0.05) (Fig. 2h–j).
Fig. 2.
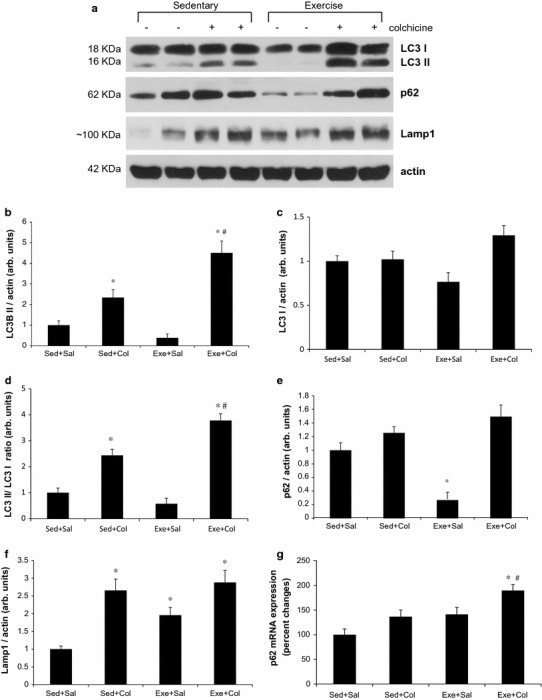
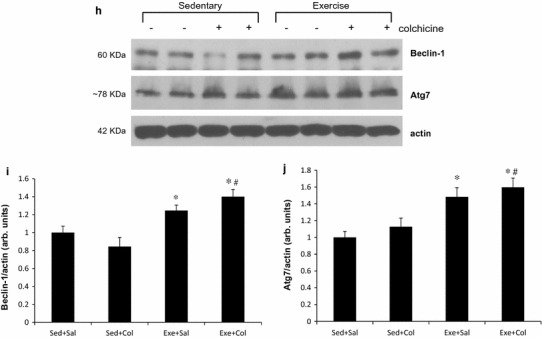
Eight-week swimming exercise training increases autophagic flux in mouse skeletal muscle. Representative immunoblot images of LC3, p62, LAMP1 or actin (a). LC3 II/actin (b), LC3 I/actin (c), LC3 II/LC3 I ratio (d), p62/actin (e) and LAMP1/actin (f) ratios were quantitated via densitometry from 8 mice per treatment conditions. Example immunoblot of Beclin-1/Atg6, Atg7 or actin (h). Quantification of Beclin-1/actin (i) and Atg7/actin (j); values are means ± SE; (n = 8) * p < 0.05 vs. sed+sal, # p < 0.05 vs. sed+col. Expression of p62 mRNA in triceps muscles of mice was measured following 8-week of swimming exercise training. mRNA levels are relative to the GAPDH (n = 4 per group) (g)
To see the involvement of mitochondrial dynamics with autophagy/mitophagy, we evaluated the expressions of four proteins, which are associated with mitochondrial fusion and fission following an 8-week swimming training in skeletal muscle of mice through immunoblotting. There is no significant difference of MFN2 between the Sed+Sal and the Sed+Col groups. Two days of colchicine treatment did not affect MFN2 in mouse skeletal muscle. However, swimming training increased MFN2 protein levels significantly in two exercise groups (Exe+Sal and Exe+Col) compared with two sedentary control groups (p < 0.05, Fig. 3a, b). Two days of colchicine treatment increased Opa1 protein levels significantly by ~70 % compared with the Sed+Sal group (p < 0.05, Fig. 3a, c). However, synergistic effects of colchicine and exercise in Opa1 protein levels were not found. Colchicine treatment increased Drp1, mitochondrial fission protein, significantly but did not increase Drp1 protein levels further along with exercise training. An 8-week swimming training resulted in ~two-fold increase in Drp1 in triceps muscles of mice (Fig. 3d, e). However, Fis1 did not alter by either colchicine or exercise training in mouse skeletal muscle group (Fig. 3a, f).
Fig. 3.
