Abstract
A pharmacophore-hybridized strategy based on previously reported HSP90 C-terminal inhibitors was utilized to prepare 32 aryl/penta-1,4-dien-3-one/amine hybrids. Among them, a silicon-containing compound 1z exhibited remarkable broad-spectrum antiproliferative effects on various human breast cancer cell lines. Through fluorescence polarization and AlphaScreen-based assays, we demonstrated that 1z specifically inhibited the HSP90 C-terminus without affecting HSP90 N-terminus. Furthermore, 1z effectively inhibited the HSP90 C-terminus without inducing heat-shock response (HSR), leading to the degradation of its client proteins HER2, pAKT, AKT, and CDK4, causing G1 arrest of MCF-7 and SKBr3 cells, and ultimately contributing to apoptosis of these cells through caspase-3, caspase-8, and caspase-9 activation. Additionally, the penta-1,4-dien-3-one linker in the hybrid, a large bulky lipophilic substitution in the aryl fragment at the 3′-site, and the presence of N-methylpiperazine as the amine fragment were identified as crucial factors that significantly contributed to the observed antiproliferative activity through structure–activity relationship (SAR) analysis. Lastly, we found that 1z exhibited superior thermostability compared to vibsanin B derivatives and good in vitro metabolic stability in simulated intestinal fluid, representing one of the few reported silicon-containing HSP90 C-terminal inhibitors.
Compound 1z, a thermostable silicon-containing aryl/penta-1,4-dien-3-one/amine hybrid kills breast cancer cells by targeting the HSP90 C-terminus without inducing heat-shock response.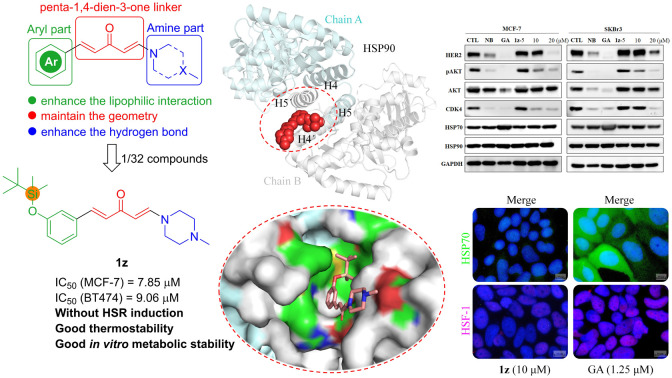
Introduction
Cancer is one of the most fatal diseases that endanger human life. There were about 19.3 million new cases of cancer worldwide in 2020, of which 10 million deaths were mostly found in developing countries, and unfortunately the number is increasing quickly.1 Breast cancer is currently the leading cause of cancer-related deaths in women.2–4 Although there are targeted therapeutic agents available for different subtypes of breast cancer, such as trastuzumab and pertuzumab for HER2-positive subtypes, olaparib for PARP inhibition, alpelisib for PI3K inhibition, and AKT inhibitors, these treatments may not fully meet the needs of all patients.5,6 Heat shock protein 90 (HSP90), an ATP-dependent molecular chaperone, is responsible for the conformational maturation, activation, and stability of more than 200 client proteins involved in signal transduction which is essential for hallmarks of cancer and drug resistance development. In eukaryotic cells, HSP90 is upregulated from its native content of 1% to 5% only under cellular stress and is continuously overexpressed in cancerous cells but not in normal cells. Thus, HSP90 inhibitors offer a promising approach to treating breast cancer by targeting the HSP90 protein and degrading its client proteins (including HER2, PI3K, and AKT, etc.), resulting in a multi-faceted attack on cancer cells.7 Geldanamycin-, radicicol-, and novobiocin-based derivatives are representative examples of HSP90 inhibitors that have shown high potential in treating various subtypes of breast cancer, and some are already undergoing clinical studies.8–13
