Abstract
We have characterized the Chlamydia trachomatis ribosomal promoter, rRNA P1, by measuring the effect of substitutions and deletions on in vitro transcription with partially purified C. trachomatis RNA polymerase. Our analyses indicate that rRNA P1 contains potential −10 and −35 elements, analogous to Escherichia coli promoters recognized by E-ς70. We identified a novel AT-rich region immediately downstream of the −35 region. The effect of this region was specific for C. trachomatis RNA polymerase and strongly attenuated by single G or C substitutions. Upstream of the −35 region was an AT-rich sequence that enhanced transcription by C. trachomatis and E. coli RNA polymerases. We propose that this region functions as an UP element.
Chlamydia trachomatis is a gram-negative obligate intracellular pathogen with a biphasic developmental life cycle (reviewed in references 16 and 20). We have been characterizing the transcriptional machinery and defining the structure of chlamydial promoters. C. trachomatis RNA polymerase (RNAP) resembles other eubacterial RNAPs in being a multisubunit enzyme containing α, β, β′, and ς subunits (3, 4, 11, 12). Three ς subunits have been identified in Chlamydia to date. ςA is a homolog of ς70, and two alternative ς subunits show sequence homology to Escherichia coli ς54 and Bacillus subtilis ς28, respectively (21). ςA shows striking amino acid sequence conservation with ς70 in subregions 2.4 and 4.2 (3, 12), which recognize the −10 and −35 promoter elements, respectively. Within these regions, specific amino acid residues that have been shown to be involved in promoter recognition are completely conserved (14).
It is not clear if this striking conservation in the ς promoter recognition domains is accompanied by conservation of promoter structure between C. trachomatis and E. coli. Consensus ς70 −10 promoter sequences can be recognized upstream of the transcription initiation sites of many C. trachomatis genes (7). Some, but not all, of these genes also have −35 promoter sequences spaced an optimal 16 to 18 bp upstream of the −10 promoter sequences (6, 8, 13, 23). However, other chlamydial genes are not preceded by recognizable ς70 promoter sequences.
In vitro transcription studies have begun to define the structures of promoters in C. trachomatis. Matthews and Sriprakash found that in vitro promoter activity was reduced or eliminated by multiple mutations in the predicted −10 and −35 regions of the plasmid countertranscript (PCT) promoter, although single substitutions had no measurable effect (15). Douglas and Hatch showed that sequences in the −35 region and adjacent AT-rich regions were important for major outer membrane protein (MOMP) P2 promoter activity in vitro (2). The predicted −10 hexamer of MOMP P2, TATCGC, differed from the E. coli −10 hexamer by the presence of C and G residues in the last three positions, but only the first two bases of this hexamer were important for promoter activity.
We have been characterizing C. trachomatis rRNA P1 by in vitro transcription with heparin-agarose-purified C. trachomatis RNAP. rRNA is highly transcribed, and rRNA P1 is likely to be a strong promoter. In E. coli, the rRNA promoters are among the strongest promoters and are very close in sequence to the consensus ς70 promoter. E. coli rRNA promoters also contain a third promoter element, the UP element, that can enhance transcription as much as 30-fold (17). Unlike −10 and −35 promoter elements, which are recognized by ς70, the UP element is recognized by the carboxy-terminal domain of the α subunit of RNAP (α-CTD). While UP elements have not been previously identified in C. trachomatis, the amino acids of α that recognize the UP element are completely conserved between E. coli and C. trachomatis (10, 11).
In our previous study, we used 5′ deletions and 5-bp substitutions of rRNA P1 to demonstrate that sequences in the approximate −10 and −35 regions were required for promoter activity with C. trachomatis or E. coli ς70 RNAP (22). This analysis also defined a region within −26 to −22 that was required for transcription by partially purified C. trachomatis RNAP but not by E. coli RNAP.
To further define the specific bases required for transcription by C. trachomatis RNAP, we performed saturation mutagenesis of rRNA P1 by constructing single base substitutions from −41 to −1. The effects of these substitutions on transcription by C. trachomatis and E. coli RNAPs were compared. Our results defined four regions that contributed to in vitro transcription by partially purified C. trachomatis RNAP.
MATERIALS AND METHODS
Reagents.
Products were obtained from the following sources and were used according to the manufacturers’ specifications. Restriction enzymes, bacterial alkaline phosphatase, and T4 DNA ligase were from Gibco BRL (Gaithersburg, Md.); T4 polynucleotide kinase was from Boehringer Mannheim Biochemicals (Indianapolis, Ind.); RNasin and RQ1 DNase were from Promega Biotech (Madison, Wis.); SP6 RNA polymerase was from Ambion (Austin, Tex.); Sequenase DNA polymerase was from United States Biochemical Corp. (Cleveland, Ohio); Thermus aquaticus DNA polymerase was from Cetus Corp. (Emeryville, Calif.); 32P-containing nucleoside triphosphates were from Amersham Corp. (Arlington Heights, Ill.); SeaPlaque and SeaKem agarose were from FMC Bioproducts (Rockland, Maine); ampicillin and gentamicin sulfate were from Sigma Chemical Co. (St. Louis, Mo.); vancomycin hydrochloride was from Abbott Laboratories (North Chicago, Ill.), and dimethyl sulfoxide was from Fisher Scientific (Pittsburgh, Pa.).
DNA manipulation.
Standard recombinant DNA methods were used for nucleic acid preparation and analysis (18). DNA was amplified by PCR as described previously (22). DNA sequencing was performed with the dideoxy-chain termination method (19) using a Sequenase kit (United States Biochemical Corp.) on double-stranded plasmid DNA.
Synthetic oligonucleotides.
