Abstract
The activity of the sporulation transcription factor ςE in Bacillus subtilis is governed by an intercellular signal transduction pathway that controls the conversion of the inactive proprotein pro-ςE to the mature and active form of the factor. Here I use immunofluorescence microscopy to show that the activation of the proprotein is associated with its progression through three patterns of subcellular localization. In the predivisional sporangium, pro-ςE was found to be associated with the cytoplasmic membrane. Next, at the stage of asymmetric division, pro-ςE accumulated at the sporulation septum. Finally, after processing, mature ςE was found to be distributed throughout the mother cell cytoplasm. The results of subcellular fractionation and sedimentation in density gradients of extracts prepared from postdivisional sporangia confirmed that pro-ςE was chiefly present in the membrane fraction and that ςE was predominantly cytoplasmic, findings that suggest that the pro-amino acid sequence is responsible for the sequestration of pro-ςE to the membrane. The results of chemical cross-linking experiments showed that pro-ςE was present in a complex with its putative processing protein, SpoIIGA, or with a protein that depended on SpoIIGA. The membrane association of pro-ςE was, however, independent of SpoIIGA and other proteins specific to B. subtilis. Likewise, accumulation of pro-ςE at the septum did not depend on its interaction with SpoIIGA. Sequestration of pro-ςE to the membrane might serve to facilitate its interaction with SpoIIGA and may be important for preventing its premature association with core RNA polymerase. The implications of these findings for the compartmentalization of ςE are discussed.
Sporulation in Bacillus subtilis involves the formation of an asymmetrically positioned septum, which partitions the sporangium into unequally sized compartments called the forespore (the small compartment) and the mother cell (20). Both compartments receive a complete chromosome but subsequently establish different programs of gene expression (for a review, see reference 29). Differential gene expression is principally governed by four sporulation-specific transcription factors: ςF and ςE, which act in the forespore and the mother cell, respectively, shortly after asymmetric division, and ςG and ςK, which appear in the forespore and the mother cell, respectively, later in development (14). The compartment-specific programs of gene expression do not, however, proceed independently of one another but are linked through intercellular pathways of signal transduction (7, 14). These pathways serve to coordinate the activation of a transcription factor in one compartment with the activity of a factor in the adjoining compartment.
Here I am concerned with the regulation of the mother cell transcription factor ςE, which is subject to temporal and spatial mechanisms of control. The ςE factor is derived from an inactive proprotein precursor called pro-ςE (11), which carries an NH2-terminal extension of 27 amino acids (17). The activation of pro-ςE is governed by an intercellular signal transduction pathway that couples proteolytic processing of the proprotein to ςF-directed gene expression in the forespore (8, 13, 14, 16). This pathway consists of the signaling protein, SpoIIR, which is produced in the forespore under the control of ςF, and SpoIIGA, a membrane-bound protein that is likely to be the proprotein-processing enzyme (4, 8, 13). The signal transduction pathway is a timing mechanism that links the processing of pro-ςE in the mother cell to the activation of ςF in the forespore (21, 38). The compartmentalization of ςE-directed gene expression is achieved by an independent mechanism that restricts pro-ςE protein to the large chamber of the sporangium (21).
Later in development, the mother cell transcription factor ςK is similarly derived from an inactive precursor (pro-ςK) whose conversion to the mature factor is under the control of (ςG-directed) gene expression in the forespore (2, 10, 15). Hence, both mother cell transcription factors are initially synthesized as inactive proproteins and rely on intercellular signal transduction pathways for their proteolytic activation. Regulated proteolysis is an emerging theme in the activation of several eukaryotic transcription factors. Thus, entry into the nucleus of the mammalian transcription factors NF-κB (18) and the sterol regulatory element-binding protein 1 (SREBP-1) (32) and the Drosophila protein cubitus interruptus (Ci) (1) is regulated at the level of proteolytic maturation of the transcription factor itself or of proteins that sequester the factors to the cytoplasm or cytoplasmic membrane.
To gain a more detailed understanding of the mechanisms that regulate the accumulation and subsequent proteolytic activation of pro-ςE, I investigated its subcellular localization by immunofluorescence microscopy and by fractionation of cell extracts. Recent work by Ju et al. (6) had indicated that the NH2-terminal 55 amino acids of pro-ςE are sufficient to direct a green fluorescent protein (GFP) fusion to the sporulation septum. In the present communication, I confirm and extend this finding by showing that ςE exhibits three distinct patterns of subcellular localization which are associated with the conversion of the transcriptionally inactive proprotein, pro-ςE, to the mature and active form of the factor. I show that pro-ςE is associated with the cytoplasmic membrane in the predivisional sporangium and selectively accumulates at the newly formed septum in the postdivisional sporangium. Following its proteolytic conversion to mature ςE via the intercellular signal transduction pathway, the active form of the transcription factor is released from the septum into the cytoplasm of the mother cell where it associates with core RNA polymerase. In addition, I present evidence that pro-ςE forms a complex with SpoIIGA but that association with the cytoplasmic membrane and sequestration to the septum do not depend on interaction with the putative processing enzyme.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Most of the Bacillus subtilis strains used in this study are isogenic with PY79 (36). B. subtilis BZ184 carries a transcriptional spoIID-lacZ fusion linked to the chloramphenicol acetyltransferase gene (28) that was inserted into the chromosome at the amyE locus. The B. subtilis AH42 and AH162 strains were constructed by transforming the spoIIGAΔ17 mutant strain (28) and the spoIIRΔ::kan mutant strain (13), respectively, with chromosomal DNA of BZ184 and selecting for chloramphenicol resistance. MO1190, the spoIIGAΔ mutant strain of B. subtilis (3), is a JH642 derivative and was a gift from P. Stragier (Institut de Biologie Physico-Chimique).
