Abstract
The acid tolerance response enables Salmonella typhimurium to survive exposures to potentially lethal acidic environments. The acid stress imposed in a typical assay for acid tolerance (log-phase cells in minimal glucose medium) was shown to comprise both inorganic (i.e., low pH) and organic acid components. A gene previously determined to affect acid tolerance, atbR, was identified as pgi, the gene encoding phosphoglucoisomerase. Mutations in pgi were shown to increase acid tolerance by preventing the synthesis of organic acids. Protocols designed to separate the stresses of inorganic from organic acids revealed that the regulators ς38 (RpoS), Fur, and Ada have major effects on tolerance to organic acid stress but only minor effects on inorganic acid stress. In contrast, the two-component regulatory system PhoP (identified as acid shock protein ASP29) and PhoQ proved to be important for tolerance to organic acid stress but had little effect against organic acid stress. PhoP mutants also failed to induce four ASPs, confirming a role for this regulator in acid tolerance. Acid shock induction of PhoP appears to occur at the transcriptional level and requires the PhoPQ system. Furthermore, induction by acid occurs even in the presence of high concentrations of magnesium, the ion known to be sensed by PhoQ. These results suggest that PhoQ can sense both Mg2+ and pH. Since phoP mutants are avirulent, the low pH activation of this system has important implications concerning the pathogenesis of S. typhimurium. The involvement of four regulators, two of which are implicated in virulence, underscores the complexity of the acid tolerance stress response and further suggests that features of acid tolerance and virulence are interwoven.
Neutralophilic bacteria such as Salmonella typhimurium prefer to live and grow at a pH near neutrality. However, S. typhimurium often encounters a variety of potentially lethal acid stress conditions both in nature and during pathogenesis (10). Acid stress is a complex phenomenon involving the combined biological effects of acidic pH and organic acids that may be present in a given environment. Severe acidic pH (e.g., pH 3) creates a situation whereby protons leak across the membrane faster than housekeeping pH homeostasis systems can remove them. The result is an intracellular acidification to levels that damage or disrupt key biochemical processes. However, even the mild acid stress of a pH 5 medium can become a severe challenge if the medium also contains 200 mM acetate. The reason for this is that even in mildly acidic environments, the protonated form of an organic acid can permeate the cell membrane and dissociate inside the cell, in which the released proton can acidify intracellular pH. After dissociation, the membrane-impermeable, ionized form of the organic acid will accumulate intracellularly, causing further cell damage.
S. typhimurium responds to acidic challenges through a complex adaptive system called the acid tolerance response (ATR), in which adaptation to mild (pH 5.8) or moderate (pH 4.4) acid conditions enables the cell to endure periods of severe acid stress (pH 3). The ATR of S. typhimurium requires the synthesis of over 50 acid shock proteins (ASPs) that can be grouped into what appear to be a variety of survival systems. Some of these systems function primarily in exponentially growing cells, while others function in stationary-phase cells. ATR systems operating in stationary phase include those that are dependent on the alternative sigma factor ς38 and others that are ς38 independent. The ς38 protein, which is encoded by rpoS, was first recognized as an important transcription factor in stationary-phase bacteria but is now acknowledged to be crucial for many stress responses (15, 19). The stationary-phase, ς38-dependent acid tolerance systems are not induced by acid, probably because ς38 levels are already elevated by entry into stationary phase. In contrast, there is a ς38-independent ATR system evident in stationary phase that does require induction by acid (2, 5, 17).
Exponentially growing cells also exhibit ς38-dependent and -independent ATR systems (16). However, while ς38-dependent acid tolerance is not induced by low pH in stationary phase, it is induced by acid shock in exponential-phase cells. Rapidly growing cells that undergo an acid shock will increase ς38 production, which will, in turn, increase the expression of a subset of ASPs. The acid shock-induced increase in ς38 occurs due to the decreased proteolytic turnover of this sigma factor, a feature of ς38 control mediated by MviA (3).
Mutants deficient in ς38 have an interesting acid-sensitive phenotype evident in log-phase cells (16). While rpoS+ cells become progressively more acid tolerant during acid shock (pH 4.4) adaptations lasting up to 90 min, rpoS mutants will induce an ATR only if adaptation does not exceed 20 min (16). Acid shock adaptation for more than 20 min will render rpoS mutants extremely acid sensitive. Thus, rpoS mutants only transiently induce an ATR. Sustained induction of the ATR is referred to as RpoS-dependent acid tolerance because of its dependence on ς38 (16). RpoS-independent systems are also involved in the ATR of log-phase cells. One such system includes a set of ASPs controlled by the major iron regulatory protein Fur (8, 14). The present report describes a second RpoS-independent system controlled by PhoPQ, a two-component regulatory system known to sense extracellular magnesium concentrations (28, 30, 32).
Multiple systems of acid tolerance may, in part, provide fail-safe redundancies that ensure survival should one system fail. However, the multifactorial nature of acid stress (i.e., the effects of acidic pH and organic acid concentration) might dictate a need for systems specific for one or the other acid stress component. If this is so, one may be able to classify specific acid response systems with respect to their utility in handling organic (weak acid) versus inorganic (low pH) acid stress. The acid stress experienced by cells exponentially growing in minimal glucose media shifted to pH 3 has been shown to involve both organic and inorganic acid components. The evidence presented indicates that RpoS is essential for surviving the organic acid stress component but two systems, one RpoS dependent and the other PhoPQ dependent, provide partially redundant protection against inorganic acid stress (i.e., low pH). In agreement with its role in inducible acid tolerance, PhoP is shown to be an acid shock protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The strains used throughout this study are listed in Table 1. Plasmids pEG9050 and pEG9071 were provided by E. Groisman (27). LB complex medium and Vogel and Bonner E minimal medium supplemented with 0.4% glucose were prepared as liquid and solid (1.5% agar) media (31). NCE-lactose medium was prepared as described by Maloy (20). N-minimal medium included 38 mM glycerol and 0.1 or 1% vitamin-free Casamino Acids (23). The following antibiotics were used at the concentrations indicated; ampicillin, 60 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 10 or 20 μg/ml for minimal and rich media, respectively.