The following single-stranded oligonucleotide primers were synthesized by Gibco BRL: M13 forward −40, 5′ GTTTT CCCAG TCACG AC; M13 reverse −40, 5′ GTTGT GTGGA ATTGT G; pGLS3′, 5′ ATAGG AGGAA TAATG; rRNA-3, 5′ CAGGG TACCA GGCCT CCGCG TTCAA GA; rRNA-4, 5′ CACGA ATTCC GCGTT CAAGA AAGG; Tx1, 5′ AAAGT AACAT CTTAT ATCAA CCTCT; Tx25, 5′ TATTA TATAG TGTCA CCTAA AT; Tx27, 5′ GGGAA GAGGG GGTGA GAG; and Tx33, 5′ CCGAA CGACC GAGCG CAGCG.
Plasmid containing the rRNA P1 and SP6 control promoters.
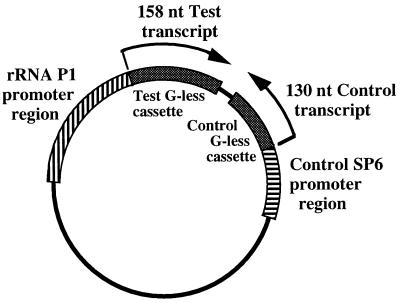
pMT589 (Fig. 1) was derived from pMT504 (22) by deletion of the lacZ and T7 promoters on a 124-bp PvuII-ApaI fragment and construction of a new SP6 control transcription template. The SP6 promoter was amplified by PCR, using pGEM-7Zf(+) as the template with M13 reverse −40 and Tx25 primers, and cloned upstream of the control G-less cassette. The G residue at +1 was replaced with a T residue to allow for transcription in the absence of GTP. Transcription with SP6 RNAP produced a 130-nucleotide control transcript.
FIG. 1.
Transcription template plasmid with rRNA P1 test promoter and SP6 control promoter. Wild-type rRNA P1 (−302 to +5), or promoter templates containing mutations, was cloned immediately upstream of the test G-less cassette in plasmid pMT589. In vitro transcription with C. trachomatis RNAP or E. coli RNAP produced a 158-nucleotide (nt) test transcript. The control promoter region contained an SP6 promoter cloned in front of the control G-less cassette. Transcription with SP6 RNAP produced a 130-nucleotide control transcript.
pMT589 also contained a longer G-less cassette with convenient upstream restriction sites for cloning a test promoter. For this study, rRNA P1 (−302 to +5), containing the wild-type sequence or a mutation, was used as the test promoter. The G residue at +1 was replaced with an A residue to allow for transcription in the absence of GTP. This substitution did not change the transcription initiation site. Transcription with C. trachomatis or E. coli RNAP produced a 158-nucleotide test transcript.
Supercoiled DNA plasmids for use in the in vitro transcription assays were prepared by using a Qiagen Plasmid Midi Kit (Qiagen Inc., Chatsworth, Calif.) according to the manufacturer’s directions. The DNA sequences of the promoter templates were determined to ascertain that there were no inadvertent mutations from the PCR and cloning steps.
Construction of rRNA P1 templates containing mutations.
A 300-bp region of rRNA P1 (−302 to +5) was generated by PCR or by the megaprimer method (see below). PCR was used if the desired mutation was located close to the transcription start site so that it could be introduced on a PCR primer. The promoter region was amplified by using pMT513 (22) as the template, rRNA-4 as the 5′ primer, and a 3′ primer containing the mutation. The 300-bp PCR product was phosphorylated, digested with EcoRI, and ligated to plasmid pMT589 previously digested with EcoRI and EcoRV.
Megaprimer method for generating promoter templates with mutations.
A modification of the megaprimer method (22) was used to introduce mutations that could not be located on a primer for one-step PCR amplification. The megaprimer was generated by PCR with pMT513 as the template, pGLS3′ as the 3′ primer, and a mutation-containing 5′ primer. The 200-bp megaprimer was electrophoresed and recovered from an agarose gel (22). The megaprimer was extended with Sequenase DNA polymerase, using 100 ng of heat-denatured, linearized plasmid pMT175 (22) as the template in a 10-μl reaction volume. Two microliters of the extended megaprimer mix was used as the template in a second PCR with 100 pmol of primers Tx33 and Tx27 and 10% (vol/vol) dimethyl sulfoxide. The 600-bp PCR product was recovered from an agarose gel, digested with EcoRI and DraI, and ligated to plasmid pMT589 previously digested with EcoRI and EcoRV.
In vitro transcription.
The method used is as described in reference 22 with slight modifications. The following reaction mixture was assembled and preincubated on ice for 30 min: 50 mM potassium acetate, 8.1 mM magnesium acetate, 50 mM Tris acetate (pH 8.0), 27 mM ammonium acetate, 2 mM dithiothreitol, 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.21 μM [α-32P]CTP (3,000 Ci/mmol), 100 μM 3′-O-methylguanosine 5′-triphosphate, Na salt (Pharmacia Biotech, Piscataway, N.J.), 18 U of RNasin, 10% glycerol, and 5 U of SP6 RNA polymerase. The DNA template (final concentration, 25 nM) and 0.25 μl of heparin-agarose-purified C. trachomatis RNAP (22) were added, and the reaction mixture was incubated at 37°C for 5 min. Heparin was added to a final concentration of 150 μg/ml, and the incubation was continued at 37°C for a further 10 min. The final reaction volume was 10 μl. The reaction was stopped by the addition of 10 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol); 7 μl of the sample was electrophoresed on an 8 M urea–6% polyacrylamide gel. Transcripts were visualized by autoradiography and quantified with a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager. The size of the transcript was determined by coelectrophoresis with an M13 sequencing ladder. A single preparation of C. trachomatis RNAP was used for the transcription reactions in this study.
In some transcription reactions, 0.003 U of E. coli ς70 RNAP (Epicentre Technologies, Madison, Wis.) was used instead of C. trachomatis RNAP. In these experiments, 3 U of SP6 RNA polymerase was used to transcribe the control SP6 promoter. E. coli RNAP containing wild-type α or a truncation of the α-CTD were reconstituted in vitro from purified subunits as described previously (10, 17).
Calculation of promoter activity.
