Abstract
G-protein coupled receptors for the pineal hormone melatonin have been partially cloned from rats. However, insufficient information about their cDNA sequences has hindered studies of their distribution and physiological responses to melatonin using rats as an animal model. We have cloned cDNAs of two rat membrane melatonin receptor subtypes, melatonin receptor 1a (MT1) and melatonin receptor 1b (MT2), using a rapid amplification of cDNA end (RACE) method. The rat MT1 and MT2 cDNAs encode proteins of 353 and 364 amino acids, respectively, and show 78–93% identities with the human and mouse counterparts. Stable expression of either rat MT1 or MT2 in NIH3T3 cells resulted in high affinity 2-[125I]-iodomelatonin (125I-Mel) binding (K d = 73.2 ± 9.0 and 73.7 ± 2.9 pM, respectively), and exhibited a similar rank order of inhibition of specific 125I-Mel binding by five ligands (2-iodomelatonin > melatonin > 6-hydroxymelatonin > luzindole > N-acetyl-5-hydroxytryptamine). RT-PCR analysis showed that MT1 is highly expressed in the hypothalamus, lung, kidney, adrenal gland, stomach, and ovary, while MT2 is highly expressed in the hippocampus, kidney, and ovary. We also performed multi-cell RT-PCR to examine the expression of mRNAs encoding MT1 and MT2 in adult GnRH neurons. MT1 was weakly expressed in male GnRH neurons, and was less expressed in the female neurons. MT2 expression was undetectable in GnRH neurons from either sex. This study delineates the gene structures, fundamental properties, and distribution of both rat melatonin receptor subtypes, and may offer opportunities to assess the physiological significance of melatonin in rats.
Electronic supplementary material
The online version of this article (doi:10.1007/s12576-008-0003-9) contains supplementary material, which is available to authorized users.
Keywords: Melatonin, Melatonin receptors, GnRH neuron
Introduction
The hormone melatonin is nocturnally synthesized and released by the pineal gland, providing a neurochemical cue of day/night cycles. Melatonin regulates a variety of physiological functions, including circadian rhythms, sleep–wake cycles, and reproduction [1]. Melatonin functions via specific high-affinity G-protein coupled membrane receptors, of which there are two receptor subtypes in mammals: melatonin receptor 1a (MT1) and melatonin receptor 1b (MT2) [2, 3].
Although cDNAs of two melatonin receptors have been cloned in humans and mice [4–7], only partial cDNA sequences have been identified in rats [4, 8, 9]. As a result, the paucity of information about complete open reading frames (ORFs), and 5′- and 3′-untranslated regions (UTRs) of rat MT1 and MT2 has hindered investigation of melatonin receptors in rats. In the post-genomic era, the ORF regions of rat membrane melatonin receptors can be deduced from the rat genome. Recently, pharmacological properties of the receptors were characterized using genomically identified ORF sequences [10]. However, genomically identified sequences lack information about UTRs that contain the signal motifs involved in post-transcriptional regulation and mRNA decay [11–13].
This study was carried out to characterize the fundamental properties of rat melatonin receptors. We cloned cDNAs including complete ORFs, and 3′- and 5′-UTRs of rat MT1 and MT2 in a rapid amplification of cDNA end (RACE) method, and identified their gene structures. Furthermore, we confirmed specific melatonin binding to the receptors, and determined the mRNA expression patterns in the central nervous system, several peripheral tissues, and GnRH neurons by RT-PCR.
Materials and methods
Chemicals
2-[125I]-Iodomelatonin (125I-Mel) was purchased from GE Healthcare Bio-Sciences (Fairfield, CT). Melatonin, 6-hydroxymelatonin, and N-acetyl-5-hydroxytryptamine (NAS) were obtained from Wako Chemicals (Osaka, Japan), and 2-iodomelatonin (I-Mel) and luzindole from TOCRIS bioscience (Bristol, UK).
Animals
All experiments were performed with the approval of the Nippon Medical School Animal Care Committee. Wistar rats, and transgenic rats that express enhanced green fluorescent protein (EGFP) under control of the GnRH promoter [14] were used in these studies. The rats had free access to water and food, and were kept under a 14-h light, 10-h dark cycle. Rats aged 2–3 months were used.
For total RNA isolation, rats were decapitated under ether anesthesia, and brains and peripheral organs were quickly removed and stored in liquid nitrogen until use.
Cell culture
NIH3T3 (cell number JCRB0615; provided from the Health Science Research Resources Bank, Osaka, Japan), Hela, and HEK293 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Sigma Chemical, St Louis, MO) supplemented with 1 mM Na pyruvate, 24 mM NaHCO3, 4 mM l-glutamine, 10% fetal bovine serum (JRH Biosciences, Lenexa, KS), 100 U/ml penicillin, and 0.1 mg/ml streptomycin without phenol red. For NIH3T3 cells stably expressing rat MT1 or MT2, 5 μg/ml blasticidin S (Invitrogen, Carlsbad, CA) was added to the culture medium. The cultures were maintained at 37°C in a water-saturated atmosphere of 95% air and 5% CO2. Cells were routinely passaged every 3–4 days, and used in experiments within 20 passages.
For experiments measuring the I-Mel binding ability and melatonin receptor mRNA expression, cells were cultured in 75-mm2 flasks for 3–4 days, then washed three times with phosphate-buffered saline (PBS: 137 mM NaCl, 8.10 mM Na2HPO4, 2.68 mM KCl, 1.47 mM KH2PO4, and 0.5 mM EDTA, pH 7.4), and pelleted by centrifugation at 300×g for 5 min. Cell pellets were frozen under liquid nitrogen and stored at −80°C until use.
Total RNA isolation
Total RNA was extracted using a SV Total RNA Isolation System (Promega, Madison, WI) following the manufacturer’s instructions. Total RNA was then treated with Turbo DNase (RNase-free DNase I; Ambion, Austin, TX), and purified. The concentration was quantified by absorption at 260 nm.