The expression vectors pDG178 and pDG150 (28) contain the entire spoIIG operon and the spoIIGB gene alone, respectively, downstream of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac promoter (35). The B. subtilis AH17 and AH16 strains carry pDG178 and pDG150, respectively, and were described previously (4). The Escherichia coli AH15 strain carries pDG150 in a TG1 background.
Growth conditions.
The B. subtilis strains were sporulated in Sterlini Mandelstam resuspension medium (27) and were harvested 90, 120, and 150 min after the induction of sporulation.
B. subtilis cells of strain AH17 (and the control strain AH16) were grown at 37°C in 50 ml of Luria-Bertani (LB) medium supplemented with kanamycin (5 μg/ml) until the optical density at 600 nm of the culture was about 0.3. At this time, the synthesis of SpoIIGA and pro-ςE (or, in the case of the control strain, pro-ςE synthesis alone) was induced with 1 mM IPTG. After 3 h, the cells were harvested by centrifugation (5,000 × g for 5 min at 4°C), washed once in 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7), and either used directly or stored at −20°C. Protoplasts of strain AH17 (and the control strain AH16) were prepared as previously described (4).
E. coli cells of strain AH15 were grown at 37°C in 50 ml of LB medium supplemented with ampicillin (100 μg/ml). When the optical density at 600 nm of the culture had reached about 0.5, the cells were induced for pro-ςE synthesis with 1 mM IPTG. After 2 h, the cells were harvested by centrifugation (5,000 × g for 5 min at 4°C), washed once in 50 mM MOPS (pH 7), and either used directly or stored at −20°C.
Antibodies, immunofluorescence microscopy, and Western blot analysis.
The mouse monoclonal antibody that binds to both pro-ςE and ςE (31) was a gift from W. Haldenwang (University of Texas) and was used at a 1:20 dilution in immunofluorescence experiments and at a 1:100 dilution in Western blot analyses. The rabbit polyclonal anti-SpoIIE antibodies were prepared by C. Webb (Harvard University) and were used at a 1:10,000 dilution in Western blot analyses. Rabbit polyclonal antibodies raised against β-galactosidase were obtained from 5′-3′ Inc., and used at a 1:1,500 dilution. The secondary antibodies (Jackson Immunolabs) were affinity-purified donkey anti-rabbit or anti-mouse antibodies conjugated either to fluorescein isothiocyanate (FITC) or to indocarbocyanine (Cy3). FITC-conjugated secondary antibodies were used at a 1:100 dilution, while Cy3-conjugated antibodies were used at a 1:200 dilution. Propidium iodide (PI; Molecular Probes) was used at a final concentration of 10 μg/ml, and 4′,6-diamidino-2-phenylindole (DAPI; Sigma) was used at a final concentration of 0.2 μg/ml. The cytoplasmic membrane was stained with the fluorescent lipophilic tracer N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino(phenyl)-hexatrienyl)pyridinium dibromide) (FM 4-64; Molecular Probes), which was applied to fixed cells at a final concentration of 1.5 μg/ml.
Immunofluorescence experiments were performed as described previously (21). Fluorescence micrographs of identical fields of cells were recorded by using a cooled charge-coupled device camera (Princeton Instruments) and a personal computer with the MetaMorph imaging system (version 3.0; Universal Imaging Corp.). DAPI images were assigned to the blue channel, PI and Cy3 images were assigned to the red channel, and FITC images were assigned to the green channel. Adobe Photoshop (version 3.0.5) was used to overlay FITC images on PI images of identical fields.
Western blot analysis was performed with alkaline phosphatase-conjugated anti-mouse immunoglobulin G (heavy and light chains) or anti-rabbit immunoglobulin G (Fc), using the ProtoBlot nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate color development system (Promega).
Rabbit polyclonal anti-SpoIIGA antibodies.
A 0.4-kbp fragment of spoIIGA was amplified from genomic DNA of B. subtilis PY79 by PCR (24) with Vent polymerase (New England Biolabs), using oligonucleotide primers (synthesized by Gibco BRL) that were complementary to nucleotides 556 through 574 in the coding sequence and to the 3′ untranslated region of spoIIGA. To facilitate subsequent cloning of the spoIIGA fragment into the expression vector pRSETA (Invitrogen), the oligonucleotide primers carried SacI and KpnI sites. AH103, the expression strain for the spoIIGA fragment under control of the T7 promoter, was created by transforming the recombinant plasmid into E. coli BL21(DE3) (30). This strain could be induced to produce a fusion protein of six NH2-terminal histidine residues via a 28-amino-acid linker region to the COOH-terminal 124 amino acids in the cytoplasmic domain of SpoIIGA.
Hexahistidine-tagged SpoIIGA was overproduced in a 200-ml culture of E. coli AH103 and purified from inclusion bodies under denaturing conditions by nickel affinity chromatography by the procedure recommended by Qiagen. Purified fractions were subjected to preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and gel slices containing hexahistidine-tagged SpoIIGA were macerated and lyophilized. Purified protein (800 μg) was sent to Immuno-Dynamics, Inc. (La Jolla, Calif.) and used to immunize two rabbits.
Protein concentration determination.
Protein concentrations were determined by the bicinchoninic acid method (26) with bovine serum albumin as a standard. The reagent kit was purchased from Pierce.
Cell extracts and subcellular fractionation.