For each promoter template, transcription with C. trachomatis RNAP (or E. coli RNAP) was normalized to a control SP6 RNAP transcript, which corrected for DNA concentration and sample handling. The promoter activity obtained with wild-type rRNA P1 was defined as 100%, and the promoter activity of each mutant promoter was normalized to this value. For each mutant promoter, three measurements of promoter activity were obtained and a mean and standard deviation were calculated.
Primer extension analysis of in vitro transcription products.
Unlabeled in vitro transcripts were synthesized as described above, with the following changes: each 10-μl reaction mixture contained 2 μl of C. trachomatis RNAP (or 0.05 U of E. coli RNAP), 400 μM CTP, and no [α-32P]CTP. Prior to primer extension analysis, RNA samples were treated with RQ1 DNase to remove the DNA template. Primer extension analysis was carried out as previously described (5) with 32P-end-labeled primer Tx27, which is complementary to the extreme 3′-end of the test G-less cassette template. Primer extension products were electrophoresed next to a Tx27-primed DNA sequence of pMT513 (22).
RESULTS
We have renumbered the positions of rRNA P1 since our previous study (22). We have shifted +1 downstream one position, as repeat primer extension studies have localized the in vitro transcription initiation site to this position (data not shown). The base at the new +1 position is a G residue. The three adjacent in vivo start sites (5) are now at positions −1, +1, and +2. The inefficiency of the C. trachomatis transcription system required that we employ a G-less transcript sequence and use transcription reactions in which no GTP was added in order to increase sensitivity and permit quantitation of in vitro transcription (22). To allow transcription in the absence of GTP, we have made a substitution of G to A at +1. We cannot rule out the possibility that this substitution altered the properties of the promoter. Nevertheless, since E. coli RNAP does not have a preference for G over A at +1 so long as the initiating nucleoside triphosphate is provided at a high concentration (reference 9 and unpublished results), we have made the assumption that C. trachomatis RNAP recognizes rRNA P1 promoters similarly with A or G at this position.
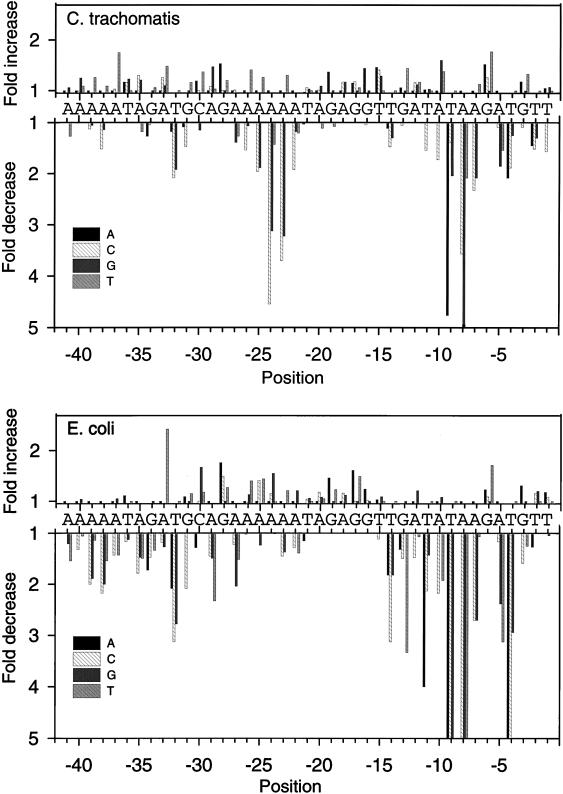
We tested the effect on in vitro transcription of single base substitutions at positions −41 to −1 of rRNA P1 (Table 1 and Fig. 2). All three substitutions were tested for 37 of 41 positions, and two substitutions were tested for each of the remaining positions. Mutations with effects on transcription clustered in several regions that were specific for C. trachomatis and E. coli RNAPs.
TABLE 1.
Promoter activities of rRNA P1 templates with single base substitutions
| Relative promoter activity ± SDa
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Position | Wild-type sequence |
C. trachomatis RNAP
|
E. coli RNAP
|
||||||
| A | C | G | T | A | C | G | T | ||
| −1 | T | 105 ± 9 | 64 ± 7 | 107 ± 17 | 100 | 118 ± 3 | 108 ± 5 | 96 ± 3 | 100 |
| −2 | T | 69 ± 3 | 66 ± 3 | 77 ± 18 | 100 | 79 ± 6 | 116 ± 2 | 120 ± 2 | 100 |
| −3 | G | 118 ± 5 | 92 ± 5 | 100 | 133 ± 14 | 131 ± 8 | 63 ± 1 | 100 | 80 ± 2 |
| −4 | T | 47 ± 3 | 53 ± 9 | 80 ± 19 | 100 | 15 ± 4 | 20 ± 5 | 34 ± 13 | 100 |
| −5 | A | 100 | 92 ± 27 | 54 ± 4 | 65 ± 4 | 100 | 86 ± 8 | 42 ± 1 | 32 ± 3 |
| −6 | G | 152 ± 30 | 126 ± 8 | 100 | 177 ± 23 | 123 ± 5 | 109 ± 0 | 100 | 171 ± 14 |
| −7 | A | 100 | 43 ± 7 | 48 ± 19 | 113 ± 15 | 100 | 37 ± 3 | 37 ± 2 | 94 ± 7 |
