Abstract
Transcription of the ferric citrate transport genes of Escherichia coli is induced by ferric citrate bound to the outer membrane receptor FecA. Additional ferric citrate-specific regulatory proteins are FecR in the cytoplasmic membrane and the FecI sigma factor in the cytoplasm. To further understand the assumed FecR-mediated signal transduction across the cytoplasmic membrane, the transmembrane topology of FecR (317 amino acids) was determined with hybrid proteins containing portions of FecR and mature BlaM β-lactamase. BlaM fused to FecR regions extending from residues 107 to 149 and residues 230 to 259 conferred high ampicillin resistance to cells, while BlaM fused to sites between residues 159 and 210 and between residues 265 and 301 conferred low resistance. Cells that synthesized FecR′-BlaM with fusion joints between residues 8 and 81 of FecR were fully sensitive to ampicillin. The ampicillin resistance of the low-resistance FecR′-BlaM hybrids was increased 2- to 10-fold by cosynthesis of plasmid-encoded GroEL GroES and SecB chaperones and in degP and ompT protease mutants, which suggested that the decreased ampicillin resistance level of these hybrids was caused by the formation of inclusion bodies and proteolytic degradation. Replacement of glycine by aspartate residues in the only hydrophobic FecR sequence (residues 85 to 100) abolished the β-lactamase activity of high-resistance FecR′-BlaM proteins, indicating that there are no other transmembrane regions in FecR that translocate BlaM into the periplasm independent of the hydrophobic sequence. All FecR′-BlaM proteins with at least 61 FecR residues complemented a fecR mutant such that it could grow on ferric citrate as the sole iron source and induced fecA-lacZ transcription independent of ferric citrate. The low resistance mediated by two FecR′-BlaM proteins in a fecA deletion mutant was increased 20-fold by transformation with a fecA-encoding plasmid. We propose that FecR spans the cytoplasmic membrane once, interacts in the periplasm with its C-terminal region with FecA occupied by ferric citrate, and transmits the information through the cytoplasmic membrane into the cytoplasm, where it converts FecI into an active sigma factor.
In Escherichia coli, citrate-mediated iron transport is catalyzed across the outer membrane by the FecA protein and the TonB, ExbB, and ExbD proteins, which presumably energize active transport across the outer membrane through the electrochemical potential of the cytoplasmic membrane. Transport of iron across the cytoplasmic membrane is mediated by an ATP binding cassette transporter, which consists of the periplasmic binding protein FecB, the hydrophobic membrane proteins FecC and FecD, and the FecE ATPase (23, 31).
The special feature of the ferric citrate transport system is the induction of the fecABCDE transport gene operon by ferric citrate, which does not have to be taken up into the periplasm or into the cytoplasm (4). By binding to the FecA outer membrane protein, ferric citrate triggers a signal that is transferred across the outer membrane, the periplasm, and the cytoplasmic membrane into the cytoplasm (10). The N terminus of FecA is located in the periplasm and is required for signaling but is dispensable for transport (16). As in transport, TonB, ExbB, ExbD, and the electrochemical potential of the cytoplasmic membrane are involved in signal transfer across the outer membrane (16). The FecR regulatory protein is required for the response of cells to ferric citrate (21, 36) and activates the FecI protein in the cytoplasm by an unknown mechanism; FecI functions as a sigma factor that directs the RNA polymerase to the promoter upstream of fecA (1, 6, 7, 22). In addition to induction of the fec transport genes by ferric citrate, transcription of the regulatory genes fecI and fecR and of the fec transport genes is repressed by iron via the Fur repressor (6, 13, 41), which binds to the promoter upstream of fecI fecR and of fecABCDE (2).
To understand the surface signaling mechanism of fec transport gene induction, it is important to know the transmembrane topology of the FecR protein. We constructed hybrid proteins between C-terminally truncated FecR derivatives and the BlaM β-lactamase and determined the fusion sites of FecR that indicated a periplasmic or a cytoplasmic location of BlaM. Furthermore, all of the FecR′-BlaM hybrid proteins that contained the fusion sites after the proposed transmembrane segment of FecR activated transcription and translation of fecA independent of ferric citrate. In addition, the ampicillin resistance of two FecR′-BlaM hybrid proteins was increased by FecA, suggesting a physical interaction of FecR with FecA.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and plasmids used in this study are listed in Table 1; also see Tables 2 to 4.
TABLE 1.
E. coli K-12 strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| 5K | hsdR mcrA ser thr lacY rpsL thi fhuA supE | This institute |