5′- and 3′- RACEs of rat melatonin receptors
The unknown cDNA sequences including 5′- and 3′-UTRs of rat melatonin receptors were analyzed by 5′- and 3′-RACEs. 5′-RACE was performed using a concatemer formation method. Total RNA isolated from male rat hypothalamus was reverse-transcribed with 5′-phosphorylated RT primers. Reaction mixtures (final volume 25 μl) contained 10 μg of total RNA, 1× RT buffer, 1 mM dNTP mixture, 5 pmol RT primer, 20 U RNasin Plus (RNase inhibitor; Promega), and 100 U ReverTra Ace (M-MLV reverse transcriptase (RTase), RNase H (–); TOYOBO, Osaka, Japan). The reaction was carried out at 42°C for 60 min, and stopped by heating at 75°C for 15 min. Subsequently, single-strand cDNA was obtained using RNase H (Takara bio, Shiga, Japan). 5′-phosphorylated single-strand cDNA was concatemerized by T4 RNA ligase (Takara bio), and two-round PCR with gene-specific primers was performed to amplify the 5′-RACE products. The first strand cDNA for 3′-RACE was synthesized from male rat hypothalamus total RNA with adapter-oligo(dT)18 primers. 3′-RACE fragments were amplified using two-round PCR with gene-specific primers and 3′-RACE adapter primers. The conditions for the first- and second-round PCRs consisted of 28 cycles of 94°C for 30 s, 60°C for 20 s and 72°C for 1 min, with an initial denaturing at 94°C for 2 min and a final elongation of 72°C for 5 min. The reaction was performed in 50 μl of reaction mixture comprising the RACE cDNAs corresponding to 1 μg of total RNA, 1× GC PCR buffer, 0.4 mM dNTP mixture, 0.4 μM forward and reverse primers, and 1.2 U LA Taq polymerase (Takara bio). The oligonucleotide primers used in RACEs are listed in Table 1. RACE products were excised, purified, and cloned into pGEM-T-Easy vectors (Promega). The complete ORFs, and 5′- and 3′-UTRs of rat MT1 and MT2 cDNAs were identified by DNA sequencing.
Table 1.
Primers used for 3′-, 5′-RACE, ORF cloning, and plasmid construction
| Gene | Direction | Primer sequences |
|---|---|---|
| 3′-RACE | ||
| Universal | ||
| RT primer | Reverse | 5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18-3′ |
| 1st PCR | Reverse | 5′-GCTGTCAACGATACGCTACGTAACG-3′ |
| 2nd PCR | Reverse | 5′-CGCTACGTAACGGCATGACAGTG-3′ |
| MT1 | ||
| 1st PCR | Forward | 5′-AAACCGGACAGCAAACCCAAACT-3′ |
| 2nd PCR | Forward | 5′-GGGCCCCACTCAACTTCATAGGTC-3′ |
| MT2 | ||
| 1st PCR | Forward | 5′-CCGAAGGAAGGCAAAGGCTGAGAG-3′ |
| 2nd PCR | Forward | 5′-CCAGAAGGGCTTTTTGTCACCAG-3′ |
| 5′-RACE | ||
| MT1 | ||
| RT primer | Reverse | 5′-pAGTAACTAGCCACGAAGA-3′ |
| 1st PCR | Forward | 5′-AAACCGGACAGCAAACCCAAACT-3′ |
| Reverse | 5′-GCCCAGGATGTCCACCACGATAGT-3′ | |
| 2nd PCR | Forward | 5′-GGGCCCCACTCAACTTCATAGGTC-3′ |
| Reverse | 5′-ACGACGGCCGCGATCTTATTTC-3′ | |
| MT2 | ||
| RT primer | Reverse | 5′-pATTAAGGCAGCTGTTGAA-3′ |
| 1st PCR | Forward | 5′-CCGAAGGAAGGCAAAGGCTGAGAG-3′ |
| Reverse | 5′-GGTGCTGGCTGTCTGGATGAAGGT-3′ | |
| 2nd PCR | Forward | 5′-CCAGAAGGGCTTTTTGTCACCAG-3′ |
| Reverse | 5′-TGACTGCAGGCTCGGTGGTAGGT-3′ | |
| ORF cloning | ||
| MT1 | Forward | 5′-GCGCGGGGCTACAGGATGATG-3′ |
| Reverse | 5′-CAGGAGGTCCGCTCCAACACTATG-3′ | |
| MT2 | Forward | 5′-GGAGCGCCCCCAAGCAGT-3′ |
| Reverse | 5′-AGGTCAAGGTGGCAGGGAAAATG-3′ | |
| Plasmid construction | ||
| MT1 | Forward | 5′-CACCATGAAGGGCAATGTCAGC-3′ |
| Reverse | 5′-TTAAACAGAGTCCACCTTTATTAAATT-3′ | |
| MT2 | Forward | 5′-CACCATGCCTGACAACAGCTCCAT-3′ |
| Reverse | 5′-CTAGAGAGCACCTTCCTGGACAG-3′ | |
Construction of NIH3T3 cells that stably express rat MT1 or MT2
Rat MT1 and MT2 cDNAs were first amplified with LA Taq polymerase, cloned into pGEM-T-Easy vectors, and sequenced. The ORFs of the receptors were then amplified from pGEM-T-Easy-rat MT1 or pGEM-T-Easy-rat MT2 vectors using the primers shown in Table 1, subcloned into pcDNA6.2-Gatway Directional TOPO vectors (Invitrogen), and confirmed by DNA sequencing. Recombinant plasmid vectors (pcDNA6.2-rat MT1 or pcDNA6.2-rat MT2) were introduced into NIH3T3 cells using a GeneJuice Transfection Reagent (Novagen, Darmstadt, Germany), according to the manufacturer’s instructions. Cells were cultured in supplemented DMEM containing 5 μg/ml blasticidin S until the resistant clones were apparent. Each colony was picked up, and the cells were dispersed and plated onto 96-well culture plates at a concentration of 0.1 cells/well. The clones were tested by the I-Mel binding assay and RT-PCR analysis, and positive clones were maintained for subsequent experiments.
I-Mel binding assay
Cell pellets were homogenized in binding buffer (50 mM Tris–HCl and 1 mM MgCl2) and briefly sonicated. The protein concentration was measured using a BCA Protein Assay Reagent Kit (Pierce, Rockford, IL). Cell lysates (1 mg/ml whole cell protein) were incubated in 200 μl binding buffer with 0.1 mg/ml bovine serum albumin and 125I-Mel for 12 h at room temperature (25°C). In saturation studies, 125I-Mel was added at concentrations ranging from 1 to 200 pM. In competition curves with melatonin and its analogues, the radioligand concentration was 100 pM. Incubation was terminated by dilution with 5 ml of ice-cold binding buffer, followed by filtration through glass-fiber filters (Whatman, Wheeling, IL). The filters were washed four times with 5 ml binding buffer. Non-specific binding was assessed in the presence of 20 μM cold melatonin.
