Abstract
The spirochete which causes Lyme disease, Borrelia burgdorferi, has many features common to other spirochete species. Outermost is a membrane sheath, and within this sheath are the cell cylinder and periplasmic flagella (PFs). The PFs are subterminally attached to the cell cylinder and overlap in the center of the cell. Most descriptions of the B. burgdorferi flagellar filaments indicate that these organelles consist of only one flagellin protein (FlaB). In contrast, the PFs from other spirochete species are comprised of an outer layer of FlaA and a core of FlaB. We recently found that a flaA homolog was expressed in B. burgdorferi and that it mapped in a fla/che operon. These results led us to analyze the PFs and FlaA of B. burgdorferi in detail. Using Triton X-100 to remove the outer membrane and isolate the PFs, we found that the 38.0-kDa FlaA protein purified with the PFs in association with the 41.0-kDa FlaB protein. On the other hand, purifying the PFs by using Sarkosyl resulted in no FlaA in the isolated PFs. Sarkosyl has been used by others to purify B. burgdorferi PFs, and our results explain in part their failure to find FlaA. Unlike other spirochetes, B. burgdorferi FlaA was expressed at a lower level than FlaB. In characterizing FlaA, we found that it was posttranslationally modified by glycosylation, and thus it resembles its counterpart from Serpulina hyodysenteriae. We also tested if FlaA was synthesized in a spontaneously occurring PF mutant of B. burgdorferi (HB19Fla−). Although this mutant still synthesized flaA message in amounts similar to the wild-type amounts, it failed to synthesize FlaA protein. These results suggest that, in agreement with data found for FlaB and other spirochete flagellar proteins, FlaA is likely to be regulated on the translational level. Western blot analysis using Treponema pallidum anti-FlaA serum indicated that FlaA was antigenically well conserved in several spirochete species. Taken together, the results indicate that both FlaA and FlaB comprise the PFs of B. burgdorferi and that they are regulated differently from flagellin proteins of other bacteria.
Lyme disease is a tick-transmitted illness caused by the spirochete Borrelia burgdorferi. It is the most prevalent vector-borne bacterial disease in the United States. In 1996, there was a 41% increase in confirmed cases over the previous year (12). B. burgdorferi is morphologically similar to other spirochetes; within the outer membrane sheath, several periplasmic flagella (PFs) wrap around the protoplasmic cell cylinder (5, 30). These PFs have an essential role in motility and cell morphology (30, 31, 63). One striking feature of B. burgdorferi and other spirochetes is their capacity to efficiently swim in a viscous gel-like medium such as connective tissue where other bacteria are slowed down or immobilized (31, 40); this invasive attribute may facilitate their passage through the extracellular matrix and cell junctions in infected tissues (40, 63).
The spirochete PF apparatus is similar to the flagellum of other bacteria; it has a filament, hook, and basal body (5, 6, 8, 14, 33, 34, 44). The PFs of most spirochetes are comprised of a class of core proteins referred to as FlaB and a class of outer layer proteins referred to as FlaA (7, 8, 14, 15, 41, 42, 46, 54, 55, 61, 69). Both FlaA and FlaB are among the most abundant cell proteins (42, 46, 55). Depending on the species, the PFs consist of one to two different FlaA proteins and two to four different FlaB proteins (8, 14, 42, 46, 54, 55, 69). FlaB proteins show sequence similarity to the flagellin proteins from other bacteria and are evidently excreted by the flagellin-specific pathway (9, 14, 54, 55). On the other hand, FlaA proteins show no homology to other proteins and are most likely excreted to the periplasm by the secA-mediated pathway (9, 35, 41, 54). In addition, there is some evidence for glycosylation of PFs. Based on lectin binding results, Brahamsha and Greenberg suggest that a minor FlaB protein may be glycosylated in Spirochaeta aurantia (8). Most recently, FlaA from Serpulina hyodysenteriae (46) has been shown by several criteria to be posttranslationally modified by glycosylation.
For over a decade, both ultrastructural analysis and biochemical isolation and characterization indicated that the PF filaments of B. burgdorferi differed from those of other spirochetes. These PFs were said to be comprised primarily of a 41-kDa FlaB protein (5, 6, 14, 16). However, we recently found a flaA homolog in B. burgdorferi which mapped in a flagellum/chemotaxis operon (fla/che) (20, 23). In addition, B. burgdorferi flaA was found to be expressed in growing cells, and the encoded protein reacted with an antiserum directed to FlaA of T. pallidum (23).
In this study, we examined whether B. burgdorferi FlaA is associated with the PFs, and we also characterized FlaA in detail. Using a new procedure to isolate the PFs, we found that FlaA purified along with the PFs. These results, along with the analysis of a spontaneously occurring PF-deficient mutant, HB19Fla−, isolated by Sadziene et al. (63), indicate that FlaA is a PF protein. We also analyzed the transcription and translation of flaA in the wild type and HB19Fla−. The results indicate that expression of both flaB (63) and flaA is likely to be controlled at the translational level. Finally, we present evidence that FlaA of B. burgdorferi is glycosylated and that it is antigenically similar to its counterparts from other spirochete species.
MATERIALS AND METHODS
Bacterial strains.
The strains of B. burgdorferi senso stricto (3) include 212 (17), HB19 and HB19Fla− (64), and B31 (39). B. burgdorferi cells were grown in BSK-II medium without gelatin (4). S. hyodysenteriae B204 (61), Treponema denticola 33520, and the PF-deficient flgE mutant HL51 were cultured as previously described (45, 62). Other spirochete cell pellets or lysates were from the following sources: Borrelia afzelii VS461, Borrelia garinii G2, and Borrelia hermsii HS1 from T. Schwan (Rocky Mountain Laboratories, Hamilton, Mont.), Treponema phagedenis Kazan 5 and T. pallidum subsp. pallidum from R. Limberger and K. Wicher (Wadsworth Center for Laboratories and Research, Albany, N.Y.), and Spirochaeta aurantia M1 from E. P. Greenberg (University of Iowa, Iowa City).
DNA and RNA manipulation.
Unless noted, all procedures for genetic manipulations were carried out according to standard methods (2). The PCR was done with PrimeZyme (Biometra) DNA polymerase (24). The amplified PCR products were cloned into pGEM-T vector (Promega) for further manipulation. Reverse transcription-PCR (RT-PCR) was performed by using the Promega RT-PCR Access system as previously reported (27, 28).
RNase protection assays were carried out according to standard procedures, using a Hybspeed RPA kit from Ambion (68). Briefly, primers to the B. burgdorferi flaA (GenBank accession no. U62900) and 16S rRNA (GenBank accession no. L40596) genes were used to amplify specific regions by PCR. The amplified regions (305-bp flaA, 354-bp rRNA) were cloned into pGEM-T, and the plasmids were then linearized with either PstI, SalI, or SphI. Antisense riboprobe was synthesized with [α-32P]UTP (Amersham) by using an in vitro transcription kit (Ambion) and purified by using a 5% polyacrylamide–8 M urea gel. Total cellular RNA was extracted and purified from 1 liter of cells, using an RNeasy Mini Kit from Qiagen. Hybridizations and RNase A and RNase T1 digestions were carried out as described for the Hybspeed RPA kit. Protected fragments were separated on 5% polyacrylamide–8 M urea denaturing gels and were analyzed both by autoradiography and quantitatively with a Molecular Dynamics PhosphorImager.
