Abstract
The Streptomyces peucetius dpsY and dnrX genes govern early and late steps in the biosynthesis of the clinically valuable antitumor drugs daunorubicin (DNR) and doxorubicin (DXR). Although their deduced products resemble those of genes thought to be involved in antibiotic production in several other bacteria, this information could not be used to identify the functions of dpsY and dnrX. Replacement of dpsY with a mutant form disrupted by insertion of the aphII neomycin-kanamycin resistance gene resulted in the accumulation of UWM5, the C-19 ethyl homolog of SEK43, a known shunt product of iterative polyketide synthases involved in the biosynthesis of aromatic polyketides. Hence, DpsY must act along with the other components of the DNR-DXR polyketide synthase to form 12-deoxyaklanonic acid, the earliest known intermediate of the DXR pathway. Mutation of dnrX in the same way resulted in a threefold increase in DXR production and the disappearance of two acid-sensitive, unknown compounds from culture extracts. These results suggest that dnrX, analogous to the role of the S. peucetius dnrH gene (C. Scotti and C. R. Hutchinson, J. Bacteriol. 178:7316–7321, 1996), may be involved in the metabolism of DNR and/or DXR to acid-sensitive compounds, possibly related to the baumycins found in many DNR-producing bacteria.
Daunorubicin (DNR) and its C-14 hydroxylated derivative doxorubicin (DXR) are clinically important antitumor anthracycline antibiotics (38) isolated from Streptomyces peucetius ATCC 29050 (3, 4, 6). Studies of DNR and DXR production in this organism (Fig. 1) have elucidated the organization and regulation of many of the biosynthetic genes (10, 13, 15, 25, 27, 32, 33, 37), as well as the mechanism of antibiotic resistance (16, 24) and the regulation of DNR and DXR biosynthesis (11, 26, 30, 41). Here we report the characterization of the dnrX and dpsY (formerly dnrY) genes situated between the dnmZ daunosamine biosynthesis and drrD self-resistance genes in the cluster of DXR biosynthesis genes (Fig. 2A). Although the deduced product of dpsY is not similar to enzymes with an established function, replacement of the wild-type copy in the 29050 strain with a mutant form resulted in the accumulation of a shunt product derived from an early intermediate of carbon chain assembly by the DNR-DXR polyketide synthase (15). Thus, DpsY is a type of polyketide cyclase heretofore unrecognized as essential for the formation of 12-deoxyaklanonic acid, the product of the DNR polyketide synthase. DnrX, in contrast, appears to govern further metabolism of DNR and DXR to as yet uncharacterized products, some of which can be hydrolyzed back to these two anthracyclines upon acid treatment of culture extracts. Replacement of the wild-type copy of dnrX in the 29050 strain with a mutant form resulted in a threefold increase in the production of DXR, the most valuable of the two antitumor drugs, but had an insignificant effect on the amount of DNR produced.
FIG. 1.
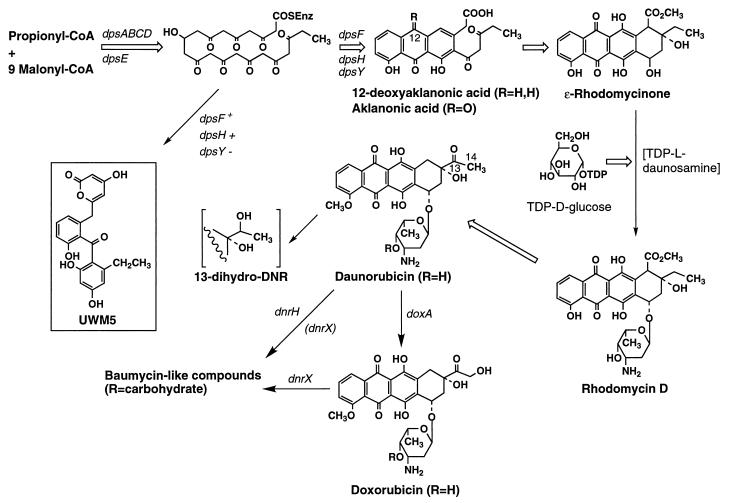
Biosynthesis of DNR and DXR in S. peucetius. Only the key intermediates and genes discussed in the text are shown. Thin and open thick arrows signify single or multiple steps, respectively. For the baumycin-like compounds, the structure of baumycin A1 is illustrated but the C-4 and C-13 positions can be modified in other types of baumycins (39).
FIG. 2.
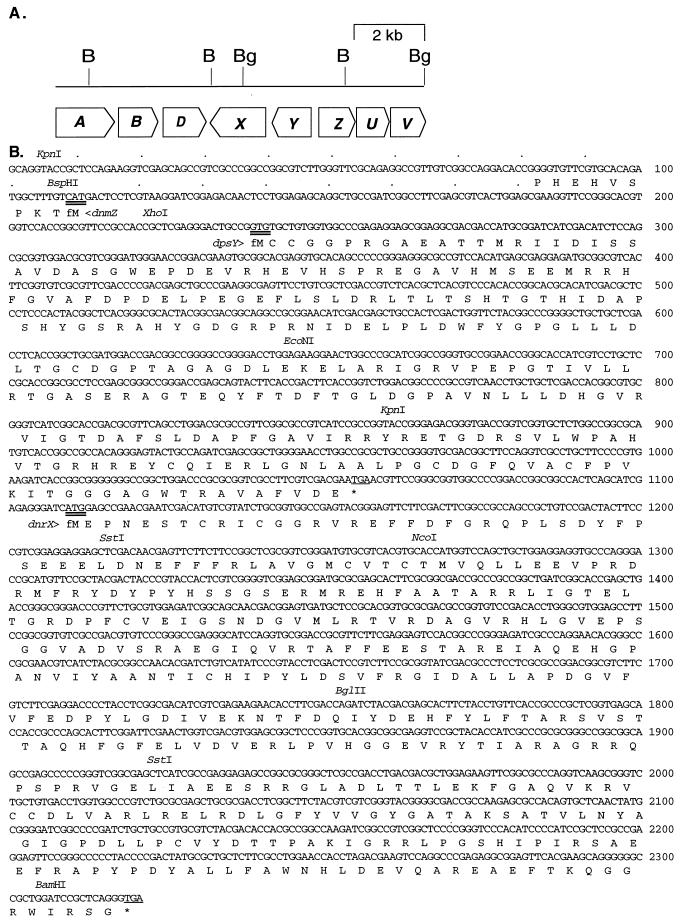
(A) Organization of the DXR biosynthesis and self-resistance genes in the region surrounding dnrX and dpsY. The drrA and drrB resistance genes are described by Guilfoile and Hutchinson (16), and dnmZ is described by Otten et al. (32). B, BamHI; Bg, BglII (B). The DNA and deduced protein sequence of the region from panel A characterized in this paper. Only the restriction stites of interest are shown. Probable translational start and stop codons are doubly and singly underlined, respectively. The fM and asterisk indicate the probable beginning and end of the open reading frame (only the first 10 amino acids of dnmZ are shown).