TABLE 1.
Strains used
| Strain | Genotype | Source |
|---|---|---|
| SF242 | oxrC (pgi)::Tn5 | C. Higgins |
| SF416 (MM197) | mgtB11::MudJ leuBCD485 | M. Maguire |
| SF530 (UK1, χ3761) | R. Curtiss III | |
| SF588 (χ4971) | UK1 fur-1 zbf-5123::Tn10 | R. Curtiss III |
| SF627 (YG7153) | hisG46 rfa+ Δada::Km | 34 |
| EG1202 | phoQ::MudJ | E. Groisman |
| SF687 | 14028S pagA1::MudJ | S. Miller |
| SF720 (EG9671) | 14028S mgtA::MudJ pmrA1::Cm | E. Groisman |
| SF732 (EG5170) | 14028S phoP5090::MudJ | E. Groisman |
| JF1899 | phoP::Tn10 | 7 |
| JF1915 | psiD19::Mud1-8 | 9 |
| JF2471 | atbR (pgi)::Tn10dTc fabF (atrB)13::MudJ | 6 |
| JF2585 | LT2 phoQ::MudJ | EG1202 × SF1 |
| JF2690 | UK1 rpoS::Ap | 16 |
| JF2731 | UK1 rpoS::Ap atbR (pgi)::Tn10dTc | JF2471 × JF2690 |
| JF2733 | UK1 atbR (pgi)::Tn10dTc | JF2471 × SF530 |
| JF2811 | LT2 atbR (pgi)::Tn10dTc atrB (fabF)::MudJ/pBF119 pgi+ | Clone pool × JF2471 |
| JF2812 | LT2 atbR (pgi)::Tn10dTc atrB (fabF)::MudJ/pBF120 pgi+ | Clone pool × JF2471 |
| JF2938 | UK1 rpoS10::MudJ | MudJ pool × SF530 |
| JF2955 | UK1 rpoS::Ap oxrC (pgi)::Tn10 | JF2690 × SF242 |
| JF3024 | UK1 ada::Km | SF627 × SF530 |
| JF3203 | UK1 phoP::Tn10 | JF1899 × SF530 |
| JF3204 | UK1 rpoS::Ap phoP::Tn10 | JF1899 × JF2690 |
| JF3264 | UK1 pmrA508ΩKm | M. Spector |
| JF3274 | UK1 mgtB11::MudJ | SF416 × SF530 |
| JF3302 | UK1 phoQ::MudJ rpoS::Ap | JF2585 × JF2690 |
| JF3303 | UK1 pagA1::MudJ | SF687 × SF530 |
| JF3439 | UK1 rpoS10::MudJ phoP::Tn10 | JF1899 × JF2938 |
| JF3529 | UK1 rpoS10::MudJ phoP::Tn10/pEG9050 (phoQ expressed from lacP) | JF3439 × pEG9050 (E. Groisman) |
| JF3530 | UK1 rpoS10::MudJ phoP::Tn10/pEG9071 (phoPQ expressed from lacP) | JF3439 × pEG9071 (E. Groisman) |
| JF3531 | UK1 pagA1::MudJ phoP::Tn10 | JF1899 × JF3303 |
| JF3547 | UK1 pagA1::MudJ pmrA1::Cm | SF720 × JF3303 |
| JF3550 | UK1 psiD19::MudA | JF1915 × SF530 |
| JF3551 | UK1 psiD19::MudA pmrA::Km | JF1915 × JF3264 |
| JF3552 | UK1 mgtB11::MudJ phoP::Tn10 | JF1899 × JF3274 |
| JF3553 | UK1 mgtB11::MudJ pmrA1::Cm | SF720 × JF3274 |
| JF3554 | UK1 psiD19::MudA phoP::Tn10 | JF1915 × JF3203 |
| JF3561 | UK1 psiD19::MudA pmrA1::Cm | SF720 × JF3550 |
| JF3594 | UK1 phoP5090::MudJ | SF732 × SF530 |
| JF3609 | UK1 phoP5090::MudJ/pEG9071 (phoPQ expressed from lacP) | JF3401 × JF3594 |
β-Galactosidase and acid tolerance assays.
β-Galactosidase was measured according to the method described by Miller (21). Standard ATR assays were conducted with strains grown overnight at 37°C in E glucose (EG) broth containing the appropriate antibiotic. A 1/100 dilution of the overnight broth was inoculated into 3 ml of EG broth (pH 7.7), and the mixture was incubated at 37°C with shaking. The cells were grown to an optical density at 600 nm (OD600) of 0.40 (2 × 108 CFU/ml), at which point cultures destined for adaptation were adjusted to pH 4.4 and incubated for 60 min. Acid challenge of unadapted and adapted cultures involved readjusting the pH to 3.1 for the indicated time. The acid stress in this assay included both organic and inorganic acids. Percent survival was calculated by dividing the number of CFUs on LB agar at time x by the number of CFUs at time zero and multiplying by 100.
The fresh medium ATR assay (organic acid free) was performed by a similar method. However, spent medium was removed by centrifugation of the culture for 3 min in a microcentrifuge. The cell pellet was resuspended in fresh EG medium preadjusted to pH 4.4 or 3.1 at 37°C. The pH of the fresh medium culture was adjusted after resuspension to correct for subtle changes in pH. When indicated, 1 mM acetic acid (Fisher Scientific, Norcross, Ga.) was added to fresh EG medium to mimic spent EG medium. Lengths of adaptation and challenge were the same as those used in the standard ATR assay described above.
Cloning of atbR.