Cell extracts were prepared from induced cultures of strains AH16 and AH17 as well as from wild-type sporangia of strain BZ184 (harvested 2 h after the induction of sporulation) by protoplasting and sonication. The cells were washed once in 20 ml of buffer consisting of 50 mM MOPS (pH 7), 1 mM EDTA, 10 mM MgCl2, and phenylmethylsulfonyl fluoride (PMSF) (20 μg/ml), suspended in 5 ml of the same buffer, and incubated with lysozyme (1 mg/ml) for 10 min at 37°C. Finally, the protoplasts were completely disintegrated by sonication (three 1-min bursts at 200 W with 2-min cooling intervals on ice). Cell debris was carefully removed by two subsequent centrifugations (each centrifugation, 5,000 × g for 20 min at 4°C). Cell extracts were then subjected to ultracentrifugation (110,000 × g for 1 h at 4°C) after which the supernatants containing the soluble proteins were withdrawn and stored on ice. The membranes were washed once in 5 ml of buffer containing 50 mM MOPS (pH 7), 1 mM EDTA, 10 mM MgCl2, and PMSF (20 μg/ml) and then suspended in 2 ml of the same buffer. Proteins (40 μg of cell extract, 40 μg of soluble proteins, 10 μg of membrane proteins) were separated by SDS-PAGE on 12% polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Immobilon-P; Millipore) for subsequent immunological detection with the anti-ςE antibody.
Cell extracts and density gradient centrifugation.
Cell extracts were prepared from wild-type sporangia of strain BZ184 (harvested 2 h after the induction of sporulation) by protoplasting, osmotic shock, and gentle shearing (9). The cells from a 50-ml sporulating culture were washed in 20 ml of buffer consisting of 20 mM potassium phosphate (pH 7.5), 15 mM MgCl2, 20% (wt/vol) sucrose, and 1 mM PMSF and resuspended in 5 ml of the same buffer. Protoplasts were prepared by the addition of lysozyme (1 mg/ml; Sigma) and incubation at 37°C. When the protoplasting efficiency was greater than 80%, as monitored by phase-contrast microscopy, the protoplasts were harvested by centrifugation (3,000 × g for 15 min at 21°C) and resuspended in 1.5 ml of a solution consisting of 50 mM Tris-HCl (pH 8), 10 mM EDTA, and 1 mM PMSF. Following the addition of RNase A (1 μg/ml) and DNase I (0.5 μg/ml), the protoplasts were disrupted by several passages through an injection needle (27 gauge). Unbroken cells and protoplasts were discarded after centrifugation (3,000 × g for 10 min at 4°C), and the cell extract was applied to the top of a sucrose gradient. The gradient had been prepared by layering 1.4 ml each of 60, 50, 40, 30, 20, and 10% sucrose in buffer consisting of 50 mM Tris-HCl (pH 8) and 10 mM EDTA (22). After ultracentrifugation (200,000 × g for 34 h at 15°C; Beckman SW41 rotor), fractions of 350 μl were collected, and their proteins were separated by SDS-PAGE on 12% polyacrylamide gels and analyzed by immunoblotting with the anti-ςE antibody and the anti-SpoIIE antiserum. Sucrose concentrations of the fractions were determined by their refractive indices.
Chemical cross-linking.
Protoplast suspensions of strains AH17 and AH16 (25 μl) were incubated with 2 μl of 50 mM ethylene glycobis(succinimidylsuccinate) (EGS; 16.1-Å spacer arm length) in the presence of 23 μl of 50 mM MOPS (pH 7). Control reaction mixtures did not contain EGS. After 2 h on ice, 12.5 μl of 5× SDS gel loading buffer was added, and the samples were heated to 100°C for 5 min. The proteins (10 μl of each reaction mixture) were separated by SDS-PAGE on 12% polyacrylamide gels and analyzed by immunoblotting with the anti-ςE antibody.
RESULTS
Pro-ςE coincides in position with the cytoplasmic membrane of the predivisional sporangium and accumulates at the newly formed septum of the postdivisional sporangium.
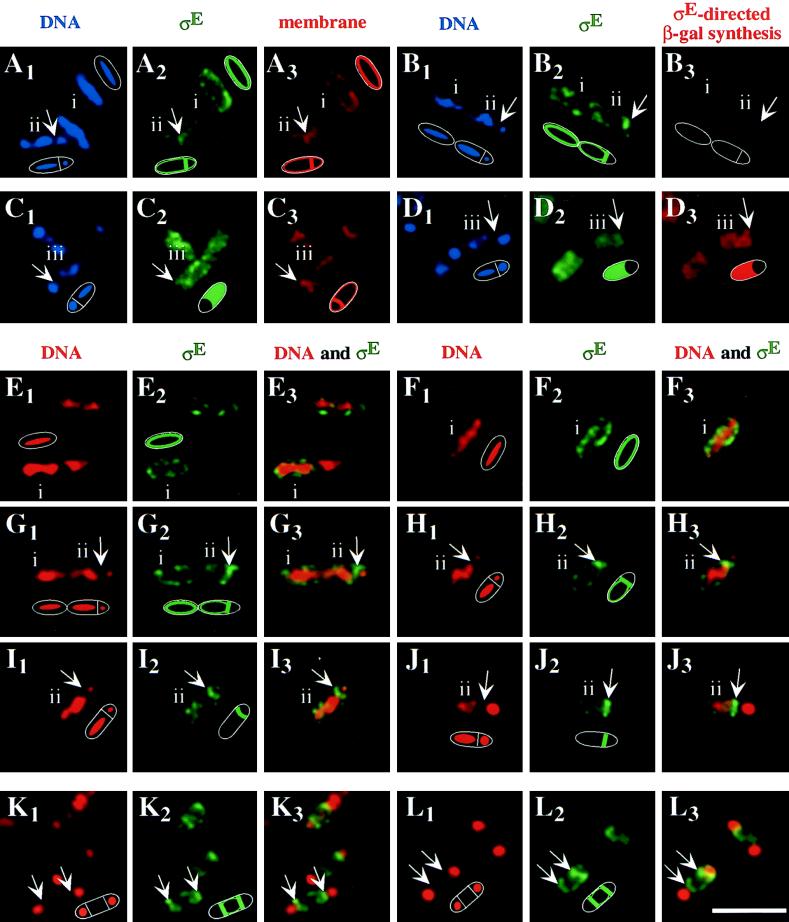
Immunofluorescence microscopy was applied to investigate the subcellular localization of pro-ςE in predivisional and postdivisional sporangia. The mouse monoclonal antibody for immunostaining of pro-ςE, a gift of W. Haldenwang (University of Texas), binds to both the inactive form and the active form of the transcription factor (31). To determine whether mature ςE was present, the sporangia were coimmunostained for the β-galactosidase product of a lacZ fusion to a gene (spoIID) under the control of ςE. The developmental stage of the sporangia was assessed by nucleoid staining. In predivisional sporangia, the nucleoid exhibits an elongated structure known as the axial filament (20, 23). Immediately following asymmetric division, one of the two chromosomes is translocated into the newly formed forespore. The forespore of early postdivisional sporangia is, therefore, characterized by a low but increasing DNA content (34). After chromosome segregation, forespores can be identified by the presence of a round and highly condensed nucleoid, in contrast to the more diffuse nucleoid in the mother cell (25). Finally, membranes were visualized with the fluorescent stain FM 4-64, a reagent that was brought to my attention by K. Pogliano (University of California at San Diego).
