Abstract
Two genes, xylP and xylQ, from the xylose regulon of Lactobacillus pentosus were cloned and sequenced. Together with the repressor gene of the regulon, xylR, the xylPQ genes form an operon which is inducible by xylose and which is transcribed from a promoter located 145 bp upstream of xylP. A putative xylR binding site (xylO) and a cre-like element, mediating CcpA-dependent catabolite repression, were found in the promoter region. L. pentosus mutants in which both xylP and xylQ (LPE1) or only xylQ (LPE2) was inactivated retained the ability to ferment xylose but were impaired in their ability to ferment isoprimeverose (α-d-xylopyranosyl-(1,6)-d-glucopyranose). Disruption of xylQ resulted specifically in the loss of a membrane-associated α-xylosidase activity when LPE1 or LPE2 cells were grown on xylose. In the membrane fraction of wild-type bacteria, α-xylosidase could catalyze the hydrolysis of isoprimeverose and p-nitrophenyl-α-d-xylopyranoside with apparent Km and Vmax values of 0.2 mM and 446 nmol/min/mg of protein, and 1.3 mM and 54 nmol/min/mg of protein, respectively. The enzyme could also hydrolyze the α-xylosidic linkage in xyloglucan oligosaccharides, but neither methyl-α-d-xylopyranoside nor α-glucosides were substrates. Glucose repressed the synthesis of α-xylosidase fivefold, and 80% of this repression was released in an L. pentosus ΔccpA mutant. The α-xylosidase gene was also expressed in the absence of xylose when xylR was disrupted.
The disaccharide isoprimeverose [α-d-xylopyranosyl-(1,6)-d-glucopyranose] is the major building block of xyloglucan, a widely distributed hemicellulose which occurs in the primary cell wall in plants (12). Xyloglucan polysaccharides contain a β-(1,4)-glucan backbone with α-(1,6)-d-xylose residues linked to about 75% of the backbone glucosyl residues. Additional α-l-fucosyl-(1,2)-β-d-galactosyl-(1,2)- branchings to position 2 of the xylosyl side chains also regularly occur, but the extent of these branchings is dependent on the species of the plant (12, 23). Xyloglucan can be degraded by cellulolytic microorganisms which produce various endoglucanases acting with different specificities on cellulose and/or hemicellulose (for a review, see reference 37). In general, treatment of xyloglucan by microbial endoglucanases yields xyloglucan fragments (hepta- to nonasaccharides) and smaller oligosaccharides, such as isoprimeverose (36). In contrast to the degradation of xyloglucan, relatively little is known about the enzymatic hydrolysis of isoprimeverose. Until now, the characterization of a genetic system implicated in the metabolism of this disaccharide has not been reported in the literature. However, a few α-xylosidases acting on xyloglucan oligosaccharides and/or isoprimeverose have been reported to exist in microorganisms and plants (22, 24, 39, 40, 43). In these studies, some of the biochemical properties of the purified enzymes were investigated. The α-xylosidases described vary considerably in molecular weight and substrate specificity. For instance, the α-xylosidase isolated from pea seedlings cleaves only the xylosidic linkage in xyloglucan oligosaccharides, whereas most of the microbial enzymes are barely active on these substrates. On the other hand, the microbial enzymes can hydrolyze smaller α-xylosides, such as isoprimeverose, p-nitrophenyl-α-d-xylopyranoside (α-p-NPX), and, in some cases, methyl-α-d-xylopyranoside. The affinity of these α-xylosidases for isoprimeverose was found to be low (apparent Km, 10 to 50 mM) (22, 39, 41, 43).
In general, lactobacilli are not degraders of polysaccharides such as cellulose or hemicellulose but readily ferment smaller carbohydrates (mono-, di-, or trisaccharides). Lactobacillus pentosus is a facultatively heterofermentative bacterium frequently associated with lactic-acid fermentation on vegetables such as cucumbers, cabbages, or olives (42). L. pentosus MD353 was originally isolated from a cucumber fermentation and was studied for its ability to ferment d-xylose. Previous studies have shown that the fermentation of d-xylose by L. pentosus involves the expression of two genes encoding d-xylose isomerase (xylA) and d-xylulose kinase (xylB) (18, 19). The transcription of the xylAB operon is induced by growth on xylose and is negatively controlled by a repressor protein (XylR) and by the trans-acting protein CcpA, a global regulator of catabolite repression (CR) in gram-positive bacteria (10, 14, 20). The repressor gene, xylR, is located upstream of the xylAB operon, and its transcription occurs from its own promoter in the absence of xylose, with the same polarity as that of xylAB. However, Lokman et al. (19) have previously shown that in the presence of xylose, xylR can be 10-fold more efficiently expressed from an unidentified promoter located upstream of the xylR gene. The size of the xylR-containing mRNA, which is inducible by xylose, was found to be large enough (>5 kb) to comprise additional open reading frames (ORFs). Preliminary sequencing results (19) revealed the presence of at least two genes: xylP, encoding a putative permease, and xylQ, encoding a protein of unknown function. In this paper we report the complete cloning and sequence analysis of the xylPQ genes and of the regulatory elements of the xylPQR operon. We also demonstrate that the xylPQ genes are involved in the metabolism of isoprimeverose rather than in xylose metabolism. This constitutes the first description of the primary structure of an α-xylosidase (XylQ) and of a putative isoprimeverose cation symporter (XylP).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and genetic elements used in this study are listed in Table 1. Escherichia coli JM109 was grown on Luria-Bertani agar or in Luria-Bertani broth. Ampicillin was added at a final concentration of 100 μg/ml. L. pentosus strains were grown at 37°C in MRS medium (Difco Laboratories, Detroit, Mich.) or M medium (19) containing 1% (wt/vol) of the indicated sugar. Erythromycin was added at a concentration of 2.5 μg/ml when necessary. The test of sugar fermentation was performed in 200 μl of M medium containing 0.5% (wt/vol) of the corresponding sugar and 0.005% bromocresol purple. Fermentation was tested by the color change of the medium from purple to yellow due to acid production. For plating, media were solidified with 1.5% agar.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| L. pentosus | ||
| MD353 | Wild-type strain | 18 |
| MD363 | Wild-type strain | 20 |
| LPE1 | MD353 ΔxylP ΔxylQ Emr Apr | This study |
| LPE2 | MD353 ΔxylQ Emr Apr | This study |
| LPE3 | MD363 ΔxylR Emr Apr | 20 |
| LPE4 | MD363 ΔccpA Emr Apr | 20 |
| E. coli JM109 | recA1 relA1 thi Δ(lac-proAB) gyrA96 hsdR17 endA1 supE44 | |
| Plasmids | ||
| pIN15E | 5.8-kb integration plasmid for Lactobacillus; Emr Apr | 20 |
| pXH50A | pUC19 with a 5-kb HindIII fragment; Apr | 19 |
| pLPA1 | pBR322 with xylP xylQ ClaI-BamHI fragment; Apr | This study |
| pLPA3 | pIN15E with xylP SpeI-MscI fragment; Emr Apr | This study |
| pLPA4 | pIN15E with xylP xylQ SpeI-BamHI fragment; Emr Apr | This study |
Emr, erythromycin resistant; Apr, ampicillin resistant.
