Abstract
Elevation of anabolism and concurrent suppression of catabolism are critical metabolic adaptations for muscular hypertrophy in response to resistance exercise (RE). Here, we investigated if RE-induced muscular hypertrophy is acquired by modulating a critical catabolic process autophagy. Male Wistar Hannover rats (14 weeks old) were randomly assigned to either sedentary control (SC, n = 10) or resistance exercise (RE, n = 10). RE elicited significant hypertrophy of flexor digitorum profundus (FDP) muscles in parallel with enhancement in anabolic signaling pathways (phosphorylation of AKT, mTOR, and p70S6K). Importantly, RE-treated FDP muscle exhibited a significant decline in autophagy evidenced by diminished phosphorylation levels of AMPK, a decrease in LC3-II/LC3-I ratio, an increase in p62 level, and a decline in active form of lysosomal protease CATHEPSIN L in the absence of alterations of key autophagy proteins: ULK1 phosphorylation, BECLIN1, and BNIP3. Our study suggests that RE-induced hypertrophy is achieved by potentiating anabolism and restricting autophagy-induced catabolism.
Keywords: Resistance exercise, Autophagy, Skeletal muscle, Hypertrophy
Introduction
Resistance exercise (RE) is a powerful intervention to enhance muscular strength as well as hypertrophy [1] and thus has been used as a potent avenue to prevent muscular atrophy [2, 3]. Although accumulating evidence has suggested that anabolism is enhanced in response to RE [4, 5], the molecular signaling nexus of hypertrophic pathways has not been clearly established yet. Nevertheless, activation of anabolic signaling axis (AKT-mTOR-p70S6K) has emerged as a potent trigger of muscular hypertrophy in response to RE [6–8]. More specifically, RE consisting of both a lengthening and shortening phase of muscular contraction (muscular mechanical stress) increases the release of growth hormones and growth factors (i.e., IGF-I), which stimulates receptor tyrosine kinases. These in turn activate AKT by PIP3 (phosphatidylinositol triphosphate) generated via PI3K activation. Then, the activated AKT relays the anabolic signaling by activating mTOR (mammalian target of rapamycin). Subsequently, the activated mTOR transmits an anabolic signaling to its downstream target molecule p70S6K, leading to protein synthesis and muscular hypertrophy [9, 10].
In addition to its role in anabolism, AKT contributes to muscular hypertrophy by inhibiting a potent catabolic process, ubiquitin–proteasome system (UPS) [11]. For example, when a transcription factor FOXO (Forkhead box O) family protein is activated, it translocates to a nucleus and promotes the expression of potent E3 ubiquitin ligases (i.e., Atrogin-1/MAFbx and MuRF1), which induce polyubiquitination on targeted proteins and subsequently consigns them to proteasomes for the final degradation process [12]. However, when activated AKT phosphorylates FOXO, FOXO’s translocation to the nucleus is interrupted, and thus catabolic potential is prevented [13].
Autophagy is another catabolic process involved in sustaining cellular homeostasis by regulating the rate of protein turnover. Interestingly, it can be strongly promoted in both denervated [14] and mechanically stressed (i.e., endurance exercise) muscle tissues [15–17], suggesting that the variation in cellular activities may be a key regulator of autophagy and a potential source of either favorable or undesired muscular adaptation, depending upon types of stresses. In this regard, endurance exercise-induced autophagy appears beneficial in that reduction in the cross-sectional area of muscle fibers by autophagic catabolism improves oxygen diffusion distance and capillary density; however, in case of RE used to promote muscular anabolism, autophagy may be an unfavorable outcome. Interestingly, the skeletal muscle of aged animal and human after RE displayed elevated autophagy [18, 19]; given that aging-mediated aberrant proteostasis (e.g., accumulation of damaged proteins and organelles) contributes to sarcopenia, it seems reasonable that the occurrence of autophagy by RE may play a potential role in maintaining healthy cellular environment of skeletal muscle undergoing aging. However, it is currently unknown whether RE promotes or inhibits autophagy or coactivates both autophagy and hypertrophy in the young-adult population. Obviously, clear investigation of the full spectrum of signaling nexus of autophagy pathways and processes linked to RE is important to elucidate the relationship between RE-induced muscular hypertrophy and autophagy.
Autophagy is accomplished by the following procedures: initiation, nucleation, elongation, maturation/fusion, and degradation by lysosomes. To induce autophagy, a master metabolic modifier AMP-activated protein kinase (AMPK) is activated, by which Unc-51 like kinase 1 (ULK1) becomes phosphorylated at Ser555. This leads to the activation of VPS34/class III PI3 kinase and subsequent recruitment of BECLIN1 for the construction of a phagophore (premature form of autophagosome). Then, the phagophore expands its membranes via interplays between autophagy (ATG) proteins such as ATG12, ATG5, ATG16, and LC3-II in conjunction with ATG7 and simultaneously encloses targeted cellular cargos, and becomes a mature autophagosome. Finally, matured autophagosomes are fused with lysosomes via lysosome-associated membrane protein (LAMP), and they are degraded by lysosomal proteases CATHEPSINs [20].
A degree of autophagy has been assessed using the ratio of LC3-II (active form) to LC3-I (inactive form), with the elevated ratio considered to be an increase in autophagy. However, this ratio can mislead the interpretation of autophagy flux because the disruption of a fusion process between autophagosomes and lysosomes can result in high levels of LC3-II levels in cytosol, instead of being increased due to upregulated autophagy flux [21]. For this reason, the levels of a key autophagy protein, p62, should also be measured because this protein is found in active autophagosomes and is degraded by lysosomes; thus, if autophagy flux is enhanced or interrupted, p62 should be diminished or accumulated, respectively [22].
Recent studies showed that mTOR activation antagonizes autophagy induction [23, 24]. The suggested mechanism is that mTOR-induced ULK1 phosphorylation at Ser757 inhibits BECLIN1 recruitment, resulting in the abolishing autophagy induction, whereas AMPK-mediated ULK1 phosphorylation at Ser555 activates autophagy induction. This implies that mTOR and AMPK competition for ULK1 phosphorylation may be a prime switch to determine the fate of metabolism (anabolism vs. catabolism).