Wild-type sporangia were harvested 90 min after the initiation of sporulation. Three classes were distinguished from the 472 sporangia; class i, 164 (35%) predivisional sporangia in which pro-ςE had not been activated; class ii, 61 (13%) postdivisional sporangia in which pro-ςE had not been activated; and class iii, 247 (52%) postdivisional sporangia that showed ςE activity.
In the majority (132 of 164) of class i sporangia that were assigned to the predivisional stage of sporulation on the basis of nucleoid staining, pro-ςE/ςE immunostaining (Fig. 1A2, B2, E2, E3, F2, F3, G2, and G3, green) was preferentially detected at the cell periphery, coincident with the location of fluorescence (red) from the membrane stain FM 4-64 (Fig. 1A3). There was little, if any, overlap of the green immunostaining of pro-ςE/ςE with the red DNA staining (Fig. 1E3, F3, and G3). The absence of coimmunostaining with antibodies against β-galactosidase (Fig. 1B3) confirmed that little or no active ςE was present in class i sporangia and, hence, that the peripheral pattern of pro-ςE/ςE immunostaining was due to pro-ςE. The remaining class i sporangia (32 of 164) showed no detectable pro-ςE/ςE immunostaining.
FIG. 1.
Immunolocalization of pro-ςE and ςE in sporulating wild-type cells and spoIIR and spoIIGA mutant cells of B. subtilis. The sporangia were harvested 90 and 150 min after the induction of sporulation and prepared for immunofluorescence microscopy as described previously (21). The arrows point to the sporulation septa and are oriented perpendicularly to the long axis of the postdivisional sporangia. Each panel includes an idealized diagram of the sporangial outline and the observed pattern of immunostaining. (A3 and C3) The cytoplasmic membranes were stained with FM 4-64 (red). (B3 and D3) The activity of ςE was visualized by immunostaining β-galactosidase (red) synthesized from a spoIID-lacZ fusion that had been inserted into the chromosome at the amyE locus. The DNA was stained either with DAPI (blue) (A1 through D1) or with PI (red) (E1 through L1). (E3 through L3) Overlays of red DNA staining and green immunostaining of pro-ςE of identical fields of cells which would result in a yellow color in case of an overlap. (A and B) Wild-type class i predivisional sporangia display a pattern of pro-ςE/ςE immunostaining (green) (A2) that coincides with the membrane stain (red) (A3) and lack ςE-directed β-galactosidase synthesis (B3). Class ii postdivisional sporangia show pro-ςE/ςE immunostaining (green) (A2) predominantly at the newly formed septum (red) (A3) and similarly lack ςE-directed β-galactosidase synthesis (B3). The arrow points to the newly formed septum which partitions the not yet fully translocated forespore nucleoid and the more elongate mother cell nucleoid (blue) (A1 and B1) of class ii sporulating wild-type cells. (C and D) Wild-type class iii postdivisional sporangia show pro-ςE/ςE immunostaining (green) (C2) and ςE-directed β-galactosidase synthesis (D3) in the mother cell, which is outlined by the membrane stain (C3). Immunostaining of pro-ςE/ςE (green) in class i predivisional (E2 and F2) and class ii postdivisional (G2 through J2) wild-type sporulating cells which lack ςE-directed β-galactosidase synthesis (not shown). In spoIIRΔ::kan mutant cells of strain AH162 (K) and in spoIIGAΔ17 mutant cells of strain AH42 (L), which fail to process pro-ςE to ςE, all the disporic sporangia display preferentially septum-associated pro-ςE staining (green) (K2 and L2). Bar, 5 μm.
A second pattern of pro-ςE/ςE immunostaining was observed in class ii sporangia that had undergone polar division but had not yet activated pro-ςE as judged, once again, by the absence of immunostaining for β-galactosidase (Fig. 1B3). In the majority (49 of 61) of such class ii sporangia, pro-ςE immunostaining (Fig. 1A2, B2, G2, G3, H2, H3, I2, I3, J2, and J3, green, arrows) was observed to be most intense at the newly formed sporulation septum (Fig. 1A3, red, arrow). A similar pattern of septal localization was reported previously by Ju et al. (6) who visualized a GFP fusion to the NH2-terminal 55 amino acids of pro-ςE. The remaining class ii sporangia (12 of 61) continued to show peripheral pro-ςE immunostaining, albeit more intense at the newly formed sporulation septum. Therefore, pro-ςE appears to be membrane associated in predivisional wild-type sporangia and to preferentially accumulate at the newly formed septum of postdivisional sporangia.