Electrophoresis, immunoblotting, and recombinant protein purification.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as previously reported (23, 28). T. pallidum and S. hyodysenteriae polyclonal anti-FlaA antisera have been previously described and were obtained from S. Norris (University of Texas Medical School, Houston) and M. Jacques (University of Montreal, Montreal, Quebec, Canada), respectively (23, 29, 46, 54). Monoclonal antibody to S. aurantia FlaA (.3E9F6) was obtained from E. P. Greenberg (8), monoclonal antibody to T. pallidum FlaA (H9-2) was obtained from J. Radolf (Southwestern Medical School, Dallas, Tex.) (50), and monoclonal antibody H9724 to B. burgdorferi FlaB (6) was obtained from A. Barbour (University of California, Irvine). Western blot assays were performed as previously described, using the Amersham ECL (enhanced chemiluminescence) system (23, 24, 28, 29). Primary antibodies were usually diluted from 1:1,000 to 10,000. For monoclonal antibody H9724, we used a dilution of 1:2000; for T. pallidum polyclonal anti-FlaA, we used a 1:5,000 dilution.
Purification of PFs and electron microscopy.
The procedure for purification of T. denticola PFs has been previously described (62). For the purification of B. burgdorferi PFs, we used two PF isolation procedures. First, the PFs were isolated by using a modified version of our previously described methods (44). One liter of logarithmic phase cells (5 × 107 cells/ml) was harvested and washed twice in 200 ml of a cold phosphate buffer (0.13 M phosphate [pH 7.4]). Outer membranes were removed by resuspending the pellet in 50 ml of 1% Triton X-100 followed by incubation at 37°C for 60 min (10). The cell cylinders were collected by centrifugation and resuspended in 40 ml of phosphate-buffered saline (PBS). To shear the PFs from the cell cylinders, the suspension was vortexed for 45 s with 1-mm-diameter glass beads (Sigma). The cellular debris was removed by centrifugation for 15 min at 15,000 × g for at 4°C. The PFs in the supernatant were collected by centrifugation at 100,000 × g for 1 h at 10°C and stored at −20°C (crude PF fraction). For further purification, the pellet was resuspended in 7 ml of PBS–5 g of CsCl and centrifuged at 50,000 × g at 15°C for 50 h. The band containing the PFs (density, 1.37 g/cm2) was collected and dialyzed overnight against an excess volume of PBS (purified PF fraction).
The second protocol for PF purification is similar to one previously described (6, 16). In brief, after three washes in PBS containing 5 mM MgCl2, the bacteria were lysed with 30 ml of 2% N-lauroylsarcosine, sodium salt (Sarkosyl; Fisher Scientific), in 10 mM Tris–1 mM EDTA, pH 8.0 (2% STEDTA), for 1 h at 37°C. The PFs were collected by centrifugation at 45,000 × g for 2 h at 25°C, and resuspended in 2% STEDTA buffer, and incubated for 10 min at 37°C as before. After centrifugation, the pellet was resuspended in 150 mM NaCl and sheared with glass beads. The suspension was centrifuged at 150,000 × g for 90 min at 25°C, and the pellet containing the PFs was collected and resuspended in PBS. The procedures used for electron microscopy have been previously described. PFs were negatively stained with 2% uranyl acetate for 1 min and examined in a JEOL 100CS microscope (62).
Purification of FlaA.
Native 38.0-kDa FlaA protein was isolated directly from SDS-polyacrylamide gels for Western blots and glycoprotein detection (2). After SDS-PAGE of the crude PFs, the appropriate protein bands were cut from the gel and transferred into a dialysis bag with transfer buffer (25 mM Tris, 0.2 M glycine, 15% methanol). The electroelution was performed at 100 V for 30 min. The electroeluted protein was collected, concentrated, and stored for further analysis.
Peptide analysis of FlaA.
FlaA was identified by peptide mass fingerprinting (38). A 5-pmol aliquot of protein was subjected to SDS-PAGE. After staining with Coomassie blue R-250, the single band was excised from the gel and digested with trypsin (60). The mixture of peptides released was subjected to salt exchange by using a 1.5-cm by 0.25-mm guard column, containing Waters Delta-Pak 300Å, C18 stationary phase, packed by Microtech Scientific (Sunnyvale Calif.). Peptides were eluted from the column directly onto a mass spectrometer sample plate with a solution of 80% acetonitrile, 20% water, and 0.1% formic acid. The masses of the tryptic peptides were measured with a Voyager DE time-of-flight (TOF) mass spectrometer (Perseptive Biosystems, Inc., Framingham, Mass.). Ionization by matrix-assisted laser desorption (MALDI) was performed with α-cyano-4-hydroxycinnamic acid as the matrix. A precision of at least ±0.4 Da was achieved by using delayed extraction and by calibrating the mass scale, using two fragments of trypsin as internal standards. Searching of the mass values against the National Center for Biotechnology Information (NCBI) database (revised 21 Dec. 1997) was performed with the MS-Fit program, available at http://prospector.ucsf.edu/htmlucsf/msfit.htm. Protein identification was confirmed by sequencing selected tryptic peptides. Sequence analysis was performed by collisional dissociation (56) in a Quattro II triple-quadrupole mass spectrometer (Micromass, Altrincham, Cheshire, England), using nanoelectrospray for sample introduction and ionization (70). Signals in the daughter ion spectra were compared with those expected from the peptide sequences matched in the mass fingerprinting experiments.
Glycoprotein detection.
Several techniques were used for glycoprotein detection. First, the Amersham ECL glycoprotein detection system was used according to the manufacturer’s instructions; this system is similar to that reported by Doig et al. (18). Briefly, proteins were first separated by SDS-PAGE and then transferred to a polyvinylidene difluoride membrane. The blot was then treated with sodium metaperiodate, followed by reaction with a biotin X hydrazine reactant. The biotinylated product was then detected by using streptavidin-horseradish peroxidase and the ECL assay. Serum glycoprotein transferrin served as a positive control. Recombinant FlaA (with a deletion of the N-terminal signal sequence; amino acids 1 to 26) and FliI proteins were overexpressed and purified as previously reported (28, 29). Second, glycosylation was detected by using a digoxigenin (DIG)-glycan differentiation kit (Boehringer Mannheim). Samples were subjected to SDS-PAGE and blotted with DIG-labeled lectins and specific anti-DIG alkaline phosphatase conjugates. Reactions were developed with 4-nitroblue tetrazolium chloride–5-bromo-4-chloro-3-indolylphosphate. The lectins which were screened and their corresponding recognition sites include the following: GNA (from Galanthus nivalis, terminal mannose); SNA (from Sambucus nigra, α-[2-6]-linked sialic acids), MAA (from Maackia amurensis, α[2-3]-linked sialic acids), PNA (Arachis hypogaea, Galβ[1-3]GalNAc), and DSA, (β[1-4]-linked oligomers of N-acetylglucosamine and terminal N-acetyllactosamine). Proteins were electrophoresed as previously described. Known positive controls provided by the manufacturer were used in all reactions and yielded expected results. Finally, native and recombinant N-glycosidase F (N-glycosidase F* designates the recombinant form) from Flavobacterium meningosepticum were used to test individual proteins while in solution as described by the manufacturer (Boehringer Mannheim). Deglycosylations were carried out with 6 mU of enzyme for 18 h at 37°C in 35 mM EDTA–35 mM sodium phosphate, pH 6.1 (67). These proteins were subsequently electrophoresed and stained by Coomassie brilliant blue or by specific lectins as described above after SDS-PAGE and blotting.
