Abstract
We investigated the association between the angiotensin I-converting enzyme (ACE) gene insertion (I)/deletion (D) polymorphism and endurance running performance in Japanese elite runners, including several Olympic athletes. The frequency of the I/I genotype was not significantly higher and the frequency of the D/D genotype was not significantly lower in elite runners compared with non-athletes. However, the frequency of the I/D genotype tended to be lower in elite runners than in non-athletes. The best performance was significantly higher for runners with the D/D genotype than for those with the I/I genotype, and the average running speed was significantly higher for those with the combined D/D + I/D genotypes than for those with the I/I genotype. There were no I/I genotypes among the five fastest marathon runners. These results suggest that the D allele of the ACE gene I/D polymorphism is associated with a high level of human endurance.
Keywords: Angiotensin I-converting enzyme, Gene polymorphism, Endurance performance, Genetic factor
Introduction
An insertion (I) or deletion (D) polymorphism in the angiotensin I-converting enzyme (ACE, or kininase II) gene was the first genetic factor reported to influence human physical performance [1]. The polymorphism constitutes a 287-bp insertion/deletion of the 17q23 chromosome and has been found to strongly affect circulatory [2] and tissue [3] ACE activity. ACE is a key factor in regulating blood pressure in the renin–angiotensin system via the production of angiotensin II.
Previous studies have reported that the I allele of the ACE gene I/D polymorphism is present at a higher frequency in British mountaineers [1], Australian and Polish rowers [4, 5], British long distance runners [6], and South African triathletes [7] than in the general population. In addition, a higher maximal oxygen consumption (aerobic capacity), VO2 max, has been reported in postmenopausal women carrying the I/I rather than the D/D genotype [8]. Conversely, the D allele was found to be favorable for (better) performance in power-oriented sports among Russian athletes and Caucasian swimmers [9–11]. However, several studies have reported no difference in the frequency of the I allele in elite endurance athletes in Australia and Korea [12, 13]. Nazarov et al. [9] found an association between the ACE gene I/D polymorphism and sporting prowess in swimmers and track and field athletes, but not in skiers or triathletes. Based on this result, these authors suggested that the association between the ACE gene I/D polymorphism and sporting prowess may only emerge in studies of elite athletes recruited from a single sporting discipline and that the use of subjects from mixed sporting disciplines may account for the absence of an association observed in some studies. In an earlier study, we demonstrated that individuals with the I/I genotype exhibited a significantly higher percentage of type I skeletal muscle than those with the D/D genotype. Conversely, the proportion of type IIb skeletal muscle was higher in D/D individuals [14]. These results support the notion that the I allele is favorable for enhanced performance in endurance sports.
The influence of the ACE gene I/D polymorphism on human physical performance has been investigated in Caucasian populations and people of African descent. In the study reported here, our aim was to identify the effect of this gene polymorphism on endurance performance in Asian people by comparing endurance performance (i.e., running speed) between genotypes.
Materials and methods
Ethical approval
This study was approved by the Ethics committee of Fukuoka University School of Medicine, and written informed consent was obtained from all subjects prior to participation.
Subjects
Thirty-seven elite long distance (over 5,000 m) runners and 335 (non-athlete) control subjects were recruited for this study. All subjects were Japanese males. The elite runners were selected by the Japanese Amateur Athletics Federation. The runners performed at the national or international level, and the group included an Olympic medalist and world record holder.
Genotyping
Peripheral blood samples were obtained from all subjects. DNA was extracted from white blood cells. The ACE gene I/D polymorphism was genotyped by PCR amplification of the relevant fragment of intron 16 of the ACE gene. The PCR products, 490 and 290 bp, respectively, were then separated by electrophoresis in 1.5% agarose gels and visualized under UV light after ethidium bromide staining [14, 15].