| AA93 | F−araD139 ΔlacU169 rpsL150 relA1 deoC1 flbB5301 ptsF25 rbsR aroB fecB::Mud1 (Ap lac) | 21 |
| BL2173 | F−hsdS gal, T7 RNA polymerase under lacUV5 control | 33 |
| WM1567 | Carries a heat-inducible T7 RNA polymerase gene on pGP1-2 | 34 |
| DH5α | Δ(lacZYA-argF)U169 endA recA1 hsR17(rK− mK+) supE44 thi-1 gyrA96 relA1 F′ φ80 ΔlacZ ΔM15 | Stratagene |
| IS176 | aroB malT tsx thi fecR lac::Tn10 | I. Kim |
| H2331 | araD139 ΔlacU169 rpsL150 relA1 deoC1 flbB5301 ptsF25 rbsR aroB Δfur | 14 |
| KS272 | Δlac-X74 galE galK rpsL ΔphoA | 32 |
| KS474 | Δlac-X74 galE galK rpsL ΔphoA degP41 | 32 |
| SG4008 | Δlac rpsL lon | This institute |
| UT5600 | azi-6 lacY1 leu-6 mtl-1 proC14 rpsL109 thi-1 trpE30 tsx67 ompP entA401 Δ(fepA-ompT) | C. F. Earhart |
| WA176 | aroB malT tsx thi fecR | 21 |
| Plasmids | ||
| pMa2-54 | ori ColE1, Apr | H. J. Fritz |
| pMc2-54 | ori ColE1, Cmr | H. J. Fritz |
| pT7-7 | ori ColE1, T7 gene 10 promoter, Apr | 34 |
| pSU18 | ori p15A, Cmr | 19 |
| pHSG576 | pSC101 derivate, Cmr | 35 |
| pJBS633 | ori ColE1 Tetr ′blaM, Neor | 5 |
| pAA70 | pT7-7 fecR Apr | 40 |
| pAK330 | pBR322 secB (under ptet control) | 17 |
| pBV25 | pMc2-54 fecIRA, Cmr | 36 |
| pIS127 | pSU18 fecR, Cmr | 21 |
| pIS135 | pHSG576 fecIR, Cmr | 21 |
| pIS1034 | pMLB1034 fecA fecA-lacZ, Apr | 21 |
| pMMO1034 | pMLB1034 fecA-lacZ, Apr | 21 |
| pMS2 | pHSG576 groEL groES, Cmr | R. Freudl |
| pSV6 | pHSG576 fec′RAB′, Cmr | 37 |
| pDW3 | pMa2-54 fecIRA′, Apr | This study |
| pDW10 | pMc2-54 fecI fecR(G91D G93D)fecA′, Cmr | This study |
| pDW20 | pJBS633 fecR, Neor | This study |
| pDW28.56 | pMLB1034 fecI fecR56-lacZ, Cmr | This study |
| pDW43 | pHSG576 fecI fecR blaM, Cmr | This study |
| pDW51.107 | pJBS633 fecR(Δ24–68)107-blaM, Neor | This study |
| pDW51.248 | pJBS633 fecR(Δ24–68)248-blaM, Neor | This study |
| pDW52.107 | pJBS633 fecR(Δ6–58)107-blaM, Neor | This study |
| pDW52.248 | pJBS633 fecR(Δ6–58)248-blaM, Neor | This study |
| pDW53.230 | pJBS633 fecR(Δ25–185)230-blaM, Neor | This study |
| pDW53.248 | pJBS633 fecR(Δ25–185)248-blaM, Neor | This study |
| pDW53.259 | pJBS633 fecR(Δ25–185)259-blaM, Neor | This study |
| pDW53.281 | pJBS633 fecR(Δ25–185)281-blaM, Neor | This study |
| pDW57.107 | pJBS633 fecR(G91D G93D)107-blaM, Neor | This study |
| pDW57.237 | pJBS633 fecR(G91D G93D)237-blaM, Neor | This study |
| pDW69 | pSU18 fecR, Cmr | This study |
| pDW71 | pSU18 fecR(Δ24–68), Cmr | This study |
| pDW72 | pSU18 fecR(Δ6–58), Cmr | This study |
| pDW73 | pSU18 fecR(Δ25–185), Cmr | This study |
| pDW102 | pHSG576 secB under ptet control | This study |
TABLE 2.
Properties of FecR′-BlaM hybrid proteins
| Plasmid | Fusion site at amino acid | Size of hybrid protein (kDa) | Ampicillin resistance (μg/ml) | Growth of WA176 | Cleavage of hybrid protein |
|---|---|---|---|---|---|
| pDW20 | 35.5 | 0 | + | + | |
| pDW26.08a | 8 | 29.9 | 0 | − | − |
| pDW26.21 | 21 | 31.5 | 0 | − | − |
| pDW26.25 | 25 | 31.8 | 0 | − | − |
| pDW26.61 | 61 | 35.8 | 0 | + | − |
| pDW26.75 | 75 | 37.4 | 0 | + | − |
| pDW26.78 | 78 | 37.7 | 0 | + | − |
| pDW26.81 | 81 | 38.0 | 0 | + | − |
| pDW26.107 | 107 | 41.0 | 200 | + | − |
| pDW26.112 | 112 | 41.5 | 50 | + | − |
| pDW26.127 | 127 | 43.2 | 100 | + | − |
| pDW26.129 | 129 | 43.5 | 50 | + | − |
| pDW26.133 | 133 | 43.9 | 100 | + | − |
| pDW26.138 | 138 | 44.5 | 50 | + | − |
| pDW26.149 | 149 | 45.7 | 50 | + | − |
| pDW26.159 | 159 | 46.8 | 0 | + | − |
| pDW26.172 | 172 | 47.9 | 5 | + | − |
| pDW26.191 | 191 | 50.4 | 5 | + | − |
| pDW26.209 | 209 | 52.4 | 0 | + | − |
| pDW26.210 | 210 | 52.5 | 0 | + | − |
| pDW26.230 | 230 | 54.8 | 100 | + | + |
| pDW26.237 | 237 | 55.8 | 100 | + | + |
| pDW26.245 | 245 | 56.4 | 200 | + | + |
| pDW26.248 | 248 | 56.8 | 100 | + | + |
| pDW26.257 | 257 | 57.8 | 100 | + | + |
| pDW26.259 | 259 | 58.0 | 50 | + | + |
| pDW26.265 | 265 | 58.7 | 10 | + | + |
| pDW26.274 | 274 | 59.7 | 5 | + | + |
| pDW26.279 | 279 | 60.2 | 5 | + | + |
| pDW26.281 | 281 | 60.4 | 5 | + | + |
| pDW26.301 | 301 | 62.7 | 5 | + | + |
In this and the following plasmids, the last number denotes the number of N-terminal amino acids of FecR fused to mature BlaM.
TABLE 4.
Expression of fecA-lacZ in E. coli AA93 Δfec
| Plasmid | Fusion site at amino acid: | β-Galactosidase activity (units)
|
|
|---|---|---|---|
| No citrate | With citrate | ||
| pHSG576 | 5 | 7 | |
| pIS135 | 6 | 502 | |
| pDW46.08 | 8 | 7 | 9 |
| pDW46.61 | 61 | 85 | 85 |
| pDW46.81 | 81 | 439 | 349 |
| pDW46.107 | 107 | 287 | 176 |
| pDW46.127 | 127 | 226 | 137 |
| pDW46.149 | 149 | 220 | 113 |
| pDW46.159 | 159 | 71 | 68 |
| pDW46.172 | 172 | 86 | 98 |
| pDW46.210 | 210 | 60 | 79 |
| pDW46.230 | 230 | 54 | 64 |
| pDW46.237 | 237 | 41 | 47 |
| pDW46.248 | 248 | 42 | 56 |
| pDW46.265 | 265 | 41 | 70 |
| pDW46.281 | 281 | 33 | 63 |
| pDW46.301 | 301 | 53 | 93 |
| pDW41 | FecR (G91D,G93D) | 81 | 65 |
Plasmid pDW3 was constructed by inserting an EcoRI-HindIII fecIRA′ fragment from pBV25 (36) into the vector plasmid pMa2-54 (30) digested with EcoRI and HindIII.