Materials.
Enzymes were purchased from Boehringer or Bethesda Research Laboratories and were used according to the specifications of the manufacturer. [α-35S]dATP (1,000 Ci/mol), [α-32P]dATP and [γ-32P]dATP (3,000 Ci/mmol), and d-[U-14C]xylose (89 mCi/mmol) were obtained from Amersham. Xyloglucan oligosaccharides were prepared as described elsewhere (35) by treatment of tamarind seed xyloglucan (Dainippon Pharmaceutical, Osaka, Japan) with an endoglucanase preparation (Maxazyme C1; Gist-Brocades, Delft, The Netherlands). Isoprimeverose was obtained by treatment of 200 mg of xyloglucan oligosaccharides with 2 mg of protein of a Driselase preparation (Sigma, St. Louis, Mo.) in 10 ml of a 50 mM sodium acetate buffer (pH 5) for 16 h at 40°C (36). The free glucose and free galactose were removed by incubating the mixture for 2 h at 37°C with 10 mg (dry weight) of Lactobacillus plantarum 80 cells grown on galactose. After centrifugation (at 10,000 × g for 10 min), the isoprimeverose was filter sterilized and the purity of the disaccharide was verified by thin-layer chromatography (TLC).
Preparation of cell extracts.
Cells in the logarithmic phase of growth were harvested by centrifugation (at 5,000 × g, at 4°C, for 10 min), washed twice with 50 mM potassium phosphate buffer (pH 6.5) containing 0.5 mM EDTA, and resuspended to 1/100 of the culture volume in the same buffer to which 1 mM of dithiothreitol was added (KPED buffer). Cells were broken by three passages through a French pressure cell (11,000 lb/in2). Cell debris was removed by centrifugation (at 20,000 × g, at 4°C, for 20 min), and the membranes were separated from the soluble fraction by centrifugation (at 100,000 × g, at 4°C, for 2 h). The membranes were washed once with KPED buffer, and the two fractions (cytosol and membranes) were stored frozen at −70°C until they were used. Protein concentration was determined by the method of Smith et al. (32) by using bicinchoninic acid.
Enzyme assays.
The conversion of d-xylose (200 mM) to d-xylulose by d-xylose isomerase was measured as described elsewhere (4). The NADH oxidase activity present in the cytosolic fraction was inhibited by incubating the reaction mixture for 15 min with KCN at a final concentration of 1 mM. d-Xylulose kinase activity was determined by measuring the appearance of ADP formed as a result of the d-xylulose kinase reaction. ADP formation was determined as described previously for the determination of acetate kinase activity (33). d-Xylulose was used at a final concentration of 1 mM. α-Xylosidase activity with α-p-NPX as the substrate was determined in 500 μl of KPED buffer (pH 6.5) at 37°C with a final α-p-NPX concentration of 5 mM. The reaction was stopped by addition of 250 μl of 1 M Na2CO3, and the optical density at 410 nm (OD410) was measured. α-Xylosidase activity with isoprimeverose (0.5 mM) as the substrate was determined by coupling the release of glucose from the disaccharide to the hexokinase–glucose 6-phosphate dehydrogenase reaction as described elsewhere (2), except that the reaction buffer was KPED (pH 6.5). α-Glucosidase activity was determined under the conditions used for the α-xylosidase activity determination, except that the substrate was p-nitrophenyl-α-d-glucopyranoside (α-p-NPG) at a final concentration of 5 mM.
d-Xylose uptake measurements.
Prior to the start of the transport experiment, xylose-growing cells were diluted to a protein concentration of 3 to 5 mg/ml in 800 μl of uptake buffer (50 mM potassium phosphate [pH 6.5] supplemented with 2 mM MgSO4). After 5 min of incubation at 37°C with gentle stirring, transport was initiated by the addition of d-[U-14C]xylose (0.4 μCi/mmol) at a final concentration of 500 μM. After 1 min, the reaction mixture was diluted into 10 ml of ice-cold 0.1 M LiCl, rapidly filtered through glass fiber filters (Whatman GF/F), and washed with 2 ml of ice-cold 0.1 M LiCl. The radioactivity on the filter was determined by liquid scintillation. The uptake of d-xylose was linear up to 2 min.
Plasmid constructions.
To construct plasmid pLPA1, genomic DNA from strain MD353 was first digested by several restriction enzymes and analyzed by Southern hybridization using different fragments from the xylR upstream sequence previously cloned in pXH50A (18) as probes. Unique DNA fragments hybridizing to the probes were identified. MD353 chromosomal DNA double digested by ClaI-BamHI was cloned into pBR322 to yield plasmid pLPA1. Plasmid pLPA4 was constructed by cloning of a 1.6-kb SpeI-BamHI fragment from pLPA1 into the XbaI-BamHI sites of the integration vector pIN15E (20). To obtain pLPA3, a digestion was performed on pLPA4 from the MscI site located in xylP to the SmaI site of the multicloning region of pIN15E. The blunt ends of the vector were ligated to give a shortened SpeI-MscI fragment of 660 bp. E. coli JM109 was used as the host for the propagation of all vectors.
DNA and RNA manipulation and nucleotide sequence analysis.
L. pentosus chromosomal DNA was isolated as described by Lokman et al. (18), and RNA was isolated as described by Pouwels et al. (28). For primer extension analysis, RNA was isolated from wild-type cells during the exponential phase of growth on xylose. To obtain the LPE1 (ΔxylPQ) and LPE2 (ΔxylQ) mutants, L. pentosus MD353 competent cells were transformed with plasmids pLPA3 and pLPA4 by electroporation, and the integrants were isolated as described previously for the ΔxylR mutant (20). Recombinant DNA procedures and transformation of E. coli were performed by standard methods (30). Nucleotide sequencing was performed by the dideoxy termination method (31). DNA fragments were isolated from agarose gels by using the GeneClean kit from Bio 101 (La Jolla, Calif.). To obtain the MunI inverse PCR fragment, genomic DNA was digested by MunI and ligated at a concentration of 2 ng/μl. The ligated DNA was precipitated in the presence of 1 μg of glycogen as the carrier and resuspended at a concentration of 30 ng/μl. One hundred nanograms of DNA and 20 pmol each of xylp5 (5′-GGCACCATATTTTTATGGAT-3′), complementary to codons 22 to 28 of xylP, and xylp3 (5′-GGAGTGAACGTTTCAGTTAT-3′), complementary to anticodons 30 to 35 of xylP, were used in the amplification reaction performed with the Expand high-fidelity PCR system (Boehringer Mannheim). The PCR fragment was sequenced by using the fmol DNA sequencing kit (Promega). Primer extension analysis was performed by annealing 1 pmol of 32P-labeled xylp5 oligonucleotide with 20 μg of total RNA, followed by synthesis of cDNA with reverse transcriptase (from Moloney murine leukemia virus; Bethesda Research Laboratories). The BLAST research and the amino acid PILEUP comparisons were performed by using the Genetics Computer Group programs through the facilities of the CAOS/CAMM center, Nijmegen, The Netherlands.