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
Escherichia coli DH5α (36) and the pUC18 (45), pSP72 (Promega, Madison, Wis.), and pSE380 (Invitrogen, Carlsbad, Calif.) plasmids were used for routine subcloning. The high-copy-number Streptomyces shuttle vector pWHM3 was from Vara et al. (43). The Streptomyces strains, other plasmids, and φC31-derived phages used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and phages
| Strain, plasmid, or phage | Genotype | Source |
|---|---|---|
| S. peucetius | ||
| ATCC 29050 | Wild type | ATCCa |
| WMH1650 | dnrX aphII, obtained after infection with phWHM281 | This work |
| WMH1655 | dpsY aphII, obtained after infection with phWHM283 | This work |
| S. lividans | ||
| TK24 | SLP1− SLP2−, streptomycin resistant | 19 |
| WMH1594 | SLP1− SLP2−, streptomycin resistant, φC31 lysogen | 23 |
| Plasmids | ||
| pFDNeoS | pUC18 containing the aphII gene | 7 |
| pWHM249 | XhoI site eliminated from the aphII gene of pFDNeoS | 23 |
| pWHM335 | Cosmid clone containing a fragment of the DNR cluster | 40 |
| pWHM279 | 3.3-kb BamHI fragment cloned from pWHM335 in pUC18 | This work |
| pWHM280 | 1.0-kb SalI fragment with the aphII gene cloned blunt ended from pWHM249 into the filled-in NcoI site of pWHM279 | This work |
| pWHM282 | 1.0-kb SalI fragment with the aphII gene cloned blunt ended from pWHM249 into the filled-in EcoNI site of pWHM279 | This work |
| pWHM346 | 1.6-kb SstI-SphI fragment cloned from pWHM279 into pSP72 | This work |
| pWHM347 | 1.6-kb EcoRI-HindIII fragment cloned from pWHM346 into pWHM3 | This work |
| pWHM226 | 3.3-kb BamHI fragment cloned from pWHM279 into pWHM3 | This work |
| Phages | ||
| KC515 | c+ attP, thiostrepton resistant, viomycin resistant | 5 |
| phWHM281 | c+ attP, viomycin resistant; 4.3-kb BamHI fragment cloned from pWHM280 into KC515 between the BamHI and BglII sites | This work |
| phWHM283 | c+ attP, viomycin resistant; 4.3-kb BamHI fragment cloned from pWHM282 into KC515 between the BamHI and BglII sites | This work |
ATCC, American Type Culture Collection.
Biochemicals and chemicals.
Thiostrepton was obtained from S. J. Lucania at Bristol-Myers-Squibb (Princeton, N.J.). ɛ-Rhodomycinone (RHO) and DNR were obtained from Pharmacia & UpJohn (Milan, Italy), and aklanonic acid was a gift from M. Gerlitz (University of Wisconsin, Madison). Restriction enzymes and other molecular biology reagents were from standard commercial sources.
Media and other growth conditions.
E. coli strains carrying plasmids were grown in Luria-Bertani medium (36) and were selected with ampicillin (100 μg ml−1). S. peucetius strains were grown on ISP4 medium (Difco Laboratories, Detroit, Mich.) containing Difco yeast extract (10% [wt/vol] per liter) for sporulation. R2YE agar medium (19) was used in transformation experiments, and R2YE agar medium, without sucrose was used for infection of S. peucetius with φC31 derivatives and for sporulation of Streptomyces lividans TK24 and the TK24 (φC31) lysogen. The minimal medium (MM) of Hopwood et al. (19) was used to screen S. peucetius recombinant clones. S. peucetius strains were grown at 30°C in R2YE liquid medium for preparation of protoplasts and isolation of chromosomal DNA. KC515 derived from φC31 and KC515-derived phages were propagated as described by Hopwood et al. (19).
Isolation and in vitro manipulation of DNA.
Plasmid DNA was isolated from bacterial cells with the Bio 101 kit (Vista, Calif.). Phage DNA was isolated with the Qiagen lambda kit (Chatsworth, Calif.). Total S. peucetius DNA was isolated by the protocol of Hopwood et al. (19). Restriction endonuclease digestions and ligation used standard techniques (36). DNA fragments for labeling and subcloning were isolated with the Qiaex (Qiagen) gel extraction kit. The conditions for phage DNA transfection and S. peucetius transformation were as described by Hopwood et al. (19).
DNA sequencing.
DNA sequencing was carried by the dideoxy chain termination method using single-stranded DNA as described previously (41).
Southern analysis.
Streptomyces chromosomal DNA was digested with restriction endonuclease enzymes for 4 h, electrophoresed in a 0.8% agarose gel overnight, and blotted to Hybond N membranes (Amersham, Chicago, Ill.) by capillary transfer (36). Labelling, hybridization, and detection were carried out with the Genius1 nonradioactive DNA labelling kit (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer’s instructions.