To create two specific mutations in the predicted transmembrane segment of the FecR protein, double nucleotide replacements were introduced into the fecR gene by the gapped-duplex DNA method (30) with the oligonucleotide 5′-GTTGCTCGACGCTGACGG-3′ (replacements are underlined). Plasmids pBV25 (36) and pDW3 were used as templates. The mutated fecR gene of the resulting plasmid, pDW10, was examined by fragment exchange with wild-type fecR in the regions not mutagenized and by sequencing of the mutagenized segment. Plasmid pDW20 was obtained by inserting the BglII-SalI fecR fragment with the ideal ribosome binding site of pAA70 (1) in the tetracycline resistance gene upstream of blaM (β-lactamase gene) of pJBS633 (5) cleaved with BamHI-SalI. fecR transcription was under the control of the tet promoter and of the phage T7 gene 10 promoter. The pDW26 series was derived from pDW20 by Bal 31 exonuclease digestion for various periods. The derivatives were blunt ended with Klenow polymerase I, cleaved with PvuII upstream of blaM, and religated. E. coli 5K was transformed with the resulting plasmids, which carry various fecR′-blaM gene fusions under the control of the tetracycline promoter (ptet) and the phage T7 gene 10 promoter and which contain an ideal ribosome binding site. pDW26.81 was constructed by deleting a PmaCI-PvuII fragment. pDW26.191 was constructed by amplifying a fecR fragment with the primers 5′-CTGGAGTATGGCATATGAATC-3′ and 5′-GCTGCGTAATATTATCC-3′; the fragment was digested with CelII and SspI and inserted in pDW20 cleaved with CelII and PvuII. Ampicillin-resistant colonies were spread as single cells on TY agar plates supplemented with increasing concentrations of ampicillin (5, 10, 25, 50, 100, 200, 400, 800, and 1,600 μg/ml).
pDW28.56 was constructed by cleavage of fecR with AviI (position 1919 of the published sequence [36]) and cloned into the XmaI site of the lac fusion vector pMLB1034 (29). pDW41 contains the CelII-PstI fecR(G91D G93D) fragment of pDW10 inserted into pIS135 cleaved with CelII and PstI.
To construct the plasmids of the pDW46 series, a blaM fragment was synthesized by PCR with the primers 5′-CTCAAGGATCTTACCGC-3′ and 5′-GAGCTCAAAAAGAGCTCCGGGGTCTGACGCTCAGTG-3′; the fragment was then digested with PstI and SacI and inserted into pIS135 cleaved with PstI and HindIII, resulting in pDW43. The plasmids of the pDW26 series were digested with HpaI (pDW26.08) or CelII and PstI, and the fecR′-blaM fragments were inserted into pDW43 cleaved with HpaI or CelII and with PstI, yielding the pDW46 series. Gene fusions on the low-copy-number plasmids (pDW46 series) were tested with 2.5, 5, 7.5, 10, 25, and 50 μg of ampicillin/ml. pDW51.107 and pDW51.248 resulted from the insertion of the XbaI-PmaCI fragment of pDW71 into pDW26.107 or pDW26.248 digested with PmaCI and XbaI. pDW52.107 and pDW52.248 contained the XbaI-PmaCI fragment of pDW72 inserted into pDW26.107 and pDW26.248, respectively.
The pDW53 series was constructed by insertion of the XbaI-PstI fragment of pDW73 into pDW26.230, pDW26.248, pDW26.259, or pDW26.281, cleaved with XbaI and PstI.
pDW57.107 and pDW57.237 contain the CelII-Cfr10I fecR(G91D G93D) fragment of pDW10 inserted into pDW26.107 and pDW26.237 digested with CelII and Cfr10I. pDW69 contains the BglII-PstI fecR′ fragment of pAA70 inserted into pIS127 digested with BamHI and PstI.
pDW71 was derived from pDW69 which was cleaved with CelII and BglI, blunt ended with Klenow polymerase I, and religated. pDW72 was derived from pDW69, which was cleaved with HpaI and BspMI, blunt ended, and religated. pDW73 resulted from a fecR fragment synthesized by PCR with the primers 5′-GGGACAGAGCTGAGCGTCCGC-3′ and 5′-CTGGAGTATGGCATATGAATC-3′; the fragment was digested with CelII and HindIII and inserted into pDW69 digested with CelII and HindIII.
pDW102 was constructed by insertion of the EcoRI-MluNI fragment (secB under tet promoter control) of pAK330 into pHSG576 digested with EcoRI and HindIII.
Media.
Cells were grown in TY medium, which contained (per liter) 8 g of tryptone (Difco Laboratories), 5 g of yeast extract, and 5 g of NaCl (pH 7). Growth on ferric citrate as the sole iron source was tested on Fec agar plates, which contained (per liter) 8 g of nutrient broth, 5 g of NaCl, 15 g of nutrient agar, 0.2 mM 2,2-dipyridyl, and 1 mM sodium citrate (pH 7). The antibiotics ampicillin (50 μg/ml), chloramphenicol (25 μg/ml for cells containing low-copy-number plasmids and 40 μg/ml for those containing high-copy-number plasmids), neomycin (50 μg/ml), and tetracycline (15 μg/ml) were added as required.
Recombinant DNA techniques.
DNA isolation from bacteria, recovery of DNA fragments from agarose gels, cloning of restriction fragments, and transformation were done by standard methods (27). All of the constructed gene fusions, deletions, nucleotide replacements, and PCR-amplified DNA fragments were sequenced by the enzymatic dideoxy chain termination method (28) with [35S]dATP (Amersham) or fluorescein-15dATP for labeling. The fluorescein-labeled reaction products were analyzed with an A.L.F. DNA sequencer (Pharmacia Biotech, Freiburg, Germany). The primer blaM (5′-CTGGTGCACCCAACTGA-3′) was complementary to codons 15 to 21 of mature β-lactamase, and other primers complementary to various regions of fecIR were also used. DNA was amplified by PCR, using the Gene Amp process with Taq polymerase (Promega, Madison, Wis.) as described previously (21).
Determination of β-galactosidase activity.