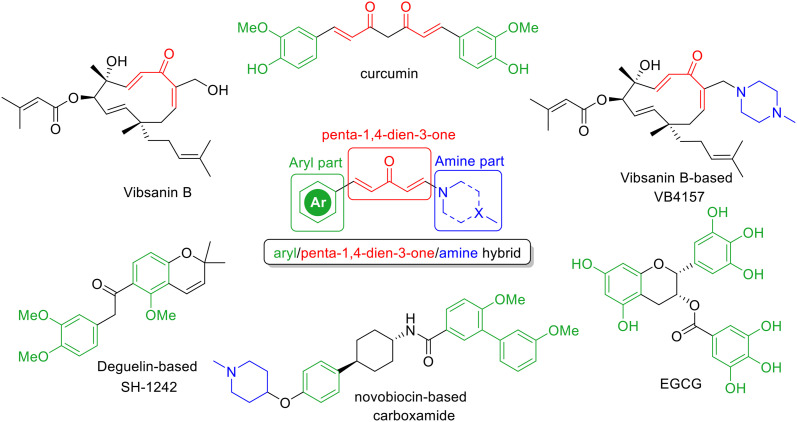
A range of structurally designed HSP90 inhibitors with remarkable properties have been reported during the past two decades. Notably, HSP90 C-terminal and HSP90 isoform-selective inhibitors have emerged as current hotspots for HSP90 inhibitors due to their outstanding functions in molecular biology.10,11,14 These inhibitors have exhibited potent antiproliferative activity against various breast cancer cell lines. Blagg et al. carried out a landmark medicinal chemistry on the natural product novobiocin and systematically developed various novobiocin-based HSP90 C-terminal inhibitors that exhibited a great potency towards HER2-positive breast cancer.8,15–29 Lee and coworkers successfully designed and synthesized a series of deguelin-based HSP90 C-terminal inhibitors, which demonstrated excellent antiproliferative activity against different subtypes of breast cancer cell lines, especially the trastuzumab-resistant HER2-positive breast cancer.5,6,30–35 McAlpine and coworkers uncovered the effects of San A-amide-based derivatives on HSP90 C-terminal interactions with its clients IP6K2, FKBP38, FKBP52, and HOP through the binding to the HSP90 N/M-terminus, which represented novel HSP90 C-terminal allosteric modulators.36–40 Very recently, a covalent inhibitor binding to Cys598 on the Hsp90 C-terminus and exhibiting antiproliferative activities against a lot of cancer cells without inhibiting ATPase activity was reported by Xu and co-workers, which gave new insights into the exploration of HSP90 C-terminal inhibition and HSP90 functions.41 Nevertheless, the limited scaffolds for HSP90 C-terminal inhibitors remain to be investigated. In a previous study, we demonstrated that the natural product vibsanin B and its analogue VB4157 showed impressive HSP90 C-terminal inhibitory activity and antiproliferative activities against various breast cancer cell lines.42 However, besides the induction of heat-shock response (HSR), the thermostability of these derivatives remains a concern; the time for 50% decomposition in boiling toluene (120 °C) of these derivatives was estimated to be shorter than 5 min due to the oxy-Cope rearrangement.42,43 To address this issue, we aimed to create a structurally simplified and thermostable scaffold inspired by vibsanin B and other well-established HSP90 C-terminal inhibitors44,45 (Fig. 1) by retaining the α,β-unsaturated ketone fragment as a linker and connecting its two ends to the frequently occurring pharmacophores of HSP90 C-terminal inhibitors to form an aryl/penta-1,4-dien-3-one/amine chimera. Thus, this article focuses on the design and synthesis of a series of aryl/penta-1,4-dien-3-one/amine hybrids as a novel generation of thermostable HSP90 C-terminal inhibitors without HSR induction.
Fig. 1. Selected HSP90 C-terminal inhibitors and the proposed hybrid.