RESULTS
Coisolation of the FlaA protein with the PFs.
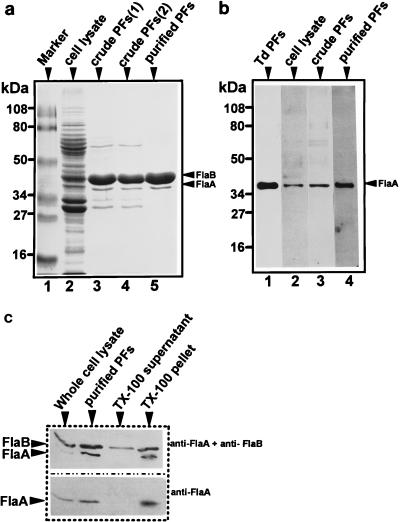
We have previously shown that the B. burgdorferi 38-kDa FlaA homolog has a primary and predicted secondary structure similar to those of its counterparts from other bacteria (23, 29). Based on this information, we hypothesized that FlaA was closely associated with FlaB and resided on the PF filament. Accordingly, we isolated the PFs and tested whether FlaA also copurified during the procedure. To minimize the dissociation of FlaA from FlaB, we used a simple protocol that avoided harsh chemical treatment. B. burgdorferi cells were first treated with Triton X-100 to remove the outer membrane sheath (10). The PFs were then isolated by shearing followed by centrifugation (crude PF preparation). Further purification was achieved by CsCl isopycnic gradient centrifugation (purified PF preparation). Electron microscopic examination of the purified PFs indicated that this preparation was relatively free of debris (Fig. 1). As shown by SDS-PAGE, both the 41- and 38-kDa proteins were the prominent protein species in both the crude and purified PFs (Fig. 2a, lanes 3 to 5). Several other proteins, of 60 kDa, 33 kDa, and 31 kDa, were detected in the crude PFs (Fig. 2a, lanes 3 and 4) but not the purified PF preparations (Fig. 2a, lane 5) and were thus likely contaminants. Immunoblotting with T. pallidum anti-FlaA confirmed that the 38-kDa protein was FlaA (Fig. 2b, lanes 2 to 4; Fig. 2c), and the 41-kDa protein corresponded to FlaB (Fig. 2c) PFs isolated from T. denticola were used as a positive control for FlaA (Fig. 2b, lane 1) (62). SDS-PAGE and Coomassie blue staining revealed that several proteins were released into the Triton X-100 supernatant fraction (not shown). However, immunoblot analysis revealed that although some FlaB partitioned into this fraction (Fig. 2c, lane 3, top), no FlaA was detected (Fig. 2c, lane 3, bottom). These results suggest that FlaA is not associated with the outer membranes and that it resided with the PFs. Because both FlaA and FlaB proteins were the major proteins detected following PF purification, they evidently are closely associated and comprise the PFs. FlaA and FlaB also copurified in crude PF preparations from strains B31 (Fig. 2a, lane 4) and HB19 (not shown), indicating that FlaA and FlaB associate with the PFs in other strains of B. burgdorferi. In cell lysates, and in both crude and purified PFs, the amount of FlaA was considerably less than that of FlaB (Fig. 2a).
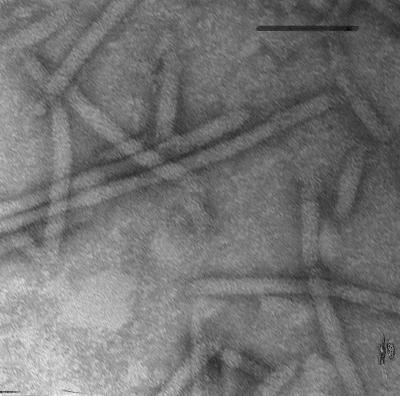
FIG. 1.
Electron microscopy of CsCl-purified PFs obtained by Triton X-100 extraction of B. burgdorferi from strain 212 negatively stained with uranyl acetate. Bar = 0.1 μm.
FIG. 2.
Analysis of PFs by SDS-PAGE and immunoblotting. (a) Cell lysates and PF preparations were subjected to SDS-PAGE (12% [wt/vol] polyacrylamide gel) and stained with Coomassie brilliant blue. Approximately 50 μg of protein was loaded in each lane. Lanes: 1, protein markers; 2, B. burgdorferi 212 cell lysate; 3, crude PFs of 212; 4, crude PFs of B31; 5, purified PFs of 212. (b) Western blotting with antiserum to T. pallidum FlaA. Approximately 10 μg of protein was loaded per lane. Lanes: 1, purified T. denticola (Td) PFs (63); 2, B. burgdorferi 212 cell lysate; crude PFs of 212; 4, purified PFs of 212. (c) Western blotting using polyclonal T. pallidum FlaA and monoclonal H9724 to B. burgdorferi FlaB during purification of 212 PFs.
Peptide analysis of FlaA.
Although the protein which purified along with the PFs reacted with an anti-FlaA antiserum, we had no direct proof that the flaA gene (20, 23) encodes that protein. In our attempts to analyze this protein by N-terminal amino acid analysis, we found that it was blocked to Edman degradation. Accordingly, purified FlaA was analyzed by mass fingerprinting in a MALDI-TOF mass spectrometer after trypsin digestion. The mass-to-charge ratios (m/z) of the protonated molecular ions corresponding to six tryptic peptides (1,065.22, 1,188.28, 1,630.63, 1,644.60, 1,786.80, and 2,063.80 Da) were searched against those of all proteins in the NCBI database with masses of 30 to 40 kDa (58,035 entries). Allowance was made for up to one missed tryptic cleavage site per peptide. Five of the peptides matched values for the protein encoded by B. burgdorferi flaA (NCBI accession no. 2688608) to within 0.24 Da. The sixth peptide, that with an m/z of 2,063.80, matched to within 0.35 Da a peptide from the same protein with the condition that both methionine residues contained by the peptide would have become oxidized. The identification of the protein encoded by flaA was then confirmed by fragmenting the doubly or triply charged ions corresponding to all six of the peptides in a triple-quadrupole mass spectrometer and matching the fragments with those expected for the peptide sequences identified in the mass fingerprinting experiment. The results from five of the six peptides were in accord with the expected sequences. One peptide, however, did not match the sequence predicted for it. This was the peptide with an m/z of 2,063.80 Da, which was also the one that matched with lowest precision in the fingerprinting experiment. We therefore judge this one match to be spurious. Accordingly, we conclude that the protein which reacts to the specific FlaA antiserum is encoded by the flaA gene previously identified.