β-Galactosidase activity was determined by the method of Miller (20) and Giacomini et al. (9). Cells were grown to the exponential growth phase in M9 medium supplemented with vitamin-free Casamino Acids (Difco Laboratories) and 0.1 mg each of tryptophan, phenylalanine, and tyrosine per ml. The media were supplemented, as indicated, with sodium citrate (1 mM) and 2,2-dipyridyl (50 μM).
Overexpression of FecR′-BlaM hybrid proteins.
The fecR′-blaM gene fusions were transcribed by the T7 RNA polymerase encoded on the chromosome of E. coli BL2173. Cells were grown in 2 ml of TY medium at 37°C to the exponential growth phase (optical density at 578 nm, 0.5), and then transcription by the RNA polymerase was induced for 1 h with 2 mM isopropyl-β-d-thiogalactoside (IPTG).
Detection of inclusion bodies.
Plasmids with fecR and fecR′-blaM fusions were transcribed by the temperature-inducible phage T7 RNA polymerase encoded by plasmid pGP1-2, which was contained in E. coli WM1576 (K38) (34). Overexpression of the FecR derivatives and labeling with [35S]methionine were performed by the method of Fischer et al. (8). Spheroplasts were prepared by suspending cells in 500 μl of ice-cold 0.2 M Tris-HCl (pH 8.0)–0.5 M sucrose. The cells were treated for 15 min on ice with 50 μl of lysozyme (5 mg/ml in 50 mM Tris-HCl [pH 8.0]), 50 μl of 5 mM EDTA, and 500 μl of 0.2 M Tris-HCl (pH 8.0)–0.5 mM EDTA. Spheroplasts were collected by centrifugation for 30 min at 15,000 × g in an Eppendorf centrifuge. The proteins in the supernatant were precipitated with 0.5 volume of 30% trichloroacetic acid for 30 min on ice, pelleted for 15 min, washed with 100 μl of acetone and 100 μl of acetone-water (1:1), and then suspended in 20 μl of sodium dodecyl sulfate (SDS) buffer. The sedimented spheroplasts were suspended in 500 μl of H2O, 3 μl of DNase (2 μg/ml), and 25 μl of 1 M MgSO4. After 30 min at room temperature, the spheroplasts were lysed by three cycles of freezing to −80°C and thawing. The lysate was centrifuged for 15 min at 600 × g, and the sediment was suspended in 20 μl of SDS-polyacrylamide gel electrophoresis (PAGE) lysis buffer. The supernatant was centrifuged at 6,000 × g and again at 14,000 × g. Proteins of the 14,000 × g supernatant were precipitated with trichloroacetic acid (final concentration, 10%) and washed with acetone and acetone-water.
SDS-PAGE.
Cells and proteins in the various cell fractions were dissolved in lysis buffer in a boiling-water bath for 2 min. Proteins were separated by SDS-PAGE (15% polyacrylamide) (18) and identified by staining with Serva Blue (Serva, Heidelberg, Germany).
Computer-assisted analysis.
Protein and nucleic acid sequences were analyzed with PC/Gene, release 6.01 (IntelliGenetics Inc., Mountain View, Calif.), Husar and Phd (EMBL, Heidelberg, Germany), and TMbase (Swiss Institute of Experimental Cancer Research, Lausanne, Switzerland) (12).
RESULTS
Determination of the transmembrane topology of the FecR protein.
To determine the arrangement of the FecR protein in the cytoplasmic membrane of E. coli, hybrid proteins between FecR and the mature TEM β-lactamase, lacking its own signal sequence, were constructed. Since only periplasmic β-lactamase confers resistance to ampicillin, it is inferred that hybrid proteins which render cells ampicillin resistant contain β-lactamase fused to sites in FecR that are exposed to the periplasm. This method was developed by Broome-Smith and Spratt (5) and has been successfully used for the determination of the transmembrane topology of a number of proteins (3, 15, 24). Plasmid pDW20 (fecR) was cleaved with SalI, digested for various periods with Bal 31 exonuclease, blunt ended, cleaved with PvuII upstream of blaM, religated, and transformed into E. coli 5K. For selection of inframe fusions, neomycin-resistant colonies were streaked on nutrient agar plates containing ampicillin (200 μg/ml). The ampicillin resistance was tested with single cells on agar plates containing ampicillin concentrations from 5 to 1,600 μg ml−1. The fusion sites between fecR and blaM were determined by DNA sequencing.
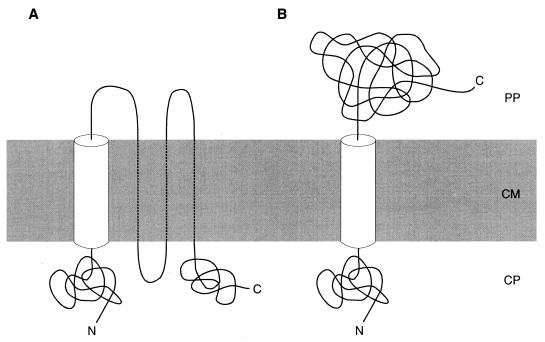
The level of ampicillin resistance conferred by the 30 isolated FecR′-BlaM hybrid proteins was tested in E. coli 5K in which the fecR-blaM genes were transcribed under the control of the tet promoter. FecR′-BlaM proteins that contained up to the first 81 amino acids of FecR conferred no ampicillin resistance (Table 2). Most hybrid proteins containing at least 107 amino acids of FecR rendered cells ampicillin resistant. When expressed in E. coli AA93 Δfec, the few ampicillin-sensitive transformants observed in E. coli 5K were resistant to 5 μg of ampicillin ml−1 (data not listed in Table 2). This result is consistent with the proposal that the hydrophobic sequence (boldface letters) RLTRRHVMKGLLLLGAGGGWQLWQSE between residues 85 and 100 spans the cytoplasmic membrane. The hydrophobic sequence is preceded by positively charged amino acids that localize the N terminus on the inside, according to the “positive-inside rule” of von Heijne (38). Hybrid proteins containing portions of FecR between residues 159 and 210 and between residues 265 and 301 conferred either no ampicillin resistance or only a low-level resistance (to 5 μg of ampicillin/ml), which is generally considered indicative of a cytoplasmic location of the fusion site. According to these data, FecR would traverse the cytoplasmic membrane four times (Fig. 1A). Increased transcription by the T7 RNA polymerase in E. coli BL2173, transformed with plasmids of the pDW26 series, of those fecR-blaM genes that conferred low resistance resulted in ampicillin resistance levels no higher than those listed in Table 2.
