Abstract
The heat shock response in Escherichia coli is mediated primarily by the rpoH gene, encoding ς32, which is specifically required for transcription of heat shock genes. A number of ς32 homologs have recently been cloned from gram-negative bacteria that belong to the gamma or alpha subdivisions of the proteobacteria. We report here some of the regulatory features of several such homologs (RpoH) expressed in E. coli as well as in respective cognate bacteria. When expressed in an E. coli ΔrpoH strain lacking its own ς32, these homologs activated the transcription of heat shock genes (groE and dnaK) from the start sites normally used in E. coli. The level of RpoH in Serratia marcescens and Pseudomonas aeruginosa cells was very low at 30°C but was elevated markedly upon a shift to 42°C, as found previously with E. coli. The increased RpoH levels upon heat shock resulted from both increased synthesis and stabilization of the normally unstable RpoH protein. In contrast, the RpoH level in Proteus mirabilis was relatively high at 30°C and increased less markedly upon heat shock, mostly by increased synthesis; this ς32 homolog was already stable at 30°C, and little further stabilization occurred upon the shift to 42°C. The increased synthesis of RpoH homologs in all these gamma proteobacteria was observed even in the presence of rifampin, suggesting that the induction occurred at the level of translation. Thus, the basic regulatory strategy of the heat shock response by enhancing the RpoH level is well conserved in the gamma proteobacteria, but some divergence in the actual mechanisms used occurred during evolution.
Induction of heat shock proteins represents a ubiquitous and homeostatic cellular response to cope with the accumulation of unfolded and misfolded proteins following exposure to heat or other stresses. In Escherichia coli, the induction occurs primarily by activating the transcription of heat shock genes by a transient increase in the cellular level of ς32 (10), encoded by the rpoH gene (8, 36). The level of ς32 during steady-state growth is very low (ca. 10 to 30 molecules per cell at 30°C [4]) due to the extreme instability (half life, 1 min) and restricted translation of rpoH mRNA (8, 37). Upon exposure to higher temperatures (e.g., 42°C), both rapid stabilization of ς32 and translational activation of rpoH mRNA result in a marked and transient increase in the ς32 level, which in turn activates the transcription of heat shock genes such as those encoding molecular chaperones (e.g., DnaK and GroEL) and ATP-dependent proteases (e.g., Lon and ClpP) (7, 8). Whereas stabilization of ς32 is thought to occur by accumulation of partially unfolded proteins that can titrate chaperones away from ς32 (2, 4), translational activation probably occurs by resolving extensive mRNA secondary structure formed under nonstress conditions (17, 37, 38).
Previous work on heat-induced synthesis of ς32–β-galactosidase fusion protein in E. coli revealed that two proximal coding regions (A and B) of rpoH mRNA are involved in translational control of ς32 synthesis (17, 37). Region A is similar to the “downstream box,” which is complementary to part of 16S rRNA (27), and acts as a positive element by enhancing translation, whereas region B is a negative element which forms an extensive secondary structure with region A. In addition, a segment of ς32 (region C; around residues 122 to 144 [18]) that corresponds to an area further downstream of rpoH was shown to be important for the DnaK/DnaJ-mediated negative feedback control of the heat shock response (28, 30, 31). Region C affects both the synthesis and stability of the fusion protein (18, 37), presumably by providing sites for interaction with DnaK (14).
Recent work in this and other laboratories led to the isolation and characterization of rpoH genes encoding ς32 (RpoH) homologs from a number of gram-negative bacteria (1, 5, 6, 16, 19–23, 25, 34). Some of them were isolated by directly selecting for clones that can complement the defects of an E. coli ΔrpoH mutant which grows only at or below 20°C (1, 19, 21). Comparison of nucleotide and amino acid sequences among these homologs revealed several conserved sequences that seemed to be of regulatory importance for the heat shock response. Specifically, both downstream box and predicted mRNA secondary structures similar to those seen in E. coli were found among the homologs from most members of the gamma but not alpha proteobacteria (1, 19, 34, 36). Besides, a stretch of 9 amino acid residues, highly conserved among all ς32 homologs but absent in other ς factors (designated the RpoH box [19, 36]), was detected within region C, implicated in the chaperone-mediated feedback control (18; see above). These interesting developments prompted us to examine the regulatory roles of ς32 homologs in the heat shock response of some of the representative bacteria.
