Abstract
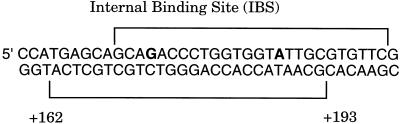
The catBCA operon of Pseudomonas putida encodes enzymes involved in the catabolism of benzoate. Transcription of this operon requires the LysR-type transcriptional regulator CatR and an inducer molecule, cis,cis-muconate. Previous gel shift assays and DNase I footprinting have demonstrated that CatR occupies two adjacent sites proximal to the catBCA promoter in the presence of the inducer. We report the presence of an additional binding site for CatR downstream of the catBCA promoter within the catB structural gene. This site, called the internal binding site (IBS), extends from +162 to +193 with respect to the catB transcriptional start site and lies within the catB open reading frame. Gel shift analysis and DNase I footprinting determined that CatR binds to this site with low affinity. CatR binds cooperatively with higher affinity to the IBS in the presence of the two upstream binding sites. Parallel in vivo and in vitro studies were conducted to determine the role of the internal binding site. We measured β-galactosidase activity of catB-lacZ transcriptional fusions in vivo. Our results suggest a probable cis-acting repressor function for the internal binding site. Site-directed mutagenesis of the IBS verified this finding. The location of the IBS within the catB structural gene, the cooperativity observed in footprinting studies, and phasing studies suggest that the IBS likely participates in the interaction of CatR with the upstream binding sites by looping out the intervening DNA.
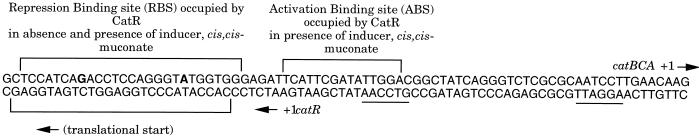
Pseudomonas putida metabolizes benzoate by way of the β-ketoadipate pathway to the tricarboxylic acid cycle intermediates succinate and acetyl coenzyme A. Growth of P. putida in the presence of benzoate leads to an induction of the catBCA operon, which encodes three enzymes of the β-ketoadipate pathway: cis,cis-muconate-lactonizing enzyme I (catB), muconolactone isomerase (catC), and catechol dioxygenase I (catA) (13). This induction requires the transcriptional regulator CatR and the pathway intermediate cis,cis-muconate (CCM) as the inducer molecule. The catR gene is divergently transcribed from the catBCA operon, and the two promoters overlap. CatR is a 32-kDa protein that negatively regulates its own expression and belongs to the LysR family of transcriptional regulators (11, 25). The DNA binding sites of LysR proteins almost invariably contain the sequence T-N11-A within an inverted repeat (9). In the absence of inducer, CatR binds to the repression binding site (RBS), which contains a G-N11-A motif within an imperfect, interrupted, inverted repeat thought to be important for specific binding by CatR (24). Binding to this site presumably allows CatR to negatively regulate its own expression. In the presence of CCM, CatR occupies an additional, adjacent downstream site designated the activation binding site (ABS) (4, 19, 22). This site is adjacent to the −35 region of the catBCA promoter. The sequences and organization of the CatR binding sites are depicted in Fig. 1.
FIG. 1.
Sequences and organization of the RBS, ABS, and IBS. The −35 and −10 consensus promoter sequences are underlined. The G and A residues of the G-N11-A motif are in boldface.
Although most LysR family members have been shown to bind only to the promoter regions of the genes that they regulate, exceptions have been reported (5). The metF gene is involved in methionine biosynthesis in Salmonella typhimurium, and its expression is positively regulated by the LysR family member MetR. MetR binding sites within the promoter region as well as the downstream binding site centered at +77 within the metF gene were shown to be required for expression (5).