AtbR was previously identified as a putative negative regulator of fabF (initially designated atrB [6]). The fabF-lac atbR strain (JF2471) grew poorly on NCE-lactose medium, whereas the fabF-lac strain grew well. This phenotype was counterintuitive with respect to AtbR being a negative regulator of fabF. Nevertheless, this characteristic provided a clue as to the identity of AtbR in that its absence interfered with lactose metabolism and served as a useful selection marker for cloning efforts. Plasmids able to complement the Lac− phenotype of JF2471 were selected from a clone pool on NCE-lactose (Ap) medium. Two distinct plasmids with this ability were isolated from JF2811 and JF2812 and designated pBF119 and pBF120, respectively. A 3.8-kb BglI fragment from pBF120 was used as a probe in hybridization experiments against BglI-digested chromosomal DNA from S. typhimurium LT2 and JF2475 (atbR::Tn10). Differences seen in the hybridization pattern of the two strains indicated that the cloned insert contained atbR. A 5.5-kb ClaI/SalI insert from pBF120 was subcloned into pMOB (TN1000 kit; Gold Biotechnologies, St. Louis, Mo.) and designated pBF215. A 2.4-kb PstI fragment was removed from pBF215 by digestion with PstI followed by religation of the plasmid. This smaller derivative, designated pBF215-1, was unable to complement the slow growth phenotype of JF2471. Restriction enzymes and T4 DNA ligase were purchased from Gibco BRL (Gaithersburg, Md.). Dideoxy-DNA sequencing of these plasmids was performed with the Sequenase version 2.0 DNA Sequencing kit (U.S. Biochemicals, Cleveland, Ohio). α-35S-dATP for sequencing was purchased from NEN Life Sciences Products (Boston, Mass.). Analyses of nucleotide and amino acid sequences were performed with the Genetics Computer Group Package (version 7).
Two-dimensional SDS-PAGE.
Two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of acid shock proteins was performed as described by Spector et al. (29) with cells labeled for 3 min with 35S-Translabel (40 μCi/ml; ICN Pharmaceuticals, Inc., Irvine, Calif.). Approximately 5 μg of protein was analyzed for each sample. Basic and acidic proteins are situated to the left and right, respectively, of each autoradiograph. The first dimension was a pH 5 to 7 isoelectric focusing gel containing 1.6% (pH 5 to 7) and 0.4% (pH 3 to 10) ampholytes (Bio-Rad Laboratories, Melville, N.Y.), and the second dimension was an SDS–11.5% polyacrylamide gel. The results presented are representative of three independent experiments.
N-terminal sequence analysis of ASP29.
ASP-29 was purified by using a modified two-dimensional gel protocol. A 0.5-liter volume of minimal E glucose medium was inoculated with a 1/100 dilution of an overnight culture of JF2690 (rpoS). At an OD600 of 0.40, a portion of the culture (3 ml) was removed for 35S-Translabel labeling. The labeled and unlabeled cultures were combined and centrifuged, and the pellets were resuspended in 1× sonication buffer (100 mM Tris [pH 7.4], 5 mM MgCl2). The sample was sonicated for five 30-s bursts. After sonication, the cellular debris was pelleted by low (12,100 × g)- and high (380,000 × g)-speed spins. The supernatant was loaded onto a Centricon-100 (Amicon) concentrator with a 100,000-molecular-weight protein cutoff. The Centricon-100 concentrator was spun at 2,800 rpm. The filtrate was then loaded onto a Centricon-30 concentrator and centrifuged at 2,800 rpm. Retentates from both centricons were processed for two-dimensional gels. After electrophoresis, the polyacrylamide gels were transferred to polyvinylidene difluoride membranes (Bio-Rad). The blots containing proteins from the Centricon-30 retentate were sent to the WISTAR Institute (Philadelphia, Pa.) for amino-terminal protein sequencing of ASP29.
RESULTS
Mutations in pgi suppress the pH 3 acid-sensitive phenotype of rpoS mutations.
The identity of a Tn10 insertion mutation (originally called atbR::Tn10) that increased the acid tolerance of unadapted S. typhimurium LT2 in a minimal glucose medium (6) was determined to be pgi, the gene encoding phosphoglucoisomerase. GenBank comparison of the nucleotide sequence obtained from pBF215-1 (obtained as described in Materials and Methods) with primer T7 revealed that atbR was 81.1% homologous over 169 bp to Escherichia coli pgi (GenBank accession no. X15196; data not shown). The pgi (atbR)::Tn10dTc insertion strain also exhibited the pgi phenotype of defective glucose metabolism.
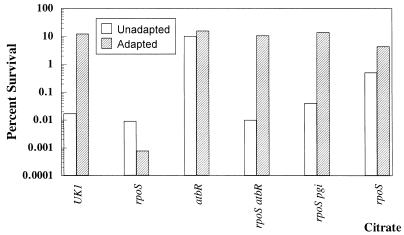
The LT2 strain originally used to analyze the pgi (atbR)::Tn10 insertion produces very little ς38, which in turn limits the ATR (16). Therefore, we suspected that the pgi mutation suppressed the acid-sensitive phenotype of rpoS mutants. To test this theory, the pgi (atbR)::Tn10 insertion was analyzed for its effect on an rpoS+ strain (UK1) and an rpoS null mutant. At pH 3, the rpoS mutant JF2690 proved to be extremely acid sensitive after 60 min of adaptation, while the rpoS pgi (atbR) mutant (JF2731) was very acid tolerant even when unadapted (Fig. 1). The effect of pgi (atbR) on the ATR of an rpoS+ strain was to increase tolerance even in unadapted cells. Subsequently, a pgi mutation obtained from the Salmonella Genetic Stock Center was also shown to suppress the acid-sensitive phenotype of an rpoS mutant (Fig. 1).
FIG. 1.
The influence of rpoS, pgi, and growth on citrate on the ATR. Cells were grown to mid-log phase (approximately 2 × 108 cells per ml) in minimal EG medium (pH 7.7). When indicated, E medium with citrate was used as the carbon source. Unadapted cell cultures were adjusted to pH 3, and samples were taken for viable counts at time zero and 60 min and after acid challenge. Adapted cultures were adjusted from pH 7.7 to 4.4 for 1 h and then acid challenged for 1 h at pH 3. Wild-type UK1 (SF530), rpoS::Ap (JF2690), atbR (JF2733), rpoS atbR (JF2731), and rpoS pgi (JF2955) cells were assayed. The data are a representative sample of triplicate experiments.