Dissociation of FlaA from FlaB after treatment with Sarkosyl.
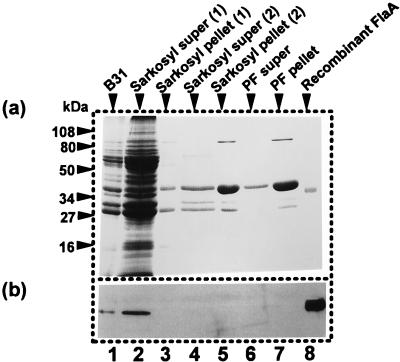
To explore why previous studies failed to detect FlaA associated with PFs, we repeated the purification procedure by using a method similar to one described by other investigators (6, 16). The major difference between these two methods is that the other method (6, 16) used Sarkosyl. We found that the use of Sarkosyl resulted in the release of most of the B. burgdorferi proteins (Fig. 3, lane 2). No FlaA was detected by Coomassie blue staining in the PF pellet after the first round of Sarkosyl lysis (Fig. 3a, lanes 3 and 5). We also tested for FlaA during PF purification by Western blot analysis. All of the FlaA dissociated from the PFs and was released into the supernatant after the first round of treatment with Sarkosyl (Fig. 3b, lane 2); no FlaA was detected in the final PF preparation (Fig. 3b, lanes 3 to 7). These results suggest that FlaA dissociated from the PFs during purification using Sarkosyl. In other experiments, FlaA was found to be associated with the PFs after the first treatment with Sarkosyl, but it dissociated after subsequent rounds of treatment (26). The above results, along with the evidence that FlaA is considerably lower in concentration than FlaB, likely account for the failure to detect FlaA in previous B. burgdorferi PF preparations. Electron microscopic examination of the PFs prepared by both methods revealed no obvious differences in structure (data not shown).
FIG. 3.
PFs of strain B31 isolated using Sarkosyl. (a) Coomassie brilliant blue staining; (b) Western blot analysis using anti-T. pallidum FlaA. The conditions for running the gel were the same as described for Fig. 2. Lanes: 1, cell lysate; 2, supernatant (super) fluid after first Sarkosyl extraction; 3, pellet after first extraction; 4, supernatant fluid after second extraction; 5, pellet after second extraction; 6, supernatant fluid after shearing; 7, pellet after shearing (crude PFs); 8, recombinant FlaA.
Defect in expression of FlaA in HB19Fla−.
Sadziene et al. isolated a spontaneously occurring nonmotile PF-deficient mutant, HB19Fla− (63). This mutant synthesized the hook-basal body structures but did not synthesize the flagellar filaments. Western and Northern blot analysis indicated that although HB19Fla− failed to produce the FlaB protein, the encoding message was still produced at a level equivalent to the wild-type level. Accordingly, Sadziene et al. suggested that FlaB could be regulated on the translational level (63). HB19Fla− is thus likely to be deficient in the last stage of PF assembly, i.e., assembly of the PF filament after hook-basal body synthesis.
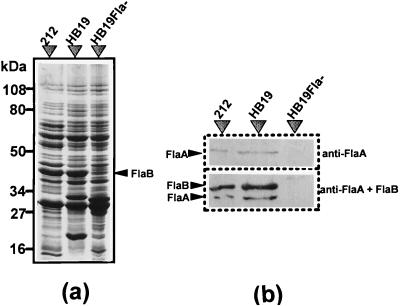
We hypothesized that like FlaB, FlaA is involved in the last stage of filament assembly. To test this hypothesis, we analyzed HB19Fla− with respect to FlaA synthesis by immunoblot analysis. SDS-PAGE and Coomassie blue staining revealed that FlaB was evident in the parental HB19 cell lysate but absent in HB19Fla− (Fig. 4a). Western blot analysis revealed that both FlaA and FlaB were also present in HB19 but absent in HB19Fla− (Fig. 4b). These results extend those of Sadziene et al. (63) and suggest that both FlaA and FlaB failed to be expressed in HB19Fla−.
FIG. 4.
Analysis of FlaA in HB19Fla− by SDS-PAGE and by immunoblotting. Approximately 10 μg of protein was loaded into each lane. (a) SDS-PAGE. Cell lysates of strains 212, wild-type HB19, and HB19Fla− were examined in a 10% polyacrylamide gel. FlaB is marked with an arrow. (b) Western blot. The whole-cell lysates of B. burgdorferi were treated with anti-T. pallidum FlaA serum (top) and anti-T. pallidum FlaA serum with anti-B. burgdorferi FlaB monoclonal antibody (bottom).
We examined whether the lack of expression of FlaA is caused by a genetic defect in flaA or failure to be transcribed. The region containing flaA from HB19Fla− was amplified by using a pair of primers derived from the adjacent genes orfA and cheA (23) and then sequenced. Comparison of the HB19Fla− region to that of strain 212 revealed 100% identity at the nucleotide level (GenBank accession no. U62900). These results suggest that flaA is intact in HB19Fla−.
We used RT-PCR analysis to test if flaA is transcribed in HB19Fla− (23, 28). Three pairs of primers spanning the adjacent and central regions were used: orfA to flaA, internal regions of flaA, and flaA to cheA (20, 23). Three specific products of the predicted sizes were successfully amplified from the mRNAs of HB19 and HB19Fla− (22). By extracting the PCR products from the gel and reamplifying with internal primers, the bands of the predicted sizes were shown to be the desired specific amplified products (22). These results, together with those obtained by Western blot analysis, suggest that flaA is transcribed but not translated in HB19Fla−.
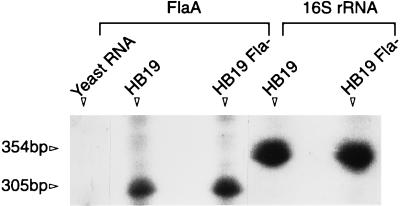
We compared the amount of flaA message in HB19Fla− to that in the wild type, using RNase protection assays. As a control, we also compared 16S rRNA messages in the two strains. There were no obvious differences between HB19 and HB19Fla− with either the flaA or 16S rRNA riboprobe (Fig. 5). The RNA protection was specific, as hybridization with yeast RNA was negative. We quantitatively compared the amounts of mRNA protected by the riboprobes with a PhosphorImager. Using four different amounts of total RNA in two different experiments, we found no significant difference in RNase protection between HB19 and HB19Fla− with either riboprobe. For example, we found that with 20 μg of total RNA and the flaA riboprobe, 11,069 cpm was protected in HB19 and 9,830 cpm was protected in HB19Fla−. These results indicate that HB19Fla− synthesized flaA message in an amount similar to the wild-type amount.
FIG. 5.
RNase protection assays of HB19 and HB19Fla−. The riboprobes to flaA and 16S rRNA were hybridized with total RNA (20 μg for flaA and 1.5 μg for 16S rRNA) from either HB19 or HB19Fla−, treated with RNase, and electrophoresed under denaturing conditions. Yeast RNA served as the control for the flaA riboprobe.
FlaA is a glycosylated protein.