Subsequently, binding sites within genes located downstream of regulated promoters were identified in the CatR-regulated pheBA operon and the ClcR-regulated clcABD operon in P. putida (18a). The presence of binding sites downstream of the regulated promoters in those systems suggests that these binding sites may be conserved regulatory elements. In this study, we report the presence of a third, low-affinity, CatR binding site designated the internal binding site (IBS) within the catB structural gene and investigate its role in the regulation of the catBCA operon. We found that the IBS negatively regulates the expression of the catBCA promoter and that this repression is CatR mediated. Our studies also suggest that occupancy of the IBS by CatR is either dependent on or facilitated by a DNA loop that entails a CatR-bound RBS plus an ABS (collectively called RBS/ABS) and the IBS as its elements.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
The Escherichia coli strains used for general cloning procedures were JM109 (30) and TG1 (Amersham, Arlington Heights, Ill.) (8). Protocols for plasmid isolation, plasmid construction, and transformation were as described previously (17). For plasmid selection, ampicillin was used at a final concentration of 75 μg/ml for E. coli and carbenicillin was used at 1,000 μg/ml for P. putida. Plasmids were introduced into P. putida by triparental mating using pRK2013 (7) as a helper plasmid. The promoter probe vectors used to monitor the in vivo expression of catB were derivatives of pKRZ-1 (24). Promoter probe studies were performed with P. putida cells grown in basal salts medium (2) as described previously (20). E. coli and P. putida were grown at 37 and 30°C, respectively.
Generation of mutant promoter probe constructs.
Subcloning of DNA fragments was done by standard procedures (17). DNA prepared for sequencing was purified by using a Qiagen Plasmid Midi kit (Qiagen Inc., Chatsworth, Calif.). Nucleotide sequencing of both DNA strands was done by using [α-35S]dATP, 7-deaza-GTP in place of dGTP, and a Sequenase kit (version 2.0; United States Biochemicals, Cleveland, Ohio) according to the manufacturer’s instructions. The oligonucleotide primers used in this study (Table 1) were purchased from Gibco-BRL, Gaithersburg, Md. Site-directed mutagenesis, including generation of +6 and +11 insertions, was performed by the overlap extension method and PCR (12). The primers consisted of complementary 25- or 54-base sequences, with the altered or inserted nucleotide(s) located in the center of the primer. The constructs were checked by DNA sequence analysis to verify the introduced mutation(s) and preclude erroneous PCR-generated mutations. Primer pairs CH1Sal-CH2 and CH1Sal-CH3 (Table 1) were used to generate 307- and 347-bp PCR products, respectively. These products were cloned into the SalI and BamHI sites of pKRZ-1 to generate constructs pCH2Z1 and pCH3Z1, respectively. Construct pCH3+6 was generated by insertion of 6 bp at a nonessential site between the catB transcriptional start site and the IBS. An additional insertion of 5 bp in the pCH3+6 construct generated construct pCH3+11, which had a total insertion of 11 bp compared to the wild type. The primer pairs used to generate the spacing mutations were +6US-+6LS and +11US-+11LS (Table 1).
TABLE 1.
Oligonucleotide primers used in this studya
| Primer | Sequence (5′-3′) |
|---|---|
| CH1 | CAGCAGCTCGGCGGCGCGAGT |
| CH1Sal | GGGTCGACCAGCAGCTCGGCGGCGCGAGT |
| CH2 | GGGTCGACGCATGGTGTGCATCGCCAGC |
| CH3 | GGGTCGACTCACTGCAGCGAACACG |
| 171GT5′a | CCATGAGCAGCATACCCTGGTGGTA |
| 171GT3′b | TACCACCAGGGTATGCTGCTCATGG |
| 173CA5′a | ATGAGCAGCAGAACCTGGTGGTATT |
| 173CA3′b | AATACCACCAGGTTCTGCTGCTCAT |
| 184TG5′a | ACCCTGGTGGTAGTGCGTGTTCGCT |
| 184TG3′b | AGCGAACACGCACTACCACCAGGGT |
| +6USa | CCGCCGCACAAGCTGGCGATGCACACAGCGACCATGAGCAGCAGACCCTGGTGG |
| +6LSb | CCACCAGGGTCTGCTGCTCATGGTCGCTGTGTGCATCGCCAGCTTGTGCGGCGG |
| +11USa | GCTGATTGAACGTATCGAGGCAATTACGAGATTGTGCATGACCTGCCGACCATTCGT |
| +11LSb | ACGAATGGTCGGCAGGTCATGCACAATCTCGTAATTGCCTCGATACGTTCAATCAGC |
| BCBC1 | CTGATTGAACGTATCGA |
| NBCBC2 | GCTTTCGATACCCGACTTGGCAAAG |
Primers with superscripts “a” and “b” are reverse complements and correspond to the published P. putida catB sequence (2) except for certain nucleotides that were changed to reflect the corrected sequence of catB. The nucleotide(s) which was altered or inserted during mutagenesis is in boldface.