Finally, a third pattern of pro-ςE/ςE immunostaining was observed in class iii postdivisional sporangia in which, as judged by immunostaining of β-galactosidase (Fig. 1D3, red), conversion of pro-ςE to mature ςE had taken place. In most (245 of 247) of these class iii sporangia, pro-ςE/ςE immunostaining (Fig. 1C2 and D2, green) was coincident with the cytoplasm of the mother cell (the position of which was revealed by nucleoid staining; Fig. 1C1 and D1, blue) and was not restricted to the septum (Fig. 1C3, red, arrow). The absence of pro-ςE/ςE immunostaining from the forespore was in agreement with the results of a previous investigation in which it was discovered that pro-ςE/ςE immunostaining becomes confined to the mother cell after the formation of the asymmetrically positioned septum (21). Because the switch in subcellular distribution was correlated with the activation of ςE, it seems likely that release from the septum into the mother cell cytoplasm was the result of the conversion of pro-ςE to mature ςE.
Pro-ςE but not ςE is predominantly membrane associated in extracts of sporulating cells.
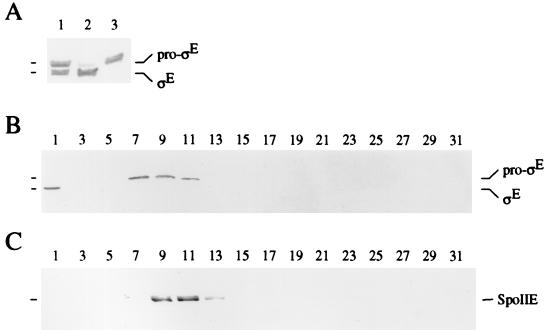
To determine if pro-ςE is indeed associated with the cytoplasmic membranes of sporulating cells, extracts of wild-type sporangia harvested 90 min after the onset of sporulation were fractionated. Protoplasts of sporangia were disintegrated by sonication, after which the cell debris was removed and the cell extract was subjected to ultracentrifugation. Cytoplasmic proteins in the supernatant and membrane proteins in the washed pellet obtained after ultracentrifugation were separated by SDS-PAGE and further analyzed in Western blots with mouse monoclonal anti-pro-ςE/ςE antibodies. Figure 2A shows that pro-ςE in the cell extract (lane 1) chiefly fractionated with the membrane pellet (lane 3), whereas somewhat more ςE was found in the soluble fraction (lane 2) than with the membranes. In a control experiment, β-galactosidase was detected exclusively in the soluble fraction (data not shown).
FIG. 2.
Pro-ςE but not ςE is membrane associated in extracts of sporulating cells. (A) Western blot of proteins before and after subcellular fractionation of an extract prepared from wild-type postdivisional sporangia (strain BZ184) that were harvested 120 min after the induction of sporulation. Lane 1, 40 μg of proteins in the extract; lane 2, 40 μg of proteins in the soluble fraction obtained after centrifugation at 110,000 × g; lane 3, 10 μg of proteins in the washed membrane fraction obtained after centrifugation at 110,000 × g. Proteins were subjected to SDS-PAGE and Western blot analysis with the monoclonal anti-ςE antibody as described in Materials and Methods. (B and C) Western blots of proteins from an extract of wild-type postdivisional sporangia after sedimentation in a sucrose density gradient. The extract was applied to the top of a sucrose density gradient of 60 to 0% and centrifuged at 200,000 × g to equilibrium. Fractions were collected from the bottom to the top of the gradient and analyzed by SDS-PAGE and subsequent Western blotting with anti-ςE antibody (B) and with anti-SpoIIE antibodies (C) as described in Materials and Methods. Fraction numbers are shown over the blots and indicate the position of the fraction in the sucrose density gradient, with fractions numbered from bottom (fraction 1) to top of the gradient.
The apparent association of pro-ςE with the cytoplasmic membrane was investigated further by sedimentation centrifugation in a sucrose gradient. Protoplasts of wild-type sporangia were broken by osmotic shock and gentle shearing. The extracts were then applied to the top of a sucrose density gradient of 60 to 0% (wt/wt) and centrifuged to equilibrium, and the fractions were analyzed by SDS-PAGE and subsequent Western blotting. Figure 2B shows that under these conditions, ςE sedimented to the bottom fraction, whereas the bulk of pro-ςE sedimented in fractions 7 (1.20 g cm−3) to 11 (1.17 g cm−3). The average density of this peak was 1.19 g cm−3, which corresponds to the density that has been determined for the cytoplasmic membrane of B. subtilis (22). SpoIIE, an integral membrane protein control which is synthesized at about the same time as pro-ςE during sporulation, sedimented mainly in fractions 9 (1.19 g cm−3) to 11 (1.17 g cm−3) (Fig. 2C). Therefore, pro-ςE but not ςE is associated with the membrane fraction of wild-type sporulating cells of B. subtilis and like the integral membrane protein SpoIIE, sediments at a density that has previously been reported for cytoplasmic membranes of B. subtilis.
Pro-ςE forms SpoIIGA-dependent complexes in cells engineered to produce the sporulation protein during growth.