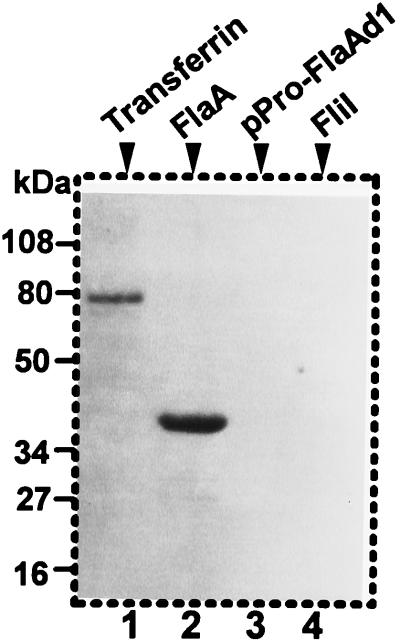
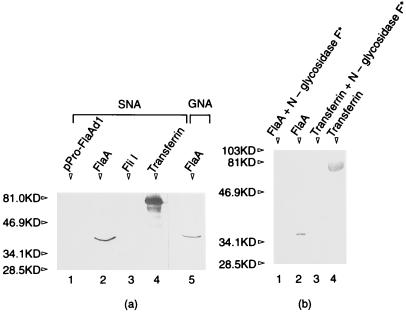
As already mentioned, we were unable to sequence the N-terminal region of the purified protein. For this reason, and because S. hyodysenteriae FlaA has been reported to be glycosylated (46), we tested if B. burgdorferi FlaA underwent a posttranslational modification. We used three different methods to test for glycosylation. First, we used the purified native FlaA protein and the highly sensitive ECL glycoprotein detection system to test for glycosylation. As expected, the positive control serum transferrin reacted strongly (Fig. 6, lane 1). In addition, we found that B. burgdorferi FlaA also yielded a strong reaction, indicating that the native protein was glycosylated (Fig. 6, lane 2). As a negative control, the cytoplasmic motility protein recombinant FliI (28) failed to react (Fig. 6, lane 4), as did the recombinant FlaA containing a deletion of the N-terminal region (29) (Fig. 6, lane 3). Also as controls, we found that reactivity was sodium metaperiodate dependent, as omitting this reagent resulted in no reactivity of FlaA and serum transferrin (not shown). Second, we tested whether FlaA reacted with five different lectins: GNA, SNA, MAA, PNA, and DSA. Only SNA and GNA gave positive reactions (Fig. 7a). SNA binds to α(2-6)-linked sialic acid, and GNA binds to terminal mannose residues. Recombinant FliI and FlaA were negative (not shown), whereas all positive controls reacted as expected (Fig. 7a, serum transferrin). Because reactivity could not be removed by pretreating proteins by boiling in 2% (wt/vol) SDS, 10 mM EDTA, or 8 M urea (not shown), the attached sugar residues are evidently covalently bound to FlaA. Finally, native and recombinant N-glycosidase F was found to remove the lectin reactivity as detected with SNA and GNA (Fig. 7b). Similar results were found when the positive control serum transferrin was treated with N-glycosidase F. These results taken together suggest that FlaA is a glycosylated protein that contains both sialic acid and mannose.
FIG. 6.
Detection of glycosylated FlaA protein by periodate oxidation followed by hydrazine detection. The proteins were subjected to SDS-PAGE (12% [wt/vol] polyacrylamide gel) and tested by a glycoprotein detection system. Lanes: 1, serum transferrin (positive control); 2, purified native FlaA; 3, recombinant FlaA with N-terminal deletion (amino acids 1 to 26); 4, purified recombinant FliI.
FIG. 7.
Lectin binding to FlaA and sensitivity to N-glycosidase F treatment. Approximately 0.5 to 1.0 μg of protein was loaded in each lane and tested as described in Materials and Methods. (a) Lectin SNA reactivity. Lanes: 1, recombinant FlaA; 2, native FlaA; 3, recombinant FliI; 4, serum transferrin; 5, lectin GNA to native FlaA. (b) Individual proteins were incubated with recombinant N-glycosidase F as described in Materials and Methods and detected with lectin SNA after electrophoresis and blotting. Lanes: 1, FlaA after enzyme treatment; 2, FlaA before enzyme treatment; 3, serum transferrin after enzyme treatment; 4, serum transferrin before treatment.
Epitopes of FlaA conserved among spirochete species.
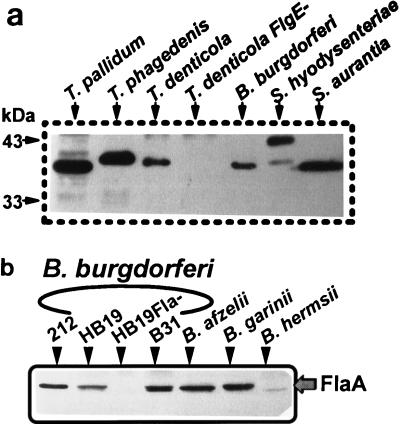
We have previously shown that anti-T. pallidum FlaA antibody reacted strongly with a 38.0-kDa protein of B. burgdorferi cell lysates (23). We investigated whether antibodies to FlaA proteins of other spirochetes reacted with B. burgdorferi FlaA. Monoclonal antibodies against S. aurantia and T. pallidum FlaA failed to react with B. burgdorferi FlaA. The polyclonal anti-S. hyodysenteriae FlaA antibody reacted positively with both T. pallidum and S. hyodysenteriae FlaA proteins in cell lysates, while the reactions with other species were inconclusive (data not shown). On the other hand, antibody specific to T. pallidum FlaA demonstrated strong reactivity to FlaA from many spirochete species as determined by Western blotting of whole-cell lysates (Fig. 8). The reactive proteins were identical in size to those reported in the literature for each FlaA of the species examined (8, 46, 54, 62). As a control, the T. denticola flgE mutant HL51, which is deficient in PF synthesis (62), failed to react with the FlaA antiserum. We also tested several Borrelia species, including B. burgdorferi senso stricto (strains 212, HB19, and B31), B. afzelii, B. garinii, and B. hermsii. All produced significant amounts of FlaA of approximately 38 kDa (Fig. 8b). B. hermsii reacted less intensely than the other species. As expected, the control HB19Fla− failed to react. These results suggest that epitopes on FlaA are evolutionarily well conserved among the spirochetes, including many species of Borrelia.
FIG. 8.
(a) Cross-reactivity of FlaA among the spirochetes. Cell lysates (approximately 10 μg of protein was loaded in each lane) from different species of spirochetes were studied by Western blotting with anti-T. pallidum FlaA antiserum. T. denticola FlgE− is a T. denticola flgE mutant. (b) Conservation of FlaA among Borrelia species, determined by Western blot analysis using anti-T. pallidum FlaA and cell lysates of various Borrelia species.