Gel shift assay and determination of CatR binding constants.
Gel shift assays and CatR binding affinity studies were performed as previously described (20) except as noted in the text. For gel shift assays, a uniformly labeled 321-bp fragment containing only the IBS (no promoter DNA) was generated by PCR (primers BCBC1 and NBCBC2). Different concentrations of purified CatR were incubated with a fixed, low DNA concentration. The reactions were run in a gel shift assay, and the KD (equilibrium dissociation constant) was calculated as the total CatR concentration that allows half-maximal DNA binding. For uniform incorporation of the label, the PCR was modified by lowering the dATP concentration to 20 μM and adding 5 μl of [α-32P]dATP (10 μCi/μl).
DNase I footprinting.
DNase I footprinting reactions were performed as previously described (21). The 321-bp DNA fragment for the DNase I footprinting reaction was generated by PCR (primers BCBC1 and NBCBC2). Cooperative binding of CatR to the IBS was examined by using a 340-bp PCR-generated fragment (primers CH1 and CH3). This fragment harbors the RBS, ABS, and IBS. The primers were end labeled with 32P as previously described (21).
β-Galactosidase assays.
The wild-type P. putida strain, PRS2000 (29), harboring the promoter probe constructs was grown in basal salts medium supplemented with either 10 mM glucose or 5 mM benzoate for 16 h at 30°C. The β-galactosidase assays were performed in triplicate, using the cell extracts as detailed previously (24), and the specific activity was determined by the method of Miller (18). The protein concentration was assayed by the method of Bradford (3), with bovine serum albumin as a standard. The activity of each construct was expressed as Miller units (nanomoles of product per minute per milligram of protein).
RESULTS
Role of the IBS in expression of the catBCA operon in vivo.
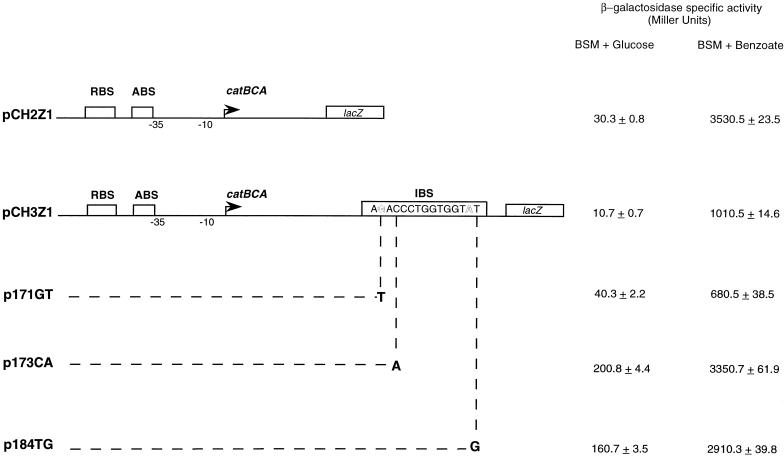
Visual inspection of the catB gene revealed a sequence that closely matched the consensus sequence for CatR binding and was designated the IBS (Fig. 2). The role of the IBS in CatR-mediated expression of the catBCA operon was examined by using transcriptional fusions to the lacZ gene (Fig. 3). CatR was supplied by the chromosomal copy of the catR gene. The results of this experiment are displayed in Fig. 3. In the presence of benzoate, the construct lacking the IBS (pCH2Z1) consistently showed about a 3.5-fold-higher level of expression than the fusion containing the IBS (pCH3Z1). The results suggest a cis-acting repressing activity for the IBS. The expression of both these constructs in the absence of benzoate was at a low basal level.