Endurance running performance
The best performance of each runner were obtained from a yearbook published by the Japanese Amateur Athletics Federation. For 36 runners, there was a best performance record for a 5,000 m race, and for 12 of the 37 runners, there was also a best performance record for a marathon (one elite runner had only a marathon record). The running speeds (m s−1) in the 5,000 m race and the marathon were calculated from the athletes’ best performances to compare endurance performance between the different genotypes.
Statistical analysis
The distributions of the I/I, I/D, and D/D genotypes in elite runners and non-athletes were compared using chi-square tests, and the odd ratios (OR) were calculated. Running speed was compared using Tukey–Kramer tests and unpaired t tests. Because the group of elite runners who had a best performance marathon record was very small, we used Scheffe’s test to compare the running speed. Additionally, Spearman rank correlation coefficients were used for the trend analysis of the association between marathon running speed and the ACE gene I/D polymorphism. A p value of <0.05 was considered to be statistically significant.
Results
The characteristics of the subjects enrolled in the study are shown in Table 1. There was no significant age difference among the genotypes in the elite runner or the non-athlete groups. Since world records are continuously improving, the relative significance of an athlete’s best record changes over time. The fact that the ages of the groups were not significantly different indicates that the athletes of each group were likely to have been around the same age when they set their records. The distribution of the ACE gene I/D polymorphism was in Hardy–Weinberg equilibrium in non-athletes [the expected genotype frequencies were: I/I, 0.46; I/D, 0.44; D/D, 0.10; χ 2 = 0.01, df = 2, not significant (NS)], but not in elite runners (expected genotype frequencies were: I/I, 0.42; I/D, 0.46; D/D, 0.12; χ 2 = 16.57, df = 2, p < 0.001).
Table 1.
Characteristics of the subjects
| Study cohort | Genotype | ||||
|---|---|---|---|---|---|
| I/I | I/D | D/D | I/I + I/D | D/D + I/D | |
| Elite runners | |||||
| n | 19 | 10 | 8 | 29 | 18 |
| Age (years) | 42 ± 10 | 42 ± 7 | 40 ± 5 | 42 ± 9 | 41 ± 6 |
| Non-athletes | |||||
| n | 155 | 146 | 34 | 301 | 180 |
| Age (years) | 63 ± 18 | 63 ± 18 | 65 ± 17 | 63 ± 18 | 65 ± 17 |
n Number of subjects, ACE angiotensin I-converting enzyme gene, I insertion, D deletion
Values are shown as mean ± standard deviation (SD)
The distribution of genotypes of the ACE gene I/D polymorphism was significantly different between elite runners and non-athletes (elite runners: I/I, 0.51; I/D, 0.27; D/D, 0.22; male non-athletes: I/I, 0.46; I/D, 0.44; D/D, 0.10; χ 2 = 19.81, df = 2, p < 0.001). The allelic frequency was similar in both groups (elite runners: I/I + I/D, 0.65; D/D + I/D, 0.35; non-athletes: I/I + I/D, 0.68; D/D + I/D, 0.32; χ 2 = 0.003, df = 1, NS). The frequency of the I/I genotype was not significantly higher (OR 1.23, 95% CI 0.62–2.42, NS), and the frequency of the D/D genotype was not significantly lower (OR 1.63, 95% CI 0.69–3.87, NS) in elite runners than in non-athletes. In contrast, the frequency of the I/D genotype tended to be lower in elite runners than in the non-athletes (OR 0.48, 95% CI 0.23–1.02, p = 0.055).