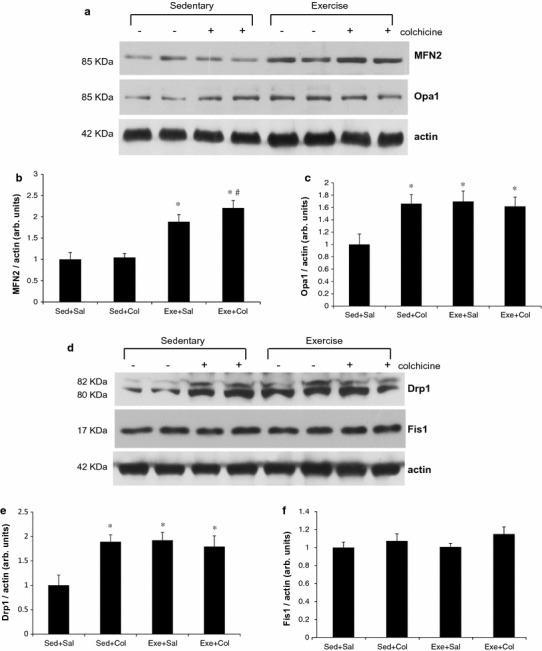
Mitochondrial dynamics proteins are increased in muscle after 8-week swimming training. Muscle homogenates were subjected to immunoblotting with MFN2 (b), Opa1 (c), Drp1 (e), Fis1 (f), or actin. Each bar represents the mean ± SE for muscles from eight mice. (n = 8) * p < 0.05 vs. sed+sal, # p < 0.05 vs. sed+col, δ p < 0.05 vs. exe+sal
We next evaluated what signaling mechanisms could be associated to the observed increase in autophagy/mitophagy following swimming training. As shown in Fig. 4, the protein levels of PINK1 and Parkin were unchanged by exercise training as well as colchicine treatment (Fig. 4b, d). PINK1 and Parkin mRNA also remained unaffected in all conditions (Fig. 4c, e). PINK1/Parkin signaling system is known to stimulate mitophagy when cells are exposed to CCCP, a mitochondrial depolarizing agent or when damaged mitochondria are accumulated in Parkinson’s disease model [21]. However, autophagy/mitophagy activated by exercise training in mouse skeletal muscle may be independent of PINK1/Parkin signaling mechanism. We observed a trend of increased Bnip3 protein levels (p = 0.075) after swimming exercise training in mouse skeletal muscle. There is no significant difference of Bnip3 mRNA and protein levels between the Sed+Sal and the Sed+Col groups (Fig. 4f, g). However, chronic exercise training resulted in an increase in Bnip3 protein concentration further than colchicine treatment alone (p < 0.05, Fig. 4f).
Fig. 4.
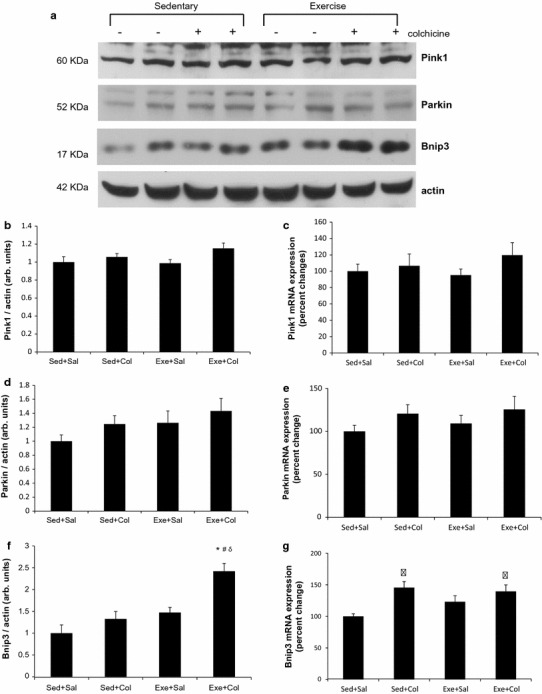
Elevated mitophagy induced by exercise training may be mediated by Bnip3 in mouse skeletal muscle. Representative immunoblot images (a) and densitometric quantification of effects of 8-week swimming training on mitophagy signaling proteins PINK1/Parkin (b, d) and Bnip3 (f). Values are means ± SE; (n = 8) * p < 0.05 vs. sed+sal, # p < 0.05 vs. sed+col. δ p < 0.05 vs. exe+sal. Expression of PINK1 (c), Parkin (e) or Bnip3 (g) mRNA in triceps muscles of mice was measured following 8-week of swimming exercise training. mRNA levels are relative to the GAPDH (n = 4 per group)
To determine whether the exercise training provides a sufficient stimulus for inducing mitochondrial biogenesis and it is altered by inhibiting mitochondrial degradation, we used the analogous method as used in the in vivo autophagic flux assay of mice treated with or without colchicine for 2 days after an 8-week swimming training. Eight weeks of the exercise training induced significant increases in PGC-1α (~50 %), succinate dehydrogenase (SDH, ~80 %) and COX IV protein (~65 %) concentrations (p < 0.05) shown in Fig. 5. We found that these elevated levels of three mitochondrial proteins were significantly down-regulated with 2 days of colchicine treatment, which inhibits mitochondrial degradation (p < 