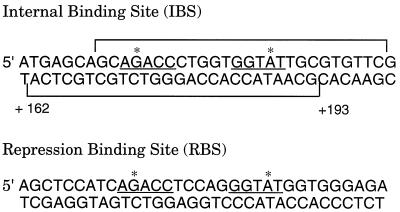
FIG. 2.
Comparison of sequences of the IBS and the RBS. The IBS sequences protected by CatR from DNase I digestion are bracketed. The interrupted inverted repeat involved in sequence specific recognition by CatR is underlined. The G and A nucleotides of the binding motif are indicated by asterisks.
FIG. 3.
Structures and in vivo activities of the catBCA constructs used in this study. Each arrow indicates the transcriptional start site (+1) of catB. All constructs harbor the RBS, the ABS, and the −35 and −10 consensus promoter sequences. Plasmid pCH2Z1 includes a part of the catB structural gene, up to the IBS, cloned upstream of the lacZ gene in pKRZ1; however, this construct lacks the IBS. Plasmid pCH3Z1 was constructed similarly; however, it extends further into the catB gene to include the IBS. Plasmids p171GT, p173CA, and p184TG are similar to pCH3Z1 except for the introduced point mutations as indicated. The β-galactosidase specific activities of each of the lacZ fusions in PRS2000 are displayed at the right.
Previous site-directed mutagenesis studies of the consensus binding sequence in the RBS had identified several nucleotides as critical for DNA binding of CatR (20). To verify that the observed decrease in activity was due to CatR-bound IBS, critical nucleotides in the IBS were altered by site-directed mutagenesis to generate the 171GT, 173CA, and 184TG mutations. In the presence of benzoate, the 171GT mutation reduced catB expression to about half of the wild-type level (compare pCH3Z1 and 171GT). There was a slight increase in the expression of the mutant in the absence of benzoate. The 173CA and 184TG mutants both demonstrated higher levels of expression of catB than the wild type in the presence of benzoate.
To determine if the differential expression of these constructs was observed throughout the growth phase, we monitored β-galactosidase activity of cells harvested at 2-h intervals from 6 to 14 h and 24 h of growth in the presence of benzoate. While the expression levels for the transcriptional fusion lacking the IBS showed a steady increase, the levels for the transcriptional fusion with the IBS increased only slightly over the course of the experiment, with the activity levels being consistently lower than those for the construct lacking the IBS (data not shown).
Gel shift assay and determination of the equilibrium dissociation constant.
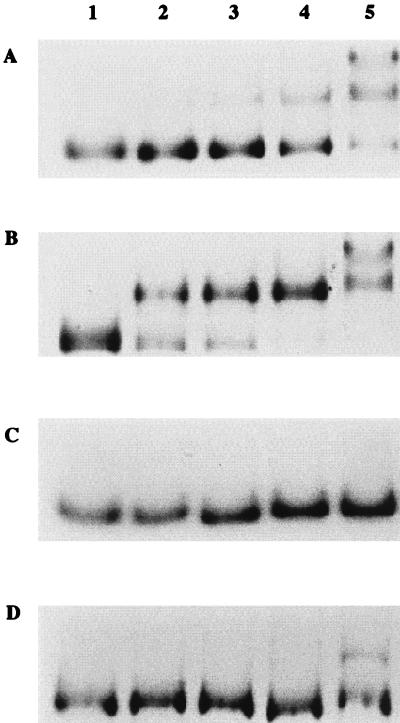
Gel shift assays were conducted to demonstrate the binding of CatR to the IBS and to determine the equilibrium dissociation constant for CatR binding to the wild-type and mutant IBS fragments (Fig. 4). These 321-bp fragments contained the IBS only. The estimated KD values were 4.9 × 10−7 M for the wild-type IBS and 1.5 × 10−7, 1.3 × 10−6, and 7.7 × 10−7 M for the three mutant IBS fragments harboring mutations 171GT, 173CA, and 184TG, respectively. These residues, as shown previously in a mutagenesis study of the RBS, are important for CatR binding (20). The 171GT alteration changes the binding site to make it look more like the consensus T-N11-A motif and increases its binding affinity for CatR. On the other hand, the 173CA and the 184TG mutations that cause a deviation of the binding site from the consensus motif result in a decrease in the binding affinity for CatR. CatR binds to the IBS with less affinity than that with which it binds to the catBCA promoter region (20). The KD of CatR for the IBS was estimated to be 4.9 × 10−7 M. Comparison of the KD values for the wild-type and mutant IBS fragments indicates that the 171GT mutant, which has a replacement of the wild-type G with the conserved T of the T-N11-A motif, has an approximately 3.5-fold-higher affinity for CatR binding than the wild type. As expected, the 173CA and the 184TG mutants showed 2.6- and 1.5-fold decreases, respectively, in binding affinity for CatR.