TLC.
TLC was performed with Merck silica gel 60 TLC plates. The solvent system was n-butanol–ethanol–water (10:1:2), and the plates were developed twice. The sugars were detected by dipping the gel into a solution of 0.5% α-naphtol–5% sulfuric acid in ethanol and heating at 120°C for 5 min.
Nucleotide sequence accession number.
The sequence, including xylP, xylQ, and the promoter-operator region, has been deposited in the EMBL/GenBank database under accession no. U89276.
RESULTS
Cloning and sequence analysis of xylP and xylQ.
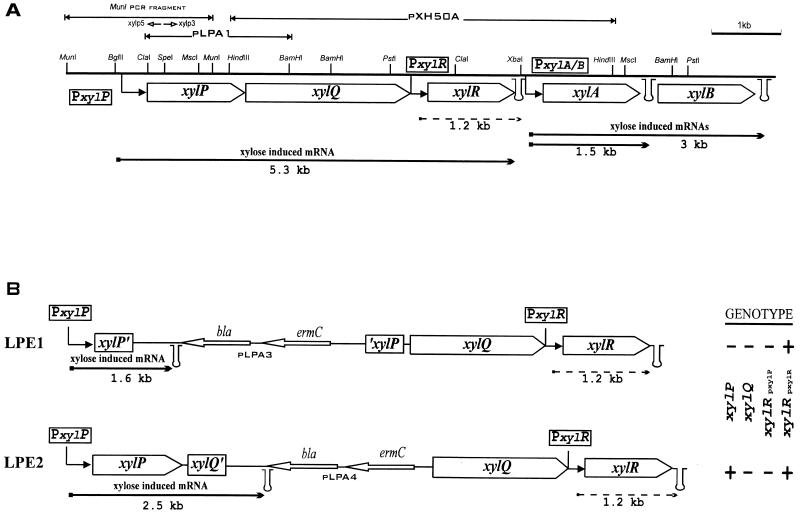
Part of the sequence of xylP and xylQ was determined by Lokman et al. (19), and the deduced amino acid sequence of xylP was used for comparison with other transport proteins by Poolman et al. (26). Here we describe the analysis of the complete xylP and xylQ region located upstream of xylR and cloned in plasmids pLPA1 and pXH50A (Fig. 1A) (19). DNA sequence analysis of these genomic fragments revealed two ORFs that were transcribed in the same direction. The first ORF, encoding a protein of 479 amino acids called XylP, started immediately after the ClaI site used for the cloning in pLPA1. This ORF contained two possible Met start codons, the first located within the ClaI site and having an ATGA motif, the second located 10 nucleotides downstream. The second ORF, encoding XylQ, a protein of 758 amino acids, probably started with a Val codon (GTG) located 27 nucleotides downstream from the translational stop of xylP and was preceded by a potential ribosome binding site (5′-GAAAGGA-3′) typical for Lactobacillus mRNAs (27). The two stop codons of xylQ (5′-TGATAA-3′) were located 5 nucleotides upstream of the −35 element of the xylR constitutive promoter. No inverted repeat that could potentially constitute a Rho-independent terminator of transcription could be identified in the sequence described here.
FIG. 1.
(A) Physical map and organization of the L. pentosus MD353 xylose regulon. The upper part shows the xylP xylQ cloning strategy. (B) Chromosomal situations of LPE1 and LPE2 integrants after integration of pLPA3 and pLPA4 plasmids. Their respective genotypes are indicated on the right (plus and minus signs indicate the presence and absence, respectively, of transcription of the genes). In both panels, arrows with right angles indicate the xylPQ and xylAB (xylose-inducible) promoters and the xylR (constitutive) promoters, and stem-loop structures indicate the putative transcriptional terminators. The sizes of the wild-type bacterial, LPE1, and LPE2 transcripts from the different promoters are given below each arrow (arrows with solid and dashed lines represent xylose-inducible and constitutive expression, respectively). The primers used for the inverse PCR, xylp5 and xylp3, are indicated by short open arrows below the MunI fragment. bla and ermC, genes for ampicillin resistance and erythromycin resistance, respectively.
Isolation of xylP upstream sequences and sequence analysis of the xylP promoter-operator region.
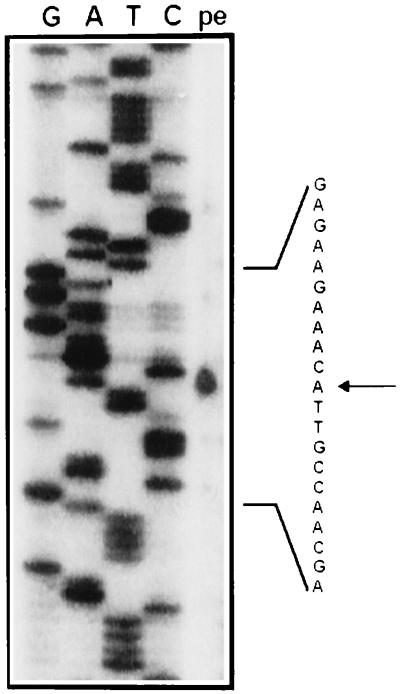
Cloning of sequences located upstream of the ClaI site was initially unsuccessful due to severe instability when either E. coli or Lactobacillus casei ATCC 393 was used as the host. However, by using inverse PCR after digestion of chromosomal DNA by MunI, a fragment of 1.9 kb, comprising 1.0 kb of xylP upstream sequences in addition to 0.9 kb of the xylP gene, could be obtained. Nucleotide sequence analysis of this PCR product showed that the first xylP ATG codon is preceded by a putative ribosome binding site similar to the one identified in the xylP-xylQ intergenic region, suggesting that the first rather than the second ATG codon is the genuine translation start site. Putative −10 and −35 consensus sequences of promoters spaced by 17 bp were found (TACTCT and TTGCAA) 144 and 167 nucleotides before the first ATG codon of xylP (Fig. 2). Primer extension analysis performed with RNA isolated from xylose-induced cells showed an RNA transcript starting 7 nucleotides downstream of the −10 element (Fig. 3). The region was also examined for sequences which might be characteristic of cis elements involved in regulation of xylPQ expression. A sequence of 27 nucleotides (Fig. 2) ending 65 bp upstream of the ATG of xylP displayed considerable homology with the palindromic xyl operator (xylO) found in the promoter regions of xylA genes of gram-positive bacteria (6). This finding indicated that the expression of xylP and xylQ could be negatively controlled by the repressor XylR. Another motif overlapping the putative −10 of the promoter (Fig. 2) showed similarity to the catabolite response element (cre) involved in general CR in gram-positive bacteria. Although this cre element showed three mismatches, at positions 7, 10, and 14, to the consensus sequence established by Weickert and Chambliss (38), this finding suggested that the xylPQ operon might be subject to CcpA-dependent CR.