In this report, we analyzed the regulatory and functional properties of ς32 homologs (RpoH) both in E. coli and in cognate bacteria. Several members of the gamma proteobacteria (E. coli, Serratia marcescens, Proteus mirabilis, and Pseudomonas aeruginosa) and one of the alpha proteobacteria (Agrobacterium tumefaciens) were chosen for this study. The ability of these rpoH homologs to complement, at least partially, the growth defect of the E. coli ΔrpoH mutant had suggested that they can be expressed to appreciable extents in E. coli and can functionally replace ς32 to varying degrees. Such expectations were fully supported by the present results. Most significantly, they revealed substantial conservation and some divergence in the regulatory strategy of ς32 homologs, particularly among the gamma proteobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
E. coli K-12 strain KY1608 (MC4100 ΔrpoH30::Kan zhf-50::Tn10), which was originally used for cloning of rpoH homologs (19), was employed to study their expression and function. Either charomid 9-36 or pTRC99A was used to express rpoH homologs from their authentic promoter(s) or trc promoter, respectively. Other strains used were S. marcescens ATCC 264, P. mirabilis PM-1, P. aeruginosa ATCC 10145, and A. tumefaciens GV3101, obtained from A. Oka, Kyoto University. λi21 nin5 was provided by H. Inokuchi; other phages and plasmids were obtained from commercial sources.
Media and chemicals.
Luria-Bertani (LB) broth (32) was generally used for growing cells. The minimal medium used was medium E supplemented with 0.5% glucose and 2 μg of thiamine per ml (32); it was further enriched with LB broth (final concentration, 1%) for some experiments. l-[35S]methionine (29.6 TBq/mmol) was obtained from American Radiochemicals, and other chemicals were obtained from Nacalai Tesque (Kyoto, Japan) or Wako Pure Chemicals (Osaka, Japan).
Radioactive labeling of proteins.
Exponentially growing cells in synthetic medium were pulse-labeled with [35S]methionine (3.7 MBq/ml), treated with 5% trichloroacetic acid, washed with acetone, dissolved in buffer containing sodium dodecyl sulfate (SDS) and EDTA, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) as described previously (35). To determine protein stability, pulse-labeled cells were further incubated with excess unlabeled methionine (200 μg/ml), harvested, and disrupted for analysis by immunoprecipitation and SDS-PAGE.
Immunoprecipitation and immunoblotting.
Immunoprecipitation of labeled proteins was carried out essentially as described previously (35) with specific antisera against E. coli ς32. Cells that produced a truncated form of ς32 were labeled with [35S]methionine and used as an internal reference to correct for differential recovery. Immunoblotting was also done as described previously (35) with a Hybond-ECL nitrocellulose membrane filter (Amersham). Quantification of protein bands following immunoprecipitation was performed with a BAS2000 BioImaging analyzer (Fuji Film, Tokyo, Japan).
Other methods.
Nucleic acid manipulation (24) and SDS-PAGE (35) were performed as described previously.
RESULTS
Induction of E. coli heat shock proteins mediated by ς32 homologs.
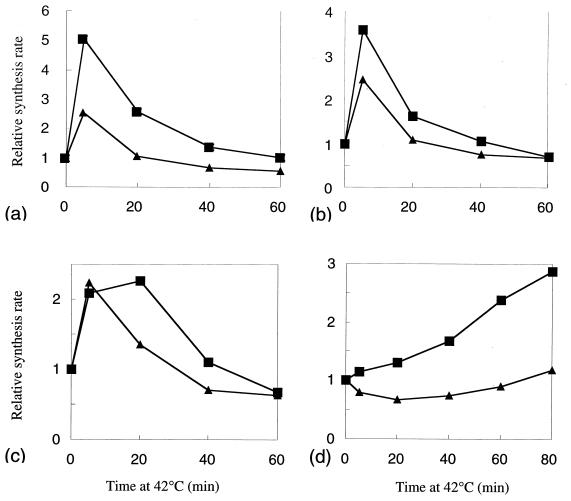
A set of E. coli (ΔrpoH) strains carrying each of the ς32 homologs on a multicopy charomid was examined by pulse-labeling cells with [35S]methionine at 30°C or after a shift to 42°C and by analyzing the synthesis rates of heat shock proteins by SDS-PAGE (10% polyacrylamide gel). The strains carrying an rpoH homolog of S. marcescens (Fig. 1b) or of P. mirabilis (Fig. 1c) exhibited heat induction of DnaK and GroEL, with maximum induction at around 5 min, much like the control strain carrying E. coli rpoH (Fig. 1a), although the induction was less pronounced with the P. mirabilis homolog. A similar result was obtained with the homolog of P. aeruginosa (data not shown). Thus, rpoH homologs from the gamma proteobacteria can support not only the basal-level synthesis of heat shock proteins but also enhanced synthesis upon exposure to high temperature. It appeared that the levels of expression and function of these homologs were appreciable though somewhat variable. In contrast, the strain carrying the rpoH homolog of A. tumefaciens (alpha subdivision) (Fig. 1d) exhibited weak and delayed induction of heat shock proteins, suggesting that the synthesis and/or function of this homolog may be restricted in E. coli.