FIG. 4.
CatR binding to the wild-type and mutant IBS. (A) Gel shift assay demonstrating binding of purified CatR in the absence of inducer to a 321-bp DNA fragment containing the wild-type IBS. Lane 1 contains free, unbound DNA. Lanes 2 to 5 contain reaction mixtures with the following concentrations of purified CatR: 1.3 × 10−7, 1.7 × 10−7, 3.4 × 10−7, and 1.3 × 10−6 M. Lanes 1 to 5 in panels B to D correspond exactly to lanes 1 to 5 in panel A except that they represent assays performed with the IBS mutant probes 171GT, 173CA, and 184TG, respectively.
DNase I footprinting and cooperative binding of CatR to the IBS.
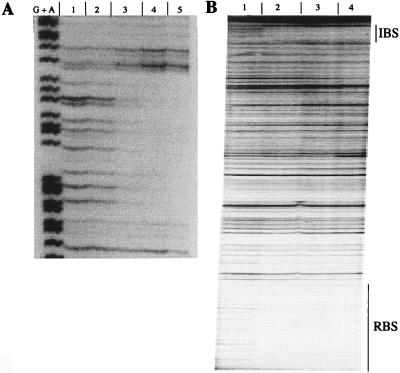
The precise location of the IBS was determined by DNase I footprinting. As shown in Fig. 5A, CatR protected a 32-bp sequence that extended from +162 to +193 relative to the catB transcriptional start site. The protection pattern was not altered in the presence of 100 μM CCM (data not shown). The IBS retains most of the imperfect, inverted repeat thought to be important for the recognition of its binding site by CatR (underlined in Fig. 2). In light of this observation, the low binding affinity of CatR for the IBS is interesting. The location of the IBS relative to the catBCA promoter region raised the possibility that the occupation of the IBS involves cooperative interaction with CatR bound at the RBS/ABS. The fact that the IBS affects in vivo expression of the catBCA operon may suggest that CatR bound to the IBS interacts with CatR bound at the promoter region. To test this hypothesis, we used a 340-bp end-labeled DNA fragment harboring both the RBS and ABS promoter sites and the IBS and subjected it to DNase I footprinting (Fig. 5B). Comparison of Fig. 5A and B shows that the IBS was occupied at over 10-fold-lower concentrations of CatR when present on a DNA fragment that also harbored the RBS and the ABS, indicating cooperative binding of CatR to the IBS. A cooperative interaction between remote binding elements suggests that CatR bound to the RBS/ABS and IBS may loop out intervening DNA.
FIG. 5.
DNase I footprinting of CatR binding to the IBS. (A) A 321-bp end-labeled DNA fragment containing the IBS but not the RBS or ABS was incubated with different amounts of CatR and subjected to digestion with DNase I. Lane G+A shows a Maxam-Gilbert reaction. Lanes 1 to 5 contain the following concentrations of CatR: 0 (free DNA), 8.5 × 10−8, 5.1 × 10−7, 1.3 × 10−6, and 5.1 × 10−6 M. The inducer did not alter the protection profile. (B) Lanes 1 to 4 contain a 340-bp end-labeled DNA fragment harboring the RBS/ABS and the IBS digested with the following concentrations of CatR present in the footprinting reactions: 0 (free DNA), 8.5 × 10−8, 3.4 × 10−7, and 8.5 × 10−7 M. The protected regions corresponding to the RBS and the IBS are indicated on the right.
Phasing dependence of IBS function.