0.05, Fig. 5). Increased PGC-1α mRNA expression by endurance exercise training was also significantly reduced with colchicine treatment (Fig. 5e). The activity of CS, a citric acid cycle enzyme, was determined to see if mitochondrial function was improved following swimming exercise training. Exercise training increased CS activity to ~40 % of the untrained control groups (p < 0.05, Fig. 5f). The quantitative PCR analysis showed that the mitochondrial-to-nuclear DNA ratio in skeletal muscle following 8-week swimming training was not significantly changed (Fig. 5g). The ratio was not significantly changed by colchicine treatment alone, either. The short-term inhibition of autophagy by colchicine treatment might not have a potent impact on changes in mitochondrial mass in skeletal muscle.
Fig. 5.
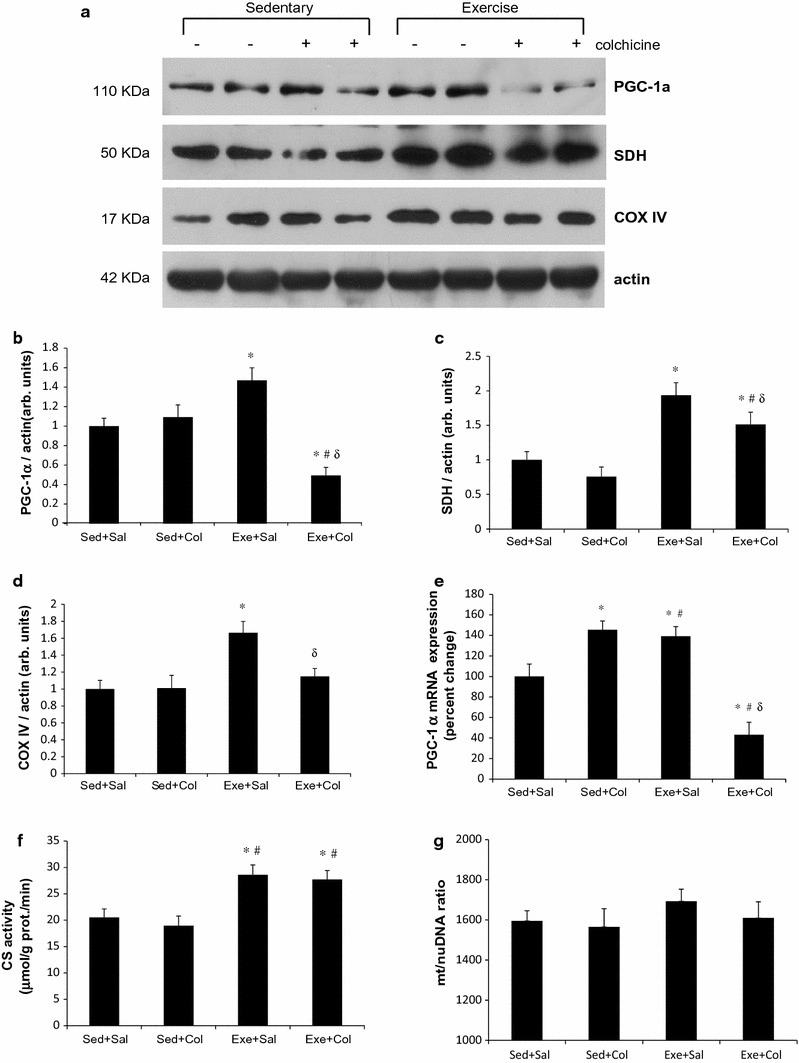
Increased mitochondrial biogenesis is abolished by inhibiting mitophagy in mouse skeletal muscle. Representative immunoblot images of PGC-1α, SDH and COX IV (a), which are indexes of mitochondrial biogenesis. Densitometric quantification of PGC-1α (b), SDH (c), COX IV (d), and CS enzyme activity (f). Values are means ± SE; (n = 8) * p < 0.05 vs. sed+sal, # p < 0.05 vs. sed+col, δ p < 0.05 vs. exe+sal. Relative mRNA expression of PGC-1α in triceps muscles of mice was measured following 8-week of swimming exercise training. mRNA levels are relative to the GAPDH (n = 4 per group) (e). Data represent mRNA levels are relative to the GAPDH (n = 4 per group). Amount of mitochondrial DNA (16s rRNA) adjusted by nuclear DNA (hexokinase 2) in triceps muscles isolated from the control and swimming trained mice (n = 4 per group) (g)
Discussion
Studies have demonstrated that both a single-bout of exercise and chronic exercise training in rodent animals resulted in activation of autophagy in skeletal muscle. However, changes in LC3-II, the most commonly used marker to monitor autophagic flux, by exercise differed among studies and resulted in two opposed interpretations. All studies showing either an increase in LC3-II (increased synthesis of autophagosomes) [14, 15, 20, 22] or a decrease in LC3-II (increased degradation of autophagosomes) [23, 24] by exercise claimed activation of autophagy. To avoid this misunderstanding, we used the autophagy flux assay with or without colchicine administration method as described previously [19], showing that LC3-II protein levels were decreased in the exercised muscles compared with those of sedentary control mice, while LC3-II proteins were more accumulated with exercise plus colchicine treatment than colchicine alone (Fig. 2). This clearly showed autophagic flux was increased in mouse skeletal muscle by endurance exercise training and it was necessary to measure LC3-II protein levels using this strategy to detect in vivo autophagy [1, 19]. A very similar pattern was also observed in the LC3 II/LC3 I ratio, which is frequently used to detect the autophagy flux, in trained muscle (Fig. 2d). However, this assay has also shown a misinterpretable result for measuring the autophagic flux without colchicine treatment. It has been indicated that p62 decreases when autophagy is induced, and accumulates when autophagy is inhibited. However, a decrease in p62 protein level is not necessarily observed, while an increase in autophagic flux can still take place [11]. In accordance with this, recent two studies showed that p62 protein levels were unchanged although the mRNA level of p62 highly upregulated after exercise [17, 18]. However, decreased p62 protein levels by exercise were found in the present study and others [14–16, 25]. p62 protein levels were significantly reduced despite transcription of p62 was not significantly altered under basal autophagy state in the present study. Treatment with colchicine resulted in only a modest further increase in p62 levels (Fig. 2a, c) indicating that p62 is less suitable marker than LC3 II for autophagic flux measurement and p62 may serve other roles besides an autophagic adapter. The validity of p62 as a specific marker of autophagic flux has been questioned due to its role in other cellular processes such as cell signaling pathways including Ras/Raf/MAPK and NF-κB, oxidative stress and tumorigenesis [26] and also conflicting results of p62 protein levels analyzed with Western blot depending on cell lysis buffer used [27].
Several studies have shown that acute exercise increases mRNA levels of MFN1/2 and Fis1 [28, 29] and protein levels of MFN1 and Drp1 [9] in skeletal muscle. Conversely, there are no changes in MFN1/2 and Drp1 mRNA [22] and MFN2 and Opa1 proteins [30] by acute exercise in mouse skeletal muscle. There has been relatively little examination of the effects of endurance exercise training on mitochondrial fusion and fission in skeletal muscle. Recently, Konopka et al. showed that 12 weeks of aerobic exercise training increases MFN1/2 and Fis1 protein levels in skeletal muscles of both young and old human subjects [31]. Perry et al. also showed that seven-session, high-intensity training increased Fis1, Drp1 and MFN1 in human skeletal muscle [9]. In the present study, despite no changes in Fis1 proteins, 8 weeks of swimming training clearly increased MFN2, Opa1 and Drp1 proteins in mouse skeletal muscle. Data from the present study and the work of others [9, 31] demonstrate that both mitochondrial fusion and fission are increased in response to endurance exercise training in skeletal muscle. This suggests that maintaining higher levels of mitochondrial fusion and fission processes during chronic exercise may result in skeletal muscle mitochondrial adaptations.