RT-PCR
Total RNA was reverse-transcribed into first-strand cDNA using oligo(dT) primer. Reaction mixtures (final volume 25 μl) contained 10 μg of total RNA, 1× RT buffer, 1 mM dNTP mixture, 1 μg oligo(dT)15 (Promega), 20 U RNasin Plus (Promega), and 100 U ReverTra Ace (TOYOBO). The reaction was carried out at 42°C for 60 min, and stopped by heating at 75°C for 15 min. cDNA was treated with 4 U of RNase H at 37°C for 30 min, and stored at −20°C until use.
PCR was performed in 25 μl of reaction mixture comprising cDNA corresponding to 200 ng of total RNA, 1× PCR buffer, 0.2 mM dNTP mixture, 0.2 μM forward and reverse primers, and 0.63 U Blend Taq polymerase (TOYOBO). PCR was performed in three steps, with a PCR cycle reaction of 95°C for 30 s, 60°C for 20 s, and 72°C for 30 s, with an initial denaturing at 95°C for 2 min and a final elongation at 72°C for 5 min. The sequences of oligonucleotide primers used in RT-PCR are shown in Table 2. To avoid amplifying any contaminating genomic DNA, the primer pairs were designed from different exons.
Table 2.
Primers used for RT-PCR and multi-cell RT-PCR
| Gene | Direction | Primer sequences | Product size (bp) |
|---|---|---|---|
| RT-PCR | |||
| MT1 | Forward | 5′-ATGGCCCTGGCTGTGCTGCGGTAAG-3′ | 316 |
| Reverse | 5′-TAAGTATAGACGTCAGCGCCAAGGGAAATG-3′ | ||
| MT2 | Forward | 5′-GGAGCGCCCCCAAGCAGTG-3′ | 390 |
| Reverse | 5′-GGATCTCCCCAAGTACCCAACCGTCAT-3′ | ||
| β-Actin | Forward | 5′-GTCCACACCCGCCACCAGT-3′ | 496 |
| Reverse | 5′-CGTCTCCGGAGTCCATCACAAT-3′ | ||
| mGAPDH | Forward | 5′-TGAAGGTCGGTGTGAACGGATTTG-3′ | 359 |
| Reverse | 5′-GGCGGAGATGATGACCCTTTTG-3′ | ||
| Multi-cell RT-PCR | |||
| MT1 | |||
| 1st PCR | Forward | 5′-ATGGCCCTGGCTGTGCTGCGGTAAG-3′ | 442 |
| Reverse | 5′-TGTGGCAAATGTAGCAGTAGCGGTTCA-3′ | ||
| 2nd PCR | Forward | 5′-CAGGCGGCGGGGAGGAAATAAG-3′ | 231 |
| Reverse | 5′-TAAGTATAGACGTCAGCGCCAAGGGAAATG-3′ | ||
| MT2 | |||
| 1st PCR | Forward | 5′-GGAGCGCCCCCAAGCAGTG-3′ | 390 |
| Reverse | 5′-GGATCTCCCCAAGTACCCAACCGTCAT-3′ | ||
| 2nd PCR | Forward | 5′-CGGGCTGCAGCGTCACCAT-3′ | 275 |
| Reverse | 5′-GTCAGCCAAGGCCAGATTCACC-3′ | ||
| GnRH | Forward | 5′-ACTGATGGCCGCTGTTGTTCT-3′ | 256 |
| Reverse | 5′-CTTCTTCTGCCCAGCTTCCTCTTCA-3′ | ||
Forward and reverse primers were designed on different exons to distinguish the amplification of cDNA templates from that of contaminating genomic DNA. To design the PCR primers, we referred to the following nucleotide sequences: AB377274 for MT1, AB377275 for MT2, NM_031144 for β-actin, NM_008084 for mGAPDH, and M12579 for GnRH
Multi-cell RT-PCR
Slice preparation and cell harvest are described in detail elsewhere [15, 16]. In brief, coronal slices (200 μm thick) containing medial septum, diagonal band of Broca, organum vasculosum of the lamina terminalis and medial preoptic area were prepared from adult male and female transgenic rats. The slice was viewed under an uplight fluorescence microscope (BX50; Olympus, Tokyo, Japan). The cytoplasmic contents were harvested from five GnRH neurons under visual control, and pooled in a thin-wall PCR tube containing RNasin Plus.
The harvested contents were heated with random hexamer primers (Promega), at 95°C for 5 min and then cooled on ice for 1 min. The reverse transcription mixture (50 μl) contained cytoplasmic contents from five cells, 1× RT buffer, 1 mM dNTP mixture, 500 ng random hexamer primers, fresh 40 U RNasin Plus, and 200 U ReverTra Ace. Reverse transcription was carried out at 30°C for 10 min and then 42°C for 45 min. After stopping the reaction by heating at 75°C for 15 min, the reaction mixture was treated with RNase H at 37°C for 30 min and stored at −80°C until use.
To confirm successful cDNA synthesis from the cytoplasmic contents of GnRH neurons, one-round PCR amplification was performed using 10 μl aliquots of the reverse transcription mixture as a template. For melatonin receptors, two-round PCR amplification was performed using 20 μl aliquots of the reverse transcription mixture as a template for the first PCR and 0.5 μl aliquots of the first PCR solution as a template for the nested PCR. The PCR conditions were 94°C for 2 min, 26 cycles for MT1 and 28 cycles for MT2 of 94°C for 30 s, 60°C for 20 s, and 72°C for 30 s, and finally 72°C for 5 min. The PCR mixture (50 μl) contained template DNA, 1× PCR buffer, 0.2 mM dNTP mixture, 0.2 μM forward and reverse primers, and 1.2 U Blend Taq polymerase. Primer sequences are shown in Table 2.
Electrophoresis
PCR products (5 μl) were separated by electrophoresis on 2% agarose gels, and visualized by ethidium bromide staining under UV irradiation. Gel images were captured using a FAS-III system (TOYOBO).
DNA sequencing
PCR products were extracted from agarose gels using a Wizard SV Gel and PCR Clean-up System (Promega), and cloned into pGEM-T-Easy vectors (Promega). Sequencing reactions were performed using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Fluorescent signals were detected using an ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
Results
Cloning and sequencing of rat MT1 and MT2 cDNAs
According to the partial cDNA sequences (accession number AF130341 for MT1 and accession number AF141863 for MT2 [9]), primers were synthesized for 3′- and 5′-RACEs to obtain the remaining sequences of rat MT1 and MT2 cDNAs. The cloned cDNA sequences of rat MT1 (accession number AB377274) and MT2 (accession number AB377275) contained 1,775 and 1,858 bp nucleotides, consisting of 206 and 115 bp of 5′-UTR, 1,062 and 1,095 bp ORF, and 507 and 648 bp 3′-UTR, respectively (Fig. 1). We identified two other forms of rat MT1 transcripts resulting from multiple polyadenylation signals (underlined in Fig. 1a).