DISCUSSION
B. burgdorferi has often been cited as being distinct from other spirochetes in having only one protein component comprising its flagellar filament (6, 14, 16, 21, 71). Two lines of experimental data have contributed to this concept. First, electron microscopy indicated that there was no obvious outer layer surrounding a filament core (5, 6, 14, 44). Second, in the analysis of purified PFs, only a single major protein band was observed with SDS-PAGE and two-dimensional gel electrophoresis (6, 16). Our recent finding of a flaA homolog in B. burgdorferi led us to reassess this notion (23). We found that both FlaA and FlaB copurified with the PFs in a gentle isolation procedure. In addition, the mutant which lacked PFs, HB19Fla−, also lacked both FlaA and FlaB. Thus, both biochemical and genetic findings strongly indicate that FlaA and FlaB comprise the PFs of B. burgdorferi. This experimental approach has been previously used to demonstrate that multiple protein species comprise the PFs of other spirochete species (48, 62).
Several reasons can account for FlaA not previously being detected as a PF protein. First, Sarkosyl was used to purify the PFs in two earlier studies (6, 16). Our results indicate that this ionic detergent completely released FlaA from the flagellar filaments, resulting in purified PFs without FlaA. Similarly, Brahamsha and Greenberg found that deoxycholate treatment during S. aurantia PF purification resulted in the loss of one of its FlaB proteins (8). As with B. burgdorferi PFs, no structural differences were noted by electron microscopy in the different PF preparations. It should be noted that the association of FlaA with FlaB of B. burgdorferi is different from that of other spirochetes, as Sarkosyl did not release FlaA from the PFs of S. hyodysenteriae during purification (26). Second, FlaA is an abundant protein in other spirochete species and is expressed at levels approximately equal to those of FlaB (7, 8, 42, 48, 54, 69). In contrast, the amount of B. burgdorferi FlaA was approximately 10% of the amount of FlaB, which can be easily missed on SDS-PAGE, especially since FlaA and FlaB have similar molecular weights. Finally, B. burgdorferi FlaA does not induce a strong immune response during infection, as do its counterparts in other spirochetal infections, or in relation to other B. burgdorferi antigens such as FlaB during Lyme disease (29). Thus, FlaA was not selected as a potential diagnostic antigen for Lyme disease.
Several lines of evidence suggest that B. burgdorferi FlaA is similar to its counterparts from other spirochetes. B. burgdorferi FlaA has 54 to 59% amino acid sequence similarity to FlaA proteins of T. pallidum, S. aurantia, and S. hyodysenteriae (23). In addition, as with the FlaA proteins of these bacteria (9, 36, 43), B. burgdorferi has an N-terminal signal peptidase I cleavage site involved in protein export. In fact, expression of intact B. burgdorferi and other spirochete FlaA proteins in E. coli results in cell death (29, 36, 57), and the N-terminal region is in part responsible for this lethality (29, 36). Western blot analysis reinforces this similarity of the FlaA proteins. Using an antiserum to T. pallidum FlaA, we found cross-reactivity in three strains of B. burgdorferi, along with B. afzelii, B. garinii, B. hermsii, S. aurantia, S. hyodysenteriae, and T. denticola (Fig. 8b). Because FlaA was found associated with the PFs, these results emphasize its evolutionary importance and likely role in spirochete motility.
It is not clear at this time whether B. burgdorferi FlaA is a flagellar outer sheath protein. Evidently, removal of FlaA during purification does not result in morphological dissociation of the PFs, as intact PFs are obtained by using Sarkosyl. Preliminary experiments using T. pallidum anti-FlaA were unsuccessful in localizing the protein by immunoelectron microscopy (26). Future experiments which involve immunoelectron microscopy and antisera specific to B. burgdorferi FlaA should allow us to determine its precise location. In the analysis of other spirochete PFs, the PF sheath tended to be associated with the region proximal to the basal body (13). Because FlaA is expressed in B. burgdorferi in a considerably smaller amount than in other spirochetes, perhaps it is found only in the area near the hook region.
Glycosylated proteins are relatively rare in bacteria. They have been associated with several archaeal flagellins, S-layer proteins, and pili (37, 66). More recently, glycoproteins have also been associated with flagella of specific species of members of the domain Bacteria (18, 53). With respect to spirochete PFs, FlaA has been shown to be glycosylated in S. hyodysenteriae (46), and Brahamsha and Greenberg reported that one of the FlaB proteins may be glycosylated in S. aurantia (8). The precise role of the carbohydrate moieties on the flagellar and PF proteins is unclear. One suggestion is that the carbohydrate residues are necessary for flagellum assembly in specific species of Archaea (37). Alternatively, the suggestion has been made that sugar residues on the flagellar glycoproteins may promote rotation of the flagella without their becoming entangled (1). One known attribute of many glycoproteins is that the carbohydrate regions are involved in increasing hydration (72). Perhaps these residues on FlaA bind water and act as a lubricant to promote PF rotation within the periplasmic space. Future experiments, which will require a sophisticated gene inactivation system, will be necessary to determine their precise function.
The spontaneously occurring PF-deficient mutant HB19Fla− has yielded significant information with respect to B. burgdorferi virulence, motility, and PF synthesis (24, 30, 31, 63, 64). We realize that construction of site-directed mutations in B. burgdorferi motility genes would be the preferable means to analyze structure and function. However, a genetic transfer system for B. burgdorferi is in the very early stages of development (59); thus, we are presently constrained in experimental design relating to genetics. With this limitation in mind, HB19Fla− has been shown to be less invasive of cultured cells than the wild type (63), and it has been shown to serve as a live attenuated vaccine for mice (64). These results suggest that motility and/or PFs are likely to be an important virulence factor for B. burgdorferi. In addition, HB19Fla− has been essential in developing a model of B. burgdorferi motility and in demonstrating that PFs in part dictate the shape of the entire cell (30, 31). Our previous studies have shown that HB19Fla− still produced many motility and chemotaxis proteins (FliI, FliG, FliM, FliN, FlhA, FlhB, and CheA) required for early flagellar assembly (22, 24). These results are consistent with the morphological data that HB19Fla− synthesizes the hook-basal body region but not the filament structure (63).
Analysis of HB19Fla− suggests that flagellar gene expression in B. burgdorferi differs from that of other bacteria. Both flaA and flaB messages were synthesized in HB19Fla−, but the encoding proteins were not detected by Western blotting (Fig. 4 and 5) (63). These results are consistent with translational control of expression of both genes. A similar conclusion was reached with respect to flaB expression in T. phagedenis (49). More recently, a flgE mutant of T. denticola was found to synthesize flaB message but not FlaB protein (47). In B. burgdorferi, flaA and flaB reside in different operons: flaB is in a monocistronic operon mapping at approximately 125 kb on the chromosome (11, 20, 65); flaA is the first of five chemotaxis and motility genes mapping on an operon at 722 kb on the chromosome (20, 23, 25). Evidently, HB19Fla− has a genetic defect that blocks expression of both FlaA and FlaB in two separate operons. In addition, cheA is downstream of flaA, and CheA but not FlaA is expressed in HB19Fla− (22). Apparently there is selective gene expression within the flaA operon. How translational control in B. burgdorferi is achieved is unclear. Perhaps antisense mRNA regulates flaA and flaB expression (19). Alternatively, RNA (58) or protein processing could be involved (32). In Salmonella typhimurium and other bacteria, the last stage of flagellum synthesis is tightly regulated on the transcriptional level (51, 52). Our results and those of others suggest that B. burgdorferi and other spirochetes adopted a different strategy to control PF filament synthesis.