Little is known regarding the molecular mechanism regulating mitophagy in response to physical exercise in skeletal muscle. The current knowledge of mitophagy came mostly from neurological or cardiac research in cultured cell/animal models from neurodegenerative (e.g., Parkinson’s disease) or cardiovascular diseases (e.g., ischemia). Briefly, mitophagy requires two steps to remove damaged mitochondria: first, induction of general autophagy, and second, priming of damaged mitochondria for selective autophagic recognition [12]. Mitochondrial priming is mediated by either the PINK1/Parkin signaling pathway or the mitophagy receptors, Nix and Bnip3 [12]. Parkin, an E3 ubiquitin ligase, is localized to the cytosol, but translocates to mitochondria with reduced membrane potential where it ubiquinates protein targets [13]. p62 then binds ubiquitinated mitochondrial proteins and LC3 on autophagosomes recruiting autophagic membranes for mitochondrial clearance. PINK1 is a serine/threonine kinase that recruits Parkin to depolarized mitochondria [32]. Bnip3 and NIX are BCL2-related proteins and localized to the outer mitochondrial membrane. Both proteins interact directly with LC3 and GABA receptor-associated protein (GABARAP) on the phagophore to tether mitochondria to form autophagosomes [33, 34]. Bnip3 is involved in hypoxia-induced mitophagy and transcriptionally regulated through HIF or FOXO3 [35]. The present study showed that the protein levels of PINK1 and Parkin were unchanged by exercise training as well as colchicine treatment. Currently, there has been insufficient evidence supporting the involvement of PINK1/Parkin with physical exercise in skeletal muscle. In contrast, swimming exercise training increased Bnip3 protein concentration further with colchicine treatment in the present study. This result is in agreement with that reported by Lira et al. in which 4-week voluntary running increased Bnip3 protein content in mouse skeletal muscle [15] and Tam et al. in which 5-month free wheel running increased Bnip3 protein levels in adult female rats [18]. These and our findings suggest that Bnip3 may be a key player for selecting mitochondria to degrade in response to exercise training in skeletal muscle. Currently, kinases and phosphatases for Bnip3 are not known [35] and more studies need to be done to elucidate the precise roles of Bnip3 in mitophagy regulation in response to exercise.
Our present study showed elevated expressions of mitochondrial biogenetic marker proteins, SDH, COX IV, and a mitochondrial biogenesis promoting factor, PGC-1α following swimming exercise training. The important aspect of our in vivo autophagic flux assay was not only able to detect increased autophagic flux but also provided an answer whether autophagy would be involved in mitochondrial biogenesis following endurance exercise training. This assay demonstrated that increased mitochondrial biogenesis markers were observed when autophagy was activated by exercise training, while exercise training-induced mitochondrial biogenesis was abolished when mitophagy was inhibited with colchicine treatment (Fig. 5). These results indicate that autophagy clearly has an effect on mitochondrial biogenesis during skeletal muscle adaptation, and mitochondrial biogenesis should be initiated under the permission of mitochondrial degradation. These results also suggest that the turnover of mitochondria is very rapid and that a short-term inhibition of autophagy is sufficient to reverse/abolish the effects of 8 weeks of exercise training and this effect is transcriptionally regulated. A failure to induce basal autophagy flux in skeletal muscle may affect the mitochondrial turnover by rapidly reducing transcriptional regulation of nuclear-encoded mitochondrial proteins and subsequent events, such as protein import and mitochondrial assembly. Instead of acute inhibition of basal autophagy, studies using gene knockdown/knockout animals of Atg5 and Atg7, essential autophagy proteins involved in autophagosome formation, also showed the importance of basal autophagy in mitochondrial function in skeletal muscle [36, 37]. These transgenic mice showed accumulated protein aggregates, abnormal mitochondria, and decreased mitochondrial function in skeletal muscle. Using an autophagy-deficient transgenic mouse model (Atg6+/- mice), a similar finding has been reported that cytochrome C and COX IV proteins were increased in WT mice skeletal muscles following 4-week voluntary wheel-running exercise, whereas mitochondrial biogenesis was blunted in muscles of Atg6 ± mice [15]. This and the present study show that autophagy serves as an important regulator in mitochondrial biogenesis during muscle adaptation in response to exercise training. The findings from the present study support the notion that mitophagy and mitochondrial biogenesis are tightly coupled and the coordinated balance between these two processes is a prerequisite for mitochondrial adaptation [38]. Several possible signaling pathways have been implicated in the crosstalk between mitophagy and mitochondrial biogenesis, and these include AMPK (AMP-activated protein kinase), CaMK (calcium/calmodulin dependent kinase) and PKD (protein kinase D1) [38]. All these molecules seem to participate in both mitophagy and mitochondrial biogenesis.