We observed a trend towards a faster 5,000 m running speed in individuals with the D/D genotype than in those with the I/I genotype, although the difference was not significant (p = 0.053). However, combined D/D + I/D individuals ran, on average, significantly faster than I/I individuals (p = 0.023; Fig. 1a), and a significant linear trend was observed for this genotype and the 5,000 m running speed (r = 0.345, p = 0.042; Fig. 1b). The marathon running speed (m s−1) for each genotype was calculated to be 5.31 (0.08) (median and interquartile range), 5.37 (0.05), and 5.46 (0.07) m s−1 for the I/I, I/D, and D/D genotypes, respectively (Fig. 2). Individuals with the D/D genotype had a significantly faster running speed than those with the I/I genotype (p = 0.005). A linear relationship was also observed between genotype and marathon running speed (r = 0.833, p = 0.006). The five marathon runners who completed the marathon within 2 h 10 min all exhibited either the D/D or I/D genotype (D/D, 4; I/D, 1; Fig. 2).
Fig. 1.
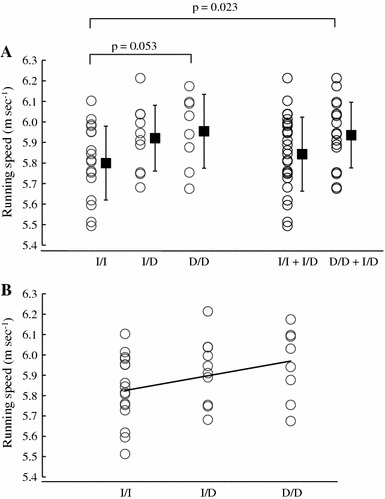
Running speed, calculated on the basis of each athlete’s best performance record in a 5,000 m race, was higher in individuals with the D/D or I/D genotype than in those with the I/I genotype (unpaired t test p = 0.023). The running speed of D/D individuals tended to be higher than that of I/I individuals (Tukey–Kramer p = 0.053). A linear trend was observed between genotype and 5,000 m running speed (Spearman rank correlation coefficients r = 0.345; p = 0.042). Data are shown as the mean and standard deviation (SD)
Fig. 2.
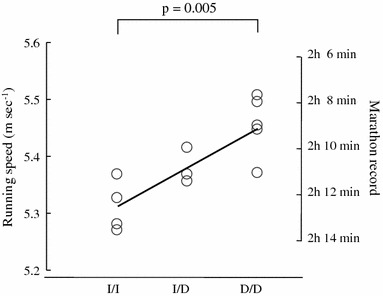
The marathon running speed (m s−1) for each genotype was 5.31 (0.08) (median and interquartile range), 5.37 (0.05), and 5.46 (0.07) m s−1 for the I/I, I/D, and D/D genotypes, respectively. Individuals with the D/D genotype had a significantly faster running speed than those with the I/I genotype (Scheffe’s test p = 0.005). A linear trend was observed between the genotype and marathon running speed (Spearman rank correlation coefficients r = 0.833, p = 0.006). The five marathon runners who ran faster than 5.41 m s−1 (i.e., completing the marathon within 2 h 10 min) all exhibited the D/D or I/D genotype (D/D, 4; I/D, 1)
Discussion
The study reported here is, to the best of our knowledge, the first investigation on whether the ACE gene I/D polymorphism influences endurance running performance in a Japanese population. We also compared running performance among the genotypes, and our analysis of the results revealed that the D allele was significantly associated with higher endurance performance.
A number of studies have found a disproportionately high frequency of the I allele of the ACE gene I/D polymorphism among elite mountaineers [1] and endurance athletes [1, 4–7, 16, 17] compared with the general population. In contrast, no association between the ACE gene I/D polymorphism and human physical performance was found in a number of large cohort studies [12, 18, 19]. Nazarov et al. [9] suggested that the inclusion of subjects from multiple sporting disciplines within a single study design may remove the association between the ACE gene I/D polymorphism and physical performance. Consequently, in our study of a Japanese sample of elite athletes, we recruited only endurance runners (and controls) to enable a simple comparison of endurance performance.
The distributions of the ACE gene genotypes were not in Hardy–Weinberg equilibrium in the elite runners enrolled in our study; however, the allelic frequency was similar to that in the control group of non-athletes. In addition, the frequency of the I/D genotype tended to be lower in the elite runners than in the non-athletes (OR 0.48, 95% CI 0.23–1.02, p = 0.055). These results demonstrate that the frequency of the ACE gene I/D polymorphism in elite runners was bimodally distributed in our sample. As such, we suggest that the D/D is also favorable to human endurance performance.