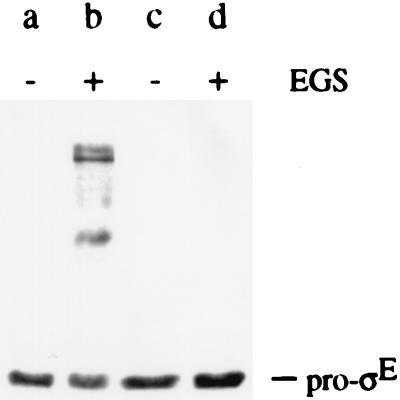
A likely candidate to confer membrane association on pro-ςE is the putative SpoIIGA protease for pro-ςE processing, which is synthesized concurrently with pro-ςE in predivisional sporangia (5) and which has been predicted to contain five membrane-spanning segments (12, 28). To test this possibility, chemical cross-linking was used to determine whether pro-ςE is present in a SpoIIGA-dependent complex. For these experiments, the association of pro-ςE with SpoIIGA was investigated in cells engineered to produce the two sporulation proteins during growth. Broken protoplasts of vegetative B. subtilis cells that had been induced to synthesize either SpoIIGA and pro-ςE or, as a negative control, pro-ςE alone were incubated in the presence or absence of the homobifunctional N-hydroxysuccinimidyl ester cross-linker EGS. Cytoplasmic and membrane proteins were then separated by SDS-PAGE and subsequently analyzed in Western blots with the mouse monoclonal antibody that binds to pro-ςE and ςE. As shown in Fig. 3, lane b, the addition of EGS caused the appearance of antibody-reactive protein complexes of molecular masses higher than that of pro-ςE only in cells that had been engineered to produce pro-ςE and SpoIIGA. EGS did not lead to the appearance of these or other immunoreactive protein complexes in cells that had been engineered to produce pro-ςE alone (Fig. 3, lane d). In the complete absence of chemical cross-linkers, no protein complexes could be detected even in the cells that had synthesized both pro-ςE and SpoIIGA (Fig. 3, lane a). In an effort to determine if the SpoIIGA-dependent protein complexes observed with pro-ςE contained SpoIIGA itself, antibodies were raised against the COOH-terminal half of SpoIIGA. Unfortunately, the antibodies raised were not of sufficiently high titer to detect SpoIIGA in Western blot analyses of extracts from B. subtilis. Therefore, pro-ςE either forms protein complexes with SpoIIGA itself or with another protein(s) whose capacity to interact with pro-ςE depends on the presence of SpoIIGA.
FIG. 3.
Pro-ςE forms SpoIIGA-dependent protein complexes. A Western blot of proteins from two strains of vegetative B. subtilis cells is shown. The B. subtilis cells had been induced for the synthesis of SpoIIGA and pro-ςE (strain AH17; lanes a and b) or for the synthesis of pro-ςE alone (strain AH16; lanes c and d). Protoplast suspensions of strains AH17 and AH1b were incubated in the absence (−) of chemical cross-linker or in the presence (+) of 2 mM EGS. Proteins were subjected to SDS-PAGE and Western blot analysis with anti-ςE antibody as described in Materials and Methods.
Association of pro-ςE with the membrane in a vegetative cell is independent of SpoIIGA and other B. subtilis-specific proteins.
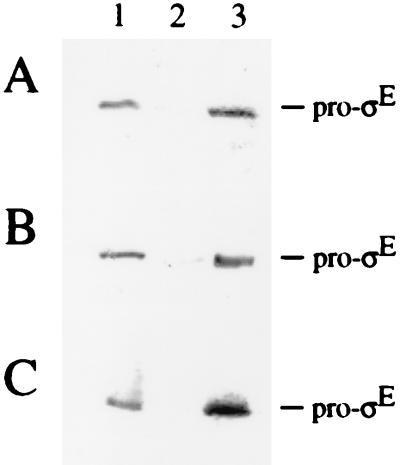
To investigate whether the membrane association of pro-ςE depends on SpoIIGA, extracts of vegetative B. subtilis cells that had been induced to synthesize either SpoIIGA and pro-ςE or, as a negative control, pro-ςE alone were fractionated. Figure 4 shows that pro-ςE in extracts of B. subtilis cells (lane 1) fractionated predominantly with the membrane pellet (lane 3) and was hardly detectable in the soluble fraction (lane 2), regardless of the presence (A) or absence (B) of SpoIIGA. When extracts of vegetative E. coli cells that had been induced to synthesize pro-ςE (Fig. 4C, lane 1) were fractionated, pro-ςE was similarly found in the membrane fraction (lane 3) and was absent from the soluble fraction (lane 2). These results show that the membrane association of pro-ςE is not only independent of SpoIIGA but it is also independent of any other protein specific to B. subtilis.
FIG. 4.
Pro-ςE is membrane associated in the absence of SpoIIGA and other sporulation-specific proteins. Western blots of proteins before and after subcellular fractionation of cell extracts. Vegetative B. subtilis cells that had been induced for the synthesis of SpoIIGA and pro-ςE (strain AH17) (A) or for the synthesis of pro-ςE alone (strain AH16) (B) and E. coli cells that had been induced for the synthesis of pro-ςE (strain AH15) (C) were used. Lane 1, 33 μg of proteins in the extract; lane 2, 33 μg of proteins in the soluble fraction obtained after centrifugation at 110,000 × g; lane 3, 8 μg of proteins in the washed membrane fraction obtained after centrifugation at 110,000 × g. Proteins were subjected to SDS-PAGE and Western blot analysis with anti-ςE antibody as described in Materials and Methods.
Accumulation of pro-ςE at the newly formed sporulation septum is independent of SpoIIR and SpoIIGA.
To analyze whether the accumulation of pro-ςE at the newly formed septum of postdivisional sporangia depends on the SpoIIR signaling protein or on a functional SpoIIGA protease, pro-ςE was immunolocalized in spoIIRΔ::kan and spoIIGAΔ17 mutant sporangia. Both of these mutant strains have impaired intercellular signal transduction pathways for pro-ςE processing and therefore fail to activate ςE, resulting in the formation of a second sporulation septum at the opposite cell pole and, ultimately, a disporic phenotype. The pattern of pro-ςE immunostaining in predivisional and postdivisional spoIIRΔ::kan (Fig. 1K2 and K3, green, arrows) and spoIIGAΔ17 mutant sporangia (Fig. 1L2 and L3, green, arrows) was similar to that observed in wild-type sporangia, in that pro-ςE localized to the membranes of predivisional sporangia (not shown) and accumulated specifically at the septa of postdivisional, disporic sporangia. Similar results were obtained with a deletion mutant lacking almost the entire spoIIGA open reading frame (not shown), which was kindly provided by P. Stragier (Institut de Biologie Physico-Chimique). Therefore, the membrane association of pro-ςE and its selective accumulation at the sporulation septum are independent of both the SpoIIR signaling protein and the SpoIIGA protease.