Currently, a very limited number of studies have investigated whether RE elicits or hinders autophagy in muscle tissues [19, 25]. Given that autophagy is associated with muscular catabolic processes, it is tempting to postulate that achievement of muscular hypertrophy would be associated with a metabolic paradigm modulation: enhancing anabolism and suppressing autophagy-mediated catabolism. In this regard, we examined the metabolic signaling pathways in hypertrophied FDP muscle by measuring an AKT-mTOR-p70S6K signaling cascade and a series of autophagy signaling pathways after 8 weeks of progressive resistance exercise. Our study demonstrated that RE elevates AKT-mTOR-p70S6K phosphorylation but suppresses AMPK phosphorylation, LC3-II/LC-I ratio, and active-CATHEPSIN L, suggesting that RE-induced autophagy interruption may contribute to muscular hypertrophy.
Materials and methods
Animals
Young adult male Wistar Hannover rats (n = 20, 14 weeks old) were maintained at a 12:12 dark–light cycle, housed at 22 °C with 50% relative humidity, and given free access to standard chow diet (Central Laboratory Animal Incorporation, Seoul, Korea) ad libitum. After 1 week of familiarization, the animals were randomly assigned to either a sedentary control group (SC, n = 10) or a resistance exercise group (RE, n = 10). Changes in body weight were recorded weekly. Rats were handled in an accredited Korea Food and Drug Administration animal facility in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International Animal Care Policies (accredited unit, Korea Food and Drug Administration: unit number 000996). All experimental procedures were approved by the Institute Animal Care and Use Committee of Korea National Sport University and performed in compliance with the guidelines.
Resistance exercise
Ladder climbing exercise was used as a resistance exercise (RE) with minimal modifications on the basis of published work demonstrating muscle hypertrophy [19, 26, 27]. Briefly, a ladder (20 cm wide, 115 cm long, 85° incline, 4 cm gap between each perch) was used (Fig. 1a), to which animals were acclimated to learn climbing activity. After 1 week of acclimation, one repetition maximum was assessed; for instance, rats began climbing a ladder with 50% of their body weight as an initial resistance, and additional resistance was gradually incremented until they reached their maximum carrying load. Total training sessions consisted of ten repetitions per session, three times a week for 8 weeks with progressive addition of weight; briefly, animals climbed a ladder with 50% (1st–2nd), 75% (3rd–4th), and 100% (5th–6th) of the previous maximal carrying weight, after which an additional 30 g of resistance was added until they reached ten repetitions. Once the animals completed each climbing trial, they rested for 2 min in the housing space provided on the top of the ladder climbing apparatus.
Fig. 1.
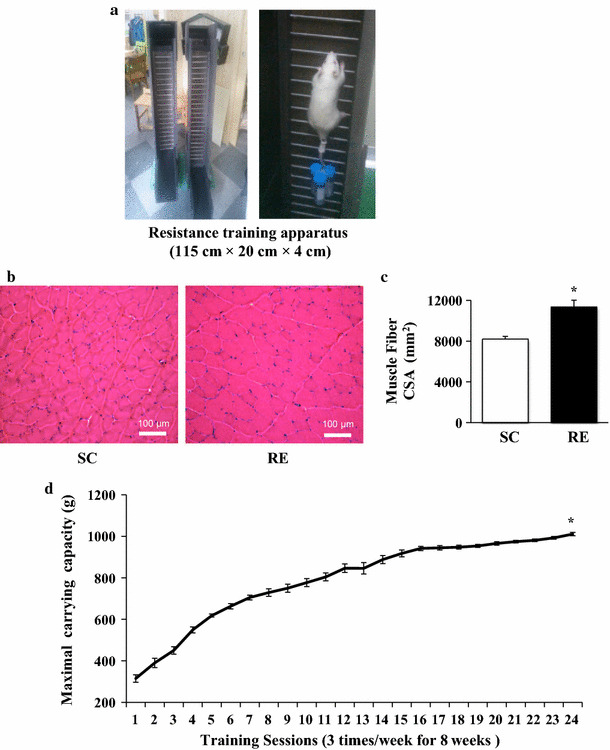
Long-term resistance exercise induces skeletal muscle hypertrophy and reinforces muscle strength. a Resistance training apparatus (width × length × gap = 115 cm × 20 cm × 4 cm). b Left Histological cross section of the flexor digitorum profundus (FDP) muscle in sedentary control group (SC), Right Histological cross section of the FDP muscle in resistance exercise group (RE). c Cross-sectional area (CSA) of FDP muscle fiber in SC and SE. Each bar represents the mean ± SE for the FDP muscles. Asterisks indicate significant differences between the two groups (*p < 0.01). d Maximal carrying capacity per each session for total training sessions. Asterisks denote significant difference in carrying capacity between the baseline and final carrying weight (*p < 0.01). The final carrying capacity (1011 g ± 8.1) was increased up to approximately 3.2-fold compared to baseline (315 g ± 17.8). Smooth marked scatters were represented as mean ± SE (n = 10). An asterisk indicates significant difference in RE groups (*p < 0.01)
Tissue extraction
Twenty-four hours after the last training session, all animals in both groups were anesthetized with sodium pentobarbiturate (40 mg/kg) by intraperitoneal injection. Once animals reached a surgical plane of anesthesia, the FDP muscles from two opposing limbs were excised: left side for histological analysis and right side for Western-blot analysis. Tissues for histological analysis were embedded with OCT freezing medium and frozen in isopentane chilled in liquid nitrogen. Other tissues were stored in cryogenic storage tubes and kept at −80 °C until needed for Western-blot analysis.
Hematoxylin and eosin (H&E) staining
FDP muscle sections (8 µm) were cut using a Cryotome (LEICA CM 1860, Germany) on a glass slide, and the tissue sections were air-dried for 20 min at room temperature and then stained with hematoxylin and eosin. Briefly, to stain nuclei, the tissue sections were submerged in Harris-Hematoxylin solution (ThermoFisher Scientific, #6765001) for 5 min using a Tissue-TEK slide stain set (VWR, #102094-050) and rinsed with tap water for 5 min. Then, the sections were submerged in 1% HCl in 70% ethanol for 1 min and then wash in tap water for 5 min. To stain cytoplasm, the tissue sections were submerged in Eosin Y solution (Sigma-Aldrich, #HT-110332) for 5 min and then dehydrated using 70, 80, and 90% ethanol for 1 min, respectively. To clear the stained tissues, the sections were submerged in 50% xylene (ThermoFisher Scientific, #X3F) diluted with 100% ethanol for 1 min, after which they were transferred to 100% Xylene and incubated for 3 min. Finally, the slides were mounted with Shandon Consul-Mounting solution (ThermoFisher Scientific, #9990440), and images were taken using an EVOS Auto inverted color microscope (Life Technologies™, #AMAFD1000). A cross-sectional area (CSA) of myofibers was assessed using ImageJ software (NIH, USA).