| −8 | A | 100 | 28 ± 4 | 11 ± 1 | 48 ± 5 | 100 | 6 ± 3 | 6 ± 4 | 9 ± 2 |
| −9 | T | 21 ± 5 | 72 ± 8 | 49 ± 5 | 100 | 8 ± 2 | 12 ± 2 | 10 ± 4 | 100 |
| −10 | A | 100 | 58 ± 7 | 160 ± 10 | 138 ± 4 | 100 | 46 ± 3 | 108 ± 8 | 52 ± 1 |
| −11 | T | 102 ± 6 | 65 ± 8 | 103 ± 7 | 100 | 25 ± 2 | 47 ± 8 | 70 ± 4 | 100 |
| −12 | A | 100 | 116 ± 11 | 112 ± 4 | 117 ± 23 | 100 | 68 ± 3 | 121 ± 3 | 94 ± 5 |
| −13 | G | 106 ± 17 | 95 ± 24 | 100 | 144 ± 25 | 76 ± 1 | 67 ± 8 | 100 | 30 ± 3 |
| −14 | T | 90 ± 4 | 68 ± 8 | 77 ± 8 | 100 | 55 ± 2 | 32 ± 2 | 55 ± 3 | 100 |
| −15 | T | 146 ± 20 | 141 ± 15 | 129 ± 9 | 100 | 103 ± 29 | 90 ± 5 | 109 ± 5 | 100 |
| −16 | G | 144 ± 13 | 96 ± 15 | 100 | Not tested | 124 ± 13 | 102 ± 6 | 100 | Not tested |
| −17 | G | 115 ± 10 | 118 ± 9 | 100 | 106 ± 14 | 161 ± 6 | 100 ± 7 | 100 | 149 ± 3 |
| −18 | A | 100 | 117 ± 26 | 117 ± 13 | Not tested | 100 | 116 ± 5 | 112 ± 9 | Not tested |
| −19 | G | 137 ± 18 | Not tested | 100 | 93 ± 21 | 146 ± 14 | Not tested | 100 | 123 ± 9 |
| −20 | A | 100 | 101 ± 15 | 109 ± 9 | 90 ± 5 | 100 | 118 ± 16 | 108 ± 7 | 105 ± 8 |
| −21 | T | 97 ± 9 | 106 ± 14 | 103 ± 17 | 100 | 87 ± 6 | 104 ± 7 | 106 ± 8 | 100 |
| −22 | A | 100 | 52 ± 6 | 85 ± 11 | 83 ± 18 | 100 | 78 ± 11 | 121 ± 3 | 72 ± 0 |
| −23 | A | 100 | 27 ± 3 | 31 ± 3 | 130 ± 8 | 100 | 69 ± 6 | 73 ± 5 | 121 ± 5 |
| −24 | A | 100 | 22 ± 3 | 32 ± 6 | 70 ± 8 | 100 | 115 ± 11 | 155 ± 4 | 99 ± 1 |
| −25 | A | 100 | 51 ± 9 | 53 ± 10 | 126 ± 18 | 100 | 141 ± 13 | 81 ± 6 | 144 ± 6 |
| −26 | A | 100 | 65 ± 4 | 94 ± 7 | 141 ± 13 | 100 | 98 ± 8 | 113 ± 16 | 140 ± 10 |
| −27 | A | 100 | 101 ± 14 | 72 ± 6 | 79 ± 7 | 100 | 82 ± 2 | 49 ± 10 | 66 ± 8 |
| −28 | G | 153 ± 19 | 100 ± 4 | 100 | 120 ± 19 | 176 ± 5 | 149 ± 10 | 100 | 127 ± 6 |
| −29 | A | 100 | 108 ± 5 | 147 ± 19 | 103 ± 5 | 100 | 69 ± 5 | 67 ± 5 | 43 ± 6 |
| −30 | C | 119 ± 7 | 100 | 87 ± 6 | 137 ± 46 | 78 ± 3 | 100 | 167 ± 9 | 118 ± 7 |
| −31 | G | 92 ± 9 | 68 ± 5 | 100 | 116 ± 15 | 109 ± 14 | 48 ± 5 | 100 | 115 ± 2 |
| −32 | T | 85 ± 15 | 49 ± 3 | 52 ± 10 | 100 | 48 ± 20 | 32 ± 4 | 36 ± 7 | 100 |
| −33 | A | 100 | 126 ± 14 | 110 ± 12 | 148 ± 23 | 100 | 85 ± 2 | 79 ± 3 | 242 ± 18 |
| −34 | G | 79 ± 7 | 96 ± 20 | 100 | 106 ± 8 | 58 ± 5 | 58 ± 5 | 100 | 75 ± 9 |
| −35 | A | 100 | 130 ± 26 | 121 ± 20 | 85 ± 17 | 100 | 56 ± 6 | 68 ± 7 | 67 ± 5 |
| −36 | T | 117 ± 15 | 116 ± 12 | 123 ± 12 | 100 | 111 ± 4 | 86 ± 2 | 89 ± 1 | 100 |
| −37 | A | 100 | 103 ± 19 | 105 ± 16 | 175 ± 8 | 100 | 70 ± 6 | 105 ± 3 | 70 ± 6 |
| −38 | A | 100 | 66 ± 7 | 88 ± 18 | 109 ± 16 | 100 | 46 ± 7 | 50 ± 5 | 65 ± 5 |
| −39 | A | 100 | 89 ± 16 | 95 ± 15 | 126 ± 26 | 100 | 50 ± 8 | 53 ± 7 | 88 ± 6 |
| −40 | A | 100 | 99 ± 20 | 125 ± 27 | 109 ± 18 | 100 | 76 ± 4 | 104 ± 8 | 95 ± 4 |
| −41 | A | 100 | Not tested | 116 ± 6 | 79 ± 4 | 100 | Not tested | 83 ± 8 | 65 ± 4 |
The promoter activity of each mutant promoter was normalized to a wild-type rRNA P1 activity of 100%. Each value is the mean of three separate measurements.
FIG. 2.
Effects of single substitutions on C. trachomatis rRNA P1 transcription by E. coli or C. trachomatis RNAPs. All three possible substitutions were tested from −41 to −1, except −41, −19, −18 and −16, where only two were tested (Table 1). The wild-type rRNA P1 sequence is shown on each graph, and changes in promoter activity resulting from the substitutions, relative to the activity of the wild-type promoter, are indicated. Decreases larger than fivefold are not illustrated as extending beyond the bottom axis.
Substitutions in the −10 region affected promoter activity.