DISCUSSION
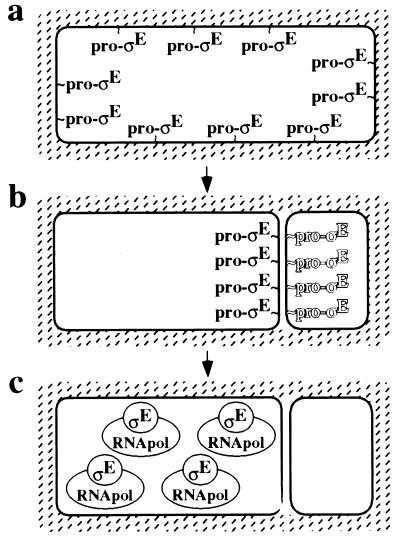
I have shown by immunofluorescence microscopy and by subcellular fractionation experiments that the mother cell-specific transcription factor ςE displays three distinct patterns of subcellular localization. As a transcriptionally inactive proprotein in predivisional sporangia, pro-ςE is preferentially associated with the cytoplasmic membrane. Next, coincident with the process of asymmetric division, pro-ςE selectively accumulates at the sporulation septum. Thus, when asymmetric division is complete, pro-ςE is situated at the septum where it awaits proteolytic conversion to active ςE via the components of the intercellular signal transduction pathway. Finally, after processing, mature ςE is freed from the septum and released into the cytoplasm of the mother cell, where it associates with core RNA polymerase and directs specific gene expression. In agreement with previous results, when mature ςE is released from the septum, it is absent from the forespore and chiefly or exclusively found in the mother cell, an observation that provides a sufficient explanation for the compartmentalization of ςE-directed gene expression (21). A model for the three patterns of subcellular localization of pro-ςE and ςE is presented in Fig. 5.
FIG. 5.
The mother cell-specific transcription factor ςE displays three patterns of subcellular localization. (a) In the predivisional sporangium, the transcriptionally inactive proprotein precursor, pro-ςE, is associated with the cytoplasmic membrane. (b) Following polar division, pro-ςE preferentially accumulates at the polar septum and possibly only on the mother cell face of the septum (as indicated by the shadowing of pro-ςE in the forespore) in a process that depends on the SpoIIIE protein (see Discussion). Finally, pro-ςE is converted to mature ςE via the components of the intercellular signal transduction pathway (not shown). (c) As a result, mature ςE is released from the septum into the cytoplasm of the mother cell where it associates with core RNA polymerase (RNApol) and directs specific gene transcription.
The conclusion that pro-ςE is associated with the cytoplasmic membrane and that this membrane association depends on the pro-amino acid sequence was independently reached in the concurrent investigation of Zhang et al. (37), who carried out subcellular fractionation experiments. That the pro-amino acid sequence also causes pro-ςE to become sequestered at the asymmetrically positioned septum is consistent with the previously reported observations of Ju et al. (6) who found that the NH2-terminal 55 amino acids of pro-ςE are sufficient to cause a fusion to GFP to become septum associated. Our present results confirm and extend the findings of Ju et al. by showing that pro-ςE is initially associated with the cytoplasmic membrane of the predivisional sporangium and that the nonrecombinant, native form of the proprotein becomes concentrated at the septum of the postdivisional sporangium.
In a parallel study on the properties of the prosequence of the late-stage mother cell-specific transcription factor ςK, Zhang et al. (37) similarly found that pro-ςK is membrane associated. Like pro-ςE, after processing, pro-ςK is released from the membrane into the cytoplasm where it associates with core RNA polymerase and directs specific gene expression. Therefore, the pro-amino acid sequences of both mother cell-specific transcription factors seem to play a similar role in promoting association with the membrane. Interestingly, however, the prosequences of pro-ςK and pro-ςE exhibit little amino acid sequence or apparent structural similarity. Whereas the prosequence of pro-ςK is highly hydrophobic (37), the prosequence of pro-ςE has been proposed to form an amphipathic alpha helix (19). Conceivably, the amphipathic alpha helix of pro-ςE could associate with the membrane itself or could provide a binding surface for interaction with another membrane-associated protein. If pro-ςE associates with the membrane by virtue of its interaction with another protein, this protein does not form a complex with pro-ςE that could be detected by chemical cross-linking and it is not specific to B. subtilis cells. Regardless of how the pro-amino acid sequences of the two proteins cause adherence to the membrane, association of the proprotein transcription factors with the cytoplasmic membrane could serve to increase the interaction with their corresponding membrane-bound processing enzymes and to help prevent their premature association with core RNA polymerase. Work by Trempy et al. (31) and Zhang et al. (37) has also shown that the pro-amino acid sequence interferes with the binding of the proprotein to core RNA polymerase.
Inasmuch as the signal transduction pathway for the proteolytic activation of ςE operates across the membranes of the forespore and the mother cell, enriching pro-ςE at the sporulation septum might be crucial to ensure its efficient conversion to mature ςE. What then is the mechanism by which pro-ςE accumulates at the newly formed sporulation septum? A likely candidate to sequester pro-ςE to the septum is the putative protease SpoIIGA, an integral membrane protein, which as shown by chemical cross-linking, either directly interacts with pro-ςE or promotes the interaction of pro-ςE with another protein(s). If SpoIIGA indeed recruits pro-ςE to the sporulation septum, then the septal localization of pro-ςE would be expected to be disrupted in a spoIIGA mutant. However, as reported here and elsewhere (6), the pattern of pro-ςE localization was unaffected in various spoIIGA mutants including a null mutant. Therefore, a protein other than SpoIIGA must be responsible for recruiting pro-ςE to the septum.