Western-blot analysis
FDP muscles were homogenized in T-PER® buffer (Thermo Scientific, #78510, Waltham, MA, USA) containing a Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, #78446, Waltham, MA, USA), using a Polytron™ PT 2500E homogenizer (Kinematica, #08-451-320, Bohemia, NY, USA). Tissue homogenates were centrifuged at 10,000 × g for 15 min for tissue protein extraction. Protein concentration was assessed using a Bradford protein assay kit (Thermo Scientific, #23236, Waltham, MA, USA). For Western-blot assays, proteins were separated by SDS-PAGE using NuPAGE® Novex® 4–12% Bis–Tris Protein gels (Thermo Scientific, # NP0322PK2, Waltham, MA, USA) and then transferred to PVDF membranes for 1 h at 30 V in a cold condition. Non-specific proteins were blocked for 60 min at room temperature in a blocking solution (5% BSA in Tris-buffered saline containing 0.1% Tween-20), and then the membranes were incubated with designated primary antibodies overnight at 4 °C. The antibodies used are as follows: AKT (9272, 1:1000), p-AKTSer473 (9271, 1:1000), mTOR (2972, 1:1000), p-mTORSer2448 (2971, 1:1000), AMPKα (2532, 1:1000), p-AMPKαThr172 (2535, 1:1000), ULK1 (8054, 1:1000), p-ULK1Ser757 (14202, 1:1000), LC3 (4108, 1:1000), p62 (5114, 1:1000), BECLIN1 (3738, 1:1000), and ATG7 (2631, 1:1000) from Cell Signaling Technology; BNIP3 (10433, 1:500), TFEB (2636, 1:500), LAMP-2 (13524, 1:500), and CATHEPSIN L (133641, 1:500) from Abcam; p-ULKSer555 (ABC124, 1:2500) from Millipore; p70S6K (SC-8418, 1:200), p-p70S6KTSer411 (SC-8416, 1:200) from Santa Cruz Biotechnology. The next day, the membranes were washed with 1X TBS-T (3 × 10 min) and then incubated with designated secondary antibodies conjugated with HRP for 1 h at room temperature: goat anti-rabbit (1148960, 1:4000), rabbit anti-goat (811620, 1:2000) from ThermoFisher Scientific; goat anti-mouse (SC-2005, 1:5000) from Santa Cruz Biotechnology. Then, the membranes were washed in TBS-T (3 × 10 min), after which immunoreactivity was detected with SuperSignal™ West Dura Extended Duration Substrate (Thermo Scientific, #34076, Waltham, MA, USA). The digital images were acquired using Odyssey®Fc Imaging System (LI-COR Biosciences, Lincoln, NE, USA), and band intensities were quantified using an ImageStudio software (LI-COR Biosciences, Lincoln, NE, USA). Each band was normalized with a loading control GAPDH.
Statistical analysis
All data were expressed as mean ± SEM. The statistical differences between the two groups were assessed using an unpaired, one-tailed Student’s t test. Statistical significance was established at p < 0.05.
Results
Progressive resistance exercise induces skeletal muscle hypertrophy and strength
To identify the effects of long-term resistance exercise training on muscle hypertrophy, we measured body weight, FDP muscle weight, and FDP muscle weight-to-body weight ratio (Table 1). While body weight and FDP muscle weight were not changed, the FDP muscle weight-to-body weight ratio was significantly increased in RE compared to SC (p < 0.05). Likewise, histological analysis showed an increase in the cross-sectional area of FDP muscle fibers (p < 0.001) (Fig. 1b, c), and the maximum weight carrying capacity in RE was 3.2-fold higher than SC (initial phase: 315 g ± 17.8, final phase: 1011 g ± 8.1, p < 0.05) (Fig. 1d).
Table 1.
Effects of 8 weeks of resistance exercise on body weight, FDP muscle weight, and ratio of FDP muscle weight to body weight
| SC (n = 10) | RE (n = 10) | p | |
|---|---|---|---|
| Body weight (g) | 449.3 ± 12.5 | 429.3 ± 8.1 | 0.197 |
| FDP muscle weight (g) | 1.74 ± 0.04 | 1.80 ± 0.05 | 0.365 |
| FDP muscle weight/body weight (mg/g) | 3.89 ± 0.08 | 4.20 ± 0.23 | 0.011* |
Values are represented as mean ± SE
FDP flexor digitorum profundus, SC sedentary control, RE resistance exercise
* p < 0.05 vs. SC
Resistance exercise potentiates anabolic signaling pathways, but constrains AMPK activation
Activation of AKT-mTOR-p70S6K anabolic signaling axis has been identified as an essential signature of muscular hypertrophy [7, 8, 28]. First, to verify if the ladder climbing is an effective mode of resistance exercise that relays anabolic signals, total and phosphorylated forms of AKT, mTOR, and p70S6K were assessed, respectively. RE resulted in a significant increase in AKTSer473 phosphorylation levels (p < 0.05); however, p-AKTSer473/AKT ratio was reduced due to a greater increase in total AKT than p-AKTSer473 (Fig. 2a, b). Similarly, RE also significantly elevated both p-mTORSer2448 and the ratio of the p-mTORSer2448 to total mTOR (p < 0.05) (Fig. 2a, c). Moreover, p-p70S6KSer411 (downstream signaling molecule of mTOR) was also augmented in RE compared to SC (p < 0.05) (Fig. 2a, d).
Fig. 2.