FIG. 1.
Induction of heat shock proteins in the E. coli ΔrpoH mutant (KY1608) harboring a charomid 9-36 that contains each of the rpoH homologs. The cells were grown in synthetic medium to mid-log phase at 30°C and shifted to 42°C at time zero. Samples were taken at intervals, pulse-labeled with [35S]methionine for 1 min, and treated with trichloroacetic acid, and whole-cell proteins were analyzed by SDS-PAGE (7.5% polyacrylamide gel). Radioactivities associated with bands of GroEL (▪) and DnaK (▴) were quantified with a BioImaging analyzer (BAS2000) and plotted against time after normalization to the zero time value. (a) E. coli; (b) S. marcescens; (c) P. mirabilis; (d) A. tumefaciens.
Transcription of E. coli heat shock genes mediated by ς32 homologs.
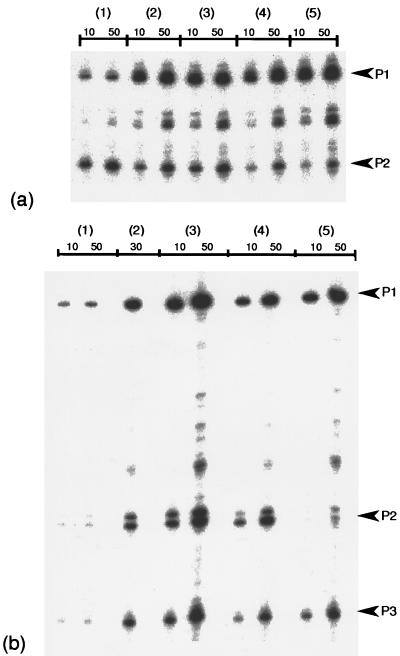
To determine the exact start sites of transcription mediated by the ς32 homologs, the same set of E. coli ΔrpoH strains but carrying each rpoH homolog under the trc promoter on a multicopy pTRC99A plasmid were grown in LB broth at 30°C. RNA was extracted from these cells and subjected to primer extension analysis to examine the groE and dnaK transcripts. Both groE and dnaK transcription mediated by these homologs were clearly initiated from sites identical to those used by E. coli ς32 (3); P1 for groE (Fig. 2a) and P1, P2, and P3 for dnaK (Fig. 2b). The plasmid carrying E. coli rpoH markedly enhanced the levels of these transcripts over the wild type (compare lanes 1 and 2). All transcripts except for the groE P2 transcript mediated by ς70 were lacking in the parental ΔrpoH mutant (reference 39 and data not shown). The relative amounts of these transcripts from individual promoters were similar among different homologs within the gamma subdivision; however, the dnaK P2 transcript was specifically reduced (five- to sixfold) in the strain carrying A. tumefaciens RpoH. The latter finding is probably due to the reduced efficiency of transcription from this particular promoter by E. coli RNA polymerase containing A. tumefaciens RpoH.
FIG. 2.
Transcription start sites of the E. coli heat shock genes (groE and dnaK) used by RpoH homologs. Cells of MC4100 (rpoH+) or KY1608 (ΔrpoH) harboring pTRC99A-rpoH that expresses each of the RpoH homologs from the trc promoter were grown in LB broth at 30°C (without isopropyl-β-d-thiogalactopyranoside [IPTG]), and RNA was extracted and hybridized with 32P-labeled primer for the 5′ ends of groE (ATTCATTGATAACTCTCCTTT) or dnaK (TAGTACCCAGGTCGATACC) transcripts. The primer extension products obtained were applied to a DNA sequencing gel (6% polyacrylamide), and relevant portions of the gels (autoradiograms) are presented. (a) groE transcripts; (b) dnaK transcripts. The amounts (in micrograms) of RNA hybridized for each lane are shown at the top, and start sites for the known promoters in E. coli (3) are indicated to the right. Lanes: 1, MC4100 (wild type); 2, KY1608 expressing RpoH (E. coli); 3, KY1608 expressing RpoH (S. marcescens); 4, KY1608 expressing RpoH (P. aeruginosa); 5, KY1608 expressing RpoH (A. tumefaciens).