Finally, the discovery that pro-ςE migrates from the cytoplasmic membrane to the sporulation septum suggests a new model for how ςE becomes confined to the mother cell. In a previous study, we (21) showed that mother cell-specific activation of ςE is, in part, controlled by a mechanism that is responsible for eliminating ςE from the forespore prior to its activation. The SpoIIIE protein was implicated in the elimination of pro-ςE/ςE from the forespore, either directly or indirectly through its effect on chromosome segregation. Conceivably, at the time of asymmetric division, pro-ςE becomes located on the mother cell face of the septum in a SpoIIIE-dependent manner (Fig. 5). Because SpoIIIE is localized in the newly formed sporulation septum, indeed in its center (33), it could be directly involved in localizing pro-ςE to the mother cell face of the septum, perhaps by creating a pore in the septum through which pro-ςE can pass. Whatever the detailed mechanism for compartmentalization of ςE, the association of pro-ςE with the sporulation septum is likely to play an intimate role both in the activation of the proprotein and in the establishment of mother cell-specific gene transcription.
ACKNOWLEDGMENTS
I thank W. Haldenwang (University of Texas) and C. Webb (Harvard University) for their gifts of antibodies, K. Pogliano (University of California at San Diego) for suggesting the use of the fluorescent lipophilic tracer FM 4-64, and P. Stragier (Institut de Biologie Physico-Chimique) for the spoIIGA deletion mutant of B. subtilis. I am grateful to B. Zhang and L. Kroos (both at Michigan State University) and W. Haldenwang for sharing results prior to publication and for helpful discussions. I thank E. Angert, N. King, and R. Losick (all at Harvard University), P. Fawcett (University of Georgia), and L. Kroos for critical reading of the manuscript.
This work was supported by NIH grant GM18568 to R. Losick. A.H. was a postdoctoral fellow of the Alexander von Humboldt Foundation.
REFERENCES
- 1.Aza-Blanc P, Ramirez-Weber F-A, Laget M-P, Schwartz C, Kornberg T B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 2.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 3.Guérout-Fleury A-M, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 4.Hofmeister A E M, Londoño-Vallejo A, Harry E, Stragier P, Losick R. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell. 1995;83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 5.Jonas R M, Weaver E A, Kenney T J, Moran C P, Jr, Haldenwang W G. The Bacillus subtilis spoIIG operon encodes both ςE and a gene necessary for ςE activation. J Bacteriol. 1988;170:507–511. doi: 10.1128/jb.170.2.507-511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju J, Luo T, Haldenwang W G. Bacillus subtilis pro-ςE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J Bacteriol. 1997;179:4888–4893. doi: 10.1128/jb.179.15.4888-4893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser D, Losick R. How and why bacteria talk to each other. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 8.Karow L M, Glaser P, Piggot P J. Identification of a gene, spoIIR, which links the activation of ςE to the transcriptional activity of ςF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kontinen V P, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 10.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 11.LaBell T L, Trempy J E, Haldenwang W G. Sporulation-specific ς factor ς29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci USA. 1987;84:1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Londoño-Vallejo J-A. Mutational analysis of the early forespore/mother-cell signalling pathway in Bacillus subtilis. Microbiology. 1997;143:2753–2761. doi: 10.1099/00221287-143-8-2753. [DOI] [PubMed] [Google Scholar]
- 13.Londoño-Vallejo J-A, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 14.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 15.Lu S, Halberg R, Kroos L. Processing of the mother-cell ς factor, ςK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margolis P, Driks A, Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 17.Miyao A, Theeragool G, Takeuchi M, Kobayashi Y. Bacillus subtilis spoVE gene is transcribed by ςE-associated RNA polymerase. J Bacteriol. 1993;175:4081–4086. doi: 10.1128/jb.175.13.4081-4086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB precursor and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 19.Peters H K, III, Carlson H C, Haldenwang W G. Mutational analysis of the precursor-specific region of Bacillus subtilis ςE. J Bacteriol. 1992;174:4629–4637. doi: 10.1128/jb.174.14.4629-4637.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogliano K, Hofmeister A E M, Losick R. Disappearance of the ςE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puohiniemi R, Simonen M, Muttilainen S, Himanen J-P, Sarvas M. Secretion of the Escherichia coli outer membrane proteins OmpA and OmpF in Bacillus subtilis is blocked at an early intracellular step. Mol Microbiol. 1992;6:981–990. doi: 10.1111/j.1365-2958.1992.tb02164.x. [DOI] [PubMed] [Google Scholar]
- 23.Ryter A. Etude morphologie de la sporulation de Bacillus subtilis. Ann Inst Pasteur (Paris) 1965;108:40–60. [PubMed] [Google Scholar]
- 24.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 25.Setlow B, Magill N, Febbroriello P, Nakhimousky L, Koppel D E, Setlow P. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol. 1991;173:6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 27.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 29.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genetics. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 30.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 31.Trempy J E, Morrison-Plummer J, Haldenwang W G. Synthesis of ς29, an RNA polymerase specificity determinant, is a developmentally regulated event in Bacillus subtilis. J Bacteriol. 1985;161:340–346. doi: 10.1128/jb.161.1.340-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Sato R, Brown M S, Jia X, Goldstein J L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 33.Wu L J, Errington J. Septal localisation of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L J, Lewis P J, Allmansberger R, Hauser P M, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 35.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 37.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-ςK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Higgins M L, Piggot P J, Karow M L. Analysis of the role of prespore gene expression in the compartmentalization of mother cell-specific gene expression during sporulation of Bacillus subtilis. J Bacteriol. 1996;178:2813–2817. doi: 10.1128/jb.178.10.2813-2817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]