Organic acids cause the pH 3 acid-sensitive phenotype of rpoS mutants.
Mutants defective in pgi do not utilize glucose, but they can grow on the citrate present in the minimal EG medium used for ATR analyses. Growth on this nonfermentable carbon source and the lack of glucose fermentation end products (organic acids) may explain why pgi mutations suppress the pH 3 acid-sensitive phenotype of rpoS mutants. Figure 1 reveals that an rpoS mutant grown on citrate as the sole carbon source did, as predicted, exhibit an acid-tolerant phenotype similar to that of the rpoS pgi mutant. However, when it was grown on glucose, the rpoS mutant could not generate an ATR.
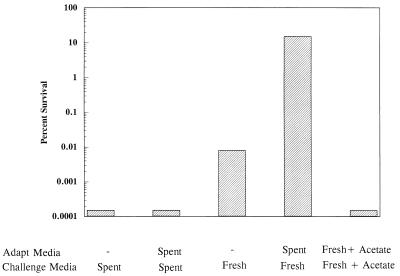
Further proof that fermentation end products were responsible for the acid-sensitive nature of rpoS mutants at pH 3 was obtained by replacing spent growth medium with fresh medium prior to pH 3 challenge. Figure 2 illustrates that when this experiment was performed, the removal of organic acids prior to acid challenge exposed an inducible acid tolerance in what previously was considered to be an acid-sensitive rpoS mutant (Fig. 2, bars 3 and 4). Adding 1 mM acetate to the fresh medium during challenge negated the fresh medium effect, proving that the acid-sensitive rpoS phenotype was due to weak acids present in growth medium (Fig. 2, bar 5). This amount of acetate is equivalent to what is produced by a mid-log culture grown on glucose. The addition of 1 mM acetate had no effect on the ability of rpoS+ cells to generate an ATR (data not shown). The results indicate that there are at least two systems involved in the log-phase ATR, i.e., an RpoS-dependent system capable of handling organic acid stress and an RpoS-independent system useless against organic acids but effective against inorganic acid stress.
FIG. 2.
RpoS-dependent systems are required for effective tolerance to organic acid stress. Cells (JF2690 [rpoS::Ap]) were grown and treated as indicated in the legend to Fig. 1. Spent, cells were grown, adapted, and challenged in the same EG medium; fresh, cells were removed from growth medium by centrifugation and resuspended in fresh EG medium already adjusted to pH 3 for challenge. In one experiment, the cells were resuspended in fresh medium made to contain 1 mM acetate (as acetic acid). The data are a representative sample of triplicate experiments.
Effects of fur and ada on organic versus inorganic acid stress survival.
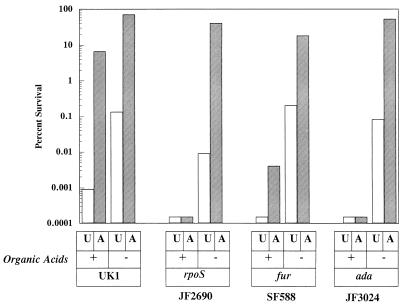
Medium exchange experiments were used to analyze two other genes (fur and ada) that when mutated cause an acid-sensitive phenotype. Fur is a global regulatory protein that senses intracellular Fe2+ and appears to sense intracellular pH independently of its ability to sense iron (8, 14). The Ada protein is involved in the adaptive response of E. coli to alkylating agents (13, 25). We have found that fur (14) and ada (Fig. 3) mutants are acid sensitive in the standard ATR assay. Using the medium exchange strategy, we decided to examine whether these genes were involved with protection against organic or inorganic acid stresses. The results indicated that an ada mutant was acid sensitive in organic acids but acid tolerant in the absence of organic acid (Fig. 3). The results with the fur mutant suggest that Fur, too, is required more for protection against organic acid stress, although a minor adaptive response is still evident in spent medium (Fig. 3).
FIG. 3.
Roles of fur and ada mutations on organic versus inorganic acid stress. Cells (UK1 [wild type], JF2690 [rpoS::Ap], SF588 [fur-1], and JF3024 [ada]) were grown and treated essentially as indicated in the legend to Fig. 1. Acid challenges (pH 3) were conducted in the presence (+ [spent medium]) or absence (− [fresh medium]) of organic acids. Results are representative of triplicate experiments. U, unadapted; A, adapted.
To this point, we have identified only genes which impact the tolerance exhibited to organic acid stress. Identification of a gene that influences inorganic but not organic acid tolerance would provide confirmation that organic and inorganic acid stresses are different and that distinct ATR systems exist for each. One such mutant was identified as described below.
ASP29 is PhoP.