Fig. 1.

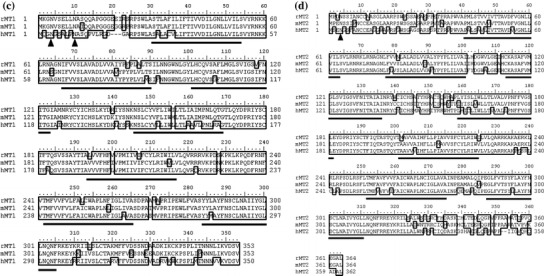
Structural analysis of rat melatonin receptor nucleotide and deduced amino acid sequences. cDNA sequences and deduced amino acid sequences of rat MT1 (a) and MT2 (b). The polyadenylation signals are underlined, and the polyadenylation sites are boxed. Shaded regions are transmembrane domains. We registered these two sequences as accession numbers AB377274 for rat MT1 and AB377275 for rat MT2. Alignments of the deduced rat MT1 (c) and MT2 (d) protein amino acid sequences with mouse and human counterparts. Identical amino acids are outlined. Solid lines and arrows indicate transmembrane domains and potential N-glycosylation sites, respectively. Genbank accession numbers referred are as follows: rat MT1 (rMT1), AB377274; mouse MT1 (mMT1), NM_008639; human MT1 (hMT1), NM_005958; rat MT2 (rMT2), AB377275; mouse MT2 (mMT2), AB377276; human MT1 (hMT2), NM_005959
These sequences exhibited the typical structures of the G-protein coupled receptor family. Two subtypes of rat receptors share 54.7% identity with each other at the amino acid level. When compared with other mammalian melatonin receptors, rat MT1 shows 84.1 and 92.6% identity (Fig. 1c), and rat MT2 exhibits 78.3% and 88.7% identity with the respective human and mouse receptors (Fig. 1d).
Genomic organization of rat MT1 and MT2
Using a BLAT alignment program [17], we mapped the rat MT1 and MT2 cDNA sequences on the rat genome version 3.4 (Fig. 2). Rat MT1 gene is located at 16q11 on rat chromosome 16 and consists of two exons (Fig. 2a). This structure is homologous to that of the human and mouse MT1 genes. Rat MT2 gene lies at 8q12 on rat chromosome 8, and consists of three exons (Fig. 2b). The 5′- and 3′-ends of the introns of both genes conform to the GT–AG rule for splicing. We newly identified mouse MT2 cDNA containing 5′- and 3′-UTRs (accession number AB377276), and mapped it on the Build 37 mouse genome assembly (Suppl. Fig. 1A). The rat MT2 gene has a homologous structure with the mouse gene. Using RT-PCR with specific primers designed on exon 2 and 3, we further confirmed that these MT2 genes were properly transcribed (Suppl. Fig. 1B). In contrast, the human MT2 gene consists of two exons (accession number BC069163, data not shown).
Fig. 2.
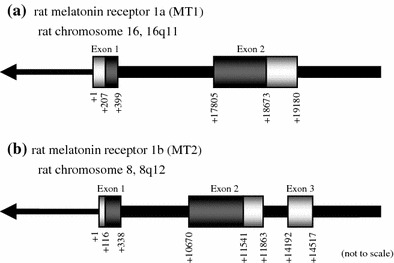
Genomic organization of rat MT1 and MT2. Schematic representation of rat MT1 (a) and MT2 (b) gene structures. cDNA sequences are mapped on rat genome version 3.4. The rat MT1 gene is located at 16q11 on rat chromosome 16, and consists of two exons. On the other hand, rat MT2 gene lies at 8q12 on rat chromosome 8, and consists of three exons. The white and gray boxes indicate the UTRs and ORFs, respectively. The arrows indicate the orientation of the chromosomes. The images are not to scale
Biochemical properties of rat MT1 and MT2
To characterize the biochemical properties of rat MT1 and MT2 using homogeneous biological materials, we constructed cell lines that stably express rat MT1 or MT2. We selected NIH3T3 cells as host cells because I-Mel binding assay and RT-PCR analysis revealed that the cells did not express mouse melatonin receptors (Suppl. Fig. 2). We constructed expression vectors of rat MT1 and MT2 from the identified sequences, and developed NIH3T3 cell lines stably transfected with either rat MT1 or MT2 (Fig. 3).
Fig. 3.
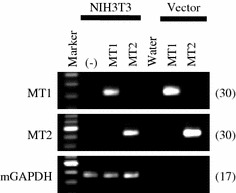
RT-PCR analysis of rat MT1 and MT2 mRNA expression in NIH3T3 cell lines stably transfected with pcDNA6.2-rat MT1 or pcDNA6.2-rat MT2. We performed RT-PCR to confirm the stable expression of rat MT1 and MT2 in transfected NIH3T3 cells. Expression of MT1 (upper panel), MT2 (middle panel), and mouse GAPDH (mGAPDH) (lower panel). “(–)” indicates samples prepared from untransfected NIH3T3 cells. 0.1 ng of pcDNA6.2-rat MT1 or pcDNA6.2-rat MT2 vectors were used as positive controls. The number on the right of each panel indicates the number of PCR cycles performed
Binding and biochemical properties were examined by stable expression of either rat MT1 or MT2 in NIH3T3 cells. Scatchard analysis of the saturation data showed that NIH3T3 cells transfected with either receptor cDNA bound 125I-Mel with high affinity (Fig. 4). The K d values of the cells expressing rat MT1 or MT2 were 73.2 ± 9.0 pM (mean ± SEM; n = 4 experiments) and 73.7 ± 2.9 pM (n = 4), respectively. The B max values were 27.1 ± 2.7 fmol/mg of whole cell protein for rat MT1 and 30.0 ± 2.4 fmol/mg for rat MT2.
Fig. 4.