Construction of the dnrX::aphII and dpsY::aphII mutants by insert-directed homologous recombination in S. peucetius.
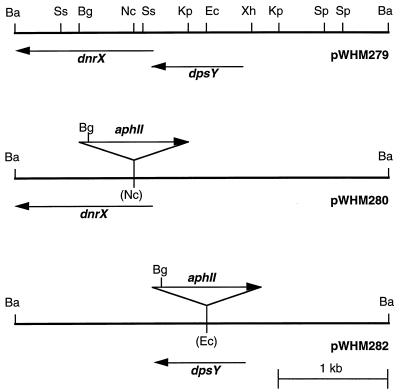
For phWHM281 (φC31 derivatives in this work are prefixed with “ph”), a 3.3-kb BamHI fragment that included the dnrX and dpsY sequences was subcloned from cosmid pWHM335 into pUC18 to create pWHM279. A 1.0-kb SalI fragment containing the aphII kanamycin-neomycin resistance gene from pWHM249 was cloned blunt ended into the unique NcoI site of pWHM279 to create pWHM280 (Fig. 3). The NcoI site is located at the beginning of the dnrX gene. A 4.3-kb BamHI fragment from pWHM280 containing a disrupted copy of the dnrX gene was subcloned into BamHI- and BglII-digested KC515 to give phWHM281. For phWHM283, the 1.0-kb SalI fragment from pWHM249 was cloned blunt ended into the unique EcoNI site of pWHM279, located within the open reading frame of the dpsY gene, to create pWHM282 (Fig. 3). The 4.3-kb BamHI fragment of pWHM282 containing the disrupted copy of dpsY was cloned into BamHI- and BglII-digested KC515 to give phWHM283. In both constructs the tsr resistance gene of KC515 was replaced by the cloned fragments. Protoplasts of S. lividans TK24 were transfected with each of the phage constructs, and the phWHM281 and -283 recombinant phages were isolated from plaques by a convenient spot test method we developed (24). The recombinant phages were characterized phenotypically as containing aphII and vph resistance genes as described previously (24), and the presence of the cloned DNA was confirmed by restriction endonuclease digestion analysis.
FIG. 3.
Insertional inactivation of the dnrX and dpsY genes. Restriction maps are shown for the BamHI fragment containing the dnrX and dpsY genes (in pWHM279), the disrupted dnrX gene (in pWHM280), and the disrupted dpsY gene (in pWHM282). Arrows indicate the relative directions of transcription of the dnrX, dpsY, and aphII genes. Restriction sites abbreviations: Ba, BamHI; Bg, BglII; Ec, EcoNI; Kp, KpnI; Nc, NcoI; Sp, SphI; Ss, SstI; and Xh, XhoI.
The S. peucetius 29050 strain was infected with phWHM281 and phWHM283 on agar plates of the R2YE medium without sucrose (5 × 107 spores and 1 × 108 to 2 × 108 phage) to allow phage multiplication and host sporulation. After 16 h the plates were overlaid with an aqueous neomycin solution to give a final concentration of 20 μg ml−1; then, after further growth for 6 days until sporulation, the resulting colonies were replica plated on MM containing neomycin. Primary neomycin-resistant clones, were isolated and their phenotype was determined after a second round of single-colony isolation. Selection for phage integration or gene replacement was carried out on MM with neomycin (10 μg ml−1) or viomycin (30 μg ml−1).
Construction of the dpsY expression plasmid.
The dpsY gene was subcloned as a 1.6-kb SstI-SphI fragment from pWHM279 into vector pSP72 to create pWHM346. A 1.6-kb EcoRI-HindIII fragment containing dpsY was cloned from pWHM346 into pWHM3 to create pWHM347.
Determination of anthracycline production.
S. peucetius strains were cultured in the APM seed and production medium (16) as described by Furuya and Hutchinson (11). The cultures were acidified with oxalic acid, heated at 60°C for 45 min, adjusted to pH 8.5, and extracted with chloroform. The extract samples were concentrated to dryness in vacuo, and the residue was dissolved in methanol and analysed by thin-layer chromatography according to Otten et al. (31). For analysis by high-performance liquid chromatography (HPLC), S. peucetius strains were inoculated into 25 ml of liquid R2YE medium with 40 μg of kanamycin sulfate ml−1 in a 300-ml flask and incubated at 30°C and 300 rpm on a rotary shaker. After 2 days of growth, 2.5 ml of this culture was transferred to 25 ml of APM production medium. After 4 to 5 days of further growth, each culture was divided into two equal portions: one was acidified with 250 mg of oxalic acid and incubated at 30°C overnight and then extracted, and the other portion was immediately extracted without acidification. Cultures were extracted with an equal volume of acetonitrile-methanol (1:1, vol/vol) at 30°C and 300 rpm for 2 h. The extract was filtered and the filtrate was analyzed by HPLC. Reverse-phase HPLC was performed with a Vydac C18 column (4.6 by 250 mm; 5-μm particle size) at a flow rate of 0.385 ml/min. The mobile phase A was 0.2% trifluoroacetic acid (Pierce Chemical Co.) in water, and mobile phase B was 0.078% trifluoroacetic acid in acetonitrile (J. T. Baker Chemical Co., Cleveland, Ohio). Elution was performed with a linear gradient from 20 to 60% phase B in phase A over 33 min and monitored with a diode array detector set at 488 nm (bandwith, 12 μm). Aklanonic acid was determined by HPLC as described by Gerlitz et al. (14).
Isolation and characterization of UWM5.