Substitutions in the −10 region had large effects on transcription by C. trachomatis RNAP and identified positions that were important for promoter activity. Some substitutions at −11, −10, −9, −8, −7, −5, and −4 had negative effects on promoter activity, while other substitutions at −10 and −6 had positive effects (Table 1 and Fig. 2). The greatest effects were seen at −8, where an A-to-G substitution caused a ninefold decrease in promoter activity, and at −9, where a T-to-A substitution decreased promoter activity fivefold. The positive effects of a G substitution at −10 and a T substitution at −6 suggested that these substitutions made the resultant promoter closer to the optimal promoter sequence. If we took the individual base(s) that produced the greatest promoter activity at each position, the sequence was A, G, or T at −11, G or T at −10, T at −9, A at −8, A or T at −7, A or T at −6, A or C at −5, and T at −4. This optimal sequence contains two potential matches to the ς70 −10 promoter element, TATAAT, from −11 to −6 and from −9 to −4.
Transcription of the same promoter templates by E. coli RNAP identified recognizable ς70 −10 promoter sequences and validated this method for identifying the locations of sequences important for promoter activity. Substitution at −14, −13, −11, −10, −9, −8, −7, −5, and −4 had negative effects, while a G-to-T substitution at −6 had a positive effect (Table 1 and Fig. 2). The greatest effect was seen at −8, where an A to C or G substitution caused an 18-fold decrease in promoter activity. The optimal E. coli −10 sequence was T at −14, G at −13, T at −11, A or G at −10, T at −9, A at −8, A or T at −7, T at −6, A or C at −5, and T at −4. Two overlapping ς70 −10 promoter elements are recognizable within this optimal sequence: a −10 hexamer that also contains an extended −10 motif, TGnTATAAT from −14 to −6, and another potential −10 hexamer extending from −9 to −4.
Single base substitutions in the extended −10 promoter motif at −14 and −13 did not affect C. trachomatis RNAP promoter activity in the context of rRNA P1 (Table 1 and Fig. 2). To confirm this finding, a 2-bp substitution at these positions (T-14C, G-13T) was tested. This substitution did not affect C. trachomatis RNAP promoter activity (data not shown).
Substitutions in the −35 region had smaller effects on C. trachomatis RNAP than substitutions in the −10 region.
We have previously shown that a 5-bp substitution from −31 to −27 decreased transcription by C. trachomatis RNAP (22). Single substitutions at −32 and −31 had negative effects, while substitutions −33, −29, and −28 had positive effects (Table 1 and Fig. 2). Substitutions at −30 had a slight positive effect. The largest effect was a T-to-C or G substitution at −32 which decreased promoter activity twofold. The optimal sequence was T at −33, T at −32, A, G or T at −31, G at −29, and A at −28, which resembles the ς70 −35 promoter element, TTGACA. The optimal −35 sequence for E. coli RNAP was T at −33, T at −32, A, G or T at −31, G at −30, A at −29, A at −28, and A at −27 (Table 1 and Fig. 2). Two overlapping potential −35 elements that resemble the E. coli consensus −35 hexamer are recognizable, from −33 to −28 and −32 to −27.
Substitutions between the −10 and −35 regions defined an AT-rich region that affected transcription by C. trachomatis RNAP but not by E. coli RNAP.
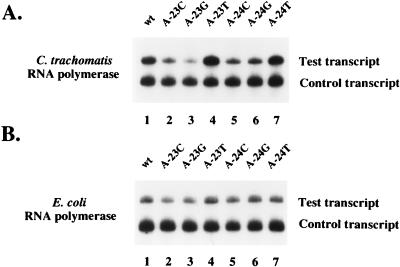
Our previous analysis suggested that sequences in the spacer region between −10 and −35 regions were important for transcription by C. trachomatis RNAP but not by E. coli RNAP (22). At positions −26 to −22, changing the wild-type sequence from AAAAA to TTTTT caused a 2-fold decrease in promoter activity, and altering it to CCCCC resulted in a 12-fold decrease in promoter activity (data not shown). Within this region, substitution of a single wild-type A residue with a C or G at −25, −24, and −23 or a C at −22 caused a negative effect on transcription (Table 1 and Fig. 2 and 3). Substitution of a C at −26 had a smaller negative effect. Substitution of a T residue at −26, −25, and 23 had a slight stimulatory effect. The greatest effect was seen at −24, where an A-to-C substitution caused a 4.5-fold decrease in promoter activity. In contrast, transcription by E. coli RNAP was slightly decreased by C or G substitutions at −23, while other substitutions in this region had no effect or a slight positive effect (Table 1 and Fig. 2 and 3). These results defined a region extending from −25 to −22 and possibly including −26, whose sequence was important for transcription by C. trachomatis RNAP. We have called this region the spacer A/T region because of its location in the spacer region between the −10 and −35 promoter elements and the sequence preference for A and T residues.
FIG. 3.
In vitro transcription of C. trachomatis rRNA P1 templates with C. trachomatis RNAP (A) or E. coli RNAP (B). The rRNA P1 templates contained the wild-type (wt) sequence (lane 1), a substitution of wild-type A at −23 with a C (lane 2), G (lane 3), or T (lane 4), or a substitution of wild-type A at −24 with a C (lane 5), G (lane 6), or T (lane 7). The shorter control transcript was transcribed by SP6 RNAP.
Substitutions upstream of the −35 region affected transcription.
We have previously shown that 5-bp substitutions from −41 to −37 and −36 to −32 decreased transcription by C. trachomatis and E. coli RNAPs (22). Some single base substitutions in this region upstream of the putative −35 hexamer affected transcription by C. trachomatis RNAP, although not as much as they affected transcription by E. coli RNAP (Table 1 and Fig. 2). Most of these deleterious substitutions were from an A to a C or G. As we will mention below, the native AT-rich sequence in this location functioned as an UP element with E. coli RNAP. Thus, it is likely that the substitutions decreased promoter activity by affecting contacts between the UP element and RNAP.
Effect of altered spacing between the −10, spacer A/T, and −35 regions.