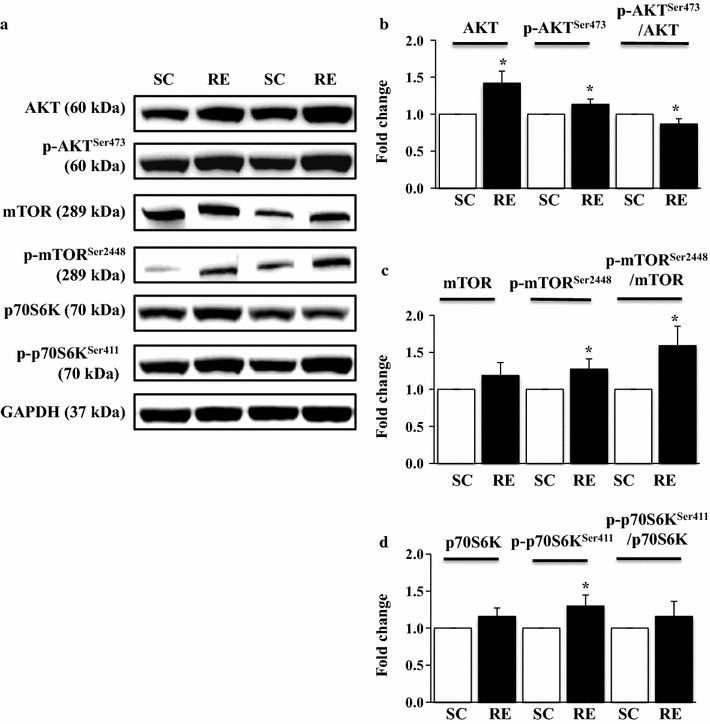
Changes in AKT-mTOR-p70S6K axis signaling pathways in response to 8 weeks of resistance exercise. a Representative Western-blot images of AKT, p-AKTSer473, mTOR, p-mTORSer2448, p70S6K, and p-p70S6KSer411 in each group. b Quantification of AKT, p-AKTSer473, and p-AKTSer473/AKT ratio. c Quantification of mTOR, p-mTORSer2448, and p-mTORSer2448/mTOR ratio. d Representative Western-blot images of p70S6K, p-p70S6KSer411, and p-p70S6KSer411/p70S6K ratio in each group. Comparisons of each target protein expression were made after normalization to GAPDH. SC sedentary control group, RE resistance exercise group. *p < 0.05 vs. SC
In contrast to mTOR, AMPK is critically involved in a catabolic process including autophagy. Surprisingly, RE substantially suppressed p-AMPKαThr172 and p-AMPKαThr172 to AMPKαThr172 ratio compared to SE, despite total AMPKα levels were upregulated in RE (p < 0.05) (Fig. 3a, b). We further examined whether RE-induced mTOR activation and AMPK deactivation modulate the activity (phosphorylation) of autophagy inductive protein ULK1 and found that neither mTOR responsive p-ULKSer757 (Fig. 3a, d) nor AMPK responsive p-ULK1Ser555 (Fig. 3a, c) was changed.
Fig. 3.
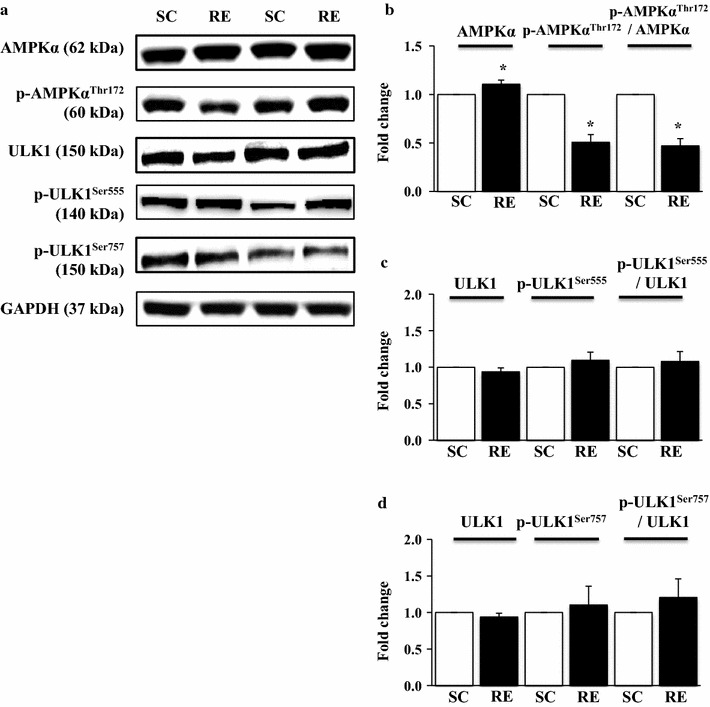
Changes in AMPKα and ULK1 signaling pathways in response to 8 weeks of resistance exercise. a Representative Western-blot images of AMPKα, p-AMPKαThr172, ULK1, p-ULK1Ser555, and p-ULK1Ser757 in each group. b Quantification of AMPKα, p-AMPKαThr172, and p-AMPKαThr172/AMPKα ratio. c Quantification of ULK1, p-ULK1Ser555, and p-ULK1Ser555/ULK1 ratio. d Representative Western-blot images of ULK1, p-ULK1Ser757, and p-ULK1Ser757/ULK1 ratio in each group. Comparisons of each target protein expression were made after normalization to GAPDH. SC sedentary control group, RE resistance exercise group. *p < 0.05 vs. SC
Resistance exercise exerts an inhibitory effect on autophagy regulation in hypertrophied skeletal muscle
To explore whether RE-induced anabolic signaling activation coincides with autophagy or inhibit it, we measured ATG proteins known to be involved in autophagy signaling. A key marker of autophagy, LC3-II was not changed, while LC3-I was significantly inclined in RE (p < 0.05), thus resulting in a decline in LC3-II/I ratio (Fig. 4a, b). p62 has been used to assess autophagy flux levels, an increase or decrease of which reflects a decline or incline of autophagy flux, respectively. Our data showed that RE displayed a remarkable elevation of p62 compared to SC (p < 0.05) (Fig. 4a, c), indicating a potential disturbance of autophagy flux. Interestingly, there were no changes for autophagy induction proteins, BECLIN1 (Fig. 4d, e) and BNIP3 (Fig. 4d, f); however, RE upregulated ATG7 known for promoting autophagosome formation (p < 0.05) (Fig. 4d, g).
Fig. 4.