ACKNOWLEDGMENTS
We thank A. Barbour, E. P. Greenberg, M. Jacques, R. Limberger, S. Norris, J. Radolf, T. Schwan, and K. Wicher for kindly sharing antisera, bacterial strains, and cell lysates, and we thank J. Kreiling for sharing her method of PF purification. We thank B. C. Pramanik for performing tandem mass spectrometric sequencing, C. Moomaw for database searching, and S. Afendis for excellent technical assistance with respect to mass spectrometric analysis. We also thank J. Radolf, R. Robinson, and D. Akins for their generous cooperation with respect to FlaA analysis.
This research was supported by Public Health Service grant AI29743 to N.W.C.
REFERENCES
- 1.Armitage J P. Tactic responses in photosynthetic bacteria. Can J Microbiol. 1988;34:475–481. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J-C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 4.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G, Hayes S F, Heiland R A, Schrumpf M E. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect Immun. 1986;52:549–554. doi: 10.1128/iai.52.2.549-554.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco D R, Champion C I, Miller J N, Lovett M A. Antigenic and structural characterization of Treponema pallidum (Nichols strain) endoflagella. Infect Immun. 1988;56:168–175. doi: 10.1128/iai.56.1.168-175.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahamsha B, Greenberg E P. A biochemical and cytological analysis of the complex periplasmic flagella from Spirochaeta aurantia. J Bacteriol. 1988;170:4023–4032. doi: 10.1128/jb.170.9.4023-4032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brahamsha B, Greenberg E P. Cloning and sequence analysis of flaA, a gene encoding a Spirochaeta aurantia flagellar filament surface antigen. J Bacteriol. 1989;171:1692–1697. doi: 10.1128/jb.171.3.1692-1697.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brusca J A, McDowall A W, Norgard M V, Radolf J D. Localization of outer surface proteins A and B in both the outer membrane and intracellular compartments of Borrelia burgdorferi. J Bacteriol. 1991;173:8004–8008. doi: 10.1128/jb.173.24.8004-8008.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens S, Delange M, Ley III H L, Rosa P, Huang W M. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J Bacteriol. 1995;177:2769–2780. doi: 10.1128/jb.177.10.2769-2780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1997. Summary of notifiable diseases, United States, 1996. Morbid. Mortal. Weekly Rep. 45(Suppl.):74.
- 13.Charon, N. W. 1998. Unpublished observations.
- 14.Charon N W, Greenberg E P, Koopman M B H, Limberger R J. Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella. Res Microbiol. 1992;143:597–603. doi: 10.1016/0923-2508(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 15.Cockayne A, Bailey M J, Penn C W. Analysis of sheath and core structures of the axial filament of Treponema pallidum. J Gen Microbiol. 1987;133:1397–1407. doi: 10.1099/00221287-133-6-1397. [DOI] [PubMed] [Google Scholar]
- 16.Coleman J L, Benach J L. Identification and characterization of an endoflagellar antigen of Borrelia burgdorferi. J Clin Invest. 1989;84:322–330. doi: 10.1172/JCI114157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson B E, MacDougall J, Saint Girons I. Physical map of the linear chromosome of the bacterium Borrelia burgdorferi 212, a causative agent of Lyme disease, and localization of rRNA genes. J Bacteriol. 1992;174:3766–3774. doi: 10.1128/jb.174.11.3766-3774.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doig P, Kinsella N, Guerry P, Trust T. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol Microbiol. 1996;19:379–387. doi: 10.1046/j.1365-2958.1996.370890.x. [DOI] [PubMed] [Google Scholar]
- 19.Franch T, Gerdes K. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol Microbiol. 1996;21:1049–1060. doi: 10.1046/j.1365-2958.1996.771431.x. [DOI] [PubMed] [Google Scholar]
- 20.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 21.Gassmann G S, Jacobs E, Deutzmann R, Gobel U B. Analysis of the Borrelia burgdorferi GeHo fla gene and antigenic characterization of its gene product. J Bacteriol. 1991;173:1452–1459. doi: 10.1128/jb.173.4.1452-1459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge Y. Molecular analysis of motility and chemotaxis genes and their organization in Borrelia burgdorferi. Ph.D. dissertation. Morgantown, W.Va: West Virginia University; 1996. [Google Scholar]
- 23.Ge Y, Charon N W. An unexpected flaA homolog is present and expressed in Borrelia burgdorferi. J Bacteriol. 1997;179:552–556. doi: 10.1128/jb.179.2.552-556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge Y, Charon N W. Identification of a large motility operon in Borrelia burgdorferi by semi-random PCR chromosome walking. Gene. 1997;189:195–201. doi: 10.1016/s0378-1119(96)00848-7. [DOI] [PubMed] [Google Scholar]
- 25.Ge Y, Charon N W. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- 26.Ge, Y., L. Corum, and N. W. Charon. 1997. Unpublished observations.