Marked increase in the level of the ς32 homolog upon heat shock.
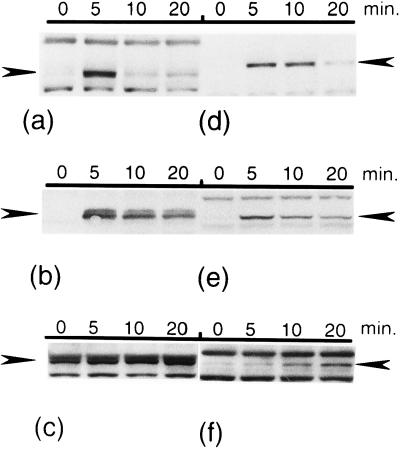
We then asked whether each of the homologs can respond to high temperature by enhancing its level as observed in E. coli. Immunoblotting of the RpoH homolog by using antiserum against E. coli ς32 was carried out with standard strains of bacteria from which the respective homologs were originally isolated. In S. marcescens, the RpoH level was very low at 30°C but was markedly and transiently elevated upon heat shock, as in E. coli (Fig. 3b). A similar response was observed in P. aeruginosa (Fig. 3d). In contrast, the RpoH level in P. mirabilis was relatively high at 30°C and only modestly enhanced upon the shift to 42°C (Fig. 3c). A similar heat-induced increase in the levels of the ς32 homologs was observed with a set of E. coli ΔrpoH strains carrying each heterologous rpoH on a single-copy λ prophage, although the amounts of homolog (particularly of P. mirabilis) produced were apparently smaller than those in the standard strains (Fig. 3e to f). We failed to determine the unequivocal response of RpoH of P. aeruginosa in E. coli or the response of A. tumefaciens RpoH, presumably because of the low expression and/or low cross-reactivities of these homologs to the antibodies used (data not shown). In spite of this limitation, these results clearly indicated that the heat-induced increase in the RpoH level is basically conserved as a primary cellular response to temperature upshift, at least within the gamma proteobacteria.
FIG. 3.
Enhancement of the RpoH level during the heat shock response. Cells were grown to mid-log phase in enriched medium E at 30°C and were shifted to 42°C. Samples were taken at the times indicated, and whole-cell proteins (5 μg/lane) were separated by SDS-PAGE (12.5% polyacrylamide gel) and immunoblotted with antiserum against E. coli ς32. Arrowheads indicate the positions of RpoH homologs. (a) E. coli MC4100; (b) S. marcescens; (c) P. mirabilis; (d) P. aeruginosa; (e) KY1608 (λi21 nin5 rpoH [S. marcescens]); (f) KY1608 (λi21 nin5 rpoH [P. mirabilis]).
Heat-induced synthesis of ς32 homologs of the gamma proteobacteria.
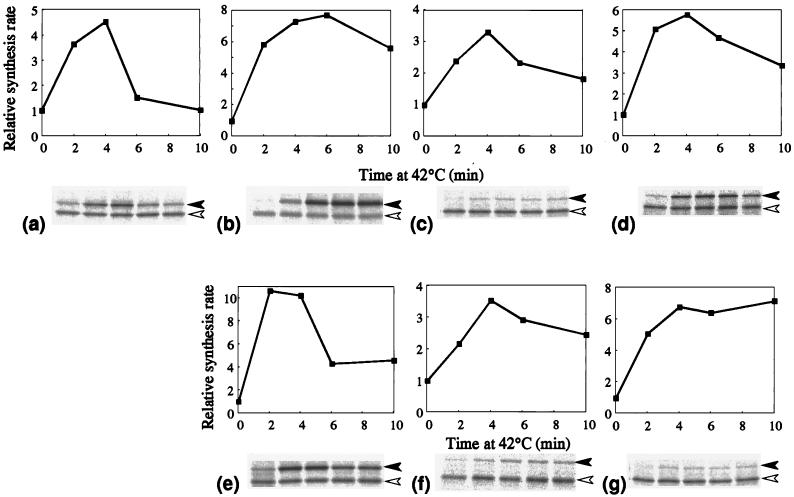
The mechanism underlying the increased RpoH level in the above strains was further investigated by examining the synthesis rate of each homolog upon heat shock. Cells were pulse-labeled with [35S]methionine before and after the temperature upshift, and RpoH proteins were analyzed by immunoprecipitation and SDS-PAGE. All three members of the gamma proteobacteria exhibited essentially identical responses, showing a marked and transient increase in synthesis rates, as in E. coli ς32, with maximum induction occurring at about 4 min. The extents of induction varied significantly and reproducibly: seven- to eightfold for S. marcescens, three- to fourfold for P. mirabilis, and five- to sixfold for P. aeruginosa (Fig. 4b to d). Quantitatively similar results were obtained with the set of E. coli ΔrpoH strains carrying the λ prophage that can express each homolog from the cognate promoter(s) (Fig. 4e to g). Thus, heat-induced synthesis of RpoH appears to be a well-conserved mechanism that contributes to the transcriptional activation of heat shock genes in the gamma proteobacteria.