The S. peucetius dpsY mutant was added to seed APM medium (12.5 ml) containing thiostrepton and neomycin (5 μg of each per ml) and incubated for 17 h. A 0.2-ml aliquot of the seed culture was transferred to 20 flasks each containing 50 ml of APM medium with thiostrepton and neomycin (5 μg of each per ml) and incubated at 30°C for 5 days. The cultures were combined and filtered, and the broth was extracted with ethyl acetate (three times with 1 liter each). The ethyl acetate-soluble fraction was evaporated under reduced pressure (<35°C) to give a residue (600 mg), which was subjected to silica gel column chromatography (Aldrich Chemicals, Milwaukee, Wis.; 1.0 by 50 cm) and eluted with CHCl3-methanol (MeOH) from 9:1 to 6:4 (vol/vol). The material eluted with CHCl3-MeOH from 7:3 to 6:4 (109 mg) was separated by gel filtration on a Sephadex LH-20 column (Pharmacia; 1.0 by 50 cm) by elution with MeOH, and a 44-mg portion of the material recovered in the fractions from 70 to 180 ml (85 mg) was purified by reverse-phase HPLC (Nova-Pak C18; Waters, Milford, Mass.; 10 by 300 mm; flow rate, 2.5 ml/min; detection with UV at 290 nm; eluent, CH3CN plus 0.008% acetic acid–H2O [3:7, vol:vol]) to yield UWM5 (25.9 mg). A colorless, amorphous residue was recovered with the following characteristics: UV (MeOH) λmax, 294 nm (ɛ 15,200) and 340 nm (sh); 1H nuclear magnetic resonance (NMR) (DMSO-d6) d 7.19 (1H, t, J = 7.9 Hz, C-9), 6.77 (1H, d, J = 7.9 Hz, C-10), 6.72 (1H, d, J = 7.9 Hz, C-8), 6.16 (1H, d, J = 2.2 Hz, C-18), 6.11 (1H, d, J = 2.2 Hz, C-16), 5.58 (1H, brs, C-4), 5.03 (1H, d, J = 1.8 Hz, C-2), 3.56 (2H, s, C-6), 2.34 (2H, dt, J = 7.4, 7.4 Hz, C-20), 1.00 (3H, t, J = 7.4, C-21); 13C NMR (DMSO-d6) d 199.3 (s, C-13), 172.1 (s, C-1), 166.9 (s, C-3), 164.1 (s, C-15), 163.5 (s, C-17), 162.2 (s, C-5), 154.4 (s, C-11), 148.0 (s, C-19), 133.6 (s, C-7), 131.5 (d, C-8), 130.7 (s, C-12), 120.7 (d, C-9), 117.2 (s, C-14), 114.5 (d, C-10), 108.5 (d, C-18), 102.2 (d, C-4), 100.2 (d, C-16), 87.7 (d, C-2), 36.5 (t, C-6), 25.9 (t, C-20), 15.1 (q, C-21). These assignments were confirmed by an HMBC NMR spectrum taken at 500 MHz (data not shown). Fast atom bombardment mass spectral data (FABMS), m/z 405 (M+Na); high-resolution FABMS, found m/z 405.0934 [calculated for C21H18O7Na (M+Na): 405.0950].
Bioconversion of aklanonic acid, RHO, and DNR.
Glycerol stock mycelium (50 ml) of the S. peucetius dpsY mutant was added to seed APM medium (12.5 ml) and incubated for 17 h. Then, 50 μl of this seed culture was transferred to 25 ml of the APM production medium and incubated at 30°C for 48 h, after which aklanonic acid, RHO, or DNR was added to a final concentration of 20 to 28, 30, or 20 μg ml−1, respectively. The cultures were further incubated for 120 h, after which they were analyzed by thin-layer chromatography as described above.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the GenBank/EMBL databases with accession number AF048833.
RESULTS
Analysis of the DNA sequences of the dnrX and dpsY genes.
The dnrX and dpsY genes were cloned in a 3.3-kb BamHI segment from a primary cosmid clone containing the DNR and DXR biosynthesis genes of S. peucetius ATCC 29050 (31, 40). Figure 2B shows the sequence of a 2,320-nucleotide (nt) portion of this segment from the first internal KpnI site and extending 12 nt past the last BamHI site, which is close to the 3′ end of the drrD gene (Fig. 2A), to complete the dnrX open reading frame (ORF). The dpsY ORF most likely begins with a GTG at nt 239 (preceded by a probable ribosome binding site, GAGG), approximately 130 nt upstream of the divergently transcribed dnmZ gene (32) (the first 10 residues of DnmZ are shown in Fig. 2B), and ends with a TGA stop codon at nt 1055. DpsY therefore should have a molecular weight of 29,320 (excluding the formylmethionine [fMet]) and an isoelectric point of 4.93. Correspondingly, the dnrX ORF should begin with an ATG at nt 1110 and end with a TGA at nt 2319; the DnrX protein should have a molecular weight of 45,176 (excluding the fMet) and an isoelectric point of 5.32. The drrA and drrB DNR and DXR resistance genes (16) and drrD, a putative resistance gene (1), lie immediately downstream of dnrX (Fig. 2A).
Comparisons of the deduced amino acid sequences of the dnrX and dpsY gene products using the Blastn and Blastp subsets (2) of the NCBI database revealed that DnrX is closely related to the following five proteins of unknown function: 65 and 66% identical, respectively, to the hypothetical products of two ORFs (GenBank no. U84349 and U84350) from different strains of Amycolatopsis orientalis; 86% identical to the hypothetical product of ORF2 from the daunorubicin-producing Streptomyces griseus strain (21); and 33% identical to the hypothetical products of two ORFs from Synechocystis sp. (GenBank no. D90901 and D90903). DnrX also is 34% identical to ORF3 from the cluster of erythromycin biosynthesis genes in Saccharopolyspora erythraea (17). This gene has recently been renamed eryBIII since it is required for the synthesis of mycarose, one of the deoxysugars of erythromycin A, and is thought to encode a C-methyltransferase (12, 12a, 22). DpsY is very similar (72% by GAP analysis [8]) only to the deduced products of ORFs from the clusters of genes for the biosynthesis of chlorotetracycline in Streptomyces aureofaciens (data not shown; 35) and oxytetracycline in Streptomyces rimosus (otcD4) (20), whose functions also are unknown.
Effect of insertional inactivation of dnrX and dpsY.