We tested the effect on promoter activity of insertions or deletions downstream (at −17) or upstream (at −27) of the spacer A/T region to determine if the spacing between the −10, spacer A/T, and −35 regions was important (Fig. 4). A 1-bp insertion or deletion at either location had minimal effects on transcription by C. trachomatis RNAP (lines B, C, F, and G). A 2-bp insertion at −27 (line E) but not −17 (line D) had a negative effect on C. trachomatis and E. coli RNAPs. The only mutation which specifically decreased transcription by C. trachomatis RNAP but not E. coli RNAP was a 2-bp deletion at −17 (line H).
FIG. 4.
Spacing mutations of C. trachomatis rRNA P1 and their effects on transcription by C. trachomatis RNAP and E. coli RNAP. The names of the spacing mutations are abbreviated so that +1 @ −17 represents a 1-bp insertion at position −17. Inserted sequences are shown in uppercase letters, and the site of deletion is marked with a Δ. The sequences are aligned by the transcription initiation site. The G residue at +1 in the native sequence was replaced by an A residue to allow for transcription in the absence of GTP. Positions that were shown by the substitution analysis to be important for C. trachomatis RNAP promoter activity are underlined in the wild-type sequence and in their new locations in the spacing mutations. The spacer A/T region is underlined twice. The promoter activity of each spacing mutation is shown for transcription by C. trachomatis RNAP (Ct) and E. coli RNAP (Ec).
To determine whether moving the spacer A/T region alone could affect promoter activity, we made spacing mutations with a deletion at −17 together with a compensatory insertion at −27. These mutations had the net effect of moving the spacer A/T region without altering the location of the sequences in the −10 and −35 regions. Shifting the spacer A/T region 1 and 2 bp downstream decreased the promoter activity of C. trachomatis RNAP to 41 and 25% of wild-type activity, respectively, but did not affect E. coli RNAP transcription (lines J and K). These results suggested that in this promoter context, the spacer A/T region could not be moved downstream without adversely affecting transcription and provided additional evidence that the spacer A/T region affected only transcription by C. trachomatis RNAP.
Substitutions that allowed transcription initiation from a new site.
Three single base changes each produced an additional C. trachomatis RNAP transcript that was faster-migrating and less abundant (Fig. 5A). The 5′ end of each extra transcript was mapped by primer extension to a site 18 downstream of the native rRNA P1 transcription start site (data not shown). These results demonstrated that the extra transcript was the result of transcription initiation at a new site, rather than premature termination. Each of the substitutions appeared to make new promoters by creating sequences that could function as −35 hexamers (T-11C) or spacer A/T regions (G-6A and G-6T) in combination with a potential −10 hexamer (TTTAAA) from the G-less cassette (Fig. 5B). E. coli RNAP only produced an extra transcript with the substitution that created a perfect −35 hexamer (T-11C), providing further evidence that the effect of the spacer A/T region is specific to C. trachomatis RNAP. These results suggest that a −35 hexamer, with the same sequence as the E. coli consensus −35 promoter element (TTGACA), and the spacer A/T region can serve as important elements for recognition by C. trachomatis RNAP.
FIG. 5.
(A) In vitro transcription of wild-type rRNA P1 and substitutions that produced an extra transcript. Lanes 1 and 5, wild-type (wt) rRNA P1; lanes 2 and 6, G-to-A substitution at −6 (G-6A); lanes 3 and 7, G-to-T substitution at −6 (G-6T); lanes 4 and 8, T-to-C substitution at −11 (T-11C). The test transcript was transcribed by C. trachomatis (Ct) RNAP (lanes 1 to 4) or E. coli (Ec) RNAP (lanes 5 to 8). The extra transcript in lanes 2, 3, 4, and 8 (marked by an asterisk) was shown to initiate 18 nucleotides downstream by primer extension (data not shown). The control transcript was transcribed by SP6 RNAP. (B) Sequences upstream of the initiation sites for the rRNA P1 transcripts shown in panel A. The sequences were aligned by the transcription initiation sites. The wild-type rRNA P1 sequence is shown in the top line, and the positions where C. trachomatis RNAP transcription was affected by substitutions are underlined. The spacer A/T region is underlined twice, and the location of a 12-bp direct repeat sequence is shown by a pair of arrows. The substituted base is shaded in each of the three mutant promoter templates. Putative −35 and −10 hexamers are underlined for T-11C, which was transcribed by both E. coli and C. trachomatis RNAPs. Proposed spacer A/T regions in G-6A and G-6T are underlined twice.
Transcription of the C. trachomatis rRNA P1 promoter was affected by deletion of the α-CTD of E. coli RNAP.
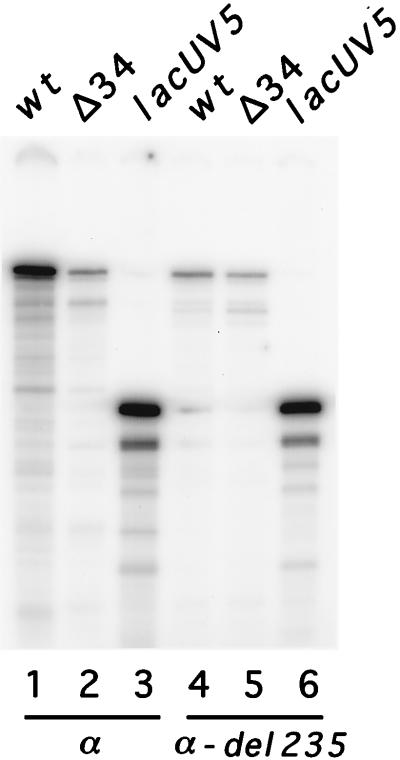
Our previous results demonstrated that the removal of AT-rich sequences upstream of the putative −35 region significantly decreased transcription by both C. trachomatis and E. coli RNAP (22). Enhancement of transcription by an AT-rich region upstream of an rRNA gene is suggestive of an UP element (17). To test whether E. coli RNAP could recognize this C. trachomatis sequence as an UP element, we compared transcription by wild-type E. coli RNAP and mutant E. coli RNAP that was unable to recognize the UP element because of deletion of the α-CTD. With wild type E. coli RNAP, transcription of a promoter template containing the proposed UP element (−302 to +5 of C. trachomatis rRNA P1) was 11-fold higher than transcription of a promoter template lacking the UP element (−34 to +5) (Fig. 6, lanes 1 and 2). The stimulation of transcription was lost when mutant E. coli RNAP lacking the α-CTD was used (lanes 4 and 5). These results show that C. trachomatis rRNA P1 contains an UP element that is recognized by E. coli RNAP. Since the residues in α responsible for UP element recognition are conserved between E. coli and C. trachomatis (10, 11), these data suggest that C. trachomatis RNAP also utilizes the rRNA P1 UP element.