To assess the role of cooperative binding in the repression effect of the catBCA operon by the IBS, we introduced 6- and 11-bp spacing alterations at nonessential sites between the ABS and the IBS. The results of this assay are depicted in Table 2. Assuming that the helix has approximately 10.5 bp per turn, addition of a half-integral turn (6 bp) impaired IBS-mediated repression, as seen by the approximately fourfold increase in the expression levels for this mutant compared to those for the wild-type IBS-containing construct. The expression levels for the 6-bp insertion mutants derived from the wild-type and the 173CA and 184TG mutant IBS-harboring constructs closely resembled levels for the IBS-deficient (wild-type) construct. It was interesting that the 6-bp insertion mutant derived from the 171GT construct showed a greater than twofold increase in expression levels, albeit the levels were lower than those for the IBS-deficient construct. Given the approximate 3.5-fold increase in the binding affinity of CatR for the 171GT mutant, it can be hypothesized that the contribution of cooperativity to the occupation of this mutant IBS, although still important, is not as consequential. Addition of approximately one helical turn in the 11-bp spacing mutations resulted in restoration of the IBS-mediated repression (Table 2). The results of the 6- and 11-bp spacing mutations strengthen the argument that cooperativity between CatR bound to the promoter and the IBS, and maintenance of the angular orientation of the IBS with respect to the catBCA promoter region, are important for this regulation.
TABLE 2.
β-Galactosidase levels in PRS2000 cells harboring the constructs with spacing alterationsa
| Construct | β-Galactosidase sp act (Miller units)
|
||
|---|---|---|---|
| Original construct | Corresponding +6 insertion mutant | Corresponding +11 insertion mutant | |
| pCH2Z1 | 3,500.9 ± 59.8 | ||
| pCH3Z1 | 1,000.8 ± 10.8 | 4,200.9 ± 53.0 | 980.4 ± 9.2 |
| p171GT | 653.5 ± 10.1 | 2,010.7 ± 14.8 | 519.5 ± 7.1 |
| p173CA | 3,330.6 ± 26.2 | 3,600.7 ± 20.9 | 3,100.7 ± 12.8 |
| p184TG | 2,890.3 ± 36.2 | 3,752.0 ± 27.3 | 3,010.8 ± 26.1 |
PRS2000 cells harboring constructs pCH3+6 and pCH3+11, with insertions of +6 and +11 bp, respectively, compared to the wild type were assayed for β-galactosidase activity after 16 h of growth in basal salts medium supplemented with 5 mM benzoate.
DISCUSSION
In this study, we investigated the role of the IBS in regulation of the catBCA operon in P. putida. The IBS reduced expression of the catBCA operon approximately three- to fourfold. Similarly located, low-affinity binding sites have also been observed in the metF gene in S. typhimurium (5) and the clcA gene and open reading frame of the clcABD and pheBA operons, respectively, in P. putida (18a). Considering the low affinity of CatR binding, the LysR consensus binding motif is surprisingly well conserved in the IBS and closely resembles that of the high-affinity RBS. This finding indicates that the consensus binding motif is not the sole important feature for high-affinity binding of LysR-type activator proteins.
Given the overlapping promoters of the catBCA operon and the catR gene, which are divergently transcribed (Fig. 1), we were interested in determining whether the IBS regulates expression of the catR gene as well. However, experiments done with catR-lacZ transcriptional fusions toward this end showed levels of activity that were too low to measure and therefore of questionable significance. Furthermore, while an IBS is present in the phe system, the lack of a divergently transcribed catR gene (14) suggests that it is unlikely that the IBS plays a role in catR regulation.