On the basis of the present findings, it is considered that endurance exercise training regulates each of these processes, mitochondrial biogenesis, fusion and fission events and autophagy/mitophagy, ensuring a relatively constant mitochondrial population. Exercise training may also have contributed to mitochondrial quality control which replaces old and/or healthy mitochondria with new and/or healthy ones in skeletal muscle. Cells having been replaced with better quality mitochondria through skeletal muscle adaptation in response to endurance exercise training might have improved mitochondrial function such as increased mitochondrial enzyme activities (e.g. citrate synthase, Fig. 5f) without further increasing mitochondrial number. Therefore, it is hypothesized that these adapted skeletal muscle cells may not require an extra same amount of mitochondria to accomplish more demanding workload in skeletal muscle, especially under moderate intensity endurance exercise training. This mitochondrial quality improvement by exercise training might provide an explanation for the unaltered mitochondrial-to-nuclear DNA ratio (mitochondrial mass) following swimming training in the present study (Fig. 5g).
In summary, taking advantage of the in vivo autophagic flux assay, the present study demonstrated that 8-week swimming exercise training increases the basal autophagy flux and expression of the autophagy/mitophagy-related genes in mouse skeletal muscle. This elevated autophagy may be related to increased mitochondrial fusion and fission events and the Bnip3 signaling pathway. The endurance exercise training induced increases in mitochondrial biogenesis marker proteins, but this effect was abolished in a short-term inhibition of basal autophagy. Our findings highlight the important role of basal autophagy in mitochondrial biogenesis/homeostasis during skeletal muscle adaptation to endurance exercise training.
Acknowledgments
The authors thank Yolanda Mathews for critical reading of the manuscript. This work was done with the sports promotion fund from Korea Institute of Sport Science, Korea Sports Promotion Foundation (KISS-13-A02002) and the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2013-332-2013S1A5A8021905).
Compliance with ethical standards
Conflict of interest
The author(s) declare that they have no competing interests.
References
- 1.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an ‘‘autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 2.Ching JK, Ju JS, Pittman SK, Margeta M, Weihl CC. Increased autophagy accelerates colchicine-induced muscle toxicity. Autophagy. 2013;12:2115–2125. doi: 10.4161/auto.26150. [DOI] [PubMed] [Google Scholar]
- 3.Peterson CM, Johannsen DL, Ravussin E. Skeletal muscle mitochondria and aging: a review. J Aging Res. 2012;2012:194821. doi: 10.1155/2012/194821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol. 2010;299:C203–C210. doi: 10.1152/ajpcell.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanza IR, Sreekumaran Nair K. Regulation of skeletal muscle mitochondrial function: genes to proteins. Acta Physiol (Oxf) 2010;199:529–547. doi: 10.1111/j.1748-1716.2010.02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, Spiegelman BM. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 7.Gómez-Sánchez R, Pizarro-Estrella E, Yakhine-Diop SM, Rodríguez-Arribas M, Bravo-San Pedro JM, Fuentes JM, González-Polo RA. Routine Western blot to check autophagic flux: cautions and recommendations. Anal Biochem. 2015;477:13–20. doi: 10.1016/j.ab.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 8.Iqbal S, Ostojic O, Singh K, Joseph AM, Hood DA. Expression of mitochondrial fission and fusion regulatory proteins in skeletal muscle during chronic use and disuse. Muscle Nerve. 2013;48:963–970. doi: 10.1002/mus.23838. [DOI] [PubMed] [Google Scholar]
- 9.Perry CG, Lally J, Holloway GP, Heigenhauser GJ, Bonen A, Spriet LL. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol. 2010;588:4795–4810. doi: 10.1113/jphysiol.2010.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez AM, Bernardi H, Py G, Candau RB. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol. 2014;307(8):R956–R969. doi: 10.1152/ajpregu.00187.2014. [DOI] [PubMed] [Google Scholar]