To examine whether the dnrX and dpsY genes were vital for the production of DNR and DXR, each gene was replaced in the wild-type strain with a mutant copy resulting from insert-directed, homologous recombination of DNA carrying an antibiotic resistance gene. The phWHM281 and phWHM283 recombinant derivatives of KC515 (5) were constructed as described in Materials and Methods to insert the aphII neomycin-kanamycin resistance gene from pWHM249 into the unique NcoI site of dnrX and the EcoNI site of dpsY (Fig. 2B), respectively. After infection of S. peucetius 29050 with each of the recombinant phages, clones resistant to neomycin and viomycin (113 from phWHM281 and 198 from phWHM283), as well as resistant to neomycin only (24 from phWHM281 and 42 from phWHM283), were identified. A large proportion of the clones were resistant to neomycin only, suggesting that many of the recombinants had resulted from double-crossover events, either following the initial single-crossover recombination producing neomycin- and viomycin-resistant strains or occurring essentially simultaneously, as observed in our previous gene disruption/replacement experiments with Streptomyces hygroscopicus (23). The neomycin-resistant WMH1650 and WMH1655 strains were chosen as representative of clones with disrupted dnrX and dpsY genes, respectively.
Southern analysis of the DNA from the WMH1650 and WMH1655 clones was performed to establish the genomic structure of each mutant in the dnrX and dpsY regions, respectively. When BglII-digested DNA from the WMH1650 and WMH1655 strains was probed with the 1.0-kb PstI-BamHI fragment of pFDNeoS containing the aphII gene, 4.9- and 4.2-kb BglII fragments between the BglII sites of the aphII gene (Fig. 3) and dnmV (Fig. 2A) were detected, confirming the presence of disrupted copies of the dnrX and dpsY genes in the chromosomes of the WHM1650 and WMH1655 strains, respectively (data not shown).
The amounts of metabolites obtained from extraction of acidified cultures are indicated in Table 2 for the WMH1650 dnrX::aphII and WMH1655 dpsY::aphII mutants. No metabolites were detected when the culture of the dpsY mutant was extracted at pH 8.5, but at pH 6, one principal metabolite was found, UWM5 (Fig. 1). Its UV, 1H, and 13C NMR and FABMS data (Materials and Methods) are consistent with the C-19 ethyl homolog of SEK43 (28, 29). The chemical shifts of most of the protons and carbons in UWM5 and SEK43 were nearly identical when their NMR spectra were obtained under the same conditions, but the signal for the C-19 methyl group of SEK43 was replaced by two new resonances at 2.34 (1H) and 25.9 (13C) ppm for the C-20–CH2 group and at 1.00 (1H) and 15.1 (13C) ppm for C-21–CH3. UWM5 arises from incomplete cyclization of the propionate-derived decaketide precursor of 12-deoxyaklanonic acid, similar to the formation of SEK43 from an acetate-derived decaketide in other systems (28, 29). Hence, DpsY appears to be a polyketide cyclase that operates with the two other cyclases, DpsF (15, 29) and DpsH (14), as part of the components of the iterative polyketide synthase that makes the aromatic portion of DNR (15). For this reason we have renamed dnrY (27) dpsY to be consistent with the nomenclature of the other 12-deoxyaklanonic acid biosynthesis genes. This deduction is consistent with the report that the S. griseus dpsY homolog appeared to govern an early step in the biosynthesis of leukaemomycin, a DNR analog (21). Since introduction of pWHM1223 containing the S. peucetius dpsABEF genes (15) along with tcmJ and tcmM, two other type II polypeptide synthase genes from Streptomyces glaucescens, into the dpsY mutant did not result in the production of aklanonic acid or its C-21 desmethyl analog, we presume that the formation of these compounds by S. lividans(pWHM1223) transformants (14) results from the action of a dpsY homolog in the latter host.
TABLE 2.
Anthracycline titers of S. peucetius 29050 (wild type), WMH1650, and WMH1655a
| Strain and compound | Titer (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| RHO | DNR | 13-dihydro-DNR | DXR | Xb | Yb | |
| 29050, no H3O+ | 0c | 6.1 | 0 | 8.8 | 6.8 | 5.6 |
| 29050 + H3O+ | 3.6 | 45.3 | 0 | 13.9 | 0 | 0 |
| WMH1650 (dnrX::aphII), no H3O+ | 0 | 35.4 | 18.0 | 41.3 | 0 | 0 |
| WMH1650 + H3O+ | 0 | 36.2 | 16.6 | 41.6 | 0 | 0 |
| WMH1655 (dpsY::aphII) | 0 | 0 | 0 | 0 | 0 | 0 |
Cultures were grown in APM medium for 120 h and worked up as described in Materials and Methods. The titers shown are typical of the values obtained in different fermentations.
X and Y, two different, unknown compounds with retention times of 35.9 and 39.3 min in the HPLC chromatogram.
No metabolite was found.
The phenotype of the WMH1650 dnrX::aphII mutant is different from that of the wild-type strain but does not lead to a clear understanding of the role of the DnrX protein. WMH1650 produced no detectable amount or only traces of RHO, as did the 29050 strain (the latter normally produces RHO [Table 3] in an amount greater than that of DNR, but did not in this experiment), and approximately threefold-more DXR than the amount isolated from the 29050 strain (Table 2). The amounts of DNR in acidified cultures of the dnrX mutant and wild-type strains were almost the same, but considerably more 13-dihydro-DNR was produced by the dnrX mutant. Apparently, more C-13 carbonyl reduction occurs when DNR is not modified by the DnrX protein. Most notable is the fact that much more DNR was obtained from the acid-treated culture of the wild-type strain than from the nonacidified one, whereas acid treatment did not increase the amounts of any of the metabolites isolated from the dnrX mutant cultures. These data suggest that a mutation blocking the function of dnrX prevents further metabolism of DNR to acid-sensitive compounds. In fact, two unknown anthracyclines, compounds X and Y, found only in the nonacidified culture of the wild-type strain, were not produced by the dnrX mutant (Table 2). Baumycin A1 is an acid-sensitive C-4′ glycoside of DNR (42) found in many DNR-producing microorganisms along with other types of baumycins (reviewed in reference 39), some of which have not been characterized. Production cultures are therefore routinely acidified before extraction to hydrolyze the baumycins and increase the recovery of DNR.