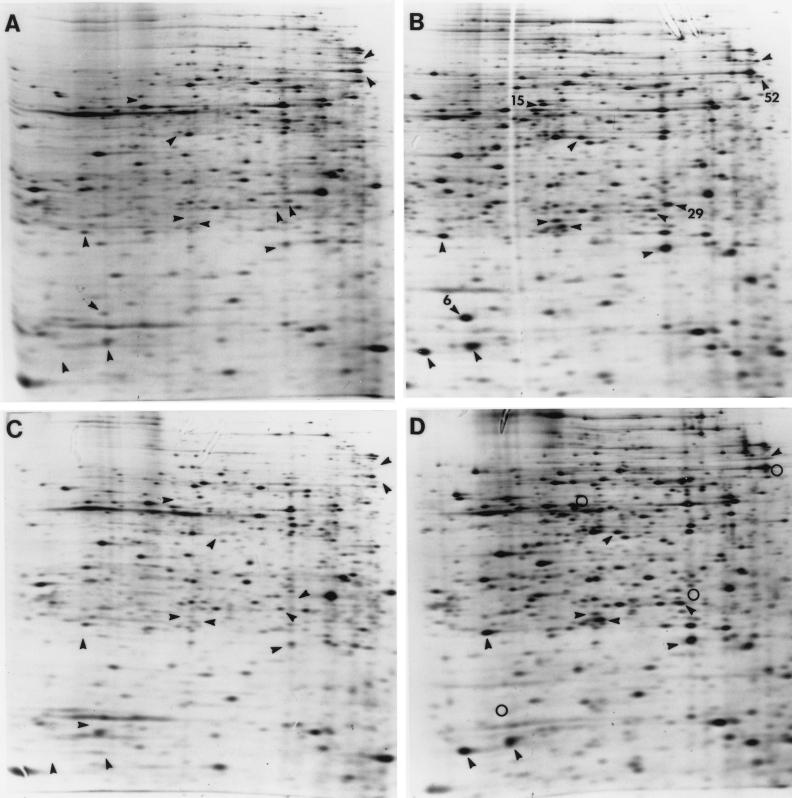
One of the ASP proteins targeted for identification was a 26-kDa protein designated ASP29 (4). Its location on two-dimensional SDS-polyacrylamide gels suggested that it was identical to a protein called spot 7 that is induced by growth in macrophages (1). Amino-terminal sequence analysis of ASP29 provided a 20 residue N-terminal sequence identical to the N terminus of the regulatory protein PhoP (data not shown). Figure 4 illustrates the position of ASP29 on a two-dimensional gel, its induction by acid shock (Fig. 4A versus B), and its absence in a phoP mutant (Fig. 4C and D). The results indicate that ASP29 is, indeed, PhoP.
FIG. 4.
PhoP controls the acid shock induction of four ASPs. Two-dimensional SDS-PAGE analysis of ASPs produced by JF2690 (rpoS) (A and B) and JF3204 (rpoS phoP) (C and D). Unadapted cells (A and C) and cells that were shocked with acid (pH 4.4) for 20 min (B and D) are shown. PhoP-dependent ASPs are indicated by numbers in panel B and by open circles in panel D. Several other ASPs are indicated by arrowheads.
PhoP- and PhoQ-dependent systems protect against inorganic but not organic acid stress.
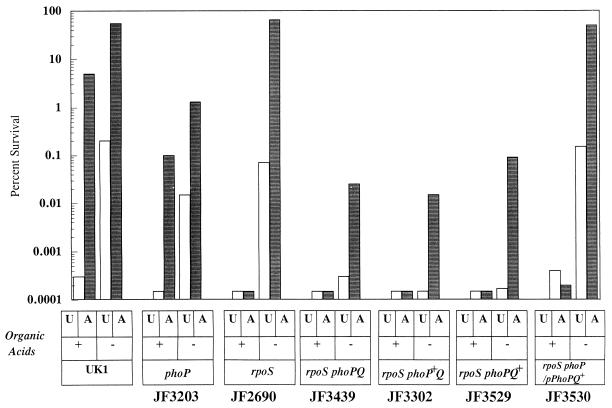
PhoP, along with PhoQ, forms a two-component system that is required for the pathogenesis of S. typhimurium (22, 30). An early study of acid tolerance suggested that PhoP was involved in the ATR (7). However, that study was unknowingly performed with an rpoS mutant background that allowed only transient induction of an ATR. To determine if PhoP was essential for acid tolerance in an rpoS+ strain, we examined the effect of phoP mutations on the ATR of UK1. As shown in Fig. 5, a mutation in phoP (JF3203) had a small but reproducible effect on acid tolerance in an rpoS+ cell, reducing it by approximately 10-fold, whether or not organic acids were present. Clearly, RpoS-dependent systems are more important than PhoP-dependent systems in protecting cells against media at pH 3 containing fermentation end products. However, when an rpoS phoP mutant was tested for inorganic acid tolerance, it was extremely acid sensitive compared to an rpoS phoP+ cell (Fig. 5 [JF3439 versus JF2690]). The data provided in Fig. 5 also illustrate that both PhoP and PhoQ are required for this system of acid tolerance. Strains that were phenotypically PhoP+ Q− (JF3302), PhoP− Q− (JF3439), and PhoP− Q+ (JF3529) all proved to be acid sensitive. In addition, acid tolerance of the phoP::Tn10 strain (PhoP− Q−) was rescued by a plasmid expressing wild-type phoPQ (JF3530).
FIG. 5.
The PhoPQ two-component signal transduction system is involved in inorganic acid tolerance. Cells (indicated below each set of bars) were grown and treated as indicated in the legends to Fig. 1 and 3. U, unadapted; A, adapted.
PhoPQ also controls the expression of another two-component regulatory system called PmrA (11, 24, 26). Some PhoPQ-dependent genes are regulated both by Mg2+ and acid (28). It has been suggested that PhoPQ controls the Mg2+-dependent expression of these genes, while PmrA controls their induction by acidic pH (26). Consequently, we questioned whether the acid-sensitive nature of phoPQ mutants could be through PhoPQ control of pmrA. Examination of an rpoS pmrA mutant revealed a normal inducible tolerance to inorganic acid (data not shown). Thus, the acid-sensitive nature of phoPQ mutants is due to a different subset of PhoPQ-regulated genes.
PhoP is required for the low-pH induction of a subset of acid shock proteins.
Shifting cells from a slightly alkaline pH (pH 7.7) to a moderately acidic pH (pH 4.4) results in the synthesis of 51 ASPs. The regulatory systems that sense environmental shifts in pH and control the expression of these ASPs are, of course, crucial to the ATR. The alternative sigma factor ς38 is required for the synthesis of 8 ASPs, while Fur is required for a separate set of 8 proteins. Since PhoP proved to be an ASP and phoP mutants are acid sensitive, it was reasonable to suspect that PhoPQ might also regulate another ASP subset. Figure 4 illustrates that a set of four ASPs, different from the previously mentioned sets, are missing from a phoP mutant. As shown above, ASP29 is PhoP, but the identities of the others, i.e., ASP6, -15, and -52, remain unknown. Thus, 20 of 51 identified ASPs fall into one of three different regulatory groups. It is interesting to note that the acid shock induction of these PhoP-dependent ASPs was not dependent on PmrA, the PhoPQ-controlled regulator suspected of sensing pH (data not shown).