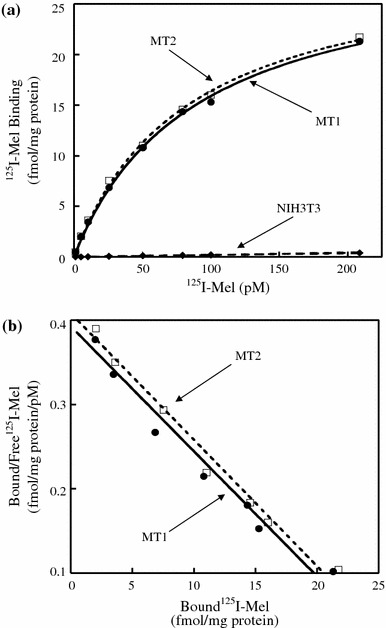
Scatchard analysis of specific 125I-Mel binding to cell lysates prepared from NIH3T3 cells stably expressing either rat MT1 or MT2. a 125I-Mel saturation binding to rat MT1 or MT2 in NIH3T3 cells. Cell lysates were incubated with various concentrations of 125I-Mel (1–200 pM). Non-specific binding was measured in the presence of 20 μM melatonin. Specific 125I-Mel binding is defined as total binding minus non-specific binding. b Scatchard plots of saturation binding to rat MT1 and MT2. The K d, B max, and Hill’s values depicted are 67.6 pM, 26.5 fmol/mg of whole cell protein, and 1.01 for rat MT1, and 66.7 pM, 27.2 fmol/mg, and 1.01 for rat MT2, respectively. Data shown are representative of four experiments. The K d, B max, and Hill’s values of four experiments are 73.2 ± 9.0 pM (mean ± SEM), 27.1 ± 2.7 fmol/mg, and 1.01 ± 0.02 for rat MT1, and 73.7 ± 2.9 pM, 30.0 ± 2.4 fmol/mg, and 1.00 ± 0.01 for rat MT2, respectively
The biochemical characteristics for inhibition of specific 125I-Mel binding in transfected NIH3T3 cells were next examined for rat MT1 and MT2 (Fig. 5; Table 3). The rank order of inhibition by melatonin and its analogues for rat MT1 was similar to that found for rat MT2. The rank order of inhibition was I-Mel > melatonin > 6-hydroxymelatonin > luzindole > NAS.
Fig. 5.
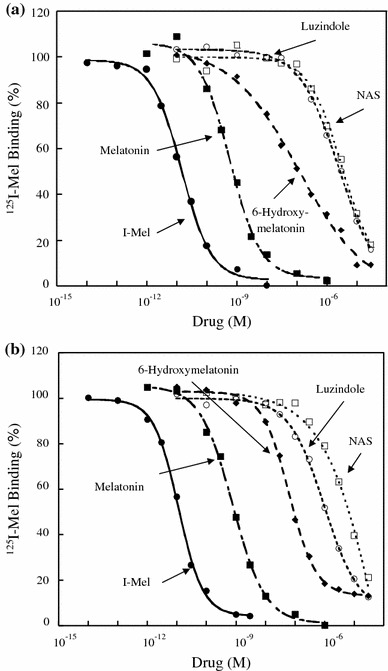
Competition curves for inhibition of specific 125I-Mel binding by melatonin and its analogues in NIH3T3 cells stably expressing either rat MT1 or MT2. Cell lysates were incubated with 100 pM 125I-Mel and various concentrations of 2-iodomelatonin (I-Mel), melatonin, 6-hydroxymelatonin, luzindole, or N-acetyl-5-hydroxytryptamine (NAS). Non-specific binding was determined in the presence of 20 μM melatonin. Competition binding with various ligands in NIH3T3 cells transfected with either rat MT1 (a) or MT2 (b). The data shown are mean values of four experiments for each drug. The K i values are listed in Table 3
Table 3.
Competition of various ligands for specific 125I-Mel binding in NIH3T3 cells transfected with either rat MT1 or MT2 cDNA
| Compound | K i | Ratio (MT2/MT1) | |
|---|---|---|---|
| MT1 | MT2 | ||
| I-Mel | 6.22 ± 0.57 pM | 4.81 ± 0.42 pM | 0.77 |
| Melatonin | 290 ± 31 pM | 378 ± 23 pM | 1.31 |
| 6-Hydroxymelatonin | 118 ± 68 nM | 31.7 ± 4.9 nM | 0.27 |
| Luzindole | 1.15 ± 0.19 μM | 433 ± 138 nM | 0.38 |
| NAS | 1.65 ± 0.42 μM | 4.98 ± 2.84 μM | 3.00 |
The K i values are mean ± SEM of four experiments for each compound
I-Mel 2-iodomelatonin, NASN-acetyl-5-hydroxytryptamine
Distribution of rat MT1 and MT2 mRNAs in rat brain subregions and peripheral organs
We determined the distribution of MT1 and MT2 mRNAs in rat brain subregions, and peripheral organs by RT-PCR (Fig. 6). MT1 mRNA was widely expressed and was strongly expressed in the hypothalamus, lung, kidney, adrenal gland, stomach, and ovary. The intensity of MT1 signals was faint in the cerebral cortex, hippocampus, and cerebellum but, using 36 cycles of PCR amplification, dense bands were observed (data not shown). We could also observe the wide distribution of MT2 mRNA, although a relatively large number of PCR cycles (37 or 38 cycles) were required to detect the expression. High expression of MT2 was observed in the hippocampus, kidney, and ovary.
Fig. 6.
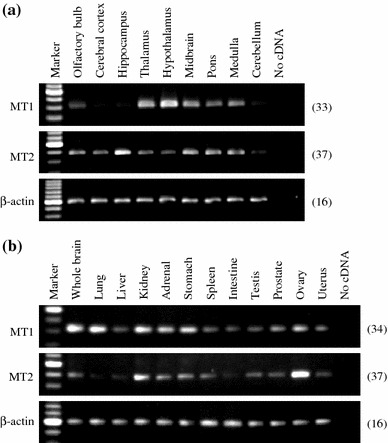
RT-PCR analysis of MT1 and MT2 mRNA expression in various brain subregions and peripheral organs. Total RNA isolated from various brain subregions and peripheral organs was subjected to RT-PCR using specific primers for MT1, MT2 and β-actin cDNAs. a Expression of MT1 (upper panel), MT2 (middle panel), and β-actin (lower panel) in rat brain subregions. b Expression of MT1 (upper panel), MT2 (middle panel) and β-actin (lower panel) in rat whole brain and peripheral organs. The number on the right of each panel indicates the number of PCR cycles performed. Similar patterns of expression were observed in three different samples
Expression of rat MT1 and MT2 mRNAs in adult rat GnRH neurons
Multi-cell RT-PCR was performed to examine the expression of mRNAs encoding MT1 and MT2 in adult male and female rat GnRH neurons (Fig. 7). The amount of template cDNA corresponding to two GnRH neurons was subjected to RT-PCR with specific primers for MT1 and MT2 cDNAs. For GnRH, the amount of cDNA corresponding to one cell was used. Positive bands of MT1 appeared in 10.6% of the reactions for male GnRH neurons, and 2.2% for female GnRH neurons. No positive bands of MT2 were detected. GnRH was positive in all reactions.
Fig. 7.