FIG. 2.
Nucleotide (nt) sequence of the promoter-regulatory region of the xylPQR operon. Open boxes, −10 and −35 elements of the promoter; boldface italic letters, putative regulatory elements (cre and xylO); stars, potential ribosome binding site; open vertical arrow, +1 transcription start; horizontal arrow, sequence complementary to the primer xylp5 used in the primer extension experiment. The beginning of the deduced amino acid sequence of the xylP gene is depicted below the nucleotide sequence. Direct repeated or inverted repeated sequences in the promoter region are underlined with arrows.
FIG. 3.
Primer extension analysis of xylose-induced RNA transcript from L. pentosus MD353. The MunI PCR-amplified fragment was used as the template for the sequencing reaction. Lane pe contains the primer extension reaction with the kinase-reacted xylp5 primer. G, guanine; A, adenosine; T, thymine; C, cytosine. The start of the transcript is indicated by an arrow.
XylP structural features and amino acid sequence similarity.
Hydropathy analysis of the XylP protein revealed 12 hydrophobic segments of 19 residues (or longer) likely to span the cytoplasmic membrane (data not shown). The deduced amino acid sequence of XylP demonstrated similarity, ranging from 28 to 32% identical amino acids, with XynC of Bacillus subtilis (8), GusB of E. coli (16) (Swiss-Prot database accession no. P30868), and three other E. coli hypothetical proteins: a protein encoded by ORFf479, present in the genomic region from 81.5 to 84.5 min (3), and two proteins encoded by ORFf723 and ORFf481, present in the genomic region from 87.2 to 89.2 min (25). These membrane proteins have recently been designated the GusB subgroup of the GPH family of translocators (26).
Inactivation of xylP and/or xylQ by chromosomal integration.
The sequence homology found between XylP and translocators of the GPH family suggested that XylP might be a xylose transporter. To assess the roles of XylP and XylQ in xylose transport and metabolism, xylP was disrupted and/or xylQ transcription from the xylP promoter was inhibited. The inactivation of these two genes was achieved by using a strategy of chromosomal integration based on the use of a plasmid harboring a temperature-sensitive replicon (pIN15E). L. pentosus MD353 was transformed with plasmid pLPA3, carrying xylP of which both the 5′ and 3′ ends were truncated, or with pLPA4, in which xylP was truncated only at its 5′ end and which contained 250 bp of xylQ downstream from its 3′ end (first BamHI site). The integration of these plasmids in the chromosome resulted in strains LPE1 and LPE2, respectively. The genetic structure of the xyl locus of LPE1 and LPE2 is shown in Fig. 1B. Briefly, integration of pLPA3 led to disruption of xylP in LPE1, with a concomitant polar effect on xylQ and xylR expression from the xylP promoter. Integration of pLPA4 also led to a polar effect on xylQ and xylR expression from the xylP promoter, but in this case, a complete xylP gene was restored after integration.
Growth characteristics of LPE1 and LPE2 on xylose.
The growth characteristics of LPE1 and LPE2 on xylose are summarized in Table 2. Inactivation of xylP and xylQ did not result in the absence of growth on xylose and transport of xylose. Although the activities of d-xylose isomerase and d-xylulose kinase were decreased in the disruption mutants compared to the activities in the wild-type strain, the mutants showed increases in their growth rates and rates of xylose uptake. The growth of LPE1 and LPE2 mutants on xylose was also characterized by a longer lag period (by about 1 day) compared to that of the wild-type strain. These observations challenged the putative role of XylP as a xylose transporter and suggested that xylose uptake might be catalyzed by another as yet unidentified transport system, distinct from XylP. These results showed, furthermore, that XylP and XylQ are not essential for xylose fermentation.
TABLE 2.
Characteristics of wild-type L. pentosus and LPE1 and LPE2 mutants grown on M medium plus 1% d-xylosea
| Strain | Growth parameter
|
Enzyme activity in cell extracts (nmol/min/mg of protein)
|
Transport activityb (nmol/min/mg [dry wt]) | ||
|---|---|---|---|---|---|
| Lag period (days) | Growth rate (h−1) | d-Xylose isomerase | d-Xylulose kinase | ||
| MD353 | 2–3 | 0.07 | 52 ± 9 | 320 ± 33 | 1.1 ± 0.1 |
| LPE1 | 3–4 | 0.13 | 22 ± 3 | 182 ± 47 | 1.9 ± 0.2 |
| LPE2 | 3–4 | 0.14 | 21 ± 5 | 252 ± 53 | 2.3 ± 0.1 |
All experiments were performed in triplicate.
Performed with d-[U-14-C]xylose.
Analysis of XylQ sequence.
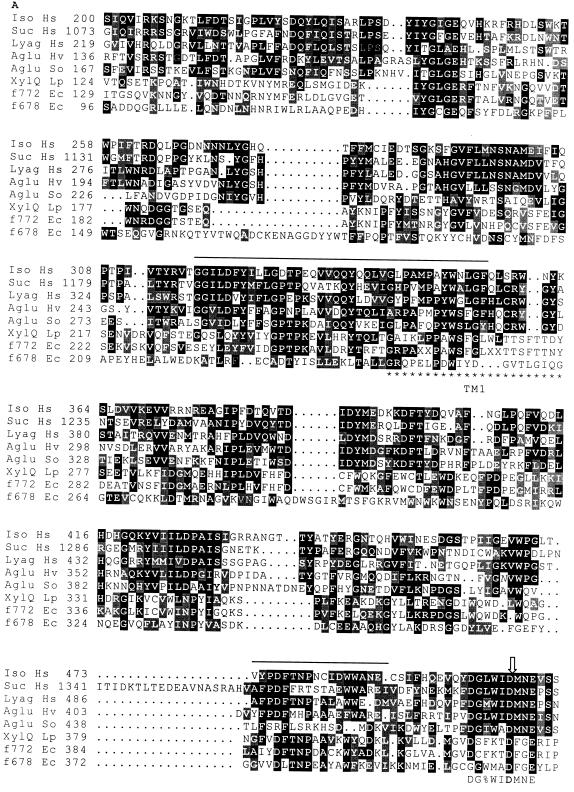
The Swiss-Prot protein sequence database was searched for entries showing similarity to XylQ with the BLAST computer program (1). XylQ showed, throughout its entire length, strong sequence homology (46%) to an E. coli 88.1-kDa protein of unknown function (encoded by ORFf772, presumably transcriptionally coupled to ORFf479, which is mentioned above) and local homology with another hypothetical E. coli protein of 77.3 kDa, of unknown function (encoded by ORFf678 and presumably transcriptionally coupled with ORFf481 and ORFf723, mentioned above). In addition, the results of the computer search revealed that the predicted amino acid sequence of XylQ contained four stretches, ranging from 18 to 38 residues, which showed striking similarity to regions highly conserved among members of family 31 of glycosyl hydrolases, described by Henrissat and Bairoch (11). However, no significant homology could be found between the prokaryotic XylQ, f772, and f678 amino acid sequences and those of the members of family 31 when the entire amino acid sequences were considered. Family 31 of glycosyl hydrolases is a very diverse group of eukaryotic α-glucosidases which were shown to share two conserved domains of about 200 and 300 residues. These two domains are connected and flanked by linker regions which can vary greatly in length and function, such as signal-anchor sequences of secretion pathways or putative glycosylation sites (15, 16). We assumed that these linker regions, specific for eukaryotic functions, were unlikely to be found in the prokaryotic proteins XylQ, f772, and f678. Therefore, we performed an alignment of XylQ, f772, and f678 sequences with the conserved domains of family 31 α-glucosidases from which the linker regions were omitted. Figure 4 shows the sequence comparisons. We found that XylQ and the two E. coli hypothetical proteins (f772 and f678) shared similar domains and that one of the amino acid clusters revealed by the BLAST search overlapped with one of the PROSITE signatures of family 31. Furthermore, the aspartic residue of the sucrase-isomaltase complex (15, 29) and of the human lysosomal α-glucosidase (13), shown to be essential for catalytic activity, appeared to be conserved in the three prokaryotic proteins. Finally, hydropathy analysis of XylQ indicated the presence of two putative transmembrane helices. The first hydrophobic segment was 23 residues (residues 254 to 276) and was separated by 210 residues from the second (residues 486 to 505), which was 20 residues.