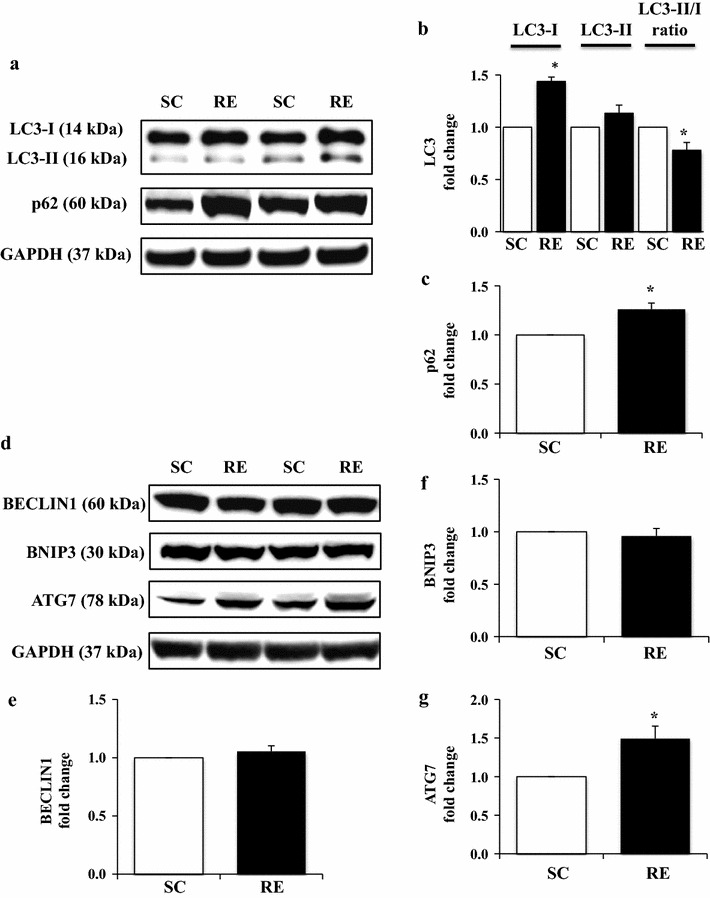
Changes in autophagy flux-related proteins (LC3 and p62) and autophagy induction-related proteins (BECLIN1, BNIP3, and ATG7) in FDP muscles after 8 weeks of resistance exercise training. a Representative Western-blot images for LC3 and p62 in each group. b Quantification of LC3-I, LC3-II, and LC3-II/I ratio. c Quantification of p62. d Representative Western-blot images for BECLIN1, BNIP3, and ATG7 in each group. e Quantification of BECLIN1. f Quantification of BNIP3. g Quantification of ATG7. Comparisons of each target protein expression were made after normalization to GAPDH. SC sedentary control group, RE resistance exercise group. *p < 0.05 vs. SC
Resistance exercise does not alter TFEB and LAMP-2, but prevents CATHEPSIN L maturation in hypertrophied skeletal muscle
To examine if RE-mediated inhibition of autophagy (i.e., reduced LC3-II/I ratio, increased p62, no changes for BECLIN1 and BNIP3) parallels with retardation of autophagy maturation, we assessed levels of lysosome-related proteins. No changes for TFEB and LAMP-2 were observed in FDP muscles between RE and SC (Fig. 5a–c). Interestingly, however, we observed that pro-CATHEPSIN L levels were accumulated; whereas, an active (cleaved) form of CATHEPSIN L was significantly declined in FDP muscles in response to RE, compared to SC (p < 0.05) (Fig. 5a, d).
Fig. 5.
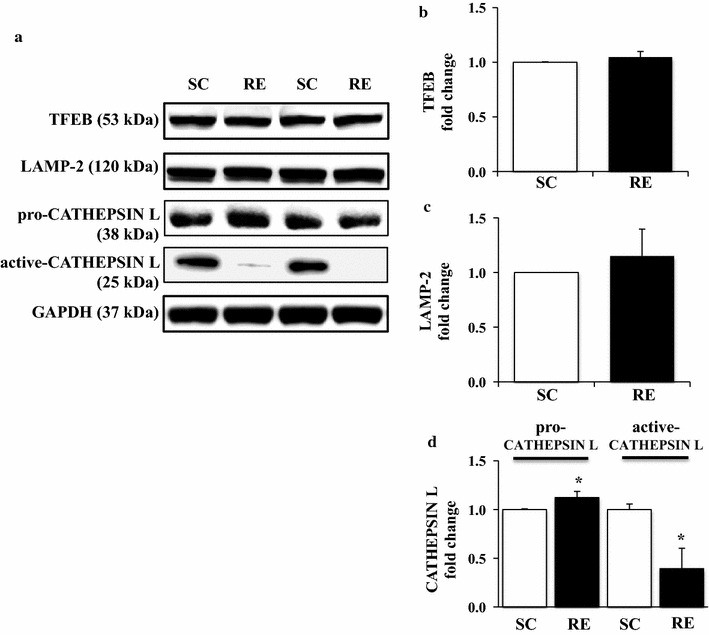
Changes in the expression level of both lysosomal biogenesis markers (TFEB, LAMP-2) and degradation marker (pro/active CATHEPSIN L) in FDP muscles in response to 8 weeks of resistance exercise training. a Representative western blot images of TFEB, LAMP-2, and pro/active CATHEPSIN L in each group. b Quantification of TFEB. c Quantification of LAMP-2. d Quantification of pro/active CATHEPSIN L. Comparisons of each target protein expression were made after normalization to GAPDH. SC sedentary control group, RE resistance exercise group. *p < 0.05 vs. SC
Discussion
In the present study, we examined RE-induced muscular hypertrophy and its signaling pathways, assessed molecular signaling nexus of autophagy, and explored possible association between hypertrophy and autophagy. Three main findings emerged from this study. First, 8 weeks of long-term RE promotes significant hypertrophy in FDP muscle as evidenced by the increased muscle weight-to-body weight ratio, enlarged cross-sectional area of muscle fibers, and a concurrent increase in muscular strength. Second, while RE enhances the activity of anabolism-related proteins: AKT, mTOR, and p70S6K, it dampens the activity of AMPK. Third, despite increased mTOR phosphorylation and reduced AMPK phosphorylation, RE does not interrupt autophagy induction (no changes in mTOR responsive ULK1Ser757 nor AMPK responsive ULK1Ser555), but does reduce levels of the active form of CATHEPSIN L. Taken together, our data suggest that RE mediates muscular hypertrophy by enhancing anabolic signaling activities and simultaneously limiting lysosomal degradation flux rather than negatively modulating the autophagy induction process.