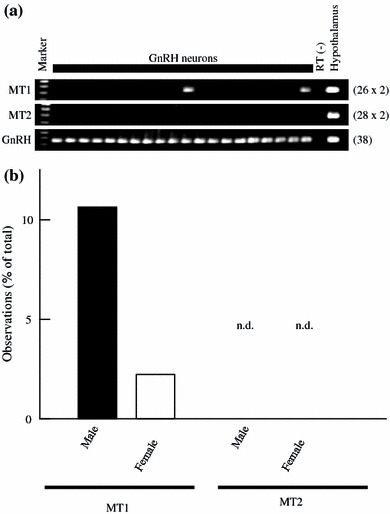
Multi-cell RT-PCR analysis of mRNAs encoding rat MT1 and MT2 in adult male and female rat GnRH neurons. Cytoplasmic contents harvested from five GnRH neurons were pooled and reverse-transcribed to generate cDNA. The amount of cDNA corresponding to two GnRH neurons was examined with each of the primer pairs for rat MT1 and MT2. For GnRH transcripts, the amount of cDNA corresponding to one GnRH neuron was used. a Representative gel images of MT1 (upper panel), MT2 (middle panel), and GnRH (lower panel) mRNA expression in adult male rat GnRH neurons. The gels show the presence of MT1 and GnRH. MT2 expression was not detectable. “RT (–)” indicates cytosol from GnRH neurons treated without reverse transcriptase. The number on the right of each panel indicates the number of PCR cycles performed. b Expression patterns of rat MT1 and MT2 mRNAs in adult male and female rat GnRH neurons. The appearance of positive bands in the multi-cell RT-PCR experiments is shown as a percentage of the total reactions for MT1 and MT2 (n = 47, 235 GnRH neurons for male; n = 45, 225 GnRH neurons for female). The bands for MT2 were not detectable (n.d.) in GnRH neurons. GnRH was positive in all reactions
Discussion
In the present study, full-length cDNAs encoding two subtypes of melatonin receptors were cloned from rat hypothalamus. The deduced proteins share high amino acid identity with other mammalian melatonin receptor counterparts, and contain motifs typical of the G protein-coupled receptor family.
We mapped the cDNA sequences on the rat genome, and found that the rat MT1 and MT2 genes are comprised of two and three exons, respectively. This structure of the rat MT1 gene is homologous to that of other mammalian MT1 genes [4, 6]. On the other hand, rat MT2 gene has an unusual structure. The last exon of MT2 gene does not contain any ORFs. In general, mRNA where the stop codon is not located on the last exon is rapidly degraded in the cytoplasm through a pathway referred to as a nonsense-mediated decay [18]. The unusual structure of rat MT2 gene is not likely to be an artifact of RACE cloning because (1) the same structure is conserved in mouse (Fig. 2, Suppl. Fig. 1A), (2) the intron boundaries of both rat and mouse MT2 genes obey the GT-AG rule, and (3) RT-PCR analysis with specific primers designed on exon 2 and 3 revealed that the transcripts are appropriately spliced (Suppl. Fig. 1B). In contrast, the human MT2 gene has a usual structure comprised of two exons, while the hamster MT2 gene lacks functional ORFs [19], suggesting that MT2 expression may be regulated in a species-specific manner.
Stable expression of rat melatonin receptor cDNAs in NIH3T3 cells showed that they have high affinities for 125I-Mel with mean K d values of 73.2 pM for rat MT1 and 73.7 pM for rat MT2. In addition, K i values for melatonin competition with specific 125I-Mel binding are characteristic of a high affinity melatonin receptor. The biochemical profiles of I-Mel and various melatonin analogues revealed similar profiles with other membrane melatonin receptors [5, 20–22].
RT-PCR analysis revealed that both rat melatonin receptor mRNAs are expressed in a wide variety of tissues although they have their own preferential expression sites. Their wide distribution may delineate the diversity of responses to melatonin within the body.
MT2 is involved in several physiological functions. However, studies using rodents lacking functional MT2 genes [7, 19] and analyses of MT1 allelic variations in ewes [23, 24] suggest that MT1 is a predominant receptor in melatonin-regulated reproductive pathways. GnRH neurons are central regulators of the reproductive axis. Several lines of evidence suggest that melatonin regulates GnRH neuronal activity and GnRH secretion through afferent neurons modulating the GnRH neuronal system [25–27]. However, direct regulatory actions of melatonin on GnRH neurons are still unknown. Our previous study showed that melatonin directly modulates GABAA receptor currents in adult rat GnRH neurons in a sexually dimorphic manner [28]. In male GnRH neurons, melatonin augments the currents, suggesting a preferential expression of MT1, because MT1 and MT2 differentially modulate GABAA receptor functions [29]. We found that MT1 mRNA expression in adult GnRH neurons exhibits a sexually dimorphic pattern (Fig. 7). This result is consistent with our previous findings. Furthermore, our recent analysis demonstrated that blocking GnRH receptor activation with the GnRH antagonist, cetrorelix, induces the expression of MT1 mRNA in immortalized GnRH-secreting neurons (GT1-7 cells). Native GnRH neurons have a functional self-stimulatory GnRH system [30]. GnRH secretion is a sexually dimorphic physiological event. Therefore, the different profiles of GnRH release between males and females may contribute to the sexually dimorphic pattern of MT1 expression in adult rat GnRH neurons via an autocrine GnRH pathway.