The reliance of PhoPQ-dependent gene expression on acidic pH, magnesium, and PmrA.
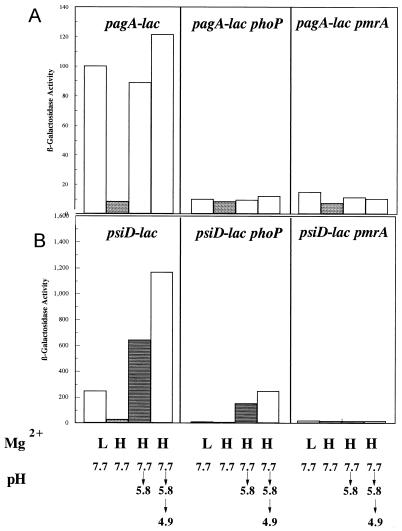
The current model for PhoPQ regulation holds that the membrane-bound sensor kinase PhoQ senses extracytoplasmic Mg2+. Under high Mg2+ concentrations, PhoQ does not phosphorylate PhoP so that PhoP is not active as a DNA-binding protein (30). PhoQ is not generally considered a pH sensor. As noted above, it has been proposed that the pH control of a subset of PhoP-regulated genes is due to the PmrAB two-component system, which itself can be activated by PhoP (26). Thus, the pH and Mg2+ controls over PhoP-dependent genes are thought to be separate. Loss of PhoP should eliminate the Mg2+ control of these genes but not their regulation by pH. Conversely, a pmrA mutation should abrogate pH control but not Mg2+ regulation. Previous studies indicating that PhoP does not sense pH were done under fairly mild pH conditions (pH 6). We retested the model under more acidic conditions, achieved in a stepwise manner (pH 7.7 to 5.8 to 4.9) thought to mimic more closely what Salmonella might experience during pathogenesis. Using a pagA-lacZ fusion as a reporter for PhoP activity, we found that pagA was induced in a PhoP-dependent fashion by acidic pH, even in the presence of repressing concentrations of Mg2+ (Fig. 6A). Since PhoP can increase pmrAB transcription but is not essential for pmrAB expression, the results were contrary to what was expected, which was that phoP should have had little effect on acidic pH control of this target gene. When a pmrA mutation was tested, it too proved to be essential for the acid induction of pagA-lacZ at high Mg2+ concentrations as well as for the low Mg2+ response.
FIG. 6.
The effects of Mg2+, acid, phoP, and pmrA on the expression of pagA and psiD (pmrC). Cells were grown in N medium (pH 7.7) supplemented with 1% vitamin-free Casamino Acids to mid-log phase (2 × 108 cells/ml) in either 10 mM (H) or 10 μM (L) MgSO4. As indicated below each bar, the cells were adapted at pH 5.8 for 60 min or underwent a stepwise adaptation at pH 5.8 for 30 min followed by 30 min at pH 4.9. The results are representative of triplicate experiments. (A) pagA-lac (JF3303), pagA-lac phoP (JF3531), and pagA-lac pmrA (JF3547) cells; (B) psiD-lac (JF3550), psiD-lac phoP (JF3554), and psiD-lac pmrA (JF3561) cells.
Because of these findings, other PhoP-dependent genes were analyzed for the effects of phoP and pmrA mutations on pH and Mg2+ regulation. In contrast to pagA, a phoP mutation did not eliminate the pH 4.9 acid induction of psiD, now known to be pmrC (10a), but did severely reduce it (Fig. 6B). This result is similar to a previous report which examined the expression of the PhoP-dependent gene pbgP under less acidic conditions (26). However, unlike pbgP, PmrA was important for both the Mg2+ and pH control of psiD (pmrC) (Fig. 6B).
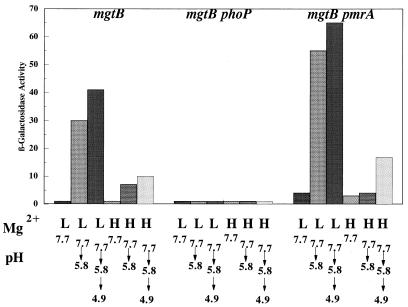
The mgtB gene, encoding a magnesium transport system, is known to be regulated by Mg2+ and is PhoPQ dependent (28). Under the conditions shown in Fig. 7, induction of the mgtB-lacZ fusion was found to require both low Mg2+ and an acidic pH shift. Low magnesium levels alone did not induce this gene in the time frame of this experiment. Consequently, mgtB can also be considered an acid-inducible gene. As shown in Fig. 7, the acid and low-Mg2+ response of mgtB proved to be PhoP dependent but, unlike that of psiD (pmrC), was PmrA independent. One possible explanation for these results is that PhoQ senses pH in addition to Mg2+ and under acidic conditions will phosphorylate PhoP even in the presence of excess Mg2+.
FIG. 7.
The effects of Mg2+, acid, phoP, and pmrA on the expression of mgtB. Cells were grown in N medium (pH 7.7) supplemented with 1% vitamin-free Casamino Acids to mid-log phase (2 × 108 cells/ml) in either 10 mM (H) or 10 μM (L) MgSO4. As indicated below each bar, the cells were adapted at pH 5.8 for 60 min or underwent a stepwise adaptation at pH 5.8 for 30 min followed by 30 min at pH 4.9. The results are representative of triplicate experiments. Results for mgtB-lac (JF3274), mgtB-lac phoP (JF3552), and mgtB-lac pmrA (JF3551) cells are shown.