Results and discussion
Design and synthesis of aryl/penta-1,4-dien-3-one/amine hybrids 1
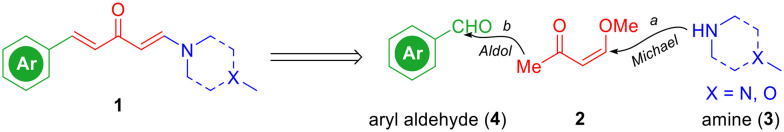
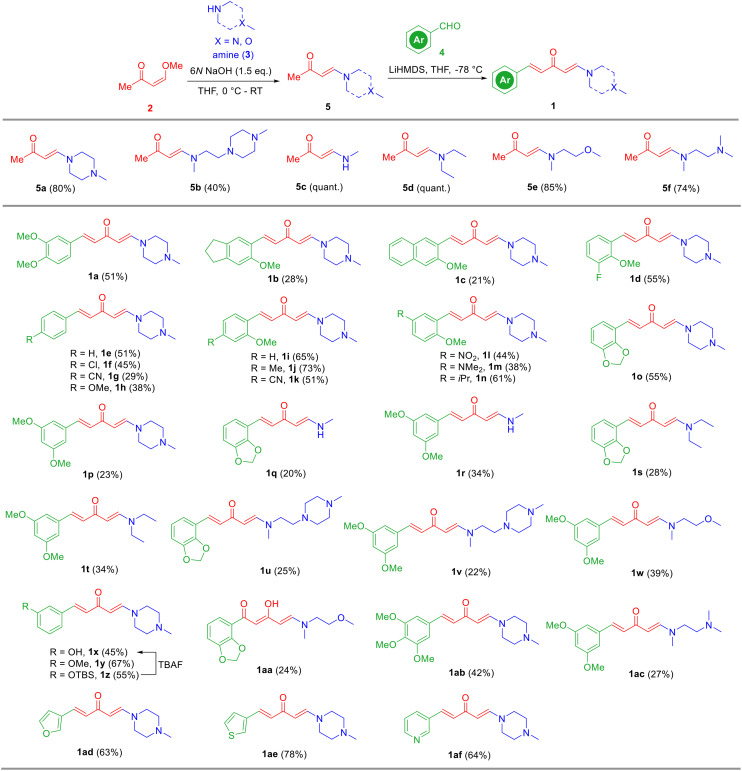
Inspired by previously published HSP90 C-terminal inhibitors (Fig. 1), the aryl groups (like the green parts in curcumin,44 SH-1242, carboxamide, EGCG, and others10), the α,β-unsaturated ketone fragments (like the red parts in curcumin, vibsanin B, VB4157, and others10), and the tertiary amines (like the blue parts in VB4157, carboxamide, and others10) are key pharmacophores and frequently presented. We envisioned that a new scaffold for HSP90 C-terminal inhibitors might be readily obtained through the combination of these key fragments. Therefore, we used the α,β-unsaturated ketone fragment as a linker and connected its two ends to the aryl part and amine part to form an aryl/penta-1,4-dien-3-one/amine chimera. The synthetic route to aryl/penta-1,4-dien-3-one/amine hybrids 1 features a two-step facile transformation including Michael addition and aldol reaction of enone 2 with amines 3 and aryl aldehydes 4 (Scheme 1). Namely, enamines 5 were readily prepared through the Michael addition of amines 3 to enone 2 in the presence of 6 N NaOH (Scheme 1, path a). Subsequently, aldol reaction of enamines 5a–5f with a series of aryl aldehydes 4 were realized by a strong base lithium hexamethyldisilazide (LiHMDS) (Scheme 1, path b). As a result, 32 hybrids (1a–1af) bearing various aryls and amines were successfully prepared in 20–78% yields by this strategy (Scheme 2).
Scheme 1. Retrosynthetic analysis of aryl/penta-1,4-dien-3-one/amine hybrids 1.
Scheme 2. Synthesis of aryl/penta-1,4-dien-3-one/amine hybrids 1.