In conclusion, in this study, we have characterized several fundamental properties of both rat melatonin receptor subtypes, which may provide opportunities for further investigation of the physiological potential of melatonin, using rats as an animal model.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1. Genomic organization of mouse MT2, and RT-PCR analysis of rat and mouse MT2 mRNAs using specific primers designed on exon 2 and exon 3.(A) Schematic representation of mouse MT2 gene structure. We newly determined a mouse MT2 cDNA sequence containing 5′- and 3′-UTRs, which we registered as accession number AB377276. Mouse MT2 cDNA is mapped onto Build 37 of the mouse genome assembly. Mouse MT2 gene is located at 9A2 on mouse chromosome 9, and consists of three exons. The white and gray boxes indicate UTRs and ORFs, respectively. The arrow indicates the orientation of the mouse chromosome. The image is not to scale. (B) RT-PCR analysis of MT2 mRNAs in rats (a) and mice (b). To exclude the possibility that the unusual MT2 gene structures were due to any artifacts of RACE cloning, we performed RT-PCR using primers designed on exon 2 and 3. Primer sequences used in this analysis are as follows: 5′-CAATTGGCTGGGACAGAAGT-3′ (forward), and 5′-TGGGAGTGCACGTAAGTCAG-3′ (reverse) for rat MT2; 5′-CCCATGAGAAGTCATTTTCC-3′ (forward), and 5′-TTGCTTGTTATCGTGATGCT-3′ (reverse) for mouse MT2. (+), total RNA reverse-transcribed with RTase; (−), without RTase; M, 100 bp ladder marker; Hy, hypothalamus. (PPT 82 kb)
Supplemental Figure 2. I-Mel binding ability and expression of mouse melatonin receptor mRNAs in NIH3T3 cells.(A) Specific I-Mel binding ability in the presence of 100 pM 125I-Mel in HEK293, Hela, and NIH3T3 cells. The data are presented as means ± SEM (n = 4). (B) RT-PCR analysis of mouse MT1 and MT2 mRNA expression in NIH3T3 cells and mouse hypothalamus. Expression of mouse MT1 (Upper panel), mouse MT2 (Middle panel), and mGAPDH (Lower panel). Primer sequences used are as follows: 5′-TACACTGGCCTTCATCCTCATCTTTACCA-3′ (forward), and 5′-TAACTAGCCACGAACAGCCACTCTG-3′ (reverse) for mouse MT1; 5′-GGAGCGCCCCAAGCAGTG-3′ (forward), and 5′-CCTCCCCAAGGACCCAACCGTCAC-3′ (reverse) for mouse MT2. The number on the right of each panel indicates the number of PCR cycles. NIH3T3 cells express neither MT1 nor MT2. The same results were obtained in four separately prepared samples. (PPT 89 kb)
Acknowledgments
We are grateful to the Health Science Research Resources Bank for donating NIH3T3 cells. We also thank Dr. Yuko Wada-Kiyama for helpful advice. This work was supported in part by JSPS Grants-in-Aid for Scientific Research (18590070, 18590226, 19790181) and a MEXT Grant-in-Aid for Scientific Research on Priority Area (1686210).
Abbreviations
- 125I-Mel
2-[125I]-Iodomelatonin
- DMSO
Dimethyl sulfoxide
- EGFP
Enhanced green fluorescent protein
- GABA
γ-Aminobutyric acid
- GnRH
Gonadotropin-releasing hormone
- I-Mel
2-Iodomelatonin
- mGAPDH
Mouse glyceraldehyde-3-phosphate dehydrogenase
- MT1
Melatonin receptor 1a
- MT2
Melatonin receptor 1b
- NAS
N-Acetyl-5-hydroxytryptamine
- ORF
Open reading frame
- PT
Pituitary pars tuberalis
- RACE
Rapid amplification of cDNA end
- SCN
Suprachiasmatic nucleus
- UTR
Untranslated region
References
- 1.Reiter RJ. Pineal melatonin: cell biology of its synthesis and of its physiological interactions. Endocr Rev. 1991;12:151–180. doi: 10.1210/edrv-12-2-151. [DOI] [PubMed] [Google Scholar]
- 2.von Gall C, Stehle JH, Weaver DR. Mammalian melatonin receptors: molecular biology and signal transduction. Cell Tissue Res. 2002;309:151–162. doi: 10.1007/s00441-002-0581-4. [DOI] [PubMed] [Google Scholar]
- 3.Witt-Enderby PA, Bennett J, Jarzynka MJ, Firestine S, Melan MA. Melatonin receptors and their regulation: biochemical and structural mechanisms. Life Sci. 2003;72:2183–2198. doi: 10.1016/S0024-3205(03)00098-5. [DOI] [PubMed] [Google Scholar]
- 4.Reppert SM, Weaver DR, Ebisawa T. Cloning and characterization of a mammalian melatonin receptor that mediates reproductive and circadian responses. Neuron. 1994;13:1177–1185. doi: 10.1016/0896-6273(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Godson C, Mahle CD, Weaver DR, Slaugenhaupt SA, Gusella JF. Molecular characterization of a second melatonin receptor expressed in human retina and brain: the Mel1b melatonin receptor. Proc Natl Acad Sci USA. 1995;92:8734–8738. doi: 10.1073/pnas.92.19.8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roca AL, Godson C, Weaver DR, Reppert SM. Structure, characterization, and expression of the gene encoding the mouse Mel1a melatonin receptor. Endocrinology. 1996;137:3469–3477. doi: 10.1210/en.137.8.3469. [DOI] [PubMed] [Google Scholar]
- 7.Jin X, von Gall C, Pieschl RL, Gribkoff VK, Stehle JH, Reppert SM, Weaver DR. Targeted disruption of the mouse Mel1b melatonin receptor. Mol Cell Biol. 2003;23:1054–1060. doi: 10.1128/MCB.23.3.1054-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong KJ, Niles LP. Induction of GDNF mRNA expression by melatonin in rat C6 glioma cells. NeuroReport. 2002;13:473–475. doi: 10.1097/00001756-200203250-00023. [DOI] [PubMed] [Google Scholar]
- 9.Poirel VJ, Masson-Pevet M, Pevet P, Gauer F. MT1 melatonin receptor mRNA expression exhibits a circadian variation in the rat suprachiasmatic nuclei. Brain Res. 2002;946:64–71. doi: 10.1016/S0006-8993(02)02824-X. [DOI] [PubMed] [Google Scholar]
- 10.Audinot V, Bonnaud A, Grandcolas L, Rodriguez M, Nagel N, Galizzi JP, Balik A, Messager S, Hazlerigg DG, Barrett P, Delagrange P, Boutin JA. Molecular cloning and pharmacological characterization of rat melatonin MT1 and MT2 receptors. Biochem Pharmacol. 2008;75:2007–2019. doi: 10.1016/j.bcp.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 12.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 13.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, Ui-Tei K, Watanabe M, Sakuma Y. Characterization of voltage-gated calcium currents in gonadotropin-releasing hormone neurons tagged with green fluorescent protein in rats. Endocrinology. 2003;144:5118–5125. doi: 10.1210/en.2003-0213. [DOI] [PubMed] [Google Scholar]