Surprisingly, we found that endurance performance was better in D/D + I/D individuals than in I/I individuals. Furthermore, all elite runners who had completed marathons within 2 h 10 min were either D/D or I/D. Only about 50 Japanese runners before 2009 and 15 runners in the last 5 years have completed marathons within 2 h and 10 min. In addition, only one Japanese runner has run a marathon within this time in 2009. The result that the most outstanding runners in our sample were D/D or I/D suggests that the D allele is favorable for human endurance performance.
Interestingly, Amir et al. [20] also suggested that the D allele was more prevalent in Israeli elite endurance athletes, although Zoossmann-Diskin [21] subsequently proposed that the Jewish populations of Israel come from many countries and that the relatively distinct gene pools may have affected the results of Amir et al.’s study. Because all participants in our study were Japanese, our results were not affected by ethnicity bias. It remains unclear just why the D allele may be associated with higher endurance performance, but our results suggest that this allele may confer some advantage for endurance running. Consequently, we present several speculative possibilities.
One possibility is that the D allele is related to better cardiac function during exercise. Previous studies have suggested that the D allele is strongly associated with exercise-induced left ventricular (LV) growth [22–25]. Greater LV wall thickness and LV mass may contribute to high endurance running performance due to higher cardiac function during exercise. In accordance with this possibility, the results of our previous study demonstrated that exercise-induced improvement in cardiac function was greater in individuals with the D/D or I/D genotype than in those with the I/I genotype [26].
Second, it may be that the D/D genotype confers less stress during exercise than the I/I genotype. In a previous study, we examined the I/D polymorphism in relation to the double product (DP) [27], which is the product of heart rate and systolic blood pressure and a commonly used index of myocardial oxygen consumption during exercise [28]. We found that at the lactate threshold, the DP was lower in D/D individuals [27]. Rankinen et al. [18] showed that D/D homozygotes have a lower blood lactate level during exercise at 60 and 80% maximum oxygen consumption than I/I homozygotes. Based on the results of our previous study, we were also able to determine that the blood lactate concentration during exercise tended to be lower in D/D individuals than in I/I individuals [29]. High blood lactic acid levels have been reported to cause a decreased power output. The D/D genotype may therefore prevent a decrease in power output due to lower blood lactic acid concentration during exercise. In this way, D/D individuals may be able to perform exercise with lower stress than I/I individuals.
Third, the D allele may reduce body weight gain caused by physical training. The I allele is associated with greater fat storage during physical training [30]. Fat is an important energy source for skeletal muscle; however, the majority of elite long distance runners have a low percentage of body fat mass. A heavier body mass consumes more energy with movement, so a low body fat mass is efficient for endurance running.
Finally, ACE may affect energy supply and fatigue. Bradykinin is one of the mediators that increases glucose uptake in skeletal muscle due to GLUT4 translocation [31]. On the other hand, bradykinin is inactivated by ACE. Long distance running, such as completing a marathon, requires a great deal of glucose in the skeletal muscles and brain, particularly in the latter since glucose is the only energy source for the brain. Hypoglycemia due to the depletion of glucose causes central fatigue and a decreased power output [32]. The D allele, which is associated with higher ACE activity, may not facilitate glucose uptake in skeletal muscle as well as the I allele, which would be advantageous in conserving glucose and preventing central fatigue due to hypoglycemia.
The main limitation of the current study is the small number of elite runners. Although these athletes were reliable elite-level runners, as attested by their running performance, the statistical power of the study is relatively low.