FIG. 4.
Alignments of similar regions of primary structure of isomaltase (Iso Hs) and sucrase (Suc Hs) from the human pro-sucrase-isomaltase complex (Swiss-Prot database accession no. P14410), the human lysosomal α-glucosidase (Lyag Hs; accession no. P10253), the Schwanniomyces occidentalis glucoamylase (Aglu So; P22861), the barley putative α-glucosidase (Aglu Hv) (34), and the three prokaryotic polypeptides L. pentosus XylQ (XylQ Lp) and the E. coli hypothetical proteins f772 (f772 Ec; P31434) and f678 (f678 Ec; P32138). Identical amino acids are shown in white letters on a solid ground, and similar amino acids are shown in white letters on a shaded ground. Stretches of amino acids revealed by the BLAST research are overlined. The PROSITE signatures (PDOC00120) of family 31 of glycosyl hydrolases are shown below the alignment, as follows: %, any one of L, I, V, or M; $, either F or Y; #, either S or T; !, either S or A. The open arrow points to the aspartic acid residue involved in the active site of the human lysosomal α-glucosidase. Stars under the sequence mark the putative transmembrane helices of XylQ. Numbers in front of the lines are amino acid positions of the proteins.
Fermentation of various sugars with an α-d-glycosyl linkage.
The similarity observed between the xylQ gene product and several α-glucosidases with specificity towards α-1,6 and α-1,4 linkages prompted us to compare LPE1 and LPE2 mutants with wild-type bacteria for their ability to ferment mal- tose [α-d-glucopyranosyl-(1,4)-d-glucopyranose], isomaltose [α-d-glucopyranosyl-(1,6)-d-glucopyranose], and sucrose [α-d-glucopyranosyl-(1,2)-d-fructofuranose]. Since the expression of xylQ is induced by xylose, we investigated the likelihood that xylQ might encode an α-xylosidase instead of an α-glucosidase by testing fermentation on methyl-α-d-xylopyranoside, isoprimeverose, and xyloglucan oligosaccharides, which are typical substrates for α-xylosidases. For the last substrate, we used a mix of xyloglucan oligosaccharides from tamarind seed (composed of 13% XXXG, 9% XLXG, 28% XXLG, and 50% XLLG) as described elsewhere (35). The nomenclature, according to Fry et al. (7), is as follows: G is β-d-glucopyranose (reducing glucose), X is α-d-xylopyranosyl-(1,6)-β-d-glucopyranosyl- (1,4)-, and L is β-d-galactopyranosyl-(1,2)-α-d-xylopyranosyl-(1,6)-β-d-glucopyranosyl-(1,4)-. The wild-type strain could ferment all substrates in 24 h except methyl-α-d-xylopyranoside and xyloglucan oligosaccharides, which were not fermented at all. For these two substrates, the fermentation ability was checked for a longer period, but it remained negative after 96 h of incubation. Similar results were obtained with the LPE1 and LPE2 mutants except that they could not ferment isoprimeverose even after 96 h, suggesting that the xylPQ operon is involved in isoprimeverose metabolism.
Identification and cellular location of XylQ enzyme activity.
To characterize the enzymatic activity of XylQ, the wild-type and LPE1 and LPE2 mutant strains were grown on xylose medium to induce the expression of XylQ. Cells were harvested at the exponential phase of growth, and α-xylosidase and α-glucosidase activities were measured in the cytosolic and membrane fractions of the three strains with the chromogenic substrates α-p-NPX and α-p-NPG, respectively (Table 3). α-Glucosidase activity was present in both the membrane and cytosolic fractions of the wild-type strain and was not significantly different in the LPE1 and LPE2 mutants. α-Xylosidase activity was detected only in the membrane fraction of the wild-type strain. The inactivation of xylQ in both the LPE1 and LPE2 mutants resulted in the loss of this membrane-associated α-xylosidase activity.
TABLE 3.
α-Glycolytic enzymes present in wild-type L. pentosus and LPE1 and LPE2 mutants grown on M medium plus 1% d-xylosea
| Strain | Enzyme activity (nmol/min/mg of protein)
|
|||
|---|---|---|---|---|
| α-p-NPX
|
α-p-NPG
|
|||
| Cytoplasmic fraction | Membrane fraction | Cytoplasmic fraction | Membrane fraction | |
| MD353 | 0 | 52 ± 5 | 464 ± 24 | 44 ± 5 |
| LPE1 | 0 | 0 | 461 ± 14 | 46 ± 5 |
| LPE2 | 0 | 0 | 465 ± 10 | 43 ± 8 |
All experiments were performed in triplicate.
Substrate specificity of XylQ.