- 15.Hiraizumi Y, Nishimura I, Ishii H, Tanaka N, Takeshita T, Sakuma Y, Kato M. Rat GnRH neurons exhibit large conductance voltage- and Ca2+-activated K+ (BK) currents and express BK channel mRNAs. J Physiol Sci. 2008;58:21–29. doi: 10.2170/physiolsci.RP013207. [DOI] [PubMed] [Google Scholar]
- 16.Yin C, Ishii H, Tanaka N, Sakuma Y, Kato M. Activation of A-type γ-amino butyric acid receptors excites gonadotrophin-releasing hormone neurones isolated from adult rats. J Neuroendocrinol. 2008;20:566–575. doi: 10.1111/j.1365-2826.2008.01697.x. [DOI] [PubMed] [Google Scholar]
- 17.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 19.Weaver DR, Liu C, Reppert SM. Nature’s knockout: the Mel1b receptor is not necessary for reproductive and circadian responses to melatonin in Siberian hamsters. Mol Endocrinol. 1996;10:1478–1487. doi: 10.1210/me.10.11.1478. [DOI] [PubMed] [Google Scholar]
- 20.Nonno R, Pannacci M, Lucini V, Angeloni D, Fraschini F, Stankov BM. Ligand efficacy and potency at recombinant human MT2 melatonin receptors: evidence for agonist activity of some mt1-antagonists. Br J Pharmacol. 1999;127:1288–1294. doi: 10.1038/sj.bjp.0702658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nonno R, Lucini V, Pannacci M, Mazzucchelli C, Angeloni D, Fraschini F, Stankov BM. Pharmacological characterization of the human melatonin Mel1a receptor following stable transfection into NIH3T3 cells. Br J Pharmacol. 1998;124:485–492. doi: 10.1038/sj.bjp.0701860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witt-Enderby PA, Dubocovich ML. Characterization and regulation of the human ML1A melatonin receptor stably expressed in Chinese hamster ovary cells. Mol Pharmacol. 1996;50:166–174. [PubMed] [Google Scholar]
- 23.Pelletier J, Bodin L, Hanocq E, Malpaux B, Teyssier J, Thimonier J, Chemineau P. Association between expression of reproductive seasonality and alleles of the gene for Mel1a receptor in the ewe. Biol Reprod. 2000;62:1096–1101. doi: 10.1095/biolreprod62.4.1096. [DOI] [PubMed] [Google Scholar]
- 24.Notter DR, Cockett NE, Hadfield TS. Evaluation of melatonin receptor 1a as a candidate gene influencing reproduction in an autumn-lambing sheep flock. J Anim Sci. 2003;81:912–917. doi: 10.2527/2003.814912x. [DOI] [PubMed] [Google Scholar]
- 25.Glass JD, Knotts LK. A brain site for the antigonadal action of melatonin in the white-footed mouse (Peromyscus leucopus): involvement of the immunoreactive GnRH neuronal system. Neuroendocrinology. 1987;46:48–55. doi: 10.1159/000124795. [DOI] [PubMed] [Google Scholar]
- 26.Revel FG, Saboureau M, Masson-Pevet M, Pevet P, Mikkelsen JD, Simonneaux V. Kisspeptin mediates the photoperiodic control of reproduction in hamsters. Curr Biol. 2006;16:1730–1735. doi: 10.1016/j.cub.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 27.Lincoln GA, Maeda KI. Reproductive effects of placing micro-implants of melatonin in the mediobasal hypothalamus and preoptic area in rams. J Endocrinol. 1992;132:201–215. doi: 10.1677/joe.0.1320201. [DOI] [PubMed] [Google Scholar]
- 28.Sato S, Yin C, Teramoto A, Sakuma Y, Kato M. Sexually dimorphic modulation of GABAA receptor currents by melatonin in rat gonadotropin-releasing hormone neurons. J Physiol Sci. 2008;58:317–322. doi: 10.2170/physiolsci.RP006208. [DOI] [PubMed] [Google Scholar]
- 29.Wan Q, Man HY, Liu F, Braunton J, Niznik HB, Pang SF, Brown GM, Wang YT. Differential modulation of GABAA receptor function by Mel1a and Mel1b receptors. Nat Neurosci. 1999;2:401–403. doi: 10.1038/8062. [DOI] [PubMed] [Google Scholar]
- 30.Krsmanovic LZ, Martinez-Fuentes AJ, Arora KK, Mores N, Navarro CE, Chen HC, Stojilkovic SS, Catt KJ. Autocrine regulation of gonadotropin-releasing hormone secretion in cultured hypothalamic neurons. Endocrinology. 1999;140:1423–1431. doi: 10.1210/en.140.3.1423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Genomic organization of mouse MT2, and RT-PCR analysis of rat and mouse MT2 mRNAs using specific primers designed on exon 2 and exon 3.(A) Schematic representation of mouse MT2 gene structure. We newly determined a mouse MT2 cDNA sequence containing 5′- and 3′-UTRs, which we registered as accession number AB377276. Mouse MT2 cDNA is mapped onto Build 37 of the mouse genome assembly. Mouse MT2 gene is located at 9A2 on mouse chromosome 9, and consists of three exons. The white and gray boxes indicate UTRs and ORFs, respectively. The arrow indicates the orientation of the mouse chromosome. The image is not to scale. (B) RT-PCR analysis of MT2 mRNAs in rats (a) and mice (b). To exclude the possibility that the unusual MT2 gene structures were due to any artifacts of RACE cloning, we performed RT-PCR using primers designed on exon 2 and 3. Primer sequences used in this analysis are as follows: 5′-CAATTGGCTGGGACAGAAGT-3′ (forward), and 5′-TGGGAGTGCACGTAAGTCAG-3′ (reverse) for rat MT2; 5′-CCCATGAGAAGTCATTTTCC-3′ (forward), and 5′-TTGCTTGTTATCGTGATGCT-3′ (reverse) for mouse MT2. (+), total RNA reverse-transcribed with RTase; (−), without RTase; M, 100 bp ladder marker; Hy, hypothalamus. (PPT 82 kb)
Supplemental Figure 2. I-Mel binding ability and expression of mouse melatonin receptor mRNAs in NIH3T3 cells.(A) Specific I-Mel binding ability in the presence of 100 pM 125I-Mel in HEK293, Hela, and NIH3T3 cells. The data are presented as means ± SEM (n = 4). (B) RT-PCR analysis of mouse MT1 and MT2 mRNA expression in NIH3T3 cells and mouse hypothalamus. Expression of mouse MT1 (Upper panel), mouse MT2 (Middle panel), and mGAPDH (Lower panel). Primer sequences used are as follows: 5′-TACACTGGCCTTCATCCTCATCTTTACCA-3′ (forward), and 5′-TAACTAGCCACGAACAGCCACTCTG-3′ (reverse) for mouse MT1; 5′-GGAGCGCCCCAAGCAGTG-3′ (forward), and 5′-CCTCCCCAAGGACCCAACCGTCAC-3′ (reverse) for mouse MT2. The number on the right of each panel indicates the number of PCR cycles. NIH3T3 cells express neither MT1 nor MT2. The same results were obtained in four separately prepared samples. (PPT 89 kb)