FIG. 4.
Induction of RpoH synthesis upon heat shock. Cells were grown at 30°C and shifted to 42°C as in the experiment in Fig. 3. Samples were taken at the times indicated and pulse-labeled with [35S]methionine for 30 s. Whole-cell proteins were immunoprecipitated with antiserum against ς32, subjected to SDS-PAGE, and visualized with a BioImaging analyzer (BAS2000). Portions of the gels are presented below each graph; each lane corresponds to the time at which the sample was taken. Solid and open arrowheads indicate ς32 homologs and truncated E. coli ς32 (reference), respectively. Synthesis rates were calculated after correction for the recovery and normalized to the zero time value. (a) E. coli MC4100; (b) S. marcescens; (c) P. mirabilis; (d) P. aeruginosa; (e) KY1608 (λi21 nin5 rpoH [S. marcescens]); (f) KY1608 (λi21 nin5 rpoH [P. mirabilis]); (g) KY1608 (λi21 nin5 rpoH [P. aeruginosa]).
Heat-induced synthesis of ς32 homologs occurs at the level of translation.
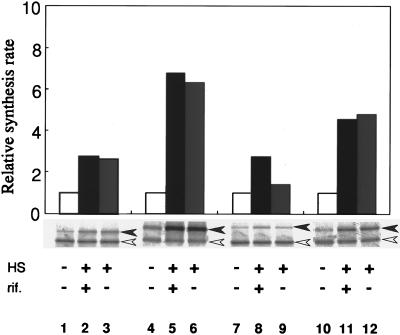
To address the question whether the increased synthesis of ς32 homologs occurs at the translational rather than the transcriptional level, we asked if the synthesis of homologs increases upon temperature upshift even in the presence of rifampin, which specifically inhibits RNA synthesis. The previous results of similar experiments with E. coli gave strong support to the translational control model (17). When cells of S. marcescens, P. mirabilis, and P. aeruginosa were grown at 30°C and shifted to 42°C in the presence and absence of rifampin, increased synthesis of RpoH was observed even in the presence of rifampin (Fig. 5). Rifampin is known to inhibit RNA synthesis (by 95%) within 3 min in E. coli under similar conditions (17), and no increase in the rpoH mRNA levels occurred in any of the strains examined (data not shown). These results therefore suggested strongly that the synthesis of RpoH is repressed under nonstress conditions and is markedly derepressed at the level of translation upon heat shock, as had previously been demonstrated in E. coli (8, 12, 17, 29, 37). The data are also consistent with the conserved secondary structure of rpoH mRNA of the gamma proteobacteria predicted from sequence analyses (19).
FIG. 5.
Induction of RpoH synthesis in the presence and absence of rifampin. Log phase cells grown in enriched medium E at 30°C were divided into three aliquots. After 1 min, rifampin was added to one aliquot (final concentration, 200 μg/ml) and shaken for 1 min; two aliquots (with and without rifampin) were shifted to 42°C, while the third (without rifampin) was kept at 30°C. After incubation for 3 min, all aliquots were pulse-labeled with [35S]methionine for 1 min and whole-cell proteins were isolated and analyzed by immunoprecipitation and SDS-PAGE as in the experiment in Fig. 4. Radioactivities were quantified, corrected for the recovery, and normalized to the 30°C control. HS, heat shock at 42°C; rif, rifampin. Lanes: 1 to 3, E. coli MC4100; 4 to 6, S. marcescens; 7 to 9, P. mirabilis; 10 to 12, P. aeruginosa.
Heat-induced stabilization of ς32 homologs.