The nucleotides targeted for site-directed mutagenesis in this study were chosen on the basis of a previous study of the RBS (20). The 171GT mutation generated a consensus LysR T-N11-A binding motif (9) and a perfect inverted repeat. Therefore, it was predicted to improve the IBS by increasing its binding affinity for CatR. The 173CA and 184TG mutations both interrupt the G-N11-A motif and inverted repeat (AGACC-N7-GGTAT) and were therefore predicted to have a contrary effect resulting in a decreased binding affinity for CatR. The expected effects on CatR binding affinity were confirmed by the estimation of equilibrium dissociation constants for the wild type and the IBS mutants. These alterations in the binding affinity of CatR for the IBS were biologically relevant since in vivo studies showed that an increase in the binding affinity of CatR resulted in a concomitant increase in repression of the catBCA operon; conversely, a lowering of the binding affinity of CatR for the IBS relieved repression of the catBCA operon considerably. One exception to this observation is that slightly elevated levels of expression were seen under noninducing conditions for the 171GT reporter (Fig. 3). One would predict slightly lower expression levels for this particular mutant. This finding may somehow reflect the fact that under noninducing conditions CatR bound to the IBS is capable of stimulating transcription to a small degree. These data confirmed that the repression from the IBS was due to bound CatR and not a polar effect on transcription. However, they do not rule out the possibility that the point mutations affect mRNA stability of the transcripts.
DNA looping mediated by protein-DNA and protein-protein interactions is a mechanism that is ubiquitously utilized by both prokaryotes and eukaryotes to modulate transcription in response to various environmental factors (1, 6, 10, 16). The location of the IBS with respect to the CatR binding sites in the promoter and the apparent effect of mutations in the IBS on the transcriptional activity of the promoter suggested that DNA looping may account for the observed regulatory activity. This possibility was explored in two ways: by DNase I footprinting and by using spacing mutations. Comparison of DNase I footprinting studies done with the IBS alone and in the presence of the RBS/ABS on the same fragment of DNA indicates that the occupation of the IBS by CatR is facilitated in the latter case as would be expected of a cooperative interaction. Since cooperativity was seen with a linear fragment used in the footprinting study, it appears that supercoiling is not necessary to elicit loop formation. Results obtained for spacing mutations indicate that insertion of 6 bp (corresponding to a half-integral turn of DNA) impaired repression, whereas insertion of 11 bp (corresponding to an integral turn of DNA) restored repression. This finding demonstrates the need for maintenance of phasing between the promoter and the IBS in a manner consistent with the requirement for the binding sites to be on the same face of the helix. The most plausible explanation for the observed results would be that the DNA or CatR bound to the DNA at the promoter region of the catBCA operon, through formation of a DNA loop, interacts with CatR bound to the IBS. This interaction results in impaired transcriptional activation from the catBCA promoter despite the occupation of the RBS/ABS.
The precise physiological significance of the IBS-mediated repression is not known. Analogous regulation has been reported in the case of certain other catabolic operons that have metabolizable inducers such as the arabinose, galactose, rhamnose, and maltose operons (15, 23, 27, 28). Given the biodegradative nature of the catBCA (and the pheBA) operon, a similar explanation could be proposed to explain the relevance of the low-level repression mediated by CatR bound to the IBS. The IBS, being a low-affinity binding site for CatR, is presumably the last of the three CatR binding sites to be occupied. This occupation likely occurs at high CCM levels, i.e., when benzoate is plentiful and being rapidly metabolized. As in the case of the arabinose operon (26), such a regulatory mechanism would arguably confer a means for fine-tuning transcriptional activity. Turning on a promoter to a high initial transcriptional rate would allow rapid initial adaptation in the presence of a metabolizable substrate. Lowering the expression levels after the optimal enzyme levels have been attained would prevent wasteful expenditure of energy by keeping expression levels to a rate that is sufficient for efficient substrate utilization. It is interesting that transcriptional repression is not observed in the case of the metF gene, which belongs to an anabolic operon, despite the presence of a similarly positioned MetR binding site.
ACKNOWLEDGMENTS
We thank David Schlictman and Bill Hendrickson for critical reading of the manuscript and for helpful discussions during the course of the study.
This work was supported by Public Health Service grant ES04050-12 from the National Institute of Environmental Health Sciences.