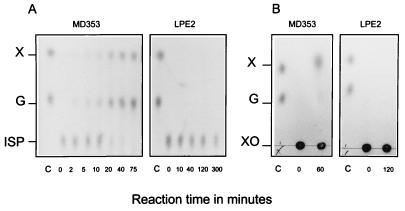
To confirm that XylQ was able to hydrolyze isoprimeverose, the membrane fractions of the wild-type bacteria and the LPE2 mutant were incubated with the disaccharide. The reaction mixtures were analyzed by TLC (Fig. 5A). Only the membrane fraction from the wild-type bacteria could hydrolyze isoprimeverose into equimolar amounts of glucose and xylose. Isoprimeverose was not hydrolyzed by the LPE2 mutant membrane fraction, even after 5 h of incubation. Similar results were obtained with the LPE1 mutant membrane fraction (data not shown). The substrate specificity of XylQ was examined by conducting similar reactions with maltose, isomaltose, sucrose, methyl-α-d-xylopyranoside, and xyloglucan oligosaccharides. Although α-glucosidase activity with α-p-NPG could be detected in the membrane fraction of each strain (see Table 3), no glucose could be released from maltose and sucrose and only a small amount of glucose could be liberated from isomaltose in 5 h of incubation time (≈5% of the amount of isomaltose). However, the levels of isomaltase activity were similar in the wild-type bacteria and the LPE1 and LPE2 mutants, indicating that this partial hydrolysis of isomaltose did not result from XylQ activity (data not shown). Methyl-α-d-xylopyranoside was not hydrolyzed either (data not shown), but liberation of 25 to 50 nmol of xylose from 2.5 mg of xyloglucan oligosaccharides, equivalent to 1.8 μmol based on the average molar weight (1,400 g/mol) of the xyloglucan oligosaccharide mix, could be detected after 1 h of incubation with the membrane fraction of the wild-type bacteria (Fig. 5B). This activity was not present in the membrane fraction of the LPE2 mutant. However, a high concentration of xyloglucan oligosaccharides (above 5 mM) was required to detect the formation of xylose, and the amount of xylose liberated by the membrane fraction of the wild-type bacteria from this substrate was not increased after 1 h, suggesting that the enzyme recognized only some of the α-xylosidic linkages. Thus, among the substrates examined, only α-p-NPX and isoprimeverose were efficiently hydrolyzed by XylQ.
FIG. 5.
TLC analysis of the products of hydrolysis of isoprimeverose (A) or xyloglucan oligosaccharides (B) by membrane fractions of wild-type L. pentosus MD353 and the LPE2 mutant. Lane C, xylose and glucose control (5 nmol of each). X, xylose; G, glucose; ISP, isoprimeverose; XO, xyloglucan oligosaccharides. The reactions were performed in 50 μl of KPED buffer (pH 6.5) at 37°C. The reaction mixtures contained 10 μg of membrane proteins and either 60 nmol of isoprimeverose or 1.8 μmol of xylogucan oligosaccharides. At each time point, 1/10 of the reaction mixture was loaded on the TLC plates.
Kinetic parameters for the hydrolysis of α-p-NPX and isoprimeverose by the membrane fraction of the wild-type bacteria were determined. In these experiments, isoprimeverose activity was evaluated by measuring the glucose released from the disaccharide, as described in Materials and Methods. The apparent Km and Vmax values of α-p-NPX and isoprimeverose were 1.3 mM and 54 nmol/min/mg of protein, and 0.2 mM and 446 nmol/min/mg of protein, respectively.
Regulation of α-xylosidase activity.
As described above, the promoter region of the xylPQ operon contained a cre-like sequence overlapping the −10 of the promoter and a putative xylO operator sequence between the promoter and the start codon of xylP. To verify the control by the repressor XylR and the influence of glucose on xylPQ expression, α-xylosidase activity was measured in the membrane fractions of several L. pentosus wild-type and mutant strains grown in the presence of either glucose, glucose plus xylose, or xylose. The results of these experiments are shown in Table 4. The repression mediated by glucose was approximately fivefold, and about 80% of this repression could be released by disruption of the ccpA gene. As expected, deletion of the xylR gene resulted in α-xylosidase activity when cells were grown in the absence of xylose, although five- to eightfold glucose repression could still be detected. Finally, a small but significant level of α-xylosidase activity could be detected in the ccpA mutant grown on glucose (1 to 2% of the activity found for the wild type grown on xylose).
TABLE 4.
Regulation of α-xylosidase activity in L. pentosusa
| Strain | Genotype | α-Xylosidase activity (nmol/min/mg of protein)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| α-p-NPX
|
Isoprimeverose
|
||||||||
| Gb | XGc | Xd | Re | Gb | XGc | Xd | Re | ||
| MD353 | Wild type | 0 | 10.1 ± 1.3 | 51.4 ± 0.8 | 5.1 | 0 | 78 ± 10 | 389 ± 22 | 5.0 |
| LPE1 | MD353 ΔxylP ΔxylQ | 0 | 0 | 0 | 0 | 0 | 0 | ||
| LPE2 | MD353 ΔxylQ | 0 | 0 | 0 | 0 | 0 | 0 | ||
| MD363 | Wild type | 0 | 9.4 ± 1.2 | 48.8 ± 1.0 | 5.2 | 0 | 79 ± 10 | 380 ± 34 | 4.8 |
| LPE3 | MD363 ΔxylR | 14.3 ± 0.6 | 14.9 ± 3.1 | 84.1 ± 3.1 | 5.6 | 68 ± 8 | 94 ± 3.0 | 787 ± 40 | 8.4 |
| LPE4 | MD363ΔccpA | 0.6 ± 0.1 | 45.0 ± 3.0 | 47.6 ± 0.3 | 1.0 | 6 ± 1 | 254 ± 14 | 322 ± 12 | 1.3 |
All experiments were performed in triplicate.
Cells were grown in M medium containing 0.5% glucose (G) and harvested at an OD600 of 2.5.
Cells were grown in M medium containing 0.5% glucose plus 1% xylose (XG) and harvested at an OD600 of 2.5.
Cells were grown in M medium containing 1% xylose (X) and harvested at an OD600 of 2.5.
The factor of repression R corresponds to the quotient of nonrepressed α-xylosidase activity (X) and α-xylosidase activity in cells grown in XG.
DISCUSSION
The initial aim of this study was to assess the possible role(s) of xylPQ in xylose transport and metabolism, which was suggested by the induction of the xylPQR operon and the homology of XylP to a family of cation symporters. Surprisingly, our data showed that inactivation of xylP and xylQ did not result in the absence of xylose fermentation and that disruption of xylP did not abolish or reduce the rate of xylose uptake, making a role for XylP in transport unlikely. However, inactivation of xylQ resulted in the loss of a membrane-associated α-xylosidase activity and of isoprimeverose fermentation. The determination of the α-xylosidase activity in the membrane fraction of the L. pentosus ΔxylR mutant and the L. pentosus ΔccpA mutant grown in the presence of xylose, glucose, or xylose plus glucose allowed us to demonstrate that the expression of xylPQ is negatively controlled by XylR and is subject to CcpA-dependent CR. Whether the complex formed between CcpA and its corepressor (Ser-P) HPr binds to the cre-like element identified in the promoter region of the xylPQ operon is not yet known.