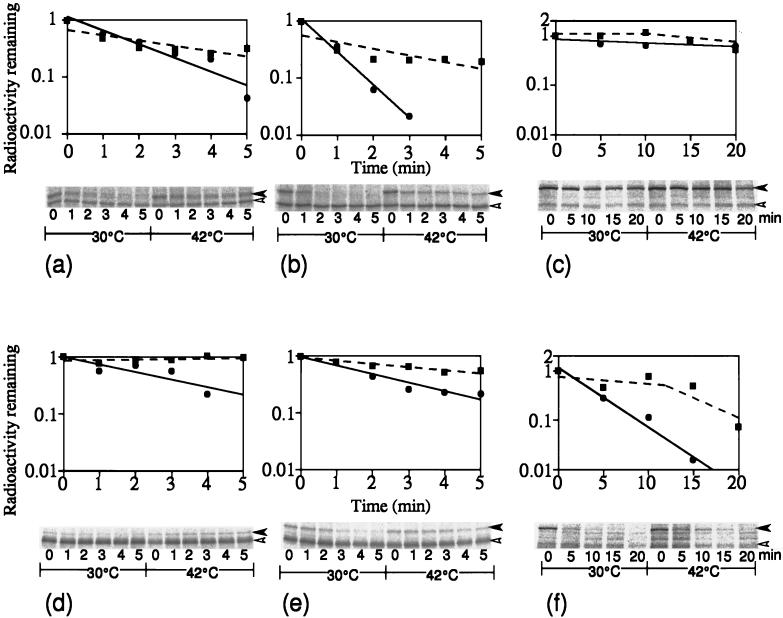
Finally, we examined the stability of ς32 homologs before and after temperature upshift by pulse-chase experiments. The homologs in S. marcescens and P. aeruginosa were found to be unstable at 30°C, with half lives of 0.7 and 2.5 min, respectively (Fig. 6b and d), much like E. coli ς32 (half life, 1.3 min) (Fig. 6a), and were markedly stabilized upon shift to 42°C. In contrast, RpoH of P. mirabilis was found to be quite stable (half life, 40 min) at 30°C and slightly more stabilized upon shift to 42°C (Fig. 6c). This agreed well with the constitutively high RpoH level at 30°C, with a modest increase upon heat shock (Fig. 3c). When these RpoH homologs were expressed in E. coli, quite different results were obtained: whereas RpoH of S. marcescens showed significantly slower degradation at 30°C (Fig. 6e), that of P. mirabilis showed much faster degradation, particularly at 30°C (half-life, 3 min) (Fig. 6f). These results indicate that although the instability of RpoH under nonstress conditions and rapid stabilization upon temperature upshift appear to be conserved, the actual half-lives and extents of stabilization can vary considerably, an extreme example being P. mirabilis RpoH, which virtually lost the mechanism of controlling its stability during the heat shock response, at least under the set of conditions used.
FIG. 6.
Stability of RpoH homologs at 30°C and after the shift to 42°C. Log-phase cultures grown in enriched medium E at 30°C were divided into two portions, and each was pulse-labeled with [35S]methionine for 30 s and chased for 1 min with excess unlabeled methionine; one was kept at 30°C, and the other was shifted to 42°C. Samples were taken at the times indicated and subjected to analysis by immunoprecipitation and SDS-PAGE, and the radioactivities associated with RpoH bands were visualized and quantified as in the experiment in Fig. 4. Panels a to f are the same as in Fig. 4. Portions of the gels are presented below each graph; the left and right halves represent data obtained at 30 and 42°C, respectively. Each lane corresponds to the time at which the sample was taken. Symbols: •, 30°C; ▪, 42°C.
DISCUSSION
The ς32 homologs of the gamma subdivision analyzed here exhibit 65 to 80% identity in amino acid sequence to E. coli ς32 and ca. 45% identity between E. coli and the alpha proteobacteria (19). As expected, the sequence similarity was particularly high for the conserved regions 2.4 and 4.2, which are involved in promoter recognition, and the regions 2.1 and 2.2, which are involved in binding to core RNA polymerase (9). Such high conservation is consistent with the fact that these homologs can at least partly replace ς32, presumably by catalyzing specific transcription from heat shock promoters when expressed in E. coli (1, 16, 19–23, 34). In line with the structural and functional similarities, all RpoH homologs tested, including that of A. tumefaciens, were able to induce the DnaK and GroE heat shock proteins to appreciable although variable extents (Fig. 1). Furthermore, they were shown to initiate transcription from start sites identical to those used by E. coli ς32, although the efficiency was low in certain cases (Fig. 2). These results suggest that each RpoH homolog can recognize the cognate heat shock promoters with sequences similar, if not identical, to those of E. coli. Indeed, the dnaK promoter of A. tumefaciens, previously thought to have a novel consensus sequence characteristic of heat shock genes of the alpha proteobacteria (26), was found to be recognized by its own RpoH (20). Similarly, the dnaK heat shock promoter of Caulobacter crescentus or Bradyrhizobium japonicum (alpha subdivision) was recently shown to be recognized by the cognate ς32 homolog in vitro (34) or in vivo (in E. coli) (15). Thus, the apparent difference in consensus heat shock promoters between E. coli and the alpha proteobacteria appears to be explained by subtle difference in promoter specificity among ς32 homologs.