Biological evaluation of aryl/penta-1,4-dien-3-one/amine hybrids
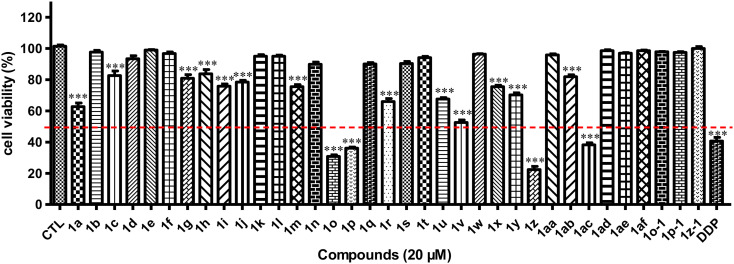
After obtaining the hybrids 1, the inhibitory ratio of hybrids 1a–1af against the proliferation of MCF-7 human breast cancer cell lines was then tested at a dose of 20 μM. The results demonstrated that hybrids 1o, 1p, 1z, and 1ac exhibited significant inhibitory effects on the growth of MCF cell lines at a concentration of 20 μM, with all inhibitory rates above 50% (***p < 0.001, Fig. 2). Further determination of their half-inhibitory concentrations (IC50) against the growth of MCF cell lines revealed that compound 1z (IC50 = 7.85 μM) exhibited greater potency than the other three compounds (IC50 values ranging from 12.86 to 16.71 μM) and was more potent than the positive control cisplatin (DDP) and novobiocin (NB, IC50 = 205 μM, Fig. S1†) but less potent than geldanamycin (GA) (Table 1). Subsequently, we assessed the inhibitory effects of 1z on the growth of five distinct breast cancer cell lines (HCC1806, MDA-MB-231, MDA-MB-468, SKBr3, and BT474) and observed its wide range of antiproliferative activities against these breast cancer cell lines (IC50 values ranging from 8.16 to 12.14 μM).
Fig. 2. Cell viability of MCF-7 cells after treating with hybrids 1 (20 μM), DMSO (CTL), and DDP (20 μM) [***p < 0.001 vs. CTL, n = 3].
Antiproliferative activities of hybrids against breast cancer cell lines (IC50, μM)a.
| Compd | MCF-7 | HCC1806 | MDA-MB-231 | MDA-MB-468 | SKBr3 | BT474 |
|---|---|---|---|---|---|---|
| 1o | 12.86 ± 1.05 | 13.20 ± 0.74 | 8.14 ± 0.66 | >20 | ND | ND |
| 1p | 14.20 ± 1.04 | 14.80 ± 0.70 | 9.13 ± 0.68 | >20 | ND | ND |
| 1z | 7.85 ± 0.09 | 10.10 ± 1.21 | 11.70 ± 1.60 | 8.16 ± 0.07 | 12.14 ± 1.05 | 9.06 ± 0.07 |
| 1ac | 16.71 ± 1.03 | 14.70 ± 1.90 | 12.90 ± 2.21 | >20 | ND | ND |
| GAb | 0.13 ± 0.01 | 0.12 ± 0.01 | 0.15 ± 0.02 | 0.033 | 0.033 | 0.027 |
| DDPb | 15.66 ± 1.03 | 2.14 ± 0.02 | 32.57 ± 1.72 | 2.31 ± 0.05 | 4.34 ± 0.27 | 11.03 ± 0.78 |
Data were determined as mean ± SD from three independent experiments.
Positive control.