This is the first report which describes the primary structure of an α-xylosidase specific for isoprimeverose. This is also the first demonstration that a gene locus, xylPQRAB, which comprises genes involved in the metabolism of both d-xylose and isoprimeverose has been found in bacteria (the characterization of XylP as an isoprimeverose transporter will be described elsewhere). Since L. pentosus is frequently associated with lactic-acid fermentation on vegetables, the ability of this bacterium to utilize isoprimeverose as an energy source may not be casual. The hydrolysis of isoprimeverose by XylQ to glucose and xylose, and the subsequent catabolism of the xylose moiety by XylA and XylB, may represent a straightforward and major pathway of d-xylose assimilation from the plant cell wall in L. pentosus. However, L. pentosus is unable to ferment xyloglucan oligosaccharides and therefore is unable to liberate isoprimeverose from these substrates. Nevertheless, hydrolysis of xyloglucan might be achieved by fibrolytic microorganisms which can be associated with the fermentation of plant materials. Indeed, previous studies have shown that many microorganisms, such as gram-negative bacteria, yeasts, molds, and lactic acid bacteria, can be involved in the natural fermentation of vegetables (5). Because of the complex array of microorganisms present during fermentation, the various polymers of the plant primary cell wall might undergo a significant breakdown, resulting in the liberation of isoprimeverose. A small amount of free xylose might be present as a result of hemicellulose hydrolysis and could mediate induction of xylPQ expression.
Our data indicate that XylQ is an enzyme which has a stringent substrate specificity and a high affinity for isoprimeverose (apparent Km, 0.2 mM). Although a measurable amount of xylose could be released from a mix of xyloglucan oligosaccharides, the rate of hydrolysis was lower than that for isoprimeverose and required a higher concentration of the substrate. It is possible that the enzyme recognized with a low affinity the isoprimeverose unit (X) located at the nonreducing end of the nona-, octa-, or heptasaccharides and then cleaved the α-xylosidic linkage. A similar activity has been demonstrated for the α-xylosidase from pea seedlings (24) and from Aspergillus niger (22). Particularly interesting is the observation that the L. pentosus α-xylosidase shares some features with the glycosyl hydrolase family 31. Our results demonstrate that, unlike the members of this family, XylQ does not hydrolyze the α-glycosidic bonds in maltose (α-1,4), isomaltose (α-1,6), and sucrose (α-1,2) but hydrolyzes only isoprimeverose. However, the only structural difference between isoprimeverose and isomaltose is a hydroxy methyl group at the C-6 position of the nonreducing end of the disaccharide. Based on this observation, it is conceivable that two enzymes hydrolyzing very similar sugars may display some preservation of their structural domains. This view is supported by the additional finding that in XylQ an aspartic acid residue which has previously been characterized as belonging to the active site in the intestinal sucrase-isomaltase complex from mammals (15, 29) and the human lysosomal α-glucosidase (13) is conserved (see Fig. 4A). Together, these observations strongly suggest that the hydrolysis of isoprimeverose by XylQ might follow a catalytic mechanism similar to that involved in the hydrolysis of maltose and isomaltose by the α-glucosidases of family 31. It is important to note that XylQ is not similar to the isomaltase enzymes of bacterial origin, which are mostly grouped within family 13 (dextran α-glucosidases and α-amylases) or family 15 (glucoamylases). The reason why a bacterial α-xylosidase is more homologous to eukaryotic isomaltases than to prokaryotic isomaltases is not immediately clear. This may indicate that the genes involved in the utilization of isoprimeverose in bacteria and isomaltose in eukaryotes could have evolved from a common ancestral genetic system. Unfortunately, since the primary structures of the α-xylosidases described earlier (22, 24, 39, 40, 43) are not yet available, it is not known whether they are structurally similar to XylQ.
Another interesting feature of the L. pentosus α-xylosidase is its membrane association. The hydrophobicity profile of XylQ does not reveal the presence of a putative signal sequence at the N terminus of the protein, nor does it show the presence of anchor sequences at one of the two extremities of the protein. It is therefore unlikely that α-xylosidase would be translocated across the membrane. Moreover, the two putative transmembrane helices located within XylQ are also conserved in the eukaryotic α-glucosidases. Since the eukaryotic enzymes were shown not to span the cytoplasmic membrane, the putative transmembrane segments of XylQ are presumably involved in the folding of the enzyme rather than in its attachment to the membrane. The possibility exists that XylQ might be associated with the cytoplasmic membrane through interaction with other membrane proteins. It should be noted that the membrane fraction of L. pentosus was obtained after passage through a French pressure cell and therefore was composed of inside-out vesicles. The measurement of α-xylosidase activity in the membrane fraction of the wild-type bacteria indicates that the substrates were directly accessible to the enzyme (XylQ located inside the cytoplasmic membrane, corresponding to the outside of the vesicles). Based on these observations, we conclude that XylQ is an α-xylosidase which most likely hydrolyzes isoprimeverose intracellularly. Strong support for this view, indeed, is the presence of a gene encoding a putative transporter (xylP) upstream of xylQ.
Finally, it is not yet known whether the E. coli hypothetical proteins f772 and f678 are α-xylosidases or α-glucosidases and whether the f479-f772 and f723-f470-f678 gene clusters are involved in the utilization of isoprimeverose. However, the ability to utilize isoprimeverose has certainly not been investigated for most bacterial species, and it seems probable that other examples of a xylPQ system will be found.
ACKNOWLEDGMENTS
We thank Gerrit Beldman, who kindly provided us with the xyloglucan oligosaccharides.
This work was supported by a grant from the EC (BIO2-CT92-0137).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bergmeyer H U. Methods of enzymatic analysis. VI. Deerfield Beach, Fla: VCH Publishers; 1985. [Google Scholar]
- 3.Burland V D, Plunkett III G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organisational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 4.Callens M, Kerstens-Hilderson H, van Opstal O, de Bruine C K. Catalytic properties of d-xylose isomerase from Streptomyces violaceoruber. Enzyme Microb Technol. 1986;8:696–700. [Google Scholar]
- 5.Daeschel M A, Andersson R E, Fleming H P. Microbial ecology of fermenting plant materials. FEMS Microbiol Rev. 1987;46:357–367. [Google Scholar]
- 6.Dahl M K, Degenkolb F R, Hillen W. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J Mol Biol. 1994;243:413–424. doi: 10.1006/jmbi.1994.1669. [DOI] [PubMed] [Google Scholar]
- 7.Fry S C, York W S, Albersheim P, Darvill A, Hayashi T, Josseleau J-P, Kato Y, Pérez Lorences E, Maclachlan G A, McNeil M, Mort A J, Reid J S G, Ulrich Seitz H, Selvendran R R, Voragen A G J, White A R. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant. 1993;89:1–3. [Google Scholar]
- 8.Hastrup, S. Personal communication.