TABLE 3.
Anthracycline titers of the S. peucetius WHM1655 dpsY::aphII mutant transformed with pWHM3 or pWHM347a
| Strain | Titer (μg/ml)
|
|||
|---|---|---|---|---|
| RHO | DNR | 13-dihydro-DNR | DXR | |
| 29050 | 4.2 | 17.2 | 0b | 8.2 |
| WMH1655(pWHM3) | 0 | 0 | 0 | 0 |
| WMH1655(pWHM347) | 0 | 23.9 | 8.7 | 24.2 |
Cultures were grown in APM medium for 96 h and worked up as described in Materials and Methods. The titers shown are averages obtained from three independent fermentations.
No metabolite was found.
Bioconversion of aklanonic acid, RHO, and DNR by the dpsY::aphII mutant.
To confirm the above implication that dpsY governs one of the earliest steps in DXR biosynthesis, bioconversion experiments were carried out with three of its precursors as described in Materials and Methods. Aklanonic acid, RHO, and DNR were all converted to the end products of the pathway (Fig. 1) in the APM production medium (data not shown). This establishes that all of the genes for the steps beyond the formation of aklanonic acid are functioning in the dpsY mutant.
Expression of dpsY and dnrX in S. peucetius strains.
The ability of the wild-type dpsY gene to complement the dpsY::aphII mutation was tested in the WMH1655 strain. pWHM347 containing dpsY was made as described in Materials and Methods. This plasmid and the pWHM3 vector as a control were each introduced by transformation into the S. peucetius 29050 and WMH1655 dpsY::aphII mutant strains. The presence of the plasmids in the transformants was confirmed by analysis of the reisolated DNA by restriction enzyme digestion. Culture extracts from three transformants of each variant were analyzed for anthracycline production by HPLC. Although pWHM347 restored DNR and DXR production in the WMH1655 dpsY::aphII mutant, the levels of these and other metabolites were atypical in comparison with those of the wild-type strain (Table 3). RHO was not found, and more DNR and especially DXR were produced. Considerably more 13-dihydro-DNR was found in the culture extracts of the transformants. Such behavior is characteristic of the phenotype of the dnrX mutant (Table 2) and could be caused by a polar effect of the dpsY::aphII mutation on expression of the dnrX gene. This belief was confirmed by observing that introduction of the dnrX and dpsY genes together (as pWHM226) into the dpsY mutant resulted in a phenotype like that of the wild-type strain (data not shown). The dpsY and dnrX genes therefore appear to be a single operon. (Mutation of the dpsY gene in an industrial strain by insertion of the aphII gene at a different location than in WMH1655 did not appear to have a polar effect on expression of dnrX.) No differences in the production of anthracycline metabolites were observed when pWHM3 and pWHM347 were each introduced into the wild-type strain (data not shown).
To complement the dnrX::aphII mutation of WMH1650, pWHM226 containing the 3.3-kb BamHI fragment with the wild-type dpsY and dnrX genes cloned into the BamHI site of pWHM3 was introduced by transformation into the WMH1650 strain, as was pWHM3 itself. HPLC analysis of extracts of the nonacidified cultures of representative transformants showed the reappearance of the two unknown, acid-sensitive compounds X and Y, while extracts of the control strain containing pWHM3 did not contain these unknown compounds (data not shown). 13-Dihydro-DNR was also not found in transformants containing pWHM226. These data confirm that the disappearance of the baumycin-like products and appearance of 13-dihydro-DNR in the WMH1650 strain is due to the disruption of dnrX.
DISCUSSION
The discovery that dpsY is an essential part of the multicomponent S. peucetius DNR-DXR polyketide synthase was unexpected since this gene is not clustered with the rest of the dps genes, and aklanonic acid is made by S. lividans TK24 and Streptomyces coelicolor CH999 containing only the DpsF cyclase gene in addition to the genes for the chain assembly (DpsABCD) and ketoreduction (DpsE) enzymes (14, 34). (The dnrG oxidase for conversion of 12-deoxyaklanonic acid to aklanonic acid is not necessary in these heterologous hosts [14, 34].) Recent evidence that the dpsH gene is also important (14) along with the information about dpsY reported here leads us to propose that DpsF catalyzes only the dehydration and cyclization associated with formation of the first ring, which is consistent with its strong sequence similarity to the cyclase-dehydratase (aromatase) enzymes of other iterative polyketide synthases (15). DpsH and DpsY appear to be more important for the second and third cyclizations, which result in the formation of the tricyclic 12-deoxyaklanonic acid (Fig. 1) because the presence of either enzyme suppresses (14) or avoids (vide supra) the formation of incompletely cyclized shunt products like SEK43 and UWM5. Chloro- (35) and oxytetracycline (20) biosyntheses are the only other systems that utilize dpsY homologs, yet neither of these gene clusters have dpsH homologs. The polyketide synthase genes in the oxytetracycline producer also appear to be broken up into different, nonadjacent subclusters (20). It is not clear which steps in oxy- and chlorotetracycline biosynthesis parallel the one governed by DpsY since even though the tetracyclic frameworks of anthracyclines and tetracyclines are different, the folding and cyclization patterns leading to tricyclic intermediates are identical among the three systems.
The role of dnrX is more of an enigma than that of dpsY. DnrX homologs in three other bacterial species are also associated with antibiotic production. For instance, in A. orientalis, these homologs are purported to be involved in the synthesis of the sugar components of the glycopeptide antibiotic chloroeremomycin (GenBank no. U84349 and U84350). Although the eryBIII gene (formerly ORF3) of S. erythraea governs a step in mycarose biosynthesis (12, 12a, 22, 44), it is not clear how this information can be related to a function for dnrX, especially if it encodes some type of methyltransferase. The dnrX gene clearly is not required for daunosamine biosynthesis or attachment to RHO to form rhodomycin D (Fig. 1), yet the acid sensitivity of the anthracycline antibiotics made by the dnrX+ strain is consistent with the presence of carbohydrate-derived moieties, as in the case of baumycin A1 (42; reviewed in reference 39). Whatever their nature, the increased production of DXR by the dnrX mutant (Table 2) implies that such compounds may not be good substrates for the DoxA CYP450 hydroxylase, which converts DNR to DXR and is thought to be ineffective with baumycin-like compounds (9). But it is unlikely that these uncharacterized metabolites are simply DXR glycosides, analogous to baumycin A1, because they should have been converted to DXR upon acidification of the wild-type culture. In this regard, the dnrH gene has also been implicated in the formation of baumycin-like compounds and its disruption increased the yield of DNR considerably, whereas the DXR titer was raised only slightly (37). Comparison of the effects of acid treatment of cultures of the dnrH and dnrX strains grown in two different media by HPLC analysis revealed only slight differences in the profile of acid-sensitive metabolites produced, apart from the distinctly different amounts of RHO and DXR produced by these mutants. Hence, although we have demarcated the roles of dnrH and dnrX in Fig. 1, dnrX probably affects more steps than the one indicated. The lack of adequate information prevents further speculation about the role of dnrX and the reason for the threefold increase in DXR production in the dnrX mutant.
ACKNOWLEDGMENTS
We thank Iain Hunter for unpublished sequence data about the oxytetracycline biosynthesis genes and Peter Leadlay for unpublished information.
This research was supported by grants from Pharmacia & Upjohn and, in part, by the National Institutes of Health (CA64161). Y.D.-K. was supported by a Promotion of Science for Young Scientists fellowship from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ali, A., and C. R. Hutchinson. Unpublished results.
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, Spalla C. 14-Hydroxydaunomycin, a new antitumor antibiotic from Streptomyces peucetius var. caesius. Biotechnol Bioeng. 1969;11:1109–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 4.Cassinelli G, Orezzi P. Daunomicina: un nuovo antibiotico ad attivita’ citostatica. Isolamento e proprieta’. G Microbiol. 1963;11:167–174. [Google Scholar]
- 5.Chater K F. Streptomyces phages and their application to Streptomyces genetics. In: Queener S E, Day L E, editors. The bacteria. 9. Antibiotic-producing Streptomyces. Orlando, Fla: Academic Press; 1986. pp. 119–158. [Google Scholar]
- 6.D’bost M, Ganter P, Maral R, Ninet L, Pinnert S, Preud’Homme J, Werner G H. Un novel antibiotique a proprietes cytostatiques: la rubidomycine. C R Acad Sci Agric Bulg. 1963;257:1813–1815. [PubMed] [Google Scholar]
- 7.Denis F, Brzezinski R. An improved aminoglycoside resistance gene cassette for use in gram-negative bacteria and Streptomyces. FEMS Microbiol Lett. 1991;81:261–264. doi: 10.1016/0378-1097(91)90224-x. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickens M L, Strohl W R. Isolation and characterization of a gene from Streptomyces sp. strain C5 that confers the ability to convert daunomycin to doxorubucin on Streptomyces lividans TK24. J Bacteriol. 1996;178:3389–3395. doi: 10.1128/jb.178.11.3389-3395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippini S, Solinas M M, Breme U, Schluter M B, Gabellini D, Biamonti G, Colombo A L, Garofano L. Streptomyces peucetius daunorubicin biosynthesis gene, dnrF: sequence and heterologous expression. Microbiology. 1995;141:1007–1016. doi: 10.1099/13500872-141-4-1007. [DOI] [PubMed] [Google Scholar]
- 11.Furuya K, Hutchinson C R. The DnrN protein of Streptomyces peucetius, a pseudo-response regulator, is a DNA-binding protein involved in the regulation of daunorubicin biosynthesis. J Bacteriol. 1996;178:6310–6318. doi: 10.1128/jb.178.21.6310-6318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaisser S, Böhm G A, Cortés J, Leadlay P F. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol Gen Genet. 1997;256:239–251. doi: 10.1007/s004380050566. [DOI] [PubMed] [Google Scholar]
- 12a.Gaisser, S., G. A. Böhm, N. Dhillon, M.-C. Raynal, J. Cortes, and P. F. Leadlay. Analysis of eryBI, eryBIII and eryBVII from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet., in press. [DOI] [PubMed]
- 13.Gallo M A, Ward J, Hutchinson C R. Cloning of the dnrL and dnrM genes from the daunorubicin-producing Streptomyces peucetius. Microbiology. 1996;142:269–275. doi: 10.1099/13500872-142-2-269. [DOI] [PubMed] [Google Scholar]
- 14.Gerlitz M, Meurer G, Wendt-Pienkowski E, Madduri K, Hutchinson C R. The effect of the daunorubicin dpsH gene on the choice of starter unit and cyclization pattern reveals that type II polyketide synthases can be unfaithful yet intriguing. J Am Chem Soc. 1997;119:7392–7393. [Google Scholar]
- 15.Grimm A, Madduri K, Ali A, Hutchinson C R. Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase. Gene. 1994;151:1–10. doi: 10.1016/0378-1119(94)90625-4. [DOI] [PubMed] [Google Scholar]
- 16.Guilfoile P G, Hutchinson C R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci USA. 1991;88:8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haydock S F, Downson J A, Dhillon N, Roberts G A, Cortes J, Leadlay P F. Cloning and sequence analysis of genes involved in erythromycin biosynthesis in Saccharopolyspora erythraea: sequence similarities between EryG and a family of S-adenosylmethionine-dependent methyltransferases. Mol Gen Genet. 1991;230:120–128. doi: 10.1007/BF00290659. [DOI] [PubMed] [Google Scholar]
- 18.Hopwood D A, Keiser T, Wright H M, Bibb M J. Plasmids, recombination chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983;129:2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 20.Hunter I S, Hill R A. Tetracyclines. In: Strohl W R, editor. Biotechnology of antibiotics. 2nd ed. New York, N.Y: Marcel Dekker; 1997. pp. 659–682. [Google Scholar]
- 21.Krugel H, Schumann G, Hanel F, Fiedler G. Nucleotide sequence analysis of five putative Streptomyces griseus genes, one of which complements an early function in daunorubicin biosynthesis that is linked to a putative gene cluster involved in TDP-daunosamine formation. Mol Gen Genet. 1993;241:193–202. doi: 10.1007/BF00280217. [DOI] [PubMed] [Google Scholar]
- 22.Leadlay, P. F. Personal communication.