Resistance exercise is a common exercise modality promoting muscular hypertrophy and strength development by enhancing anabolic processes [29, 30]. Using animals, several studies have demonstrated a ladder climbing as a potent resistance exercise that elicits muscular hypertrophy [19, 26, 27]. Consistent with these studies, our data also present that 8 weeks of resistance exercise (ladder climbing) significantly increased cross-sectional area of FDP muscle and muscle weight in conjunction with improved muscular strength (weight-carrying capacity). In general, an increased anabolic state over catabolic state is an important feature of muscular hypertrophy; thus, elevated phosphorylation levels of AKT-mTOR-p706SK signaling axis have been considered as key markers reflecting enhanced anabolism in muscles undergoing hypertrophy [7, 8]. In this context, our data also show that RE elevates phosphorylation levels of AKT, mTOR, and p706SK. Although the ratio of total protein to phosphorylated proteins was reduced, we presume that, if the absolute levels of enzyme’s phosphorylation are significantly raised, the enhanced enzyme activities (p-AKTSer473 and p-p70S6KSer411) may serve as a potent mediator of anabolism.
AMPK is an essential kinase critically involved in maintaining metabolic homeostasis by enhancing glucose uptake, fatty acid oxidation, and mitochondrial biogenesis [31–33]. AMPK also functions as a potent autophagy initiator in various stress conditions such as starvation and hypoxia [34, 35], and emerging evidence has revealed that endurance exercise-induced AMPK activation is linked to autophagy promotion [35]. However, given that heavy RE is known to augment protein synthesis and suppress protein degradation (i.e., ubiquitin–proteasome pathway), whether autophagy is coactivated or repressed during RE is poorly understood. Also, a question of how AMPK is regulated in response to chronic RE remains unclear. Our data show that chronic RE upregulates expression of AMPK proteins, but dampens AMPK phosphorylation levels. In contrast to our results, others showed no changes [36] or increases [37] in AMPK phosphorylation following acute RE; however, it should be noted that these studies utilized an electrical stimulation to generate maximal isometric contraction as a means to mimic RE. Thus, it seems unreasonable to directly compare our data with these results and deduce any conclusion. Nevertheless, we do not exclude the possibility that AMPK activity (phosphorylation) levels will change in an exercise duration or recovery period-dependent fashion, inferring that in our present study, AMPK phosphorylation data acquired 24 h post-exercise may display variable outcomes compared to the studies above. To circumvent this variability, further time-dependent studies are necessary.
Regarding muscular hypertrophy, our finding appears to indicate that RE-induced AMPK suppression may prevent muscular catabolism caused by autophagy and thereby contribute to sustaining anabolism, as inhibition of AMPK is linked to the failure of autophagy [38]. To prove our postulation, we further examined an AMPK downstream target ULK1. To our surprise, p-ULKSer555 levels were not altered, despite suppressed p-AMPKThr172 levels. Therefore, we next examined another ULK1 phosphorylation site at Ser757 because elevation of p-ULKSer757 levels by mTOR suppresses autophagy induction [39], but we did not observe any significant changes in p-ULKSer757 levels either. Our results suggest that neither AMPK nor mTOR does not exert modulatory effects on autophagy initiation process; however, based upon recent studies showing an autophagy modulation independent of mTOR [16, 40], it may be possible that RE affects autophagy in an AMPK or mTOR-independent fashion. Regarding this result, we still do not exclude a possibility that we might overlook probable changes in autophagy-inductive processes, which occur in a time-dependent manner because we sacrificed animals 24 h after the last exercise session. For example, unlike hypertrophy signaling responses typically observed 24–48 h post exercise, autophagy responses in the heart and skeletal muscle after endurance exercise peak within 2 h post exercise and begin to reduce after 3–4 h post exercise [17]. For this reason, it is recommended that future studies examining RE-induced autophagy adaptation should consider a proper time frame that precisely displays autophagic responses.
Measurement of LC3 turnover (LC3-I, LC3-II, and LC3-II/I ratio, respectively) is a key element for assessing autophagy levels. Our finding shows that the LC3-II/I ratio was reduced following RE due to substantial upregulation of LC3-I with lack of LC3-II conversions. One question is why does RE not significantly alter LC3-II but does increase LC3-I. This question can be answered by our data showing ULK1 needed in the autophagy induction was not activated, implying that LC3-II formation was limited in the beginning step of autophagy. Alternatively, the increased LC3-I levels in our study may be due to enhanced autophagy flux because LC3-II attached to autophagosomal membrane facing cytosol can be recycled to LC3-I via ATG4 [41] and thus may accumulate in proportion to an increase in autophagy. To confirm the possibility of the latter, we assessed p62 levels since it has been identified as an important indicator of autophagy flux in many studies [42–44]. p62 possesses a ubiquitin-binding motif and LC3-interacting region motif, both of which are critical for p62 to incorporate ubiquitinated toxic molecules and to tether them to autophagosomes via conjoining LC3-II, and it is then degraded with autophagosomes by lysosomes. Thus, alterations in p62 levels in response to autophagic stresses imply changes in the rate of autophagy flux [22]. In our study, p62 levels were significantly elevated in FDP muscles responding to RE. Therefore, we can exclude the possibility that an increase in LC3-I may be due to accelerated autophagy flux.
Unlike our study, recent studies showed contrasting results that RE decreases autophagy by reducing LC3-II without changes in LC3-I in human vastus lateralis muscle [18] and in rat gastrocnemius muscles [19] after exercise. We believe that a snapshot of complex processes of autophagy at different recovery periods (24 vs. 48 h), exercise duration (1 day acute bout of RE vs. 8 or 9 weeks), and age (young vs. old) may limit the chance of reaching a universal consensus of autophagy mechanisms among studies. Nonetheless, a consensus appears to converge on the possibility that RE reduces catabolism induced by autophagy in response to RE.
To further support an inhibitory effect of RE on autophagosomal degradation, we measured a key lysosomal protease, CATHEPSIN L, and provided plausible postulation that RE may constrain a lysosomal degradation process since the active form of CATHEPSIN L was dramatically reduced by RE. This reduction may limit autophagy-mediated proteolysis in lysosomes and thus contribute to muscular hypertrophy, as a recent study showed that CATEHPSIN L overexpression is linked to atrophy of skeletal muscles [45].
BECLIIN1, BNIP3, and ATG7 are critical autophagy proteins involved in induction and formation of autophagosomes [46–48]. In our study, we show that BECLIIN1 and BNIP3 remain unchanged in response to RE, indicating that RE does not promote autophagy. Interestingly, ATG7 was significantly elevated following RE. This is surprising because recent research reports autophagy promotion upon ATG7 overexpression in the heart, but our data show an inverse relationship between autophagy and ATG7. Currently, no evidence has been found to explain why ATG7 is upregulated in hypertrophied muscle in the absence of autophagy. Nevertheless, our speculation is that, due to RE-induced lysosomal proteolysis inhibition, the middle-stream of autophagy machinery (i.e., ATG7 and LC3-I) may be upregulated via a feed-forward mechanism. Apparently, this speculation should be explored by future studies.