- 9.Hastrup S. Analysis of Bacillus subtilis xylose regulon. In: Cramson A T, Hoch J A, editors. Genetics and biotechnology of Bacilli. New York, N.Y: Academic Press; 1988. pp. 79–84. [Google Scholar]
- 10.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli LacI-GalR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 11.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hensel A. Xyloglucane-Struktur, Genese und Funktionen einer weit verbreiteten Stoffgruppe. Pharm Unserer Zeit. 1993;22:228–234. doi: 10.1002/pauz.19930220426. [DOI] [PubMed] [Google Scholar]
- 13.Hermans M M P, Kroos M A, van Beeumens J, Oostra B A, Reuser A J J. Human lysosomal α-glucosidase. Characterization of the catalytic site. J Biol Chem. 1991;266:13507–13512. [PubMed] [Google Scholar]
- 14.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 15.Hunziker W, Spiess M, Semenza G, Lodish H F. The sucrase-isomaltase complex: primary structure, membrane-orientation, and evolution of a stalked, intrinsic brush border protein. Cell. 1986;46:227–234. doi: 10.1016/0092-8674(86)90739-7. [DOI] [PubMed] [Google Scholar]
- 16.Kinsella B T, Hogan S, Larkin A, Cantwell B A. Primary structure and processing of the Candida tsukubaensis α-glucosidase. Eur J Biochem. 1991;202:657–664. doi: 10.1111/j.1432-1033.1991.tb16420.x. [DOI] [PubMed] [Google Scholar]
- 17.Liang W J. The glucuronide transport system of Escherichia coli. Ph.D. thesis. Cambridge, England: University of Cambridge; 1992. [Google Scholar]
- 18.Lokman B C, van Santen P, Verdoes J, Krüse J, Leer R J, Posno M, Pouwels P H. Organization and characterization of three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol Gen Genet. 1991;230:161–169. doi: 10.1007/BF00290664. [DOI] [PubMed] [Google Scholar]
- 19.Lokman B C, Leer R J, van Sorge R, Pouwels P H. Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol Gen Genet. 1994;245:117–125. doi: 10.1007/BF00279757. [DOI] [PubMed] [Google Scholar]
- 20.Lokman B C, Heerikhuisen M, Leer R J, van den Broek A, Borsboom Y, Chaillou S, Postma P W, Pouwels P H. Regulation of expression of the Lactobacillus pentosus xylAB operon. J Bacteriol. 1997;179:5391–5397. doi: 10.1128/jb.179.17.5391-5397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo M, Seki T, Mitsuishi Y, Shoun H, Nakahara T. Purification and characterization of an intracellular α-d-xylosidase II from Penicillium wortmannii IFO 7237. Biosci Biotechnol Biochem. 1996;60:341–343. doi: 10.1271/bbb.60.341. [DOI] [PubMed] [Google Scholar]
- 22.Matsushita J, Kato Y, Matsuda K. Purification and properties of an α-d-xylosidase from Aspergillus niger. J Biochem. 1985;98:825–832. doi: 10.1093/oxfordjournals.jbchem.a135341. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa K, Hayashi T, Okamura K. Conformational analysis of xyloglucans. Int J Biol Macromol. 1990;12:218–222. doi: 10.1016/0141-8130(90)90036-a. [DOI] [PubMed] [Google Scholar]
- 24.O’Neil R A, Albersheim P, Darvill A D. Purification and characterization of a xyloglucan oligosaccharide-specific xylosidase from pea seedlings. J Biol Chem. 1989;264:20430–20437. [PubMed] [Google Scholar]
- 25.Plunkett G, III, Burland V, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993;15:3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poolman B, Knol J, van der Does C, Henderson P J F, Liang W J, Leblanc G, Pourcher T, Mus-Veteau I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol Microbiol. 1996;19:911–922. doi: 10.1046/j.1365-2958.1996.397949.x. [DOI] [PubMed] [Google Scholar]
- 27.Pouwels P H, Leer R J. Genetics of lactobacilli: plasmids and gene expression. Antonie Leeuwenhoek. 1993;64:85–107. doi: 10.1007/BF00873020. [DOI] [PubMed] [Google Scholar]
- 28.Pouwels P H, van Luijk N, Leer R J, Posno M. Control of replication of the Lactobacillus pentosus plasmid p353-2: evidence for a mechanism involving transcriptional attenuation of the gene coding for the replication protein. Mol Gen Genet. 1994;242:614–622. doi: 10.1007/BF00285285. [DOI] [PubMed] [Google Scholar]
- 29.Quaroni A, Semenza G. Partial amino acid sequences around the essential carboxylate in the active sites of the intestinal sucrase-isomaltase complex. J Biol Chem. 1976;251:3250–3253. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki K, Nakalima H, Imahori K. Acetate kinase from Bacillus stearothermophilus. Methods Enzymol. 1982;90:179–180. doi: 10.1016/s0076-6879(82)90124-0. [DOI] [PubMed] [Google Scholar]
- 34.Tibbot B K, Skadsen R W. Molecular cloning and characterization of a gibberellin-inducible, putative α-glucosidase gene from barley. Plant Mol Biol. 1996;30:229–241. doi: 10.1007/BF00020110. [DOI] [PubMed] [Google Scholar]
- 35.Vincken J P, de Keizer A, Beldman G, Voragen A G J. Fractionation of xyloglucan fragments and their interaction with cellulose. Plant Physiol. 1995;108:1579–1585. doi: 10.1104/pp.108.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincken J P. Enzymic modification of cellulose-xyloglucan networks. Ph.D. thesis. Wageningen, The Netherlands: Agricultural University of Wageningen; 1996. [Google Scholar]
- 37.Warren R A J. Microbial hydrolysis of polysaccharides. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 38.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa K, Yamamoto K, Okada S. Purification and characterization of an intracellular α-d-xylosidase I from Aspergillus flavus MO-5. Biosci Biotechnol Biochem. 1993;57:1275–1280. doi: 10.1271/bbb.57.1275. [DOI] [PubMed] [Google Scholar]
- 40.Yoshikawa K, Yamamoto K, Okada S. Purification and characterization of an intracellular α-d-xylosidase II from Aspergillus flavus MO-5. Biosci Biotechnol Biochem. 1993;57:1281–1285. doi: 10.1271/bbb.57.1281. [DOI] [PubMed] [Google Scholar]
- 41.Yoshikawa K, Yamamoto K, Okada S. Classification of some α-glucosidases and α-xylosidases on the basis of substrate specificity. Biosci Biotechnol Biochem. 1994;58:1392–1398. doi: 10.1271/bbb.58.1392. [DOI] [PubMed] [Google Scholar]
- 42.Zanoni P, Farrow J A E, Phillips B A, Collins M D. Lactobacillus pentosus (Fred, Peterson, and Anderson) sp. nov., nom. rev. Int J Syst Bacteriol. 1987;37:339–341. [Google Scholar]
- 43.Zong N, Kamiyama Y, Yasui T. Substrate specificity of Bacillus α-d-xylosidase. Agric Biol Chem. 1989;53:2129–2139. [Google Scholar]