- 12.Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 2013;27:4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LoVerso F, Carnio S, Vainshtein A, Sandri M. Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy. 2014;10(11):1883–1894. doi: 10.4161/auto.32154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vainshtein A, Tryon LD, Pauly M, Hood DA. Role of PCG-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am J Physiol Cell Physiol. 2015;308:C710–C719. doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam BT, Pei XM, Yu AP, Sin TK, Leung KK, Au KK, Chong JT, Yung BY, Yip SP, Chan LW, Wong CS, Siu PM. Autophagic adaptation is associated with exercise-induced fibre-type shifting in skeletal muscle. Acta Physiol. 2015;214(2):221–236. doi: 10.1111/apha.12503. [DOI] [PubMed] [Google Scholar]
- 19.Ju JS, Varadhachary AS, Miller SE, Weihl CC. Quantitation of “autophagic flux” in mature skeletal muscle. Autophagy. 2010;6:929–935. doi: 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grumati P, Coletto L, Schiavinato A, Castagnaro S, Bertaggia E, Sandri M, Bonaldo P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7:1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamart C, Naslain D, Gilson H, Francaux M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am J Physiol Endocrinol Metab. 2013;305:E964–E974. doi: 10.1152/ajpendo.00270.2013. [DOI] [PubMed] [Google Scholar]
- 23.Kim YA, Kim YS, Song W. Autophagic response to a single bout of moderate exercise in murine skeletal muscle. J Physiol Biochem. 2012;68:229–235. doi: 10.1007/s13105-011-0135-x. [DOI] [PubMed] [Google Scholar]
- 24.Luo L, Lu AM, Wang Y, Hong A, Chen Y, Hu J, Li X, Qin ZH. Chronic resistance training activates autophagy and reduces apoptosis of muscle cells by modulating IGF-1 and its receptors, Akt/mTOR and Akt/FOXO3a signaling in aged rats. Exp Gerontol. 2013;48:427–436. doi: 10.1016/j.exger.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Pagano AF, Py G, Bernardi H, Candau RB, Sanchez AM. Autophagy and protein turnover signaling in slow-twitch muscle during exercise. Med Sci Sports Exerc. 2014;46(7):1314–1325. doi: 10.1249/MSS.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 26.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–1705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 28.Cartoni R, Léger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Dériaz O, Zorzano A, Gobelet C, Kralli A, Russell AP. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J Physiol. 2005;567:349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding H, Jiang N, Liu H, Liu X, Liu D, Zhao F, Wen L, Liu S, Ji LL, Zhang Y. Response of mitochondrial fusion and fission protein gene expression to exercise in rat skeletal muscle. Biochim Biophys Acta. 2010;1800:250–256. doi: 10.1016/j.bbagen.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Picard M, Gentil BJ, McManus MJ, White K, St Louis K, Gartside SE, Wallace DC, Turnbull DM. Acute exercise remodels mitochondrial membrane interactions in mouse skeletal muscle. J Appl Physiol. 2013;115:1562–1571. doi: 10.1152/japplphysiol.00819.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konopka AR, Suer MK, Wolff CA, Harber MP. Markers of human skeletal muscle mitochondrial biogenesis and quality control: effects of age and aerobic exercise training. J Gerontol A Biol Sci Med Sci. 2014;69:371–378. doi: 10.1093/gerona/glt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas RL, Gustafsson AB. Mitochondrial autophagy—an essential quality control mechanism for myocardial homeostasis. Circ J. 2013;77:2449–2454. doi: 10.1253/circj.CJ-13-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17(24):3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Palikaras K, Tavernarakis N. Mitochondrial homeostasis: the interplay between mitophagy and mitochondrial biogenesis. Exp Gerontol. 2014;56:182–188. doi: 10.1016/j.exger.2014.01.021. [DOI] [PubMed] [Google Scholar]