In conclusion, our results suggest that the I allele of the ACE gene I/D polymorphism is not a strong genetic factor affecting human endurance performance in Japanese elite runners. However, the D allele of the ACE gene I/D polymorphism appears to be associated with enhanced performance in endurance running. Further studies are required to understand the mechanism of action involved. Because this study focused only on Japanese elite runners, future studies are also required to test whether this association is also present in non-athletes.
Acknowledgments
The authors are deeply grateful to the people who kindly participated in this research as subjects, and to Mitsu Funakoshi, who provided statistical analysis support. This work was supported by Grants-in-Aid from The Ministry of Education, Science, Sports and Culture of Japan (Grant No. 15300229, Tokyo, Japan), and The Fukuoka University Institute for Physical Activity, as well as a Fukuoka University Global FU Program grant.
References
- 1.Montgomery HE, Marshall R, Hemingway H, Myerson S, Clarkson P, Dollery C, Hayward M, Holliman DE, Jubb M, World M, Thomas EL, Brynes AE, Saeed N, Barnard M, Bell JD, Prasad K, Rayson M, Talmud PJ, Humphries SE. Human gene for physical performance. Nature. 1998;393:221–222. doi: 10.1038/30374. [DOI] [PubMed] [Google Scholar]
- 2.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danser AH, Schalekamp MA, Bax WA, van den Brink AM, Saxena PR, Riegger GA, Schunkert H. Angiotensin-converting enzyme in the human heart. Effect of the deletion/insertion polymorphism. Circulation. 1995;92:1387–1388. doi: 10.1161/01.cir.92.6.1387. [DOI] [PubMed] [Google Scholar]
- 4.Gayagay G, Yu B, Hambly B, Boston T, Hahn A, Celermajer DS, Trent RJ. Elite endurance athletes and the ACE I allele the role of genes in athletic performance. Hum Genet. 1998;103:48–50. doi: 10.1007/s004390050781. [DOI] [PubMed] [Google Scholar]
- 5.Cieszczyk P, Krupecki K, Maciejewska A, Sawczuk M. The angiotensin converting enzyme gene I/D polymorphism in polish rowers. Int J Sports Med. 2009;30:624–627. doi: 10.1055/s-0029-1202825. [DOI] [PubMed] [Google Scholar]
- 6.Myerson S, Hemingway H, Budget R, Martin J, Humphries S, Montgomery HE. Human angiotensin I-converting enzyme gene and endurance performance. J Appl Physiol. 1999;87:1313–1316. doi: 10.1152/jappl.1999.87.4.1313. [DOI] [PubMed] [Google Scholar]
- 7.Collins M, Xenophontos SL, Cariolou MA, Mokone GG, Hudson DE, Anastasiades L, Noakes TD. The ACE gene and endurance performance during the South African Ironman Triathlons. Med Sci Sports Exerc. 2004;36:1314–1320. doi: 10.1249/01.MSS.0000135779.41475.42. [DOI] [PubMed] [Google Scholar]
- 8.Hagberg JM, Ferrell RE, McCole SD, Wilund KR, Moore GE. VO2 max is associated with ACE genotype in postmenopausal women. J Appl Physiol. 1998;85:1842–1846. doi: 10.1152/jappl.1998.85.5.1842. [DOI] [PubMed] [Google Scholar]
- 9.Nazarov IB, Woods DR, Montgomery HE, Shneider OV, Kazakov VI, Tomilin NV, Rogozkin VA. The angiotensin converting enzyme I/D polymorphism in Russian athletes. Eur J Hum Genet. 2001;9:797–801. doi: 10.1038/sj.ejhg.5200711. [DOI] [PubMed] [Google Scholar]