FIG. 1.
Alternative topologies of FecR in the cytoplasmic membrane showing four membrane-spanning segments (A) and one membrane-spanning segment (B).
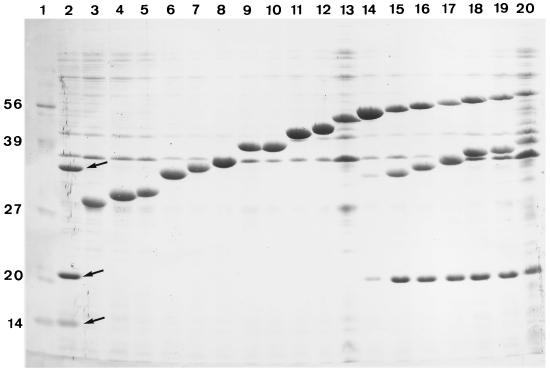
Since FecR contains no additional hydrophobic sequences that suggest further transmembrane segments, the amounts of the FecR-BlaM proteins were estimated by SDS-PAGE to determine whether the lack of ampicillin resistance was caused by low synthesis or degradation of the hybrid proteins. Each of the fecR-blaM genes was selectively and highly transcribed by the T7 RNA polymerase in E. coli BL2173 transformed with 18 of the 30 fecR-blaM genes. The FecR′-BlaM proteins showed electrophoretic mobilities that corresponded to the calculated molecular masses (Fig. 2, lanes 3 to 20). From the FecR protein (35.5 kDa), two fragments of 20 and 15 kDa were observed (lane 2); this agreed with the previously determined cleavage between residues 181 and 182, which results in fragments of 20.2 and 15.3 kDa (40). Cleavage of the FecR′-BlaM proteins did not occur unless the FecR portion was at least 230 residues long (lane 14). FecR230-BlaM conferred a high ampicillin resistance, as did five other cleaved FecR-BlaM proteins, whereas some uncleaved FecR-BlaM proteins with shorter FecR portions and some cleaved FecR-BlaM proteins with longer FecR portions conferred no or very low-level ampicillin resistance, indicating no correlation between the ampicillin resistance levels and susceptibility to cellular proteolysis.
FIG. 2.
Stained gel after SDS-PAGE of FecR′-BlaM hybrid proteins carrying fusions at residues 8, 21, 25, 61, 75, 81, 127, 133, 159, 172, 209, 230, 237, 241, 259, 274, 281, and 301 of FecR (lanes 3 to 20, respectively). The proteins were synthesized in E. coli BL2173 in which the hybrid genes were transcribed by the phage T7 RNA polymerase, whose synthesis was induced by 2 mM IPTG. Lane 1, standard proteins of 14.3, 20.1, 26.6, 39.2, 55.6, and 85.2 kDa. Lane 2, FecR (35.5 kDa) and its 20.2-kDa N-terminal and 15.3-kDa C-terminal degradation fragments (arrows). FecR230-BlaM and the larger hybrid proteins were proteolytically cleaved.
To determine FecR′-BlaM protein concentrations under the same conditions as those at which ampicillin resistance was tested, the hybrid proteins synthesized in E. coli 5K fecR-blaM transformants were analyzed by SDS-PAGE. Much less of the proteins were synthesized when the gene was under the control of the tet promoter than after transcription by the T7 RNA polymerase, but the FecR-BlaM proteins were still the most dominant proteins in the cells (data not shown). After fractionation of disrupted cells by differential centrifugation for 15 min at 600, 7,000, and 14,000 × g, most of FecR and the FecR-BlaM proteins were in the 600 × g fraction, which indicates the formation of inclusion bodies (data not shown). The fraction of FecR-BlaM proteins correctly inserted in the cytoplasmic membrane increased with the increase in the total protein but was not strictly proportional to the amount of FecR-BlaM synthesized.
To avoid the formation of inclusion bodies, the fecR-blaM genes, together with the fecI gene, were cloned into the low-copy-number vector pHSG576 (five copies per cell [35]). Transcription from the fecI promoter and translation with the natural ribosome binding site yielded very low ampicillin resistance levels (5 μg/ml) conferred by the FecR′-BlaM proteins that rendered cells highly resistant when overexpressed (Table 2). Transfer of the fecR-blaM plasmids (pDW46 series listed in Table 4) into E. coli H2331 Δfur to circumvent iron repression increased ampicillin resistance two- to fourfold, but the differences in the resistance of the strains remained as listed in Table 2 (data not shown). In particular, the FecR′-BlaM hybrids that conferred low-level resistance (5 μg of ampicillin/ml) when highly expressed (Table 2) and no resistance when moderately expressed remained sensitive when expressed in the Δfur strain. Derepression of fecR-blaM transcription gave no hints why certain transformants were only weakly resistant.
Increase of FecR-BlaM activity in protease-negative and chaperone-overproducing cells.
The E. coli 5K fecR-blaM transformants that were resistant to 5 μg of ampicillin/ml were clearly not fully sensitive, suggesting that secretion of BlaM into the periplasm by the FecR fragments had occurred (Table 2). The corresponding fecR-blaM transformants of E. coli AA93 Δfec all showed resistance to 5 μg of ampicillin/ml, and none were fully sensitive (except E. coli AA93 carrying fecR8-blaM to fecR81-blaM and fecR). Therefore, we examined whether ampicillin resistance could be increased in cells transformed with plasmids encoding chaperones or in mutants lacking certain proteases. Three of the low-resistance FecR′-BlaM proteins and, as controls, a high-resistance FecR′-BlaM (FecR107-BlaM) and a sensitive FecR-BlaM (FecR78-BlaM) (encoded on plasmids of the pDW26 series) were synthesized in E. coli 5K together with plasmid-encoded GroEL GroES (pMS2) or SecB (pDW102). Most transformants grown at 28°C displayed a twofold-higher resistance in the presence of overexpressed GroEL GroES and overexpressed SecB (Table 3). The same results were obtained with GroEL GroES at 37°C. Cells grown at 42°C exhibited a fourfold-reduced resistance compared to cells grown at 28 and 37°C, and this resistance was not increased by the chaperones. DnaK did not increase ampicillin resistance (data not shown).