Contrary to our results, a recent study reported that muscular hypertrophy, despite suppressed levels of AKT and mTOR phosphorylation, concurs with elevated autophagy in aged rats following 9 weeks of RE [19]. Because this is the only comparable research that investigates hypertrophy and autophagy using the same exercise modality (ladder climbing), we cannot explain the conflicting observations between the two studies. However, different ages (young: 6-month-old in our study vs. old: 22-month-old in Lou’s study) may elicit disparate results possibly because aged muscle may contain more dysfunctional or damaged proteins, which need to be actively removed and recycled by accelerated autophagy in parallel with enhanced anabolic processes rather than only by elevated anabolic processes as displayed in the young rats depicted in our study.
Collectively, as summarized in Fig. 6, our study demonstrates that long-term resistance training promotes muscular hypertrophy and muscular strength by potentiating an anabolic signaling pathway and concurrently interfering in autophagy, especially in the lysosomal proteolysis step of autophagy without altering an autophagy-inductive process.
Fig. 6.
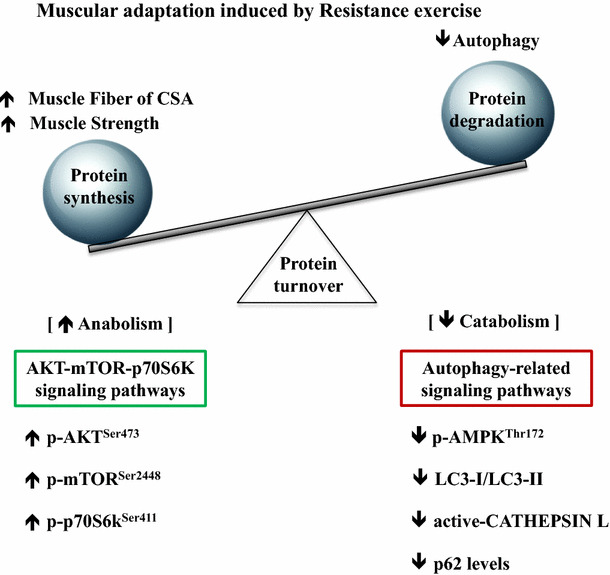
Summary of RE-induced muscular adaptation. RE-induced increase in muscle fiber cross-sectional area and strength is caused by modulating the rate of protein turnover, with protein synthesis predominating protein degradation. RE enhances an anabolic signaling nexus of hypertrophy pathways (AKT-mTOR-p70s6k) but suppresses a catabolic process, autophagy, which was evidenced by reduced AMPK phosphorylation levels, a declined LC3-I/LC3-II ratio, diminished active CATHEPSIN levels, and increased p62 levels. Our data suggest that although showing only a limited snapshot of metabolic change 24 h after the last bout of RE, sustained anabolic signaling activation may restrict autophagy-related catabolic processes, contributing to an increase in muscle fiber sizes
Acknowledgments
This project was supported by a grant from the University of West Florida though the Office of Research and Sponsored Programs (#164336) and UWF Florida Research Fellowship to YL (CF6672).
Author contributions
I.K. and Y.J. collected and analyzed data; I.K., Y.J., Y.C.J. and Y.L. interpreted results; I.K. and Y.J. prepared the table and figures; I.K. and Y.L. drafted the manuscript; I.K., Y.J., J-Y. C., Y.C.J., and Y.L. edited and revised the manuscript; I.K., Y.J., J-Y. C., J.Y.C., and Y.L. approved the final version of the manuscript; I.K. and Y.L. developed the conception and design of research.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17(2):162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Adams GR, et al. Combined isometric, concentric, and eccentric resistance exercise prevents unloading-induced muscle atrophy in rats. J Appl Physiol. 2007;103(5):1644–1654. doi: 10.1152/japplphysiol.00669.2007. [DOI] [PubMed] [Google Scholar]
- 3.Corazza AV, et al. Phototherapy and resistance training prevent sarcopenia in ovariectomized rats. Lasers Med Sci. 2013;28(6):1467–1474. doi: 10.1007/s10103-012-1251-8. [DOI] [PubMed] [Google Scholar]
- 4.Karagounis LG, et al. Contraction-induced changes in TNFalpha and Akt-mediated signalling are associated with increased myofibrillar protein in rat skeletal muscle. Eur J Appl Physiol. 2010;109(5):839–848. doi: 10.1007/s00421-010-1427-5. [DOI] [PubMed] [Google Scholar]
- 5.Coffey VG, Hawley JA. The molecular bases of training adaptation. Sports Med. 2007;37(9):737–763. doi: 10.2165/00007256-200737090-00001. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez AM, et al. Intramuscular anabolic signaling and endocrine response following resistance exercise: implications for muscle hypertrophy. Sports Med. 2016;46(5):671–685. doi: 10.1007/s40279-015-0450-4. [DOI] [PubMed] [Google Scholar]
- 7.Joassard OR, et al. Regulation of Akt-mTOR, ubiquitin-proteasome and autophagy-lysosome pathways in response to formoterol administration in rat skeletal muscle. Int J Biochem Cell Biol. 2013;45(11):2444–2455. doi: 10.1016/j.biocel.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Blaauw B, et al. Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J. 2009;23(11):3896–3905. doi: 10.1096/fj.09-131870. [DOI] [PubMed] [Google Scholar]
- 9.Ogasawara R, et al. The order of concurrent endurance and resistance exercise modifies mTOR signaling and protein synthesis in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2014;306(10):E1155–E1162. doi: 10.1152/ajpendo.00647.2013. [DOI] [PubMed] [Google Scholar]
- 10.Paturi S, et al. Effects of aging and gender on muscle mass and regulation of Akt-mTOR-p70s6k related signaling in the F344BN rat model. Mech Ageing Dev. 2010;131(3):202–209. doi: 10.1016/j.mad.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Keller J, et al. Supplementation of carnitine leads to an activation of the IGF-1/PI3K/Akt signalling pathway and down regulates the E3 ligase MuRF1 in skeletal muscle of rats. Nutr Metab (Lond) 2013;10(1):28. doi: 10.1186/1743-7075-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smiles WJ, et al. Modulation of autophagy signaling with resistance exercise and protein ingestion following short-term energy deficit. Am J Physiol Regul Integr Comp Physiol. 2015;309(5):R603–R612. doi: 10.1152/ajpregu.00413.2014. [DOI] [PubMed] [Google Scholar]
- 14.O’Leary MF, et al. Denervation-induced mitochondrial dysfunction and autophagy in skeletal muscle of apoptosis-deficient animals. Am J Physiol Cell Physiol. 2012;303(4):C447–C454. doi: 10.1152/ajpcell.00451.2011. [DOI] [PubMed] [Google Scholar]
- 15.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, et al. Cardiac kinetophagy coincides with activation of anabolic signaling. Med Sci Sports Exerc. 2016;48(2):219–226. doi: 10.1249/MSS.0000000000000774. [DOI] [PubMed] [Google Scholar]