The previous sequence analysis of rpoH homologs detected both putative downstream box and mRNA secondary structure among members of the gamma (but not the alpha) subdivision (19). Consistent with such predictions, the synthesis of all RpoH homologs of the gamma subdivision examined was similarly induced upon temperature upshift, and the induction apparently occurred at the level of translation (Fig. 5). Thus, the translational control involving rpoH mRNA secondary structure appears to be a conserved mechanism for heat shock regulation of ς32 homologs in the gamma proteobacteria. The extents of heat-induced synthesis varied among these homologs, between seven- to eightfold (S. marcescens) and three- to fourfold (P. mirabilis) (Fig. 4b to d). Since quantitatively similar differences were found when these homologs were expressed in E. coli (Fig. 4e to g), the differential extents of induction observed might reflect the differential stability of the rpoH mRNA secondary structure itself. As for the alpha subdivision, a heat-induced increase in the RpoH level was observed in C. crescentus (22, 34), B. japonicum (21), and A. tumefaciens (20), but the increase appears to occur primarily at the level of transcription.
All the RpoH homologs of the gamma subdivision tested except P. mirabilis were found to be unstable at low temperature (nonstress conditions), with some variation in half-life (0.7 to 2.5 min), the most unstable one being RpoH of S. marcescens (Fig. 6). In contrast, RpoH of P. mirabilis was quite stable at 30°C (half-life, 40 min) and showed little further stabilization upon the shift to 42°C. Interestingly, when either S. marcescens or P. mirabilis RpoH was expressed in E. coli, the apparent half-life became much like that of E. coli ς32 (1.3 min). This suggests that, in contrast to the above cases of translational induction (Fig. 4), interplay with certain cellular factors responsible for degradation of RpoH such as ATP-dependent proteases (11, 13, 33) or DnaK-DnaJ-GrpE chaperones (28, 30, 31) rather than the characteristics of RpoH itself explains the observed difference in stability between the respective bacteria. However, the actual involvement of such factors in the degradation of RpoH in S. marcescens and P. aeruginosa as well as of P. mirabilis RpoH expressed in E. coli remained to be investigated.
In summary, the results presented here and those reported by others indicate a strong conservation, at least among the alpha and gamma proteobacteria, of heat-induced elevation of cellular RpoH levels as a means of activating the transcription of the heat shock genes, much like in E. coli. However, there are differences in the actual strategies used to modulate the RpoH levels, perhaps reflecting the different ecological niche of each bacterial species. Among the gamma proteobacteria tested, P. mirabilis seemed to require higher levels of RpoH and heat shock proteins even under nonstress conditions, and this presumably brought about stable RpoH during evolution. Besides, a highly parasitic Haemophilus influenzae and endosymbiont Buchnera aphidicola, which may not require strong heat shock responses in their living environments, appear to lack rpoH mRNA secondary structures (5, 25) and perhaps translational control of RpoH synthesis. Further work on the regulatory mechanisms of RpoH homologs may provide new insights into the nature and significance of the heat shock response in eubacteria.
ACKNOWLEDGMENTS
We are grateful to Eliora Ron for helpful discussion, to A. Oka for bacterial strains, and to Masako Nakayama and Mayumi Ueda for technical assistance.