TABLE 3.
FecR-BlaM activity in protease mutants and chaperone-overproducing cells
| Hybrid protein | Activitya in strain:
|
|||||
|---|---|---|---|---|---|---|
| KS272 (degP+) | KS474 (degP) | UT5600 (ompT) | 5K | 5K(pMS2) (groEL groES) | 5K(pDW102) (secB) | |
| FecR78-BlaM | 0 | 0 | 0 | 0 | 0 | 0 |
| FecR107-BlaM | 200 | 800 | 800 | 200 | 200 | 400 |
| FecR159-BlaM | 5 | 10 | 10 | 0 | 10 | 10 |
| FecR191-BlaM | 10 | 50 | 25 | 5 | 10 | 10 |
| FecR210-BlaM | 5 | 50 | 10 | 5 | NDb | ND |
| FecR265-BlaM | 10 | 25 | 50 | 10 | 25 | 25 |
The values denote the highest ampicillin concentrations (micrograms per milliliter) in nutrient agar plates on which the cells formed single colonies at 28°C.
ND, not determined.
fecR-blaM transformants of degP and ompT protease mutants displayed 2- to 10-fold-higher ampicillin resistance levels than did the protease-containing parent cells (Table 3). The ampicillin resistance of a lon protease mutant carrying the fecR-blaM genes listed in Table 3 was not increased. With proteases, as with the chaperones, ampicillin resistance was increased only in the fecR-blaM E. coli 5K and E. coli AA93 transformants that showed resistance to at least 5 μg of ampicillin/ml. The ampicillin-sensitive transformants in the protease mutants that synthesized BlaM fused to the N-terminal cytoplasmic FecR portion in front of the hydrophobic segment remained fully sensitive.
Evidence against additional transmembrane segments in FecR.
If FecR contains transmembrane segments in addition to the hydrophobic sequence between residues 85 and 100, BlaM could be secreted into the periplasm by FecR-BlaM proteins lacking residues 85 to 100. Residues 25 to 185 were deleted in the high-resistance proteins FecR230-BlaM, FecR248-BlaM, and FecR259-BlaM and in the low-resistance protein FecR281-BlaM (Table 1, pDW53 series). All transformants of E. coli 5K were completely sensitive to ampicillin, which suggests that there is no transmembrane region beyond residue 186 of FecR that secretes BlaM independent of residues 85 to 100. To confirm this conclusion, Gly-91 and Gly-93 were replaced by Asp in the high-resistance proteins FecR107-BlaM and FecR237-BlaM. E. coli 5K carrying the resulting plasmids pDW57.107 and pDW57.237 were fully sensitive to ampicillin. Sensitivity was not caused by the lack of the proteins since, as will be shown below, a FecR protein containing the double amino acid replacement displayed regulatory activities.
Regulatory activities of FecR-BlaM hybrid proteins.
E. coli WA176 fecR does not grow on minimal agar with ferric citrate as the sole iron source (Fec plates) (39) since fecR contains a stop codon giving rise to a truncated FecR of only 18 N-terminal amino acids (21). All FecR′-BlaM proteins with at least 61 FecR residues restored growth of E. coli WA176 fecR on Fec plates (Table 2); this indicates that the fec transport genes were expressed when the FecR protein had a minimal size. To study the regulatory activity of the FecR′-BlaM proteins, some of the fecR-blaM fusion genes were transformed into E. coli AA93 Δfec, which carries a plasmid-encoded fecA-lacZ gene fusion. Of the genes required for the induction of fec transport gene transcription, fecA was carried on the fecA-lacZ plasmid, fecI was carried on the fecR-blaM plasmids, and tonB, exbB, and exbD were on the chromosome.
Transcription and translation of fecA-lacZ occurred when FecR contained 61 or more residues (Table 4). Expression of fecA-lacZ was independent of ferric citrate in the growth medium and was higher with shorter FecR portions in the FecR′-BlaM proteins than with longer FecR portions. In contrast, expression of fecA-lacZ in cells carrying wild-type fecR occurred only in the presence of ferric citrate (Table 4, pIS135). The somewhat lower fecA-lacZ expression in the presence of ferric citrate in cells containing the fecR-blaM genes than in the absence of ferric citrate may be caused by an improved iron supply, which results in a partial repression of fecA-lacZ and fecR-blaM transcription by Fe2+-Fur. However, iron uptake was not mediated by the ferric citrate transport system, which is lacking in strain AA93, but by an undefined low-affinity system, which takes advantage of the solubilization of iron by citrate. Apparently, N-terminal FecR fragments fused to BlaM activated the FecI sigma factor in the absence of the inducer. Results similar to those of the fecR-blaM fusions were obtained with fecR-lacZ gene fusions, of which the smallest active hybrid protein contained 56 N-terminal FecR residues and transcribed fecR-lacZ independently of ferric citrate. In E. coli DH5α Δlac, fecR56-lacZ on pDW28.56 is under the control of the fecI gene promoter located upstream of fecR. Since fecI fecR transcription is repressed by Fe2+-Fur, 10 U of β-galactosidase was measured under iron-replete conditions in nutrient broth and 49 U was obtained under iron-depleted conditions in nutrient broth supplemented with 0.2 mM dipyridyl.
FecR(G91D G93D), which, according to the ampicillin sensitivity of the FecR107-BlaM and FecR237-BlaM hybrid proteins, remains in the cytoplasm and does not insert into the cytoplasmic membrane, also increased fecA-lacZ expression independently of ferric citrate (Table 4).
To corroborate the importance of the FecR N terminus in FecI-mediated fecA transcription, residues 6 to 58 or residues 24 to 68 of FecR were deleted. E. coli WA176 transformed with pDW72 (FecRΔ6–58) or pDW71 (FecRΔ24–68) could not grow on Fec plates. No β-galactosidase was synthesized in E. coli IS176 fecR lacZ(pMMO1034 fecA-lacZ) transformed with either pDW71 or pDW72, showing that the intact N terminus of FecR is required for induction of fecA transcription and translation. The N-terminal region was not essential for FecR insertion into the cytoplasmic membrane, since the same deletions introduced into the high-resistance protein FecR107-BlaM reduced the ampicillin resistance of E. coli 5K from 200 to only 100 μg of ampicillin/ml. The deletions in FecR248-BlaM reduced resistance from 100 to 50 μg of ampicillin/ml [FecR(Δ24–68)248-BlaM] and to 25 μg ampicillin/ml [FecR(Δ6–58)248-BlaM].