Molecular modelling
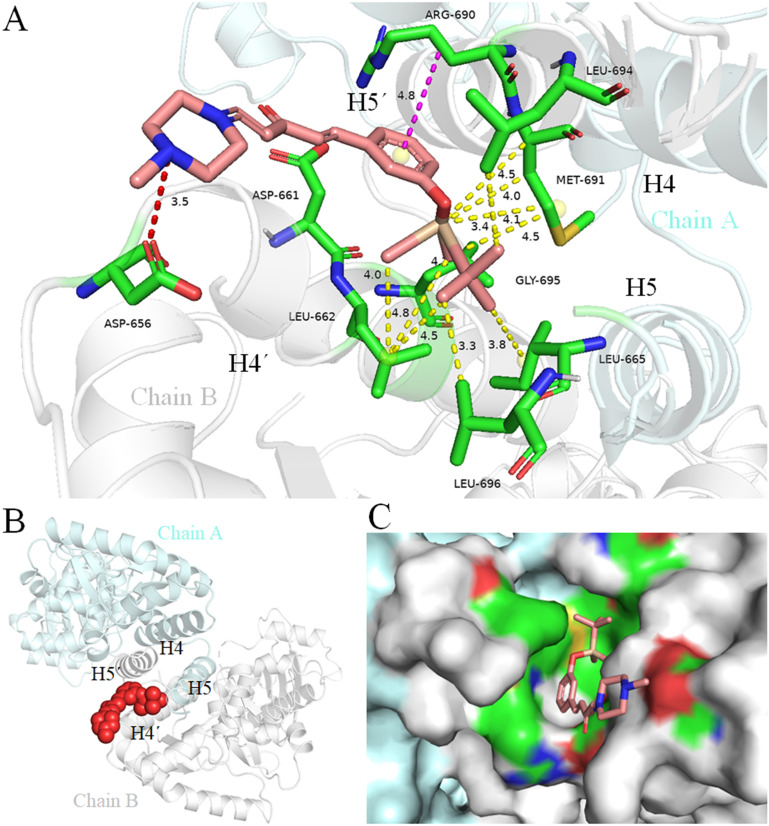
To gain more insights into the correlation of 1z with the HSP90 C-terminus, we conducted molecular simulation experiments (Fig. 3). The blind docking results demonstrated that 1z can access the HSP90 C-terminal dimerization region comprising the pocket formed by the H4, H5, H4′, and H5′ helices46,47 with an unfolded E,E-conformation. In detail, the N atom of N-methylpiperazine in the amine fragment of 1z forms a robust salt bridge with the amino acid residue ASP656, while the aryl fragment of 1z establishes significant lipophilic interactions with amino acid residues in H4′, H5′, and H5. Specifically, there is a π–δ interaction between ARG690 and the aromatic ring of 1z, alkyl–alkyl interactions involving LEU662, MET691, LEU694, and GLY695 from H4′/H5′ as well as LEU695 and LEU696 from H5 with the tert-butyldimethyl silyl (TBS) fragment in 1z, which contributed to the lipophilic interactions.48 These interactions provide a valuable support for the superior performance of 1z compared to other derivatives.
Fig. 3. Predicted binding mode of 1z in the HSP90 C-terminus. (A) Interactions between 1z (pink) with the amino acids (green) of H4′, H5′, and H5 in the HSP90 C-terminus (PDB 3Q6M), chain A (cyan), chain B (gray), salt bridge: red-dotted line; π–δ interaction: purple-dotted line; alkyl–alkyl interactions: yellow-dotted line. (B) A global view of binding mode; 1z is depicted in a red space filling model. (C) Solid surface map of the interaction pocket with compound 1z; red, blue, and green colored regions correspond to negatively charged, positively charged, and neural areas, respectively.
Structure–activity relationships
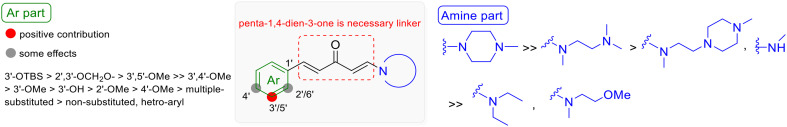
Based on the aforementioned information, the SARs for the antiproliferative activities of aryl/penta-1,4-dien-3-one/amine hybrids 1 against breast cancer cells could be summarized as follows (Fig. 4): (i) the presence of a penta-1,4-dien-3-one linker which most likely maintained the E,E-conformation of the hybrids is necessary for activity, as observed in 1ovs.1o-1, 1pvs.1p-1, and 1zvs.1z-1 (Fig. 2 and Scheme S1†); (ii) substