Abstract
Mycoplasma capricolum subsp. capripneumoniae belongs to the so-called Mycoplasma mycoides cluster and is the causal agent of contagious caprine pleuropneumonia (CCPP). All members of the M. mycoides cluster have two rRNA operons. The sequences of the 16S rRNA genes of both rRNA operons from 20 strains of M. capricolum subsp. capripneumoniae of different geographical origins in Africa and Asia were determined. Nucleotide differences which were present in only one of the two operons (polymorphisms) were detected in 24 positions. The polymorphisms were not randomly distributed in the 16S rRNA genes, and some of them were found in regions of low evolutionary variability. Interestingly, 11 polymorphisms were found in all the M. capricolum subsp. capripneumoniae strains, thus defining a putative ancestor. A sequence length difference between the 16S rRNA genes in a poly(A) region and 12 additional polymorphisms were found in only one or some of the strains. A phylogenetic tree was constructed by comparative analysis of the polymorphisms, and this tree revealed two distinct lines of descent. The nucleotide substitution rate of strains within line II was up to 50% higher than within line I. A tree was also constructed from individual operonal 16S rRNA sequences, and the sequences of the two operons were found to form two distinct clades. The topologies of both clades were strikingly similar, which supports the use of 16S rRNA sequence data from homologous operons for phylogenetic studies. The strain-specific polymorphism patterns of the 16S rRNA genes of M. capricolum subsp. capripneumoniae may be used as epidemiological markers for CCPP.
rRNA sequences are, in general, believed to show low variability between and within species or subspecies. Heterogeneities in 16S rRNA genes have been reported but only to a minor extent. Nevertheless, both macroheterogeneities and microheterogeneities are known to exist. Macroheterogeneities involving large insertions ranging from 50 to several hundred nucleotides have been observed, e.g., in the archaeon Pyrobaculum aerophilum (6) and in the (eu)bacteria Campylobacter helveticus (29), Desulfotomaculum australicum (36), and a spore-forming Bacillus species (43). The first two are examples of species with split-gene formations of the rRNA genes, and the insertions are defined as intervening sequences. Therefore, although the intervening sequence is present in the structural gene and is even represented in the primary transcript, it is absent in the mature 16S rRNA molecule and does not contribute to the structure of the gene product. The last two species, however, have unusually long extensions of helices 6 and 49, respectively, according to Van de Peer et al. (54), which are positioned within the hypervariable regions V1 and V5, respectively, following the nomenclature of Gray et al. (11). These idiosyncrasies will cause problems only if they are not present in all of the rrn operons and when PCR based sequencing is used for determination of the nucleotide sequence. The resulting extra characters will be removed from the alignment which is used for inferring the tree and therefore do not constitute phylogenetic insignia. Microheterogeneities are probably by far more common than macroheterogeneities, and they are likely to be reported more frequently when we start looking for them. Clayton et al. recently observed that slightly different 16S rRNA sequences were deposited into the data banks for different strains belonging to the same species (8). Sequencing errors as the only plausible explanation for this variability were ruled out by the authors. Instead, most of the differences were believed to be real and to be caused by intraspecific variations. However, microheterogeneities in the form of nucleotide differences between the rrn operons, so-called polymorphisms, within a species and the extent to which they occur are not known. Examples of species where polymorphisms have been identified are Haloarcula marismortui (33), Bacillus sporothermodurans (42), and members of the class Mollicutes (16, 39–41, 44, 45).
About 175 species have been recognized within the class Mollicutes, and discoveries of new species are constantly being reported. The trivial name “mollicutes” will be used herein to avoid confusion with members of the genus Mycoplasma. The mollicutes have a small genome with a low G+C content and lack a cell wall, but they are phylogenetically related to gram-positive bacteria with a low G+C content in their genomes. Phylogenetic analysis of the mollicutes based on 16S rRNA sequences (55) has, together with other data, resulted in a revised taxonomy of this class, which is now composed of eight genera (53). The tree of the mollicutes revealed five distinct groups, of which one was named the spiroplasma group (55). A cluster of mycoplasmas within the spiroplasma group, which is of particular importance in veterinary medicine, is the so-called Mycoplasma mycoides cluster. All the members of the M. mycoides cluster are closely related, and some of them are difficult to differentiate by conventional techniques. Analysis of rRNA sequences also showed that M. putrefaciens is related to the members of the M. mycoides cluster (55). The following six mollicutes (9) denoted as species, subspecies or strains are included in the classical M. mycoides cluster: Mycoplasma capricolum subsp. capripneumoniae, Mycoplasma capricolum subsp. capricolum, Mycoplasma mycoides subsp. capri, Mycoplasma mycoides subsp. mycoides type LC, Mycoplasma mycoides subsp. mycoides type SC, and Mycoplasma sp. bovine serogroup 7. The M. mycoides cluster can be subdivided into the M. capricolum species group and the M. capri species group (41). M. capricolum subsp. capripneumoniae, formerly Mycoplasma sp. strain F38 (26), which belongs to the M. capricolum species group, causes contagious caprine pleuropneumonia (CCPP). CCPP is a goat disease of great concern in Africa and Asia (26, 30) and is included in the B list of communicable animal diseases of the Office International des Epizooties (22). CCPP was first described at the end of the last century (20, 51) and was shown to be caused by M. capricolum subsp. capripneumoniae in 1976 by MacOwan and Minette (31). More than 30 countries have declared that they have detected CCPP, but the organism has been isolated from goats in only 11 countries (48). A diagnostic method for CCPP based on PCR of the 16S rRNA genes from M. capricolum subsp. capripneumoniae and restriction enzyme analysis of the PCR product has been developed (45). The members of the M. mycoides cluster have two rRNA operons, designated rrnA and rrnB (7), and the above diagnostic method for CCPP was based on a polymorphism (nucleotide difference between the two 16S rRNA genes in the same strain) which was found to be unique for M. capricolum subsp. capripneumoniae. This polymorphism can therefore be easily used as a molecular marker for this subspecies, because it is localized in a restriction site for PstI (45). This unique polymorphism has been shown to be present in 16 strains of M. capricolum subsp. capripneumoniae from different parts of the world, but it was not present in 39 strains representing other species or subspecies of the M. mycoides cluster (4, 45).
The sequences of the 16S rRNA genes from both rRNA operons were recently determined for all members of the M. mycoides cluster, and several polymorphisms (or microheterogeneities) were identified (41). The strains F38T and 4/2LC of M. capricolum subsp. capripneumoniae were found to be unique among the members of the M. mycoides cluster, because they had the largest number of polymorphisms (15 and 17, respectively). M. mycoides subsp. mycoides type SC, the causal agent of contagious bovine pleuropneumonia, was found to have eight polymorphisms and a sequence length difference of two adenosines, whereas the other members of the M. mycoides cluster had only one, two, or three polymorphisms and sequence length variations were not found (41). Other mollicutes from ruminants have also been shown to have only few polymorphisms (44). Furthermore, the polymorphism patterns in the 16S rRNA genes were not identical for the two strains F38T and 4/2LC, even though they represent the same subspecies. Thus, M. capricolum subsp. capripneumoniae is unique among the mollicutes, which so far have been analyzed for the presence of polymorphisms in the 16S rRNA genes, which also supports the current classification of M. capricolum subsp. capripneumoniae into at least a separate subspecies (26). In the present study, we therefore determined the sequences of both 16S rRNA genes from 20 M. capricolum subsp. capripneumoniae strains from different geographical origins to investigate if polymorphisms can be used as epidemiological markers. The variations in the polymorphism patterns were found to be surprisingly great within M. capricolum subsp. capripneumoniae and could therefore be used to study molecular evolution within this subspecies.
MATERIALS AND METHODS
M. capricolum subsp. capripneumoniae strains, growth conditions, and sample preparations.
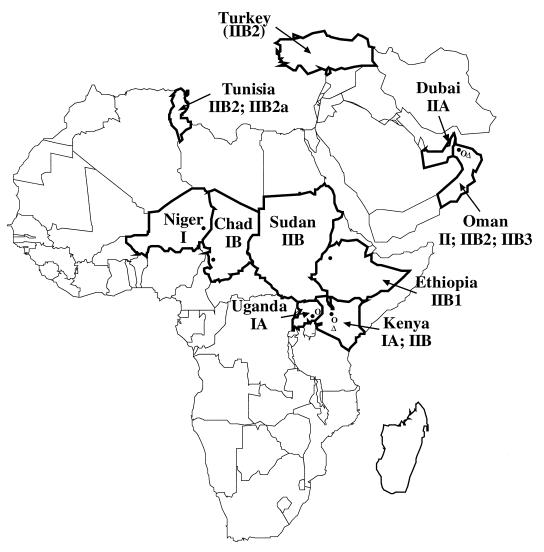
The M. capricolum subsp. capripneumoniae strains used in this work are listed in Table 1, and the geographical distribution of the strains is shown in Fig. 1. All strains were grown in Hayflick’s medium supplemented with pyruvate (50). Samples for PCR and DNA sequencing were prepared as described previously (41).
TABLE 1.
M. capricolum subsp. capripneumoniae strains from which the 16S rRNA genes have been sequenced earlier or in this worka
| Strain | Country of origin | Symbol in Fig. 1 | Yr of isolation | Polymorphism pattern | Accession no. of rrnA/rrnBb | Reference to strain |
|---|---|---|---|---|---|---|
| F38T | Kenya | 1976 | IIB | U26042/M94728 | 31 | |
| G1943/80 | Kenya | 1980 | IA | ID to AF009837/AF009845 | 5 | |
| Baragoi | Kenya | • | 1995 | IA | ID to AF009837/AF009845 | 4 |
| G280/80 | Kenya | ○ | 1980 | IA | ID to AF009837/AF009845 | 5 |
| G94/83 | Kenya | ▵ | 1983 | IA | ID to AF009837/AF009845 | 5 |
| M74/93 | Uganda | • | 1993 | IA | ID to AF009837/AF009845 | 3 |
| M79/93 | Uganda | ○ | 1993 | IA | AF009837/AF009845 | 3 |
| M79/93 (sample) | Uganda | 1993 | IA | ID to AF009837/AF009845 | 3 | |
| 9231 | Ethiopia | • | 1982 | IIB1 | AF009834/AF009842 | 49 |
| 89110bis | Sudan | 1981 | IIB | ID to U26042/M94728 | 15 | |
| 8789 | Chad | • | 1987 | IB | AF009831/AF009839 | 27 |
| 95043 | Niger | • | 1995 | I | AF009835/AF009843 | CIRAD-EMVTc |
| Gabés | Tunisia | 1980 | IIB2 | ID to AF009830/AF009838 | 37 | |
| KD | Tunisia | 1980 | IIB2 | ID to AF009830/AF009838 | 37 | |
| 7/1a | Oman (Turkey) | • | 1988 | IIB2 | AF009830/AF009838 | 5, 23 |
| 4/2LC | Oman | ○ | 1988 | IIB3 | U26051/U26052 | 7, 23 |
| 19/2 | Oman | ▵ | 1988 | IIB3 | ID to U26051/U26052 | 23 |
| 9081 | Oman | 1990 | II | AF009832/AF009840 | CIRAD-EMVT | |
| 91106/550/1 | Dubai | 1991 | IIA | AF009833/AF009841 | CIRAD-EMVT | |
| 91106/550/2 | Dubai | 1991 | IIA | ID to AF009833/AF009841 | CIRAD-EMVT | |
| GL102 | 1980 | IIB2a | AF009836/AF009844 | 37 |
Strain M74/93 was isolated from a sheep. M79/93 (sample) was not cultured and is therefore not regarded as a separate strain. Strain GL102 was obtained from passage 102 of strain Gabés. The 16S rDNA sequences of both operons of the strain F38 (U26042/M94728) and 4/2LC (U26051/U26052) were retrieved from GenBank.
The accession numbers of the nucleotide sequences of the 16S rRNA genes of the rrnA and rrnB operon are given as N/N, respectively. Boldface type indicates accession numbers of nucleotide sequences determined in this work. ID, identical. Nucleotide sequences from the two operons were deposited to GenBank for only one strain of each polymorphism pattern type.
CIRAD-EMVT: Culture Collection at Dépertement d’élevage et de médicine véterinaire, Centre de coopération internationale en recherche agronomique pour le dévelopment.
FIG. 1.
Geographical origins of the M. capricolum subsp. capripneumoniae strains which have been analyzed. The affected countries are shown with bold boundaries. When the site of isolation is known, it is indicated with a symbol (Table 1).
In vitro amplification and DNA sequencing of the 16S rRNA genes.
The 16S rRNA genes from both rRNA operons were amplified by seminested PCR. The reverse primers were biotinylated for magnetic separation of the strands. One outer primer pair was complementary to the universal regions U1 and U8 (11) and was used to amplify the 16S rRNA genes of both rRNA operons (21, 41). The other outer primer pair was complementary to the flanking regions of the 16S rRNA gene of the rrnB operon and was used for specific amplification of the gene from this operon (41). Thereafter, the respective product was used in a seminested amplification as described previously (21, 41). The biotinylated amplicons from the 16S rRNA genes were immobilized onto streptavidin-coated superparamagnetic beads (Dynabeads M280; Dynal AS, Oslo, Norway), and single strands suitable for bidirectional DNA sequencing were obtained by magnetic separation (18, 40). The sequencing reactions, performed by the method of Sanger et al. (47) with bacteriophage T7 DNA polymerase, were carried out automatically (17, 38, 40, 41). Detailed protocols and descriptions of primers for solid-phase 16S rDNA sequencing have been published (21, 38, 40, 41, 44).
Evaluation of sequence data.
The sequence of the 16S rRNA gene of the rrnA operon was deduced from the sequences obtained by PCR with general primers and with rrnB-specific primers (41). A secondary-structure model of the 16S rRNA molecule transcribed from the rrnB operon of M. capricolum subsp. capripneumoniae was constructed by modification of the Postscript file of the 16S rRNA molecule of M. capricolum subsp. capricolum (12) retrieved from the Ribosomal Database Project (32). The polymorphisms of the different M. capricolum subsp. capripneumoniae strains were introduced into the model as described previously (41).
Two kinds of phylogenetic trees were inferred from the rRNA sequences. One tree was based on mutations and was inferred from the consensus sequences by comparative analysis of the sequence differences compiled in Table 2. By this procedure, all of the polymorphisms in the 16S rRNA genes which were found to be common to all strains constituted the microheterogeneity pattern identical to that of the ancestor. Thereafter, the exceptionally found polymorphisms were used to extrapolate the evolutionary lineages originating from this hypothetical ancestor. One evolutionary event (i.e., one polymorphism or sequence truncation) was given a value of 1. The second tree was derived from a corrected distance matrix. The one-parameter model of Jukes and Cantor (24) was used to correct the matrix at single locations, assuming equal frequencies and identical substitution rates of all nucleotides of the individual 16S rRNA sequences. The dendrogram was computed by using the tree-building method of Saitou and Nei (46).
TABLE 2.
Mutational events in the 16S rRNA genes of different strains of M. capricolum subsp. capripneumoniaea
| Positionb | SRGc | Mutation in line I
|
Ancestor | Mutation in line II
|
Resulting base paird | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IB | IA | I | II | IIA | IIB | IIB1 | IIB2 | IIB2a | IIB3 | ||||
| 94 (111) | 5 · — (e) | G/G | G/G | G/G | G/G | G/G | G/G | G/A | G/A | G/A | G/A | G/A | |
| 180 (187) | 2 · — (c) | A/G | |||||||||||
| 232 (238) | 2 · — (f) | G/G | G/G | G/G | G/G | G/G | G/A | G/G | G/G | G/G | G/G | G/G | |
| 404 (411) | 6 · — (e) | A/A | A/A | A/A | A/A | C/A | C/A | C/A | C/A | C/A | C/A | C/A | |
| 444–449 (449–455) | — · — | A/– | A/– | A/– | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | |
| 452 · 467 (458 · 464) | 1 · 1 (a) | G/A | G · U/A · U | ||||||||||
| 509 · 528 (516 · 535) | 5 · 6 (e) | T/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | U · A/C · A |
| 493 · 538 (500 · 545) | 5 · 5 (e) | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/T | C/C | C/C | C/C | G · C/G · U |
| 672 · 702 (680 · 710) | 2 · 2 (c) | C/T | C · G/U · G | ||||||||||
| 684 (692) | 5 · — (e) | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/C | T/C | T/T | |
| 709 (717) | 2 · — (c) | C/T | |||||||||||
| 844 (859) | 1 · — (a) | C/T | |||||||||||
| 557 · 871 (564 · 886) | 2 · 4 (b) | A/C | U · A/U · C | ||||||||||
| 875 · 896 (890 · 910) | 4 · 6 (e) | G/G | G/A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G · C/A · C |
| 873 · 897 (888 · 912) | 4 · 5 (a) | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | T/C | C/C | U · U/U · C |
| 1060 (1079) | 6 · — (e) | A/G | |||||||||||
| 1068 · 1079 (1087 · 1098) | 4 · 4 (a) | C/T | G · C/G · U | ||||||||||
| 1067 · 1080 (1086 · 1099) | 3 · 3 (c) | G/G | G/G | G/G | G/G | G/A | G/A | G/A | G/A | G/A | G/A | G/A | U · G/U · A |
| 1146 (1166) | 5 · — (c) | G/A | |||||||||||
| 1144 · 1151 (1164 · 1172) | 2 · 2 (a) | T/C | G · U/G · C | ||||||||||
| 1238 · 1255 (1259 · 1276) | 4 · 4 (a) | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | A/G | C · A/C · G |
| 1297 (1317) | 3 · — (f) | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | T/C | |
| 1403 · 1459 (1422 · 1478) | 3 · 2 (c) | G/G | G/G | G/G | G/G | G/G | G/G | T/G | T/G | T/G | T/G | T/G | U · U/G · U |
| 1417 · 1446 (1436 · 1465) | 2 · 2 (b) | C/T | U · C/U · U | ||||||||||
Polymorphic positions are in boldface type. The nucleotides in the polymorphic positions are given as N/N for rrnA and rrnB, respectively.
Nucleotide position according to the 16S rRNA gene of the rrnB operon of M. capricolum subsp. capripneumoniae F38 (41). The number of the corresponding position of the 16S rRNA sequence of E. coli is given in parentheses (11). Boldface indicates the position of the actual polymorphism in a pair.
Substitution rate groups (SRG) of Van de Peer et al. (54). The numbers denote subsets with the following relative rate limits, from high to low variability: 1, >10+0.575; 2, 10+0.075 to 10+0.575; 3, 10−0.425 to 10+0.075; 4, 10−0.925 to 10−0.425; 5, <10−0.925; 6, absolutely conserved positions (54). Boldface indicates the variability of the actual polymorphic position in a pair. A nonpaired position is marked with a dash. Letters within parentheses denote intervals of nucleotide conservation of the actual positions within the mollicutes (a, <50%; b, 50 to 65%; c, 65 to 80%; d, 80 to 90%; e, 90 to 95%; f, 95 to 100%) as determined from consensus sequences compiled by using the different percentage values as cutoff values for displaying the main nucleotide residue which is present at the actual position.
The base pairs are given as N · N rrnA/N · N rrnB, with the alternative residues of the actual polymorphism in boldface type.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA genes from the rrnA and the rrnB operons of the M. capricolum subsp. capripneumoniae strains have been deposited in GenBank (National Center for Biotechnology Information, Bethesda, Md.) under the accession numbers listed in Table 1.
RESULTS AND DISCUSSION
Nucleotide sequence heterogeneity in 16S rRNA genes.
Despite the reports of variability within 16S rRNA genes (see Introduction), the extent to which microheterogeneities occur has not been extensively studied. This is partially due to the limitations in the detection techniques commonly used for rDNA sequencing. In the present study, we found that the 16S rRNA genes can differ by >1% between the operons within an individual strain of M. capricolum subsp. capripneumoniae. We have shown for M. capricolum subsp. capripneumoniae that the interoperon variation in, for instance, strains 4/2LC and GL102 is 1.1% as calculated from data in Table 2. We have also observed a strain-to-strain variation for a specific operon of up to 0.26% between several of the strains (Table 2).
The importance of determining the intraspecific variability in 16S rRNA genes for developing reliable phylogenetic hypothesis has recently been pointed out (8, 41). The usefulness of characterizing microheterogeneities is also demonstrated in this work, emphasizing the importance of sequencing the 16S rRNA genes of all operons (if more than one is present) and investigating the strain-to-strain variations in the 16S rRNA genes. It is, of course, also extremely important to deposit sequences in GenBank under the correct species name and strain designation. Consequently, a bacterial DNA or protein sequence should preferably not be accepted in a data bank without a recognized strain designation.
Nucleotide sequences of the 16S rRNA genes of M. capricolum subsp. capripneumoniae.
The 16S rRNA gene sequences of both operons from 20 strains of the species M. capricolum subsp. capripneumoniae listed in Table 1 were subjected to bidirectional solid-phase rDNA sequencing, which has previously been shown to give very accurate data with the possibility of detecting heterogeneity, e.g., in viral populations and in 16S rRNA genes (see, e.g., references 16, 21, 28, 38, 40–42, and 44). Ten different 16S rRNA polymorphism patterns were found among the 20 M. capricolum subsp. capripneumoniae strains. Furthermore, a sequence length variation of one adenosine in a poly(A) region was found between the 16S rRNA genes of the two rRNA operons in nine of the strains. The poly(A) region is situated between nucleotides 444 and 449 and is not homologous to the poly(A) region with sequence length variations found in M. mycoides subsp. mycoides SC (between positions 1264 and 1270) reported previously (41). Length variations between the 16S rRNA genes of the members of the M. mycoides cluster have so far been observed only within poly(A) regions. These sequence length variations are probably caused by a process known as replication slippage (57).
A primary isolate of a mycoplasma may represent several clonal variants of a particular strain. A sample was therefore also analyzed for clonal variants with respect to 16S rRNA gene sequences among the M. capricolum subsp. capripneumoniae isolates. The 16S rRNA sequences of both operons obtained from a sample from Uganda were compared with those of the cloned strain M79/93 originating from this sample. No sequence differences were observed, which indicates that in a particular sample, only one 16S rRNA clone variant is normally present. The fact that the two nucleotide alternatives in a polymorphism were always present in a 1:1 ratio also supports this observation.
The hypothetical ancestor of the M. capricolum subsp. capripneumoniae strains and two lines of descent.
The 24 evolutionary events were compiled and compared as detailed in Table 2. Eleven polymorphic positions were found to be common to all 20 strains, thus defining a hypothetical ancestor from which the M. capricolum subsp. capripneumoniae strains have evolved. The sequences of the two 16S rRNA genes of the hypothetical ancestor can be derived from Table 2. The remaining 12 polymorphisms and the length variation which occurred in only some of the strains most probably arose due to later evolutionary events. The polymorphisms in the 16S rRNA genes of the M. capricolum subsp. capripneumoniae strains (Table 2) were converted into a cladogram (Fig. 2) by comparative analysis. Both the length polymorphism and a nucleotide polymorphism were treated as one event. The horizontal lines are proportional to the number of events. The tree shown in Fig. 2 was rooted by the 11 ancestral evolutionary events and revealed two major lines of descent.
FIG. 2.
Phylogenetic tree based on mutational events of the 16S rRNA genes of the M. capricolum subsp. capripneumoniae strains. The tree was constructed by comparative analysis and with the 11 polymorphisms of the putative ancestor used as the root. Two major lines of descent (I and II) can be seen. The positions and types of the polymorphisms are shown on the axis.
Evolutionary line I contained the 9 M. capricolum subsp. capripneumoniae strains which shared the synapomorphy (444–449)A/− (Table 2; Fig. 2), situated in a poly(A) segment of the molecule in a region which is characterized by bilaterally bulged residues (Fig. 3). It remains to be shown whether the presence or absence of a sixth adenosine in this region of the 16S rRNA molecule will affect the function of the mature small ribosomal subunit from the strains of line I. The strains of line I showed a very low interstrain variability, and only two additional polymorphic positions, 509T/C and 875G/A were found, thus bifurcating into sublines IA and IB (Table 2; Fig. 2). Strain 95043 from Niger differed from the hypothetical ancestor only by the truncation of one adenosine in the 16S rRNA gene of the rrnB operon.
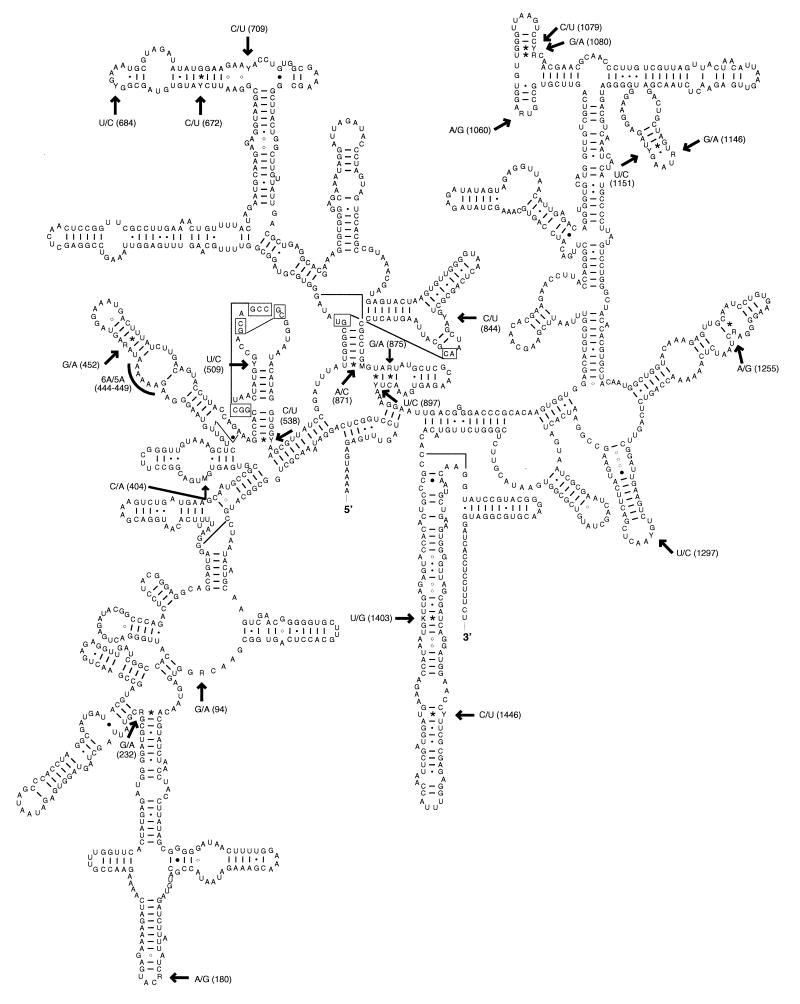
FIG. 3.
Secondary-structure model of the 16S rRNA molecule from the rrnB operon from M. capricolum subsp. capripneumoniae. All identified polymorphisms are indicated with arrows. They are numbered, and their composition (Table 2) is given according to the letter code of the International Union of Biochemistry. The nucleotide residues of the rrnA and rrnB operons are written at the arrows as N/N, respectively. The segment containing the sequence length variation between rrnA and rrnB is denoted by an arc. This model has been adapted from the secondary-structure model of the 16S rRNA molecule of M. capricolum subsp. capricolum described by Gutell et al. (13, 14).
The 11 M. capricolum subsp. capripneumoniae strains of evolutionary line II shared the two polymorphisms 404C/A and 1080C/A (Table 2; Fig. 2). The earliest member of line II was M. capricolum subsp. capripneumoniae 9081 from Oman, and line II branched into the two sublines IIA and IIB with a fork of three additional branches at the node of IIB (Fig. 2). This line is by far the most heterogeneous, and the evolutionary drift seems to be more pronounced for the 16S rRNA genes in the strains of line II than in those of line I. The two lines are separated by only three mutational events but can nevertheless be recognized as true phylogenetic entities, since none of the polymorphisms present in only some of the strains were shared among M. capricolum subsp. capripneumoniae strains of the two lines (Table 2). This observation can therefore be regarded as a consistency measure for the two lines, and it also indicates that sequence homogenization due to gene conversion has not occurred as was observed for the tuf genes of Salmonella typhimurium (1).
The 16S rRNA gene sequences of the five M. capricolum subsp. capripneumoniae strains, which were used in a recent study on the DNA relatedness of strains from the M. capricolum species group (5), have also been determined in this work. Although the hybridization values were very similar for all these strains (87 to 89%), the 16S rRNA sequence data clearly showed that two of the strains (F38T and 7/1a) belonged to line II and three of the strains (G1943/80, G280/80, and G94/83) belonged to line I (Fig. 2; Table 2).
Polymorphisms as epidemiological markers and diagnostic targets.
All members of the M. mycoides cluster are closely related as judged by biochemistry (10, 41), serological reactions (25, 35), DNA-DNA reassociation studies (2), and 16S rRNA sequence analysis (41). It is therefore important to identify polymorphisms in the 16S rRNA genes, because they can be used in diagnostic applications to distinguish between closely related species (45). The polymorphisms may also be useful as epidemiological markers. Two major lines of descent have been defined for M. capricolum subsp. capripneumoniae strains in this work. Representatives of line I were found only in Africa, whereas representatives of line II were found in both Africa and Asia (Fig. 1). A representative of line I, strain 95043, was isolated in Niger in 1995, and a strain of subline IB (Fig. 2; Table 2) was isolated in the neighboring country of Chad. Several strains of subline IA were isolated in Kenya and Uganda. The occurrence of the same type in neighboring countries, such as IA in Kenya and Uganda and IIB in Kenya and Sudan, might well be explained by trade and other movement of animals across these borders. An area of eastern Uganda and western Kenya is, for instance, populated by the same tribe, and the border is not well respected.
Strains of line II were found in countries bordering maritime routes (Mediterranean sea: Turkey, Tunisia; Indian Ocean: Kenya, Oman; Red Sea: Ethiopia, Sudan). Strain 9081, with a mutation pattern of line II, was isolated in Oman in 1990 and differed from the hypothetical ancestor by two polymorphisms in positions 404 and 1080. The polymorphism type pattern of subline IIA is the closest to that of line II and differs by one extra polymorphism in position 232. Strains of this type were isolated in Dubai in 1991. The polymorphism type pattern of the subline IIB differed from that of line II by the two polymorphisms in positions 94 and 1403. Strains of this type have been isolated in Kenya in 1976 (strain F38) and Sudan in 1989. Strains of types IIB1 and IIB2 differed from the strains of type IIB by one polymorphism in positions 538 and 684, respectively. A strain of type IIB1 was isolated in Ethiopia in 1992, and representatives of type IIB2 were isolated in Tunisia 1980 and Oman 1988. The Oman strain 7/1a originated from a goat which was imported from Turkey. Strains with a polymorphism pattern of type IIB3 differed from type IIB by two polymorphisms in positions 1255 and 1297. Representatives of type IIB3 were isolated in Oman.
Three types of M. capricolum subsp. capripneumoniae were found in Oman, namely, II, IIB2, and IIB3. A plausible explanation for this would be the extensive importation of goats from several countries in the Arabic peninsula for the religious feasts. These animals are normally slaughtered, but some might be kept alive and can thus infect indigenous herds. It is also noteworthy that two countries which were most likely to have CCPP-infected animals in the 19th century, Algeria (51) and South Africa (20), might have had strains of type IIB2. Algeria has a border with Tunisia, where type IIB2 was found in 1980, and South Africa imported Angora goats, which most probably brought CCPP from Turkey in 1881. The Turkish goat imported to Oman in 1988 was also found to be of type IIB2. In general, no correlation could be seen between the polymorphism pattern and the time of isolation of the strains.
Secondary structure analysis of the polymorphic positions and their implications.
The positions of the 24 mutational events in the 16S rRNA molecule of M. capricolum subsp. capripneumoniae are shown in the secondary-structure model in Fig. 3. The nucleotide substitution rates according to a recently published variability map of the 16S rRNA molecule (54) are included in Table 2, showing the variability in each of the polymorphic positions. For comparison, consensus 16S rRNA sequences from an alignment of the mollicutes (32, 39) were computed by using different cutoff values for which a residue in a certain position was present. These percentages are included in Table 2, indicating that positions of high, intermediate, and low variability found in (eu)bacteria (54) also seem to hold for the variability in the 16S rRNA genes of mycoplasmas. Of the 11 polymorphisms of the ancestor, 6 (positions 180, 452, 672, 844, 1151, and 1446) were located in the high-variability groups 1 and 2 as defined by Van de Peer et al. (54). In contrast, 6 of the 12 microheterogeneities found in only some of the M. capricolum subsp. capripneumoniae strains (i.e., positions 94, 404, 509, 538, 684, and 897) belonged to group 5 or 6 of low variability. Moreover, frequently observed polymorphisms situated in stems were found predominantly in positions of relatively high variability (positions 452, 672, 1080, 1151, 1403, and 1446), while rarely found microheterogeneities often were situated in highly conserved locales in the 16S rRNA molecule (positions 538, 871, 875, 897, and 1255). It is noteworthy that all but 875 of the rarely found polymorphisms in sites of low variability occur in the starting or ending base pair of a stem (positions 538, 871, 897, and 1255). The reason for this is not known, but the base pairing at these positions might be rather flexible, at least in these mycoplasmas. This observation indicates that polymorphisms shared among all strains occur in rather variable positions while rarely found polymorphisms are present in more highly conserved positions of the 16S rRNA molecule of M. capricolum subsp. capripneumoniae.
A total of 13 polymorphisms are situated in stems (Fig. 3). A compensatory mutation in the opposite position to stabilize the actual stem in the other molecule was never observed. However, the general alternatives to noncanonical base pairing (13, 54) were followed in most cases. Therefore, a guanosine, a uridine, and an adenosine residue in one strand can have C or U, A or G, and U or C, respectively, as their counterparts. Moreover, the polymorphisms 509T/C, 875G/A, and 1255A/G indicated that cytidines can tolerate A or G as pairing nucleotides and that a C · G base pair can be substituted with a C · A pair, in certain positions. Therefore, a C · A pair might not necessarily affect the stability of a stem (54), and plausible explanations are a protonated C or a tautomeric configuration of A or C in these positions, which have been suggested in previous reports (19, 54). Interestingly, over 40 polymorphisms have been localized to stem regions of the 16S rRNA molecule of mycoplasmas (39, 41, 44, also see above) and without a compensatory mutation in the corresponding nucleotide position. This finding is contradictory to the polymorphisms in the 16S rRNA genes of the recently described B. sporothermodurans (42), where most of the microheterogeneities found in stems were associated with compensatory mutations in the complementary nucleotide position.
All polymorphisms of the M. capricolum subsp. capripneumoniae strains were checked against the corresponding positions of the 16S rRNA mutation database which provides a list of mutated positions in Escherichia coli (52). Only position 897 in M. capricolum subsp. capripneumoniae (912 in E. coli)T/C was listed in the database. A change of a C to a U in this position in E. coli has been shown to confer resistance to streptomycin (34). The actual polymorphism 897(912)T/C is present in strain GL102, a laboratory strain originating from strain Gabés, which has been passaged 102 times in the laboratory. It has not been possible to find whether streptomycin has been used during any of the passages. Nevertheless, this transition indicated that mutations may also occur in positions of low variability (Table 2) with a significant rate in the 16S rRNA genes.
We have so far characterized over 80 microheterogeneities in the 16S rRNA genes of different Mycoplasma species isolated from different hosts (16, 39, 41, 44). A comparison of these polymorphisms with those determined in this study confirmed that microheterogeneities in the 16S rRNA genes of mycoplasmas are distributed throughout the molecule but are rarely seen in universal regions (11) of the molecule. Polymorphisms with identical nucleotide compositions and present in different species have been found in only 3 of the more than 80 positions.
Evolution of the 16S rRNA genes of M. capricolum subsp. capripneumoniae: rrnA operon versus rrnB operon.
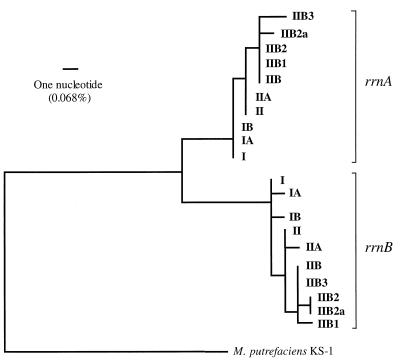
The 16S rRNA sequences of the rrnA operon were 99.73 to 99.93% similar for the different M. capricolum subsp. capripneumoniae strains. Identical values were found for the rrnB operon. The percent similarity ranged from 98.77 to 99.20% between the two operons. This means that the similarity between 16S rRNA genes within a strain is lower (11 to 17 nucleotide differences) than that between 16S rRNA genes of homologous operons in different strains (0 to 4 nucleotide differences). A phylogenetic tree was constructed from 16S rRNA sequences of the individual operons (Fig. 4). Mycoplasma putrefaciens served as the outgroup, and the branches are denoted according to the corresponding polymorphism pattern (Table 1). The phylogenetic analysis revealed two distinct entities, one of which consisted of sequences from the rrnA operon and the other consisted of sequences from the rrnB operon. A slightly higher evolutionary rate was observed for the sequences of the rrnB operon as judged from the branch lengths (Fig. 4). The tree implies that the rrnB operon has evolved by one to three further nucleotide changes. Strikingly, the topologies for the respective clade are very similar, and the line I strains form deep branches of both clades. Descendants belonging to line II form more recent branches, where M. capricolum subsp. capripneumoniae 9081 of subline II is placed intermediately in the respective operonal cluster. This shows that the 16S rRNA genes from the two operons coevolved and that both are phylogenetically informative. The tree illustrates the importance of using 16S rRNA sequences from homologous operons to infer the phylogeny when polymorphisms are present and the species to be analyzed are closely related.
FIG. 4.
Phylogenetic tree based on 16S rRNA sequences of the individual operons obtained by the neighbor-joining method (46). The tree revealed two distinct clades representing sequences of the rrnA and rrnB operons, indicating that the individual operons are monophyletic and that gene conversion has not taken place.
Conclusion.
The evolution within mycoplasmas has been reported to be unusually rapid (56). In a recent study, we suggested that members of the M. mycoides cluster could be used as a model system for molecular evolution by mapping the polymorphisms in the two 16S rRNA genes (41). In the present work, we have characterized microheterogeneities in strains of M. capricolum subsp. capripneumoniae, a subspecies of the M. capricolum species group (41) of the M. mycoides cluster (55), and have constructed two kinds of trees. One tree was based on mutational events from consensus sequences, and this tree formed two lines of descent without underlying synapomorphies (close parallelism as a result of common inherited genetic factors causing incomplete synapomorphy). In a tree based on individual operons, both 16S rRNA genes were found to evolve and to reflect similar phylogeny. Therefore, strains of M. capricolum subsp. capripneumoniae constitute a very useful model for studies of molecular evolution. M. capricolum subsp. capripneumoniae may also be a suitable subspecies for comparing the evolution of other genes with the evolution observed for the 16S rRNA genes.
ACKNOWLEDGMENTS
We are grateful to Diarmaid Hughes for valuable discussions on gene conversion. We also thank Marianne Persson for skillfully performing all PCR experiments and Joseph Tully, Henning Ernø, Gareth Jones, and Hezron Wesonga for generous supply of strains.
This work has been financially supported by grants from the Göran Gustafsson Foundation and the Swedish Engineering Science Research Council to M.U. and from the Commission of the EU (DG XII) for “Contagious caprine pleuropneumonia: distribution, evaluation of vaccines, and development of new tools” (contract IC18-CT95-0007), from the Swedish Council for Forestry and Agricultural Research, and from the Swedish International Development Cooperation Agency to K.-E.J. This study also forms a part of the EU research collaboration COST 826 on Ruminants’ mycoplasmoses.
REFERENCES
- 1.Abdulkarim F, Hughes D. Homologous recombination between the tuf genes of Salmonella typhimurium. J Mol Biol. 1996;260:506–522. doi: 10.1006/jmbi.1996.0418. [DOI] [PubMed] [Google Scholar]
- 2.Askaa G, Ernø H, Ojo M O. Bovine mycoplasmas: classification of groups related to Mycoplasma mycoides. Acta Vet Scand. 1978;19:166–178. doi: 10.1186/BF03547622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bölske G, Johansson K-E, Heinonen R, Panvuga P A, Twinamasiko E. Contagious caprine pleuropneumonia in Uganda and isolation of Mycoplasma capricolum subspecies capripneumoniae from goats and sheep. Vet Rec. 1995;137:594. [PubMed] [Google Scholar]
- 4.Bölske G, Mattsson J G, Ros Bascuñana C, Bergström K, Wesonga H, Johansson K-E. Diagnosis of contagious caprine pleuropneumonia by detection and identification of Mycoplasma capricolum subsp. capripneumoniae by PCR and restriction enzyme analysis. J Clin Microbiol. 1996;34:785–791. doi: 10.1128/jcm.34.4.785-791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet F, Saillard C, Bové J M, Leach R H, Rose D L, Cottew G S, Tully J G. DNA relatedness between field isolates of mycoplasma F38 group, the agent of contagious caprine pleuropneumonia and strains of Mycoplasma capricolum. Int J Syst Bacteriol. 1993;43:597–602. doi: 10.1099/00207713-43-3-597. [DOI] [PubMed] [Google Scholar]
- 6.Burggraf S, Larsen N, Woese C R, Stetter K O. An intron within the 16S ribosomal RNA gene of the archeon Pyrobaculum aerophilum. Proc Natl Acad Sci USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen G, Ernø H. RFLP in rRNA genes of Mycoplasma capricolum, the caprine F38-like group and the bovine serogroup 7. Zentralbl Bakteriol Suppl. 1990;20:479–488. [Google Scholar]
- 8.Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 9.Cottew G S, Breard A, DaMassa A J, Ernø H, Leach R H, Lefèvre P C, Rodwell A W, Smith G R. Taxonomy of the Mycoplasma mycoides cluster. Isr J Med Sci. 1987;23:632–635. [PubMed] [Google Scholar]
- 10.Ernø H, Stipkovits L. Bovine mycoplasmas: cultural and biochemical studies. II. Acta Vet Scand. 1973;14:450–463. doi: 10.1186/BF03547432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray M W, Sankoff D, Cedergren R J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984;12:5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutell R R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutell R R, Larsen N, Woese C R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 15.Harbi M S M A, El Tahir M S, MacOwan K J, Nayil A A. Mycoplasma strain F38 and contagious caprine pleuropneumonia in Sudan. Vet Rec. 1981;108:261. doi: 10.1136/vr.108.12.261. [DOI] [PubMed] [Google Scholar]
- 16.Heldtander M, Pettersson B, Tully J G, Johansson K-E. Sequences of the 16S rRNA genes and phylogeny of the goat mycoplasmas Mycoplasma adleri, Mycoplasma auris, Mycoplasma cottewii, and Mycoplasma yeatsii. Int J Syst Bacteriol. 1998;48:263–268. doi: 10.1099/00207713-48-1-263. [DOI] [PubMed] [Google Scholar]
- 17.Hultman T, Bergh S, Moks T, Uhlén M. Bidirectional solid-phase sequencing of in vitro-amplified plasmid DNA. BioTechniques. 1991;10:84–93. [PubMed] [Google Scholar]
- 18.Hultman T, Ståhl S, Hornes E, Uhlén M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter W N, Brown T, Anand N N, Kennard O. Structure of an adenine · cytosine base pair in DNA and its implications for mismatch repair. Nature (London) 1986;320:552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- 20.Hutcheon D. Contagious pleuropneumonia in Angora goats. Vet J. 1881;13:171–180. [Google Scholar]
- 21.Johansson K-E, Heldtander M U K, Pettersson B. Characterization of mycoplasmas by PCR and sequence analysis with universal 16S rDNA primers. In: Miles R J, Nicholas R A J, editors. Methods in molecular medicine. 104. Mycoplasma protocols. Totowa, N.J: Humana Press Inc.; 1998. pp. 145–165. [DOI] [PubMed] [Google Scholar]
- 22.Jones G E. Contagious caprine pleuropneumonia. In: Truszczynsky M, Pearson J, Edwards S, editors. The OIE manual of standards for diagnostic tests and vaccines. Paris, France: Office International des Epizooties; 1992. pp. 376–392. [Google Scholar]
- 23.Jones G E, Wood A R. Microbiological and serological studies on caprine pneumonias in Oman. Res Vet Sci. 1988;44:125–131. [PubMed] [Google Scholar]
- 24.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H H, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 25.Kanyi Kibe M, Smith G R. A study of F38-type and related mycoplasmas by mycoplasmaemia and cross-immunization tests in mice. J Hyg Camb. 1984;93:465–473. doi: 10.1017/s0022172400065062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leach R H, Ernø H, MacOwan K J. Proposal for designation of F38-type caprine mycoplasmas as Mycoplasma capricolum subsp. capripneumoniae subsp. nov. and consequent obligatory relegation of strains currently classified as M. capricolum (Tully, Barile, Edward, Theodore, and Ernø 1974) to an additional new subspecies, M. capricolum subsp. capricolum subsp. nov. Int J Syst Bacteriol. 1993;43:603–605. doi: 10.1099/00207713-43-3-603. [DOI] [PubMed] [Google Scholar]
- 27.Lefèvre P C, Breard A, Al Faroukh I, Buron S. Mycoplasma species F38 isolated in Chad. Vet Rec. 1987;121:575–576. [PubMed] [Google Scholar]
- 28.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyö E M, Uhlén M. Analysis of heterogeneous viral populations by direct DNA sequencing. BioTechniques. 1993;15:120–126. [PubMed] [Google Scholar]
- 29.Linton D, Dewhirst F E, Clewley J P, Owen R J, Burnens A P, Stanley J. Two types of 16S rRNA gene are found in Campylobacter helveticus: analysis, applications and characterization of the intervening sequence found in some strains. Microbiology. 1994;140:847–855. doi: 10.1099/00221287-140-4-847. [DOI] [PubMed] [Google Scholar]
- 30.MacOwan K J. Role of mycoplasma strain F38 in contagious caprine pleuropneumonia. Isr J Med Sci. 1984;20:979–981. [PubMed] [Google Scholar]
- 31.MacOwan K J, Minette J E. A mycoplasma from acute contagious caprine pleuropneumonia in Kenya. Trop Anim Health Prod. 1976;8:91–95. doi: 10.1007/BF02383376. [DOI] [PubMed] [Google Scholar]
- 32.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal database project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mevarech M, Hirsch-Twizer S, Goldman S, Yakobson E, Eisenberg H, Dennis P P. Isolation and characterization of the rRNA gene clusters of Halobacterium marismortui. J Bacteriol. 1989;171:3479–3485. doi: 10.1128/jb.171.6.3479-3485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montandon P E, Wagner R, Stutz E. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. EMBO J. 1986;5:3705–3708. doi: 10.1002/j.1460-2075.1986.tb04703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsson B, Bölske G, Bergström K, Johansson K-E. Analysis of caprine mycoplasmas and mycoplasma infections in goats using two-dimensional electrophoresis and immunoblotting. Electrophoresis. 1990;11:861–869. doi: 10.1002/elps.1150111016. [DOI] [PubMed] [Google Scholar]
- 36.Patel B K C, Love C A, Stackebrandt E. Helix 6 of the 16S rRNA of the bacterium Desulfotomaculum australicum exhibits an unusual structural idiosyncrasy. Nucleic Acids Res. 1992;20:5483. doi: 10.1093/nar/20.20.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perreau P, Breard A, Le Goff C. Infection Expérimentale de la chévre par les souches de mycoplasme de type F.38 (pleuropneumonie contagieuse caprine) Ann Microbiol (Paris) 1984;135A:119–124. [PubMed] [Google Scholar]
- 38.Pettersson B. Direct solid-phase 16S rDNA sequencing: a tool in bacterial phylogeny. Ph.D. thesis. Stockholm, Sweden: Royal Institute of Technology; 1997. [Google Scholar]
- 39.Pettersson, B., and K.-E. Johansson. Unpublished data.
- 40.Pettersson B, Johansson K-E, Uhlén M. Sequence analysis of 16S rRNA from mycoplasmas by direct solid-phase DNA sequencing. Appl Environ Microbiol. 1994;60:2456–2461. doi: 10.1128/aem.60.7.2456-2461.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersson B, Leitner T, Ronaghi M, Bölske G, Uhlén M, Johansson K-E. Phylogeny of the Mycoplasma mycoides cluster as determined by sequence analysis of the 16S rRNA genes from the two rRNA operons. J Bacteriol. 1996;178:4131–4142. doi: 10.1128/jb.178.14.4131-4142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersson B, Lembke F, Hammer P, Stackebrandt E, Priest F G. Bacillus sporothermodurans, a new species producing highly heat-resistant endospores. Int J Syst Bacteriol. 1996;46:759–764. doi: 10.1099/00207713-46-3-759. [DOI] [PubMed] [Google Scholar]
- 43.Pettersson, B., and F. G. Priest. Unpublished data.
- 44.Pettersson B, Uhlén M, Johansson K-E. Phylogeny of some mycoplasmas from ruminants based on 16S rRNA sequences and definition of a new cluster within the hominis group. Int J Syst Bacteriol. 1996;46:1093–1098. doi: 10.1099/00207713-46-4-1093. [DOI] [PubMed] [Google Scholar]
- 45.Ros Bascuñana C, Mattsson J G, Bölske G, Johansson K-E. Characterization of the 16S rRNA genes from Mycoplasma sp. strain F38 and development of an identification system based on PCR. J Bacteriol. 1994;176:2577–2586. doi: 10.1128/jb.176.9.2577-2586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 47.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiaucourt F, Bölske G. Contagious caprine pleuropneumonia and other pulmonary mycoplasmoses of sheep and goats. Rev Sci Tech OIE. 1996;15:1397–1414. doi: 10.20506/rst.15.4.990. [DOI] [PubMed] [Google Scholar]
- 49.Thiaucourt F, Breard A, Lefèvre P C, Mebratu G Y. Contagious caprine pleuropneumonia in Ethiopia. Vet Rec. 1992;131:585. [PubMed] [Google Scholar]
- 50.Thiaucourt F, Guérin C, Mady V, Lefèvre P C. Diagnostic de la pleuropneumonie contagieuse caprine: améliorations récentes. Rev Sci Tech OIE. 1992;11:859–865. [PubMed] [Google Scholar]
- 51.Thomas P. Rapport médical sur la maladie des chèvres désgnée par les Arabes sous le nom de Bou-Frida gui a sévi epizootiquement pendant l’hiver 1872-1873 dans le cercle de Djelfa. In: Jourdan A, editor. Rapport médical sur le Bou-Frida. Algiers, Algeria: Gouvernment général civil de L’Algérie; 1893. pp. 1–35. [Google Scholar]
- 52.Triman K L. The 16S ribosomal RNA mutation database (16SMDB) Nucleic Acids Res. 1996;24:166–168. doi: 10.1093/nar/24.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tully J G, Bové J M, Laigret F, Whitcomb R F. Revised taxonomy of the class Mollicutes: proposed elevation of a monophyletic cluster of arthropod-associated mollicutes to ordinal rank (Entomoplasmatales ord. nov.), with provision for familial rank to separate species with nonhelical morphology (Entomoplasmataceae fam. nov.) from helical species (Spiroplasmataceae), and emended descriptions of the order Mycoplasmatales, family Mycoplasmataceae. Int J Syst Bacteriol. 1993;43:378–385. [Google Scholar]
- 54.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisburg W G, Tully J G, Rose D L, Petzel J P, Oyaizu H, Yang D, Mandelco L, Sechrest J, Lawrence T G, Van Etten J, Maniloff J, Woese C R. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989;171:6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woese C R, Stackebrandt E, Ludwig W. What are mycoplasmas: the relationship of tempo and mode in bacterial evolution. J Mol Evol. 1985;21:305–316. doi: 10.1007/BF02115648. [DOI] [PubMed] [Google Scholar]
- 57.Wolfson R, Higgins K G, Sears B B. Evidence for replication slippage in the evolution of Oenothera chloroplast DNA. Mol Biol Evol. 1991;8:709–720. doi: 10.1093/oxfordjournals.molbev.a040680. [DOI] [PubMed] [Google Scholar]