Evidence that FecR interacts with FecA.
FecR248-BlaM and FecR257-BlaM conferred on E. coli 5K fec+(pDW26.248) and E. coli 5K(pDW26.257) resistance to 100 μg of ampicillin/ml, but they conferred on E. coli AA93 Δfec only resistance to 5 and 10 μg of ampicillin/ml, respectively. To examine whether this difference is caused by the lack of the FecA protein in strain AA93, E. coli AA93(pDW26.248) and E. coli AA93(pDW26.257) were transformed with pSV6fecA. Both transformants were resistant to 100 μg of ampicillin/ml. The strongly increased BlaM activity may indicate interaction between FecA and the FecR-BlaM proteins. Interaction between FecA and FecR would involve a region preceding residue 248 of FecR directly, or this region influences a FecR binding site. E. coli 5K synthesizing FecR259-BlaM was resistant to 50 μg of ampicillin/ml, and E. coli AA93 FecR259-BlaM was resistant to 5 μg/ml. Transformation with fecA increased the resistance of strain AA93 to only 10 μg/ml. The other FecR′-BlaM hybrids in E. coli AA93 were not affected by fecA.
DISCUSSION
The presence of only one hydrophobic sequence suggested that FecR consists of three topologically distinct regions extending from residues 1 to 84 in the cytoplasm, 85 to 100 within the cytoplasmic membrane, and 101 to 317 in the periplasm. Additional transmembrane regions were not obvious by inspection of the amino acid sequence and were not predicted by various computer-assisted membrane protein analysis programs (12, 26). The full ampicillin sensitivity of cells that synthesized BlaM fused to one of seven FecR sites from the N terminus up to residue 81 was consistent with this prediction. The unexpected finding was that BlaM fused to FecR residues beyond residue 107 did not confer a similar high resistance but, instead, conferred a pattern of high, low, high, low resistance. Interpretation of this finding has to take into account intracellular precipitation of the FecR′-BlaM proteins. All FecR′-BlaM proteins formed inclusion bodies, and an extensive analysis by differential centrifugation and SDS-PAGE of the amounts of protein present in the cell debris, the membrane fraction, and the soluble fraction showed that most of FecR′-BlaM was in the cell debris (inclusion bodies), regardless of whether larger or smaller amounts of hybrid proteins were synthesized. A decrease in the amount of the FecR′-BlaM proteins by using the weak natural ribosome binding site or through transcription by uninduced amounts of the T7 RNA polymerase did not prevent the formation of the inclusion bodies. Only the FecR′-BlaM proteins that conferred high-level resistance when strongly overexpressed conferred low-level resistance when they were weakly synthesized by cloning into a low-copy-number vector. Attempts to reduce the precipitation of FecR′-BlaM by overexpression of the chaperones GroEL GroES (11) and SecB (25) resulted in an increased periplasmic β-lactamase activity of all FecR′-BlaM proteins tested. Presumably, by binding of FecR′-BlaM to the chaperones, kinetic competition between insertion into the cytoplasmic membrane and precipitation was shifted in favor of insertion. Moreover, proteolysis of FecR′-BlaM decreased periplasmic β-lactamase activity since degP and ompT mutants carrying FecR′-BlaM proteins were resistant to ampicillin concentrations higher than that of the corresponding degP and ompT wild-type strains. Both the chaperones and the mutations in the proteases increased the β-lactamase activity of the FecR′-BlaM proteins that conferred only resistance to 5 μg of ampicillin/ml—and therefore would tentatively be localized in the cytoplasm—to levels which indicate a periplasmic location of BlaM. Ampicillin-sensitive cells that synthesized BlaM fused to FecR′ fragments not larger than 81 N-terminal residues remained sensitive in the presence of the overexpressed chaperones and in the absence of the DegP and OmpT proteases. These results make it likely that cytoplasmic β-lactamase did not confer the low-level ampicillin resistance.
Point mutations that replaced two glycine residues in the hydrophobic region (residues 85 to 100) of FecR′-BlaM with hydrophilic, charged aspartate residues converted ampicillin-resistant cells to ampicillin-sensitive cells, presumably by abolition of BlaM translocation into the periplasm. This result shows that there are no other FecR transmembrane segments that can translocate BlaM into the periplasm independent of the hydrophobic region. Taken together, these data are consistent with a topology model of FecR that proposes the N-terminal residues 1 to 84 in the cytoplasm, residues 85 to 100 in the cytoplasmic membrane, and residues 101 to 317 in the periplasm (Fig. 1B).
The transmembrane model of FecR agrees with the finding that cells that synthesize FecR′-BlaM proteins containing 61 or more FecR residues transcribe fecA-lacZ constitutively. This should occur only if the FecR N terminus is located in the cytoplasm, because the N-terminal FecR fragments can interact with the cytoplasmic FecI and convert FecI to an active sigma factor. BlaM fused to the FecR′ fragments does not seem to interfere sterically with FecR′ activity. Alternatively, BlaM could be proteolytically cleaved from a small proportion of FecR′-BlaM, below the detection limit of SDS-PAGE, and the free FecR′ molecules would be active. The smallest FecR fragment that supports growth on ferric citrate contained 56 N-terminal residues (FecR56-LacZ). Ferric citrate-independent transcription induction by C-terminally truncated FecR′ fragments, of which the smallest derivative contained 68 residues, has been shown previously (21). The 3′-deleted fecR genes were cloned on the same low-copy-number vector (pHSG576) as the fecR-blaM genes. The levels of constitutive fecA-lacZ transcription and translation caused by FecR′ and FecR′-BlaM were very similar, which argues against the need of a proportion of FecR′ from FecR′-BlaM to be released in order to exert activity. The largest FecR′ studied contained 273 residues, displayed activation in the absence of ferric citrate, and did not respond to ferric citrate (21).