- 23.Lomovskaya N, Fonstein L, Ruan X, Stassi D, Katz L, Hutchinson C R. Gene disruption and replacement in the rapamycin-producing Streptomyces hygroscopicus strain ATCC 29253. Microbiology. 1997;143:875–883. doi: 10.1099/00221287-143-3-875. [DOI] [PubMed] [Google Scholar]
- 24.Lomovskaya N, Hong S K, Kim S U, Fonstein L, Furuya K, Hutchinson C R. The Streptomyces peucetius drrC gene encodes a UvrA-like protein involved in daunorubicin resistance and production. J Bacteriol. 1996;178:3238–3245. doi: 10.1128/jb.178.11.3238-3245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madduri K, Torti F, Colomb A L, Hutchinson C R. Cloning and sequencing of a gene encoding carminomycin 4-O-methyltransferase from Streptomyces peucetius and its expression in Escherichia coli. J Bacteriol. 1993;175:3900–3904. doi: 10.1128/jb.175.12.3900-3904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madduri K, Hutchinson C R. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J Bacteriol. 1995;177:1208–1215. doi: 10.1128/jb.177.5.1208-1215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madduri K, Hutchinson C R. Functional characterization and transcriptional analysis of a gene cluster governing early and late steps in daunorubicin biosynthesis in Streptomyces peucetius. J Bacteriol. 1995;177:3879–3884. doi: 10.1128/jb.177.13.3879-3884.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature. 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 29.Meurer G, Gerlitz M, Wendt-Pienkowski E, Vining L C, Rohr J, Hutchinson C R. Iterative, type II polyketide synthases, cyclases and ketoreductases exhibit context dependent behavior in the biosynthesis of linear and angular decapolyketides. Chem Biol. 1997;4:433–443. doi: 10.1016/s1074-5521(97)90195-2. [DOI] [PubMed] [Google Scholar]
- 30.Otten S L, Ferguson J, Hutchinson C R. Regulation of daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J Bacteriol. 1995;177:1216–1224. doi: 10.1128/jb.177.5.1216-1224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otten S L, Stutzman-Engwall K J, Hutchinson C R. Cloning and expression of daunorubicin biosynthesis genes from Streptomyces peucetius and S. peucetius subsp. caesius. J Bacteriol. 1990;172:3427–3434. doi: 10.1128/jb.172.6.3427-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otten S L, Gallo M A, Madduri K, Liu X, Hutchinson C R. Cloning and characterization of the Streptomyces peucetius dnmZUV genes encoding a putative acyl-coenzyme A dehydrogenase, thymidine diphospho 4-keto-6-deoxyglucose-3(5)-epimerase and thymidine diphospho 4-ketodeoxyhexulose ketoreductase required for daunosamine biosynthesis. J Bacteriol. 1996;178:7316–7321. [Google Scholar]
- 33.Otten S L, Liu X, Ferguson J, Hutchinson C R. Cloning and characterization of the Streptomyces peucetius dnrQS genes encoding a daunosamine biosynthesis enzyme and a glycosyl transferase involved in daunorubicin biosynthesis. J Bacteriol. 1995;177:6688–6692. doi: 10.1128/jb.177.22.6688-6692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajgarhia V B, Strohl W R. Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis. J Bacteriol. 1997;179:2690–2696. doi: 10.1128/jb.179.8.2690-2696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan, M. J., J. A. Lotvin, N. Strathy, and S. E. Fantini. December 1996. U.S. patent 5,589,385.
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. 1 to 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Scotti C, Hutchinson C R. Enhanced antibiotic production by manipulation of the Streptomyces peucetius dnrH and dnmT genes involved in doxorubicin (adriamycin) biosynthesis. J Bacteriol. 1996;178:7316–7321. doi: 10.1128/jb.178.24.7316-7321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta S K. Inhibitors of DNA topoisomerases. In: Foye W O, editor. Cancer chemotherapeutic agents. Washington, D.C: American Chemical Society; 1995. pp. 205–217. [Google Scholar]
- 39.Strohl W R, Dickens M L, Rajgarhia V B, Woo A J, Priestly N D. Anthrcyclines. In: Strohl W R, editor. Biotechnology of antibiotics. 2nd ed. New York, N.Y: Marcel Dekker; 1997. pp. 577–657. [Google Scholar]
- 40.Stutzman-Engwall K J, Hutchinson C R. Multigene families for anthracycline antibiotic production in Streptomyces peucetius. Proc Natl Acad Sci USA. 1989;86:3135–3139. doi: 10.1073/pnas.86.9.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stutzman-Engwall K J, Otten S L, Hutchinson C R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi Y, Naganawa H, Takeuchi T, Umezawa H, Komiyama T, Oki T, Inui T. The structure of baumycins A1, A2, B1, B2, C1 and C2. J Antibiot. 1977;30:622–624. doi: 10.7164/antibiotics.30.622. [DOI] [PubMed] [Google Scholar]
- 43.Vara J A, Lewandowska-Skarbek M, Wang Y G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthetic pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber J M, Leung J O, Maine G T, Potenz R H B, Paulus T J, Dewitt J P. Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J Bacteriol. 1990;172:2372–2383. doi: 10.1128/jb.172.5.2372-2383.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13m18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]