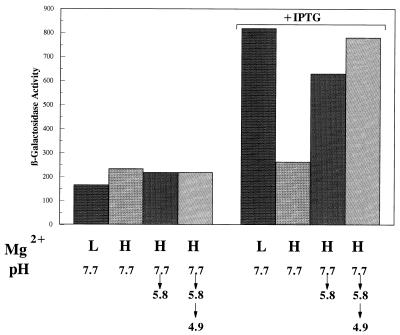
Since PhoPQ autoregulates its own expression (27), we decided to test whether phoPQ expression was itself induced by acid, as the two-dimensional gels would suggest. In order to conduct this study, we constructed a strain containing a chromosomal phoP::MudJ insertion and a plasmid in which phoPQ was placed under the control of the lac promoter (27). The results shown in Fig. 8 confirmed that when PhoPQ is produced following isopropyl-β-d-thiogalactopyranoside (IPTG) induction, the expression of phoP-lacZ is induced under low Mg2+ conditions. However, under high Mg2+ conditions, phoP was also induced by acidic pH shifts lasting no more than 1 h. The results indicate that PhoQ senses H+ in addition to Mg2+ concentrations.
FIG. 8.
The effects of Mg2+ and acid on the expression of phoP. Cells were grown as described in the legends to Fig. 6 and 7 either in the presence or in the absence of 0.2 mM IPTG to induce the plasmid-borne phoPQ operon.
DISCUSSION
The results presented confirm that acid stress is a combination of low pH (inorganic acid) and the concentration of organic weak acids present in the microbial environment. In addition, the data illustrate that there are distinct tolerance systems for each type of acid stress. The RpoS-dependent system is clearly required for protection against organic acid stress, whereas PhoPQ is used mostly against inorganic acid stress. With respect to inorganic acid stress, the data suggest that the PhoPQ and RpoS systems are somewhat redundant, although both are required for optimum acid tolerance. The minor decrease in organic acid tolerance seen in the phoP mutant probably reflects the fact that part of how these acids work is by lowering internal pH, much as inorganic acid does. Alternatively, the loss of PhoP may slightly increase permeability toward organic acids, insofar as some PhoP-regulated genes have been shown to affect the cell surface (12). The results also indicate that the iron regulatory protein Fur and the adaptive response protein Ada are required for organic but not inorganic acid tolerance. The manner in which Fur protein aids in acid tolerance is not clear. It does appear that Fur can sense acid and iron levels independently and that Fur regulates the expression of several acid shock proteins in an iron-independent manner (14). It is predicted that one or more of the Fur-dependent ASPs is involved in this protection.
The Ada protein is involved in the adaptive response of E. coli to alkylating agents in two ways, i.e., as an enzyme by removing methyl groups from O6 methyl guanine residues and as a potent transcriptional activator of its own gene and several others involved in the response (25). S. typhimurium appears to produce a defective Ada protein that has lost the ability to induce its own expression but still can activate other target genes involved in DNA repair (13, 33, 34). The fact that ada expression falls under RpoS control in E. coli suggests that one of the reasons that rpoS mutants in Salmonella are acid sensitive is because they cannot induce ada. Why an ada mutant would be sensitive to organic acids is not intuitively obvious, since organic acids probably do not cause methylation damage to DNA. However, low pH can facilitate the inappropriate methylation of DNA by S-adenosyl methionine (18). Acid shock-stimulated accumulation of RpoS may lead to the induction of Ada in Salmonella. Upon removing methyl groups from O6 methyl guanine residues, Ada may then induce other components of the Ada regulon (other than ada itself). The products of these other genes may deal with damage caused by organic acids. Confirmation of this model awaits more detailed analysis of ada expression in S. typhimurium.
Consistent with a role for PhoP in acid tolerance was the finding that PhoP is itself an acid shock protein that is required for the expression of several other ASPs. The role that acidic pH plays in the expression of PhoP-dependent genes is complex. While the acid-induced expression of some PhoP-dependent genes requires PmrA [e.g., pbgP and psiD (pmrC)], acid-induced expression of others is PmrA independent (e.g., mgtB and phoP) or requires both PhoP and PmrA (e.g., pagA). The observation that the transcription of phoPQ is induced by acid shock in a PhoPQ-dependent manner certainly suggests that PhoQ can sense pH; thus, both PhoPQ and PmrAB appear to sense acid stress. It is not apparent, at this point, whether PhoQ senses pH independently of Mg2+ or whether pH affects the interaction between Mg2+ and the Mg2+-sensing site on PhoQ. If the latter is the case, then acidic conditions could be translated by the cell as low Mg2+, possibly triggering an influx of this ion. Increasing intracellular magnesium concentrations through the acid induction of Mg2+ transport via PhoP might serve a protective function under acidic conditions. The observation that mgtB mutations do not confer an acid-sensitive phenotype (data not shown) may simply reflect a redundancy in Mg transport systems or that Mg2+ transport is not an essential component of acid tolerance.
The induction of phoPQ by low pH also has important implications in terms of virulence. Mutants defective in this system are avirulent. Since the environmental condition implicated in the control of PhoP-regulated genes is Mg2+, it has been reasoned that in vivo conditions in which Mg2+ levels are low are important in initiating the activation of this system (28, 30). However, the present studies suggest that even when Mg2+ concentrations in vivo are high, low pH environments can also trigger activation of PhoP. Therefore, the environmental cue within the macrophage that triggers the PhoPQ cascade remains undetermined but probably reflects a combination of Mg2+ and H+ ion concentrations. The following is a working model that attempts to integrate the available data concerning how Mg2+, H+, PhoPQ, and PmrAB might cooperatively regulate gene expression. The model predicts at least two basic sets of PhoP-regulated genes. In this model, one set of PhoP-regulated genes needs only PhoP phosphate (PhoP-P) for induction, while the second set requires both PhoP-P and PmrA-P. Individual members of the first set will vary in their responses to the different levels of PhoP-P achieved through PhoQ sensing different levels of Mg2+. Higher or lower H+ concentrations will influence the level of Mg2+ that is required to produce a given level of PhoP-P. Thus, genes that require little PhoP-P for induction could respond to either low pH or low Mg2+. Genes that require high PhoP-P will be induced under extremely low Mg2+ concentrations at neutral pH or under more moderate Mg2+ levels in an acidic environment.