REFERENCES
- 1.Benvenisti L, Koby S, Rutman A, Giladi H, Yura T, Oppenheim A B. Cloning and primary sequence of the rpoH gene from Pseudomonas aeruginosa. Gene. 1995;155:73–76. doi: 10.1016/0378-1119(94)00829-h. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B. Regulation of the Escherichia coli heat shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 3.Cowing D W, Bardwell J C A, Craig E A, Woolford C, Hendrix R W, Gross C A. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci USA. 1985;82:2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig E A, Gross C A. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 5.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Georghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Garvin D L, Hardies S C. Nucleotide sequence for the htpR gene from Citrobacter freundii. Nucleic Acids Res. 1989;17:4889. doi: 10.1093/nar/17.12.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 8.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: AMS Press; 1996. pp. 1382–1399. [Google Scholar]
- 9.Gross C A, Lonetto M, Losick R. Bacterial sigma factors. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- 10.Grossman A D, Erickson J W, Gross C. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 11.Herman C, Thevenet D, D’Ari R, Bouloc P. Degradation of ς32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamath-Loeb A S, Gross C A. Translational regulation of ς32 synthesis: requirement for an internal control element. J Bacteriol. 1991;173:3904–3906. doi: 10.1128/jb.173.12.3904-3906.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of Hs1VU and other ATP-dependent proteases in controlling in vivo turnover of ς32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty J S, Rudiger S, Schonfeld H-J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. Regulatory region C of the E. coli heat shock transcription factor, ς32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 15.Minder A C, Narberhaus F, Babst M, Hennecke H, Fischer H-M. The dnaKJ operon belongs to the ς32-dependent class of heat shock genes in Bradyrhizobium japonicum. Mol Gen Genet. 1997;254:195–206. doi: 10.1007/s004380050408. [DOI] [PubMed] [Google Scholar]
- 16.Naczynski Z M, Mueller C, Kropinski A M. Cloning the gene for the heat shock response positive regulator (sigma 32 homolog) from Pseudomonas aeruginosa. Can J Microbiol. 1995;41:75–87. doi: 10.1139/m95-010. [DOI] [PubMed] [Google Scholar]
- 17.Nagai H, Yuzawa H, Yura T. Interplay of two cis-acting mRNA regions in translational control of ς32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai H, Yuzawa H, Kanemori M, Yura T. A distinct segment of the ς32 polypeptide is involved in DnaK-mediated negative control of the heat shock response in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding ς32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- 20.Nakahigashi, K. Unpublished data.
- 21.Narberhaus F, Weiglhofer W, Fischer H-M, Hennecke H. The Bradyrhizobium japonicum rpoH1 gene encoding a ς32-like protein is part of a unique heat shock gene cluster together with groESL1 and three small heat shock genes. J Bacteriol. 1996;178:5337–5346. doi: 10.1128/jb.178.18.5337-5346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock ς32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahu G K, Chowdhury R, Das J. The rpoH gene encoding ς32 homolog of Vibrio cholerae. Gene. 1997;189:203–207. doi: 10.1016/s0378-1119(96)00849-9. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sato S, Ishikawa H. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J Bacteriol. 1997;179:2300–2304. doi: 10.1128/jb.179.7.2300-2304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segal G, Ron E Z. The dnaKJ operon of Agrobacterium tumefaciens: transcriptional analysis and evidence for a new heat shock promoter. J Bacteriol. 1995;177:5952–5958. doi: 10.1128/jb.177.20.5952-5958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprengart M L, Fatscher H P, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16S rRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 29.Straus D B, Walter W A, Gross C A. The heat shock response of E. coli is regulated by changes in the concentration of ς32. Nature. 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 30.Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The dnaK protein modulates the heat-shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- 31.Tilly K, Spence J, Georgopoulos C. Modulation of stability of the Escherichia coli heat shock regulatory factor ς32. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobe T, Ito K, Yura T. Isolation and physical mapping of temperature-sensitive mutants defective in heat-shock induction of proteins in Escherichia coli. Mol Gen Genet. 1984;195:10–16. doi: 10.1007/BF00332716. [DOI] [PubMed] [Google Scholar]
- 33.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Newton A. Isolation, identification, and transcriptional specificity of the heat shock sigma factor ς32 from Caulobacter crescentus. J Bacteriol. 1996;178:2094–2101. doi: 10.1128/jb.178.7.2094-2101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yano R, Nagai H, Shiba K, Yura T. A mutation that enhances synthesis of ς32 and suppresses temperature-sensitive growth of the rpoH15 mutant of Escherichia coli. J Bacteriol. 1990;172:2124–2130. doi: 10.1128/jb.172.4.2124-2130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yura T. Regulation and conservation of the heat-shock transcription factor ς32. Genes Cells. 1996;1:277–284. doi: 10.1046/j.1365-2443.1996.28028.x. [DOI] [PubMed] [Google Scholar]
- 37.Yura T, Nagai H, Mori H. Regulation of heat shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 38.Yuzawa H, Nagai H, Mori H, Yura T. Heat induction of ς32 synthesis mediated by mRNA secondary structure: a primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 1993;21:5449–5455. doi: 10.1093/nar/21.23.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y-N, Kusukawa N, Erickson J W, Gross C A, Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor ς32. J Bacteriol. 1988;170:3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]