REFERENCES
- 1.Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 2.Aldrich T L, Frantz B, Gill J F, Kilbane J J, Chakrabarty A M. Cloning and complete nucleotide sequence determination of the catB gene encoding cis,cis-muconate lactonizing enzyme. Gene. 1987;52:185–195. doi: 10.1016/0378-1119(87)90045-x. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Chugani S A, Parsek M R, Hershberger C D, Murakami K, Ishihama A, Chakrabarty A M. Activation of the catBCA promoter: probing the interaction of CatR and RNA polymerase through in vitro transcription. J Bacteriol. 1997;179:2221–2227. doi: 10.1128/jb.179.7.2221-2227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowan J M, Urbanowski M L, Talmi M, Stauffer G V. Regulation of the Salmonella typhimurium metF gene by the MetR protein. J Bacteriol. 1993;175:5862–5866. doi: 10.1128/jb.175.18.5862-5866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn T M, Hahn S, Ogden S, Schleif R F. An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci USA. 1984;81:5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, England: Cambridge University; 1984. [Google Scholar]
- 9.Goethals K, Van Montagu M, Holsters M. Conserved motifs in a divergent nod box of Arhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci USA. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn S, Hendrickson W, Schleif R F. Transcription of Escherichia coli ara in vitro: the cyclic AMP receptor requirement for PBAD induction that depends on the presence and orientation of the araO2 site. J Mol Biol. 1986;188:355–367. doi: 10.1016/0022-2836(86)90160-9. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S, Wallace J C, Brown J P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:11–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- 12.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis—a practical approach. Oxford, England: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 13.Houghton J E, Appel T M B, A J, Hughes E J, Ornston L N. Discontinuities in the evolution of Pseudomonas putida cat genes. J Bacteriol. 1995;177:401–412. doi: 10.1128/jb.177.2.401-412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasak L, Horak R, Nurk A, Talvik K, Kivisaar M. Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. J Bacteriol. 1993;175:8038–8042. doi: 10.1128/jb.175.24.8038-8042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz L, Englesberg E. Hyperinducibility as a result of mutation in structural genes and self-catabolite repression in the ara operon. J Bacteriol. 1971;107:34–52. doi: 10.1128/jb.107.1.34-52.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee N E, Davis M M. T cell receptor β-chain genes in BW5147 and other AKR tumors. J Immunol. 1988;140:1665–1675. [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 4.29–4.30. [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 18a.Parsek, M. Unpublished observations.
- 19.Parsek M R, Shinabarger D L, Rothmel R K, Chakrabarty A M. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J Bacteriol. 1992;174:7798–7806. doi: 10.1128/jb.174.23.7798-7806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsek M R, Ye R W, Pun P, Chakrabarty A M. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region: catBC promoter activation by CatR. J Biol Chem. 1994;269:11279–11284. [PubMed] [Google Scholar]
- 21.Parsek M R, Coco W M, Chakrabarty A M. Gel-shift assay and DNase I footprinting in analysis of transcriptional regulation of biodegradative genes. Methods Mol Genet. 1994;3:273–290. [Google Scholar]
- 22.Parsek M R, McFall S M, Shinabarger D L, Chakrabarty A M. Interaction of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: functional and evolutionary implications. Proc Natl Acad Sci USA. 1994;91:12393–12397. doi: 10.1073/pnas.91.26.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power J. The rhamnose genetic system in Escherichia coli K12. Genetics. 1967;55:557–568. doi: 10.1093/genetics/55.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothmel R K, Shinabarger D L, Parsek M R, Aldrich T L, Chakrabarty A M. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA-binding site by hydroxy radical footprinting. J Bacteriol. 1991;173:4717–4724. doi: 10.1128/jb.173.15.4717-4724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 26.Schleif R. Genetics and molecular biology. Reading, Mass: Addison-Wesley Publishing Company; 1985. pp. 347–366. [Google Scholar]
- 27.Schwartz M. Phenotypic expression and genetic localization of mutations affecting maltose metabolism in Escherichia coli K-12. Ann Inst Pasteur (Paris) 1967;112:673–700. [PubMed] [Google Scholar]
- 28.Soffer R L. Enzymatic expression of genetic units of function concerned with galactose metabolism in Escherichia coli. J Bacteriol. 1961;82:471–478. doi: 10.1128/jb.82.4.471-478.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheelis M L, Ornston L N. Genetic control of enzyme induction in the β-ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972;109:790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene (Amsterdam) 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]