- 17.Ogura Y, et al. Single bout of running exercise changes LC3-II expression in rat cardiac muscle. Biochem Biophys Res Commun. 2011;414(4):756–760. doi: 10.1016/j.bbrc.2011.09.152. [DOI] [PubMed] [Google Scholar]
- 18.Fry CS, et al. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci. 2013;68(5):599–607. doi: 10.1093/gerona/gls209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo L, et al. Chronic resistance training activates autophagy and reduces apoptosis of muscle cells by modulating IGF-1 and its receptors, Akt/mTOR and Akt/FOXO3a signaling in aged rats. Exp Gerontol. 2013;48(4):427–436. doi: 10.1016/j.exger.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Mammucari C, Schiaffino S, Sandri M. Downstream of Akt: foxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4(4):524–526. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 23.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15(4):406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 24.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monico-Neto M, et al. Resistance training minimizes catabolic effects induced by sleep deprivation in rats. Appl Physiol Nutr Metab. 2015;40(11):1143–1150. doi: 10.1139/apnm-2015-0061. [DOI] [PubMed] [Google Scholar]
- 26.Begue G, et al. Early activation of rat skeletal muscle IL-6/STAT1/STAT3-dependent gene expression in resistance exercise linked to hypertrophy. PLoS One. 2013;8(2):e57141. doi: 10.1371/journal.pone.0057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornberger TA, Jr, Farrar RP. Physiological hypertrophy of the FHL muscle following 8 weeks of progressive resistance exercise in the rat. Can J Appl Physiol. 2004;29(1):16–31. doi: 10.1139/h04-002. [DOI] [PubMed] [Google Scholar]
- 28.Katta A, et al. Altered regulation of contraction-induced Akt/mTOR/p70S6k pathway signaling in skeletal muscle of the obese Zucker rat. Exp Diabetes Res. 2009;2009:384683. doi: 10.1155/2009/384683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damas F, et al. A review of resistance training-induced changes in skeletal muscle protein synthesis and their contribution to hypertrophy. Sports Med. 2015;45(6):801–807. doi: 10.1007/s40279-015-0320-0. [DOI] [PubMed] [Google Scholar]
- 30.Nader GA, et al. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol. 2014;116(6):693–702. doi: 10.1152/japplphysiol.01366.2013. [DOI] [PubMed] [Google Scholar]
- 31.Peralta S, et al. Sustained AMPK activation improves muscle function in a mitochondrial myopathy mouse model by promoting muscle fiber regeneration. Hum Mol Genet. 2016;25:3178–3191. doi: 10.1093/hmg/ddw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ihsan M, et al. Regular postexercise cooling enhances mitochondrial biogenesis through AMPK and p38 MAPK in human skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2015;309(3):R286–R294. doi: 10.1152/ajpregu.00031.2015. [DOI] [PubMed] [Google Scholar]
- 33.Combes A, et al. Exercise-induced metabolic fluctuations influence AMPK, p38-MAPK and CaMKII phosphorylation in human skeletal muscle. Physiol Rep. 2015;3(9):e12462. doi: 10.14814/phy2.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwalm C, et al. Activation of autophagy in human skeletal muscle is dependent on exercise intensity and AMPK activation. FASEB j. 2015;29(8):3515–3526. doi: 10.1096/fj.14-267187. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, et al. AMPK binds to sestrins and mediates the effect of exercise to increase insulin-sensitivity through autophagy. Metabolism. 2015;64(6):658–665. doi: 10.1016/j.metabol.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Roberts MD, et al. A putative low-carbohydrate ketogenic diet elicits mild nutritional ketosis but does not impair the acute or chronic hypertrophic responses to resistance exercise in rodents. J Appl Physiol. 2016;120(10):1173–1185. doi: 10.1152/japplphysiol.00837.2015. [DOI] [PubMed] [Google Scholar]
- 37.Kido K, et al. Acute resistance exercise-induced IGF1 expression and subsequent GLUT4 translocation. Physiol Rep. 2016;4(16):e12907. doi: 10.14814/phy2.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vodicka P, et al. Assessment of chloroquine treatment for modulating autophagy flux in brain of WT and HD mice. J Huntingtons Dis. 2014;3(2):159–174. doi: 10.3233/JHD-130081. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiebel C, et al. Cannabinoid receptor 1 modulates the autophagic flux independent of mTOR- and BECLIN1-complex. J Neurochem. 2014;131(4):484–497. doi: 10.1111/jnc.12839. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 42.Lira VA, et al. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 2013;27(10):4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon I, et al. Effects of long-term resistance exercise training on autophagy in rat skeletal muscle of chloroquine-induced sporadic inclusion body myositis. J Exerc Nutrition Biochem. 2015;19(3):225–234. doi: 10.5717/jenb.2015.15090710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang D, et al. Exercise ameliorates the detrimental effect of chloroquine on skeletal muscles in mice via restoring autophagy flux. Acta Pharmacol Sin. 2014;35(1):135–142. doi: 10.1038/aps.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deval C, et al. Identification of cathepsin L as a differentially expressed message associated with skeletal muscle wasting. Biochem J. 2001;360(Pt 1):143–150. doi: 10.1042/bj3600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22(2):140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanida I, et al. The FAP motif within human ATG7, an autophagy-related E1-like enzyme, is essential for the E2-substrate reaction of LC3 lipidation. Autophagy. 2012;8(1):88–97. doi: 10.4161/auto.8.1.18339. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16(7):939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]