FIG. 6.
In vitro transcription using wild-type E. coli RNAP (α) or E. coli RNAP with a deletion of the α-CTD (α-del 235). C. trachomatis rRNA P1 templates contained an UP element (wild type [wt]; −302 to +5) or lacked an UP element (Δ34; −34 to +5). For the Δ34 template, the sequence that replaced the UP element was derived from the plasmid (sequence in Δ34 from −64 to −35: GTCGCATGCTCCTCTAGACTCGAGGAATTC. The E. coli lacUV5 promoter, which lacks an UP element (17), was used to normalize the activities of the two RNAP preparations.
DISCUSSION
We have identified four regions of C. trachomatis rRNA P1 that contributed to the promoter activity of C. trachomatis RNAP: a −10 region, a −35 region, a novel spacer A/T region just downstream of the −35 region, and an UP element. Three of these regions are analogous to the corresponding regions in E. coli promoters, but the spacer A/T region was recognized only by C. trachomatis RNAP.
Context effects limit the conclusions that can be made from the single base substitutions. This is apparent in the analyses of the effect of substitutions in the −35 region. Apparently, C. trachomatis RNAP does not require a perfect −35 hexamer in the context of the wild-type rRNA P1 promoter sequence. However, in the T-11C mutant (Fig. 5B), a −35 hexamer identical to the E. coli consensus directs C. trachomatis RNAP to recognize a new promoter even though there is an imperfect spacer A/T region. On the other hand, creation of a good spacer A/T region is sufficient to allow for transcription when the −35 sequence is imperfect (G-6A and G-6T [Fig. 5B]). It is tempting to speculate that the −35 region and the spacer A/T region work cooperatively to anchor C. trachomatis RNAP to the promoter.
The promoter sequences identified for C. trachomatis RNAP resemble the E. coli ς70 promoter structure. This is consistent with the conservation of amino acid sequence between ςA and ς70 in subregions 2.4 and 4.2 (3, 12). Most substitutions in the −10 and −35 regions of rRNA P1 that affected transcription by C. trachomatis RNAP also affected transcription by E. coli RNAP, although there were subtle differences in the recognition properties of the two enzymes. For example, C. trachomatis RNAP did not recognize the extended −10 motif in this promoter context. However, subregion 2.5 of ς70, which is the region that recognizes this motif, is well conserved between ςA and ς70 (1). The particular amino acid (E458) proposed to recognize the G residue of the TG motif is also conserved.
Our most striking result is the delineation of the spacer A/T region, which was important for transcription by C. trachomatis RNAP but not E. coli RNAP. Substitutions in this region demonstrated an AT sequence preference, and the spacing mutations showed that the location of the spacer A/T region was also important.
The role of the spacer A/T region may be explained in several ways. If the spacer A/T region can function as an additional promoter element, it may be required for specific contacts with C. trachomatis RNAP. In fact, the spacer A/T region is immediately downstream of the predicted location of the −35 promoter element, and the two regions might form an extended −35 promoter element. Alternatively, the spacer A/T region could serve as a binding site for a putative transcription factor present in the C. trachomatis RNAP preparation. A third possibility is that the preference for A or T residues in this region may reflect the energetic contribution of these sequences to promoter activity, such as with localized DNA strand melting. Finally, the spacer A/T region is part of six consecutive A residues, which could produce a nonstandard DNA structure that may affect promoter recognition by C. trachomatis RNAP.
We propose that an AT-rich region from −52 to −34 functions as an UP element to increase transcription from C. trachomatis rRNA P1. We demonstrated a fourfold increase in transcription with C. trachomatis RNAP (reference 22 and data not shown) and an 11-fold enhancement with E. coli RNAP (reference 22 and Fig. 6). The effect of the UP element on E. coli RNAP transcription was dependent on the α-CTD, strongly suggesting that the E. coli RNAP α subunit can recognize this heterologous UP element. We note that it is not yet technically possible to confirm that the C. trachomatis RNAP α-CTD makes a contact with the UP element, as this experiment will require the construction of mutant C. trachomatis RNAP lacking the α-CTD.
Interestingly, the UP element included a 12-bp sequence from −44 to −33 which was directly repeated from −29 to −18 (Fig. 5B). The downstream repeat encompasses the spacer A/T region. As the α subunit of RNAP recognizes the sequence of the UP element, it is intriguing to speculate if the α subunit can also recognize the spacer A/T region.
Our results are consistent with the analysis of C. trachomatis MOMP P2 by Douglas and Hatch (2). They reported that the first two positions of the putative −10 hexamer were important for promoter activity, and in our analysis, the first two positions of the potential −10 hexamer from −9 to −4 also showed the greatest effect with substitution; 2-bp substitutions in the AT-rich sequences upstream and downstream of the predicted MOMP P2 −35 promoter element greatly abrogated promoter activity. We propose that the AT-rich sequence upstream of the −35 promoter element of MOMP P2 is an UP element, which would explain the decreased promoter activity with deletion of sequences upstream of −39 or with a 2-bp substitution at −41 and −40. We also suggest that the AT-rich region downstream of the −35 promoter element is the equivalent of a spacer A/T region. Loss of promoter activity with a 2-bp substitution at −29 and −28 (equivalent to −26 and −25 of rRNA P1 [Fig. 7]) can be explained by alteration of the spacer A/T region from AAAA to GAAA.
FIG. 7.