- 10.Woods D, Hickman M, Jamshidi Y, Brull D, Vassiliou V, Jones A, Humphries S, Montgomery HE. Elite swimmers and the D allele of the ACE I/D polymorphism. Hum Genet. 2001;108:230–232. doi: 10.1007/s004390100466. [DOI] [PubMed] [Google Scholar]
- 11.Costa AM, Silva AJ, Garrido ND, Louro H, de Oliveira RJ, Breitenfeld L. Association between ACE D allele and elite short distance swimming. Eur J Appl Physiol. 2009;106:785–790. doi: 10.1007/s00421-009-1080-z. [DOI] [PubMed] [Google Scholar]
- 12.Taylor RR, Mamotte CD, Fallon K, van Bockxmeer FM. Elite athletes and the gene for angiotensin-converting enzyme. J Appl Physiol. 1999;87:1035–1037. doi: 10.1152/jappl.1999.87.3.1035. [DOI] [PubMed] [Google Scholar]
- 13.Oh SD. The distribution of I/D polymorphism in the ACE gene among Korean male elite athletes. J Sports Med Phys Fit. 2007;47:250–254. [PubMed] [Google Scholar]
- 14.Zhang B, Tanaka H, Shono N, Miura S, Kiyonaga A, Shindo M, Saku K. The I allele of the angiotensin-converting enzyme gene is associated with an increased percentage of slow-twitch type I fibers in human skeletal muscle. Clin Genet. 2003;63:139–144. doi: 10.1034/j.1399-0004.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Sakai T, Miura S, Kiyonaga A, Tanaka H, Shindo M, Saku K. Association of angiotensin-converting enzyme gene polymorphism with the depressor to mild exercise therapy in patients with mild to moderate essential hypertension. Clin Genet. 2002;62:328–333. doi: 10.1034/j.1399-0004.2002.620414.x. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez R, Terrados N, Ortolano R, Iglesias-Cubero G, Reguero JR, Batalla A, Cortina A, Fernandez-Garcia B, Rodriguez C, Braga S, Alvarez V, Coto E. Genetic variation in the renin-angiotensin system and athletic performance. Eur J Appl Physiol. 2000;82:117–120. doi: 10.1007/s004210050660. [DOI] [PubMed] [Google Scholar]
- 17.Scanavini D, Bernardi F, Castoldi E, Conconi F, Mazzoni G. Increased frequency of the homozygous II ACE genotype in Italian Olympic endurance athletes. Eur J Hum Genet. 2002;10:576–577. doi: 10.1038/sj.ejhg.5200852. [DOI] [PubMed] [Google Scholar]
- 18.Rankinen T, Perusse L, Gagnon J, Chagnon YC, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Angiotensin-converting enzyme ID polymorphism and fitness phenotype in the HERITAGE Family Study. J Appl Physiol. 2000;88:1029–1035. doi: 10.1152/jappl.2000.88.3.1029. [DOI] [PubMed] [Google Scholar]
- 19.Rankinen T, Wolfarth B, Simoneau JA, Maier-Lenz D, Rauramaa R, Rivera MA, Boulay MR, Chagnon YC, Perusse L, Keul J, Bouchard C. No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. J Appl Physiol. 2000;88:1571–1575. doi: 10.1152/jappl.2000.88.5.1571. [DOI] [PubMed] [Google Scholar]
- 20.Amir O, Amir R, Yamin C, Attias E, Eynon N, Sagiv M, Sagiv M, Meckel Y. The ACE deletion allele is associated with Israeli elite endurance athletes. Exp Physiol. 2007;92:881–886. doi: 10.1113/expphysiol.2007.038711. [DOI] [PubMed] [Google Scholar]
- 21.Zoossmann-Diskin A. The association of the ACE gene and elite athletic performance in Israel may be an artifact. Exp Physiol. 2008;93:1220. doi: 10.1113/expphysiol.2007.041921. [DOI] [PubMed] [Google Scholar]