The second set of PhoP-regulated genes could be regulated by a PhoP-PmrA cascade that amplifies the pH response. In this case, acidic environments (even under high Mg2+ conditions) would generate enough PhoP-P to induce the PmrA system (represented by psiD [pmrC] in Fig. 6). The PmrA system subsequently undergoes autoinduction, which would serve to amplify the pH signal. The resultant high level of PmrA-P generated either through the PmrB sensor kinase sensing pH directly or through some other signal will then fully induce the PmrA subset of PhoP-regulated genes.
The data presented underscore the complexity of acid tolerance and pH sensing systems in S. typhimurium. The number of regulatory systems involved with acid survival now numbers four, including Fur, ς38, PhoPQ, and Ada. Clearly, several more regulatory systems are involved, since there are at least 20 or so ASPs for which regulatory systems have not been revealed. The elaborate response raised against acid stress suggests that many different facets of cellular physiology are impacted by low pH and that an elaborate, interwoven regulatory network is critical for survival.
ACKNOWLEDGMENTS
We thank M. Maguire, S. Miller, E. Groisman, and M. Yamada for their generous gifts of various strains and plasmids and M. Moreno and J.-C. Giard for critically reading the manuscript. Various discussions with M. Maguire and M. Spector were especially helpful and gratefully acknowledged.
This work was supported by an award (GM48017) from the National Institutes of Health.
REFERENCES
- 1.Abshire K Z, Neidhardt F C. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J Bacteriol. 1993;175:3734–3743. doi: 10.1128/jb.175.12.3734-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearson S, Bearson B, Foster J W. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 3.Bearson S M D, Benjamin W H, Jr, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster J W. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J Bacteriol. 1991;173:6896–6902. doi: 10.1128/jb.173.21.6896-6902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster J W. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–237. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- 6.Foster J W, Bearson B. Acid sensitive mutants of Salmonella typhimurium identified through a dinitrophenol selection strategy. J Bacteriol. 1994;176:2596–2602. doi: 10.1128/jb.176.9.2596-2602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster J W, Hall H K. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990;172:771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster J W, Hall H K. The effect of Salmonella typhimurium ferric-uptake regulator (fur) mutations on iron and pH-regulated protein synthesis. J Bacteriol. 1992;174:4317–4323. doi: 10.1128/jb.174.13.4317-4323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster J W, Spector M P. Phosphate-starvation regulon of Salmonella typhimurium. J Bacteriol. 1986;166:666–669. doi: 10.1128/jb.166.2.666-669.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster J W, Spector M. How Salmonella survives against the odds. Annu Rev Microbiol. 1995;49:145–174. doi: 10.1146/annurev.mi.49.100195.001045. [DOI] [PubMed] [Google Scholar]
- 10a.Groisman, E. Personal communication.
- 11.Gunn J S, Miller S I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S I. Regulation of lipid a modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 13.Hakura A, Morimoto K, Sofuni T, Nohmi T. Cloning characterization of the Salmonella typhimurium ada gene, which encodes O6-methylguanine-DNA methyltransferase. J Bacteriol. 1991;173:3663–3672. doi: 10.1128/jb.173.12.3663-3672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall H K, Foster J W. The role of Fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol. 1996;178:5683–5691. doi: 10.1128/jb.178.19.5683-5691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengge-Aronis R. Back to log phase: ςs as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee I S, Lin J, Hall H K, Bearson B, Foster J W. The stationary-phase sigma factor ςs (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol. 1995;17:155–167. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee I S, Slonczewski J L, Foster J W. A low-pH inducible stationary phase acid tolerance response in Salmonella typhimurium. J Bacteriol. 1994;176:1422–1426. doi: 10.1128/jb.176.5.1422-1426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. J Biochem. 1972;11:3610–3617. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 19.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςs (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 20.Maloy S R. Experimental techniques in bacterial genetics. Boston, Mass: Jones and Bartlett Publishers; 1990. [Google Scholar]
- 21.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 22.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoPphoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson D L, Kennedy E P. Transport of magnesium by a repressible and a nonrepressible system in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:1091–1093. doi: 10.1073/pnas.69.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roland K L, Martin L E, Esther C R, Spitznagel J K. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saget B M, Walker G C. The Ada protein acts as both a positive and a negative modulator of Escherichia coli’s response to methylating agents. Proc Natl Acad Sci USA. 1994;91:9730–9734. doi: 10.1073/pnas.91.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soncini F C, Groisman E A. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soncini F C, Véscovi E G, Groisman E A. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soncini F C, Véscovi E G, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spector M P, Aliabadi Z, Gonzalez T, Foster J W. Global control in Salmonella typhimurium: two-dimensional electrophoretic analysis of starvation-, anaerobiosis-, and heat shock-inducible proteins. J Bacteriol. 1986;168:420–424. doi: 10.1128/jb.168.1.420-424.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Véscovi E G, Soncini F, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 31.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 32.Waldburger C D, Sauer R T. Signal detection by the PhoQ sensor-transmitter. J Biol Chem. 1996;271:26630–26636. doi: 10.1074/jbc.271.43.26630. [DOI] [PubMed] [Google Scholar]
- 33.Yamada M, Hakura A, Sofuni T, Nohmi T. New methods for gene disruption in Salmonella typhimurium: construction of an ada-deletion derivative of Salmonella typhimurium TA 1535. J Bacteriol. 1993;175:5539–5547. doi: 10.1128/jb.175.17.5539-5547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada M, Sedgwick B, Sofuni T, Nohmi T. Construction and characterization of mutants of Salmonella typhimurium deficient in DNA repair of O6-methylguanine. J Bacteriol. 1995;177:1511–1519. doi: 10.1128/jb.177.6.1511-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]