- 27.Ge Y, Old I, Saint Girons I, Charon N W. The flgK motility operon of Borrelia burgdorferi is initiated by a ς70-like promoter. Microbiology. 1997;143:1681–1690. doi: 10.1099/00221287-143-5-1681. [DOI] [PubMed] [Google Scholar]
- 28.Ge Y, Old I G, Saint Girons I, Charon N W. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus ς70 promoter. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge Y G, Charon N W. FlaA, a putative flagellar outer sheath protein, is not an immunodominant antigen associated with Lyme disease. Infect Immun. 1997;65:2992–2995. doi: 10.1128/iai.65.7.2992-2995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein S F, Buttle K F, Charon N W. Structural analysis of Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J Bacteriol. 1996;178:6539–6545. doi: 10.1128/jb.178.22.6539-6545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstein S F, Charon N W, Kreiling J A. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc Natl Acad Sci USA. 1994;91:3433–3437. doi: 10.1073/pnas.91.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 33.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hovind Hougen K. Ultrastructure of spirochetes isolated from Ixodes ricinus and Ixodes dammini. Yale J Biol Med. 1984;57:543–548. [PMC free article] [PubMed] [Google Scholar]
- 35.Isaacs R D, Hanke J H, Guzman-Verduzco L M, Newport G, Agabian N, Norgard M V, Lukehart S A, Radolf J D. Molecular cloning and DNA sequence analysis of the 37-kilodalton endoflagellar sheath protein gene of Treponema pallidum. Infect Immun. 1989;57:3403–3411. doi: 10.1128/iai.57.11.3403-3411.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaacs R D, Radolf J D. Expression in Escherichia coli of the 37-kilodalton endoflagellar sheath protein of Treponema pallidum by use of the polymerase chain reaction and a T7 expression system. Infect Immun. 1990;58:2025–2034. doi: 10.1128/iai.58.7.2025-2034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarrell K F, Bayley D P, Kostyukova A S. The archaeal flagellum: a unique motility structure. J Bacteriol. 1996;178:5057–5064. doi: 10.1128/jb.178.17.5057-5064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen O N, Podtelejnikov A, Mann M. Delayed extraction improves specificity in database searches by matrix-assisted laser desorption/ionization peptide maps. Rapid Commun Mass Spectrom. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Johnson R C, Schmid G P, Hyde F W, Steigerwalt A G, Brenner D J. Borrelia burgdorferi sp. nov.: etiological agent of Lyme disease. Int J Syst Bacteriol. 1984;34:496–497. [Google Scholar]
- 40.Kimsey R B, Spielman A. Motility of Lyme disease spirochetes in fluids as viscous as the extracellular matrix. J Infect Dis. 1990;162:1205–1208. doi: 10.1093/infdis/162.5.1205. [DOI] [PubMed] [Google Scholar]
- 41.Koopman M B H, Baats E, de Leeuw O S, Van der Zeijst B A M, Kusters J G. Molecular analysis of a flagellar core protein gene of Serpulina (Treponema) hyodysenteriae. J Gen Microbiol. 1993;139:1701–1706. doi: 10.1099/00221287-139-8-1701. [DOI] [PubMed] [Google Scholar]
- 42.Koopman M B H, Baats E, van Vorstenbosch C J A H V, van der Ziejst B A M, Kusters J G. The periplasmic flagella of Serpulina (Treponema) hyodysenteriae are composed of two sheath proteins and three core proteins. J Gen Microbiol. 1992;138:2697–2706. doi: 10.1099/00221287-138-12-2697. [DOI] [PubMed] [Google Scholar]
- 43.Koopman M B H, de Lee O S, van der Zieijst B A M, Kuster J G. Cloning and DNA sequence analysis of a Serpulina (Treponema) hyodysenteriae gene encoding a periplasmic flagellar sheath protein. Infect Immun. 1992;60:2920–2925. doi: 10.1128/iai.60.7.2920-2925.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreiling J A. Characterization of the periplasmic flagella of Borrelia burgdorferi. Ph.D. dissertation. Morgantown, W.Va: West Virginia University; 1995. [Google Scholar]
- 45.Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of a flgE mutant. J Bacteriol. 1996;178:3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Dumas F, Dubreuil D, Jacques M. A species-specific periplasmic flagellar protein of Serpulina (Treponema) hyodysenteriae. J Bacteriol. 1993;175:8000–8007. doi: 10.1128/jb.175.24.8000-8007.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Limberger, R. J. 1998. Personal communication.
- 48.Limberger R J, Charon N W. Treponema phagedenis has at least two proteins residing together on its periplasmic flagella. J Bacteriol. 1986;166:105–112. doi: 10.1128/jb.166.1.105-112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Limberger R J, Slivienski L L, Yelton D B, Charon N W. Molecular genetic analysis of a class B periplasmic flagellum gene of Treponema phagedenis. J Bacteriol. 1992;174:6404–6410. doi: 10.1128/jb.174.20.6404-6410.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukehart S A, Tam M R, Hom J, Baker-Zander S A, Holms K K, Nowinski R C. Characterization of monoclonal antibodies to Treponema pallidum. J Immunol. 1985;134:585–592. [PubMed] [Google Scholar]
- 51.Macnab R M. Genetics, structure, and assembly of the bacterial flagellum. Soc Gen Microbiol Symp. 1990;46:77–106. [Google Scholar]
- 52.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 53.Moens S, Michiels K, Vanderleyden J. Glycosylation of the flagellin of the polar flagellum of Azospirillum brasilense, a Gram-negative nitrogen-fixing bacterium. Microbiology. 1995;141:2651–2657. [Google Scholar]
- 54.Norris S J, Charon N W, Cook R G, Fuentes M D, Limberger R J. Antigenic relatedness and N-terminal sequence homology define two classes of major periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J Bacteriol. 1988;170:4072–4082. doi: 10.1128/jb.170.9.4072-4082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norris S J Treponema pallidum Polypeptide Research Group. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Microbiol Rev. 1993;57:750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papayannopoulos I A. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom Rev. 1995;14:49–73. [Google Scholar]
- 57.Parales J, Greenberg E P. Analysis of the Spirochaeta aurantia flaA gene and transcript. FEMS Microbiol Lett. 1993;106:245–252. doi: 10.1111/j.1574-6968.1993.tb05971.x. [DOI] [PubMed] [Google Scholar]
- 58.Petersen C. Control of functional mRNA stability in bacteria: multiple mechanisms of nucleolytic and non-nucleolytic inactivation. Mol Microbiol. 1992;6:277–282. doi: 10.1111/j.1365-2958.1992.tb01469.x. [DOI] [PubMed] [Google Scholar]
- 59.Rosa P, Samuels D S, Hogan D, Stevenson B, Casjens S, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. In-gel digestion of proteins for internal sequence analysis of proteins separated by polyacrylamide gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 61.Rosey E, Kennedy M J, Petrella D K, Ulrich R G, Yancey R J., Jr Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J Bacteriol. 1995;177:5959–5970. doi: 10.1128/jb.177.20.5959-5970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruby J D, Li H, Kuramitsu H, Norris S J, Goldstein S F, Buttle K F, Charon N W. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadziene A, Thomas D D, Bundoc V G, Holt S C, Barbour A G. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J Clin Invest. 1991;88:82–92. doi: 10.1172/JCI115308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadziene A, Thompson P A, Barbour A G. A flagella-less mutant of Borrelia burgdorferi as a live attenuated vaccine in the murine model of Lyme disease. J Infect Dis. 1996;173:1184–1193. doi: 10.1093/infdis/173.5.1184. [DOI] [PubMed] [Google Scholar]
- 65.Saint Girons I, Old I G, Davidson B E. Molecular biology of the Borrelia, bacteria with linear replicons. Microbiology. 1994;140:1803–1816. doi: 10.1099/13500872-140-8-1803. [DOI] [PubMed] [Google Scholar]
- 66.Stimson E, Virji M, Makepeace K, Dell A, Morris H R, Payne G, Saunders J R, Jennings M P, Barker S, Panico M. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 67.Tarentino A L, Gomez C M, Plummer T H., Jr Deglycosylation of asparagine-linked glycans by peptide: N glycosidase F. Biochemistry. 1985;24:4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 68.Tate R, Riccio A, Iaccarino M, Patriarche E J. A cysG mutant strain of Rhizobium etli pleiotropically defective in sulfate and nitrate assimilation. J Bacteriol. 1997;179:7343–7350. doi: 10.1128/jb.179.23.7343-7350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trueba G A, Bolin C A, Zuerner R L. Characterization of the periplasmic flagellum proteins of Leptospira interrogans. J Bacteriol. 1992;174:4761–4768. doi: 10.1128/jb.174.14.4761-4768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Biochem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 71.Wilson D R, Beveridge T J. Bacterial flagellar filaments and their component flagellins. Can J Microbiol. 1993;39:451–472. doi: 10.1139/m93-066. [DOI] [PubMed] [Google Scholar]
- 72.Yanagishita M. Function of proteoglycans in the extracellular matrix. Acta Pathol Jpn. 1993;43:283–293. doi: 10.1111/j.1440-1827.1993.tb02569.x. [DOI] [PubMed] [Google Scholar]