- 22.Montgomery HE, Clarkson P, Dollery CM, Prasad K, Losi MA, Hemingway H, Statters D, Jubb M, Girvain M, Varnava A, World M, Deanfield J, Talmud P, McEwan JR, McKenna WJ, Humphries S. Association of angiotensin-converting enzyme gene I/D polymorphism with change in left ventricular mass in response to physical training. Circulation. 1997;96:741–747. doi: 10.1161/01.cir.96.3.741. [DOI] [PubMed] [Google Scholar]
- 23.Nagashima J, Musha H, Takada H, Awaya T, Oba H, Mori N, Ohmiya K, Nobuoka S, Murayama M. Influence of angiotensin-converting enzyme gene polymorphism on development of athlete’s heart. Clin Cardiol. 2000;23:621–624. doi: 10.1002/clc.4960230814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fatini C, Guazzelli R, Manetti P, Battaglini B, Gensini F, Vono R, Toncelli L, Zilli P, Capalbo A, Abbate R, Gensini GF, Galanti G. RAS genes influence exercise-induced left ventricular hypertrophy: an elite athletes study. Med Sci Sports Exerc. 2000;32:1868–1872. doi: 10.1097/00005768-200011000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Hernández D, de la Rosa A, Barragán A, Barrios Y, Salido E, Torres A, Martín B, Laynez I, Duque A, De Vera A, Lorenzo V, González A. The ACE/DD genotype is associated with the extent of exercise-induced left ventricular growth in endurance athletes. J Am Coll Cardiol. 2003;42:527–532. doi: 10.1016/S0735-1097(03)00642-9. [DOI] [PubMed] [Google Scholar]
- 26.Tobina T, Kiyonaga A, Akagi Y, Mori Y, Ishii K, Chiba H, Shindo M, Tanaka H. Angiotensin I converting enzyme gene polymorphism and exercise trainability in elderly women: an electrocardiological approach. J Sports Sci Med. 2007;6:220–226. [PMC free article] [PubMed] [Google Scholar]
- 27.Kiyonaga A, Imai K, Zhang Bo, Saku K. Association between insertion/deletion polymorphism of the angiotensin I converting enzyme gene and blood pressure responses to exercise in normal controls and hypertensive patients. In: Tanaka H, Shindo M, editors. Exercise for preventing common diseases. Berlin: Springer; 1999. pp. 35–44. [Google Scholar]
- 28.Braunwald E. Control of myocardial oxygen consumption: physiologic and clinical considerations. Am J Cardiol. 1971;27:416–432. doi: 10.1016/0002-9149(71)90439-5. [DOI] [PubMed] [Google Scholar]
- 29.Tobina T, Michishita R, Zhang B, Saku K, Shindo M, Tanaka H, Kiyonaga A. Association of angiotensin I converting enzyme gene insertion/deletion polymorphism with the renin-angiotensin system and blood pressure response during a single bout of exercise. Int J Sports Health Sci. 2006;2:465–471. [Google Scholar]
- 30.Montgomery HE, Clarkson P, Barnard M, Bell J, Brynes A, Dollery C, Hajnal J, Hemingway H, Mercer D, Jarman P, Marshall R, Prasad K, Rayson M, Saeed N, Talmud P, Thomas L, Jubb M, World M, Humphries S. Angiotensin-converting-enzyme gene insertion/deletion polymorphism and response to physical training. Lancet. 1999;353:541–545. doi: 10.1016/S0140-6736(98)07131-1. [DOI] [PubMed] [Google Scholar]
- 31.Kishi K, Muromoto N, Nakaya Y, Miyata I, Hagi A, Hayashi H, Ebina Y. Bradykinin directly triggers GLUT4 translocation via an insulin-independent pathway. Diabetes. 1998;47:550–558. doi: 10.2337/diabetes.47.4.550. [DOI] [PubMed] [Google Scholar]
- 32.Newsholme EA, Blomstrand E, Ekblom B. Physical and mental fatigue: metabolic mechanisms and importance of plasma amino acids. Br Med Bull. 1992;48:477–495. doi: 10.1093/oxfordjournals.bmb.a072558. [DOI] [PubMed] [Google Scholar]