Evidence of an interaction of FecA with FecR was obtained with two FecR′-BlaM hybrids. FecR248-BlaM conferred to E. coli AA93 Δfec only 5% of the ampicillin resistance conferred to E. coli 5K, and FecR257-BlaM conferred only 10%. Transformation of E. coli AA93 with a fecA plasmid restored ampicillin resistance to 100%. The increase of β-lactamase activity could be caused by stabilization of membrane-inserted FecR′-BlaM through binding to FecA or, less likely, by an increase in the amount of membrane-inserted FecR′-BlaM through cosynthesis with FecA.
The transmembrane topology of FecR makes FecR a suitable candidate for transduction of the signal, initiated by binding of ferric citrate to FecA, across the cytoplasmic membrane into the cytoplasm.
ACKNOWLEDGMENTS
We thank A. Angerer and K. Hantke for helpful advice and K. A. Brune for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 323, project B1).
REFERENCES
- 1.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new sub-family of ς70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;19:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 2.Angerer, A., and V. Braun. Iron regulates transcription of the ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol., in press. [DOI] [PubMed]
- 3.Bowler L D, Spratt B G. Membrane topology of penicillin-binding protein 3 of Escherichia coli. Mol Microbiol. 1989;3:1277–1286. doi: 10.1111/j.1365-2958.1989.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 4.Braun V. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch Microbiol. 1997;167:325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 5.Broome-Smith J K, Spratt B G. A vector for construction of translational fusions to TEM β-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 6.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enz S, Braun V, Crosa J. Transcription of the region encoding the ferric dicitrate-transport system in Escherichia coli: similarity between promoters for fecA and for extracytoplasmic function sigma factors. Gene. 1995;163:13–19. doi: 10.1016/0378-1119(95)00380-o. [DOI] [PubMed] [Google Scholar]
- 8.Fischer E, Strehlow B, Hartz D, Braun V. Soluble and membrane bound ferrisiderophore reductases of Escherichia coli K-12. Arch Microbiol. 1990;153:329–336. doi: 10.1007/BF00249001. [DOI] [PubMed] [Google Scholar]
- 9.Giacomini A, Corich B, Ollero F J, Squartini A, Nuti M P. Experimental conditions may affect reproducibility of the β-galactosidase assay. FEMS Microbiol Lett. 1992;100:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- 10.Härle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of Escherichia coli ferric dicitrate transport system from the cell surface. EMBO J. 1995;14:1430–1438. doi: 10.1002/j.1460-2075.1995.tb07129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayer-Hartl M K, Weber F, Hartl F U. Mechanism of chaperonin action: GroES binding and release can drive GroEL-mediated protein folding in the absence of ATP hydrolysis. EMBO J. 1996;15:6111–6121. [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann H, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 13.Hussein S, Hantke K, Braun V. Citrate-dependent iron transport system in Escherichia coli K-12. Eur J Biochem. 1981;117:431–437. doi: 10.1111/j.1432-1033.1981.tb06357.x. [DOI] [PubMed] [Google Scholar]
- 14.Kammler M, Schön C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kampfenkel K, Braun V. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1993;268:6050–6057. [PubMed] [Google Scholar]
- 16.Kim I, Stiefel A, Plantör S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 17.Kumamoto C A, Nault A K. Characterization of Escherichia coli protein-export gene secB. Gene. 1989;75:167–175. doi: 10.1016/0378-1119(89)90393-4. [DOI] [PubMed] [Google Scholar]
- 18.Lugtenberg B, Meiherrs H, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the major outer membrane protein of E. coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 19.Martinez E, Bartolome B, de la Cruz F. pACY184-derived cloning vectors containing the multiple cloning site and lacZ reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Ochs M, Veitinger S, Kim I, Welz D, Angerer A, Braun V. Regulation of citrate dependent iron transport system in Escherichia coli: FecR is required for transcriptional activation by FecI. Mol Microbiol. 1995;15:119–132. doi: 10.1111/j.1365-2958.1995.tb02226.x. [DOI] [PubMed] [Google Scholar]
- 22.Ochs M, Angerer A, Enz S, Braun V. Surface signaling in transcription regulation of the ferric citrate transport system of Escherichia coli: mutational analysis of the alternative sigma factor FecI supports its essential role in fec transport gene transcription. Mol Gen Genet. 1996;250:455–465. doi: 10.1007/BF02174034. [DOI] [PubMed] [Google Scholar]
- 23.Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–2724. doi: 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prinz W A, Beckwith J. Gene fusion analysis of membrane protein topology: a direct comparison of alkaline phosphatase and β-lactamase fusions. J Bacteriol. 1994;176:6410–6413. doi: 10.1128/jb.176.20.6410-6413.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randall L L, Topping T B, Hardy S J S, Pavlov M Y, Freistroffer D V, Ehrenberg M. Binding of SecB to the ribosome-bound polypeptides has the same characteristics as binding to full-length, denatured proteins. Proc Natl Acad Sci USA. 1997;94:802–807. doi: 10.1073/pnas.94.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao J K M, Argos P. A conformational preference parameter in integral membrane proteins. Biochim Biophys Acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook F, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA. 1977;72:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silhavy T S, Bermann M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 30.Stanssens P, Opsomer C, McKeown Y M, Kramer W, Zabeau M, Fritz H J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989;17:4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staudenmaier H, Van hove B, Yaraghi Z, Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989;171:2626–2633. doi: 10.1128/jb.171.5.2626-2633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2667–2674. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier F W, Moffat B. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vector for lacZ-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 36.Van Hove B, Staudenmaier H, Braun V. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J Bacteriol. 1990;172:6749–6758. doi: 10.1128/jb.172.12.6749-6758.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veitinger S, Braun V. Localization of the entire fec region at 97.3 minutes on the Escherichia coli chromosome. J Bacteriol. 1992;174:3838–3839. doi: 10.1128/jb.174.11.3838-3839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagegg W, Braun V. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J Bacteriol. 1981;145:156–163. doi: 10.1128/jb.145.1.156-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wriedt K, Angerer A, Braun V. Transcriptional regulation from the cell surface: conformational change in the transmembrane protein FecR lead to altered transcription of the ferric citrate genes in Escherichia coli. J Bacteriol. 1995;177:3320–3322. doi: 10.1128/jb.177.11.3320-3322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann L, Hantke K, Braun V. Exogenous induction of the iron dicitrate transport system of Escherichia coli K-12. J Bacteriol. 1984;159:271–277. doi: 10.1128/jb.159.1.271-277.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]