Alignment of selected chlamydial promoters. Upstream sequences of chlamydial genes with ς70-like −10 hexamers were aligned by these putative −10 elements. Predicted −35 and −10 promoter sequences are underlined. Potential spacer A/T regions are underlined twice. In vivo transcription initiation sites are marked with a dot above the sequence. Upstream sequences of the following genes from C. trachomatis serovars L1, L2, and MoPn (mouse pneumonitis) and C. psittaci 6BC are shown (GenBank accession numbers in parentheses): L1 60-kDa CRP P1 and 15-kDa SRP (M35148), L2 MOMP P2 (M14738), MoPn S1 (M23000), L2 PCT (plasmid countertranscript) and prom7 (X07547), 6BC EUO (L13598), L2 hctA (M60902), L2 ltuA (L40822), L2 ltuB (L40838), MoPn groE (L12004), MoPn dnaK (M62819), and L2 hctB (L10193).
Analysis of other chlamydial promoters shows that many of them contain potential ς70 −10 and −35 promoter sequences (Fig. 7). A potential spacer A/T region can be identified immediately downstream of the putative −35 element in many of these promoters. The potential spacer A/T region is often part of a larger AT-rich sequence (6-8 bp). A notable exception is the dnaK promoter, which lacks an identifiable spacer A/T region but has a 5-of-6-bp match to the E. coli consensus −35 hexamer and a perfect E. coli −10 hexamer (23). This ς70-like promoter structure may be sufficient for transcription by C. trachomatis RNAP. However, for other C. trachomatis promoters, including rRNA P1, the spacer A/T region may compensate or substitute for a suboptimal −35 element.
At this time, we cannot distinguish between the likely roles of the spacer A/T region as a promoter element that interacts differently with C. trachomatis and E. coli RNAPs or as a binding site for an activator of transcription that is present in the C. trachomatis RNAP preparation. Further characterization of the spacer A/T region will require purification of C. trachomatis RNAP to homogeneity.
ACKNOWLEDGMENTS
We thank members of the Engel lab and Carol Gross for support and suggestions.
This work was supported by grants from the NIH (AI 01247 to M.T., GM 37048 to R.L.G., and AI 24436 and AI 01348 to J.N.E.).
REFERENCES
- 1.Barne K A, Bown J A, Busby S J W, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase ς70 subunit is responsible for the recognition of the ’extended −10‘ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas A L, Hatch T P. Mutagenesis of the P2 promoter of the major outer membrane protein gene of Chlamydia trachomatis. J Bacteriol. 1996;178:5573–5578. doi: 10.1128/jb.178.19.5573-5578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel J, Ganem D. A PCR-based approach to cloning sigma factors from eubacteria and its application to the isolation of a sigma70 homolog from Chlamydia trachomatis. J Bacteriol. 1990;172:2447–2455. doi: 10.1128/jb.172.5.2447-2455.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel J, Pollack J, Malik F, Ganem D. Cloning and characterization of RNA polymerase core subunits of Chlamydia trachomatis using the polymerase chain reaction. J Bacteriol. 1990;172:5732–5741. doi: 10.1128/jb.172.10.5732-5741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engel J N, Ganem D. Chlamydial rRNA operons: gene organization and identification of putative tandem promoters. J Bacteriol. 1987;169:5678–5685. doi: 10.1128/jb.169.12.5678-5685.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett K D E, Hatch T P. Sequence analysis and lipid modification of the cysteine-rich envelope proteins of Chlamydia psittaci 6BC. J Bacteriol. 1991;173:3821–3830. doi: 10.1128/jb.173.12.3821-3830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahr M J, Douglas A L, Xia W, Hatch T P. Characterization of late gene promoters of Chlamydia trachomatis. J Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahr M J, Sriprakash K S, Hatch T P. Convergent and overlapping transcripts of the Chlamydia trachomatis 7.5-kb plasmid. Plasmid. 1992;28:247–257. doi: 10.1016/0147-619x(92)90056-g. [DOI] [PubMed] [Google Scholar]
- 9.Gaal T, Bartlett M, Ross W, Turnbough C J, Gourse R. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 10.Gaal T, Ross W, Blatter E, Tang H, Jia X, Krishman V, Assa-munt N, Ebright R, Gourse R. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 11.Gu L, Wenman W M, Remacha M, Meuser R, Coffin J, Kaul R. Chlamydia trachomatis RNA polymerase α subunit: sequence and structural analysis. J Bacteriol. 1995;177:2594–2601. doi: 10.1128/jb.177.9.2594-2601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehler J E, Burgess R R, Thompson N E, Stephens R S. Chlamydia trachomatis RNA polymerase major ς subunit. J Biol Chem. 1990;265:13206–13214. [PubMed] [Google Scholar]
- 13.Lambden P R, Everson J S, Ward M E, Clarke I N. Sulfur-rich proteins of Chlamydia trachomatis: developmentally regulated transcription of polycistronic mRNA from tandem promoters. Gene. 1990;87:105–112. doi: 10.1016/0378-1119(90)90500-q. [DOI] [PubMed] [Google Scholar]
- 14.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathews S A, Sriprakash K S. The RNA polymerase of Chlamydia trachomatis has a flexible sequence requirement at the −10 and −35 boxes of its promoters. J Bacteriol. 1994;176:3785–3789. doi: 10.1128/jb.176.12.3785-3789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulder J. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schachter J. The intracellular life of Chlamydia. Curr Top Microbiol Immunol. 1988;138:109–139. [PubMed] [Google Scholar]
- 21.Stephens R S, Kalman S, Fenner C, Davis R. Chlamydia Genome Project University of California at Berkeley and Stanford University. 1997. chlamydia-www.berkeley.edu:4231 http;// chlamydia-www.berkeley.edu:4231. . [Google Scholar]
- 22.Tan M, Engel J N. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J Bacteriol. 1996;178:6975–6982. doi: 10.1128/jb.178.23.6975-6982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan M, Wong B, Engel J N. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J Bacteriol. 1996;178:6983–6990. doi: 10.1128/jb.178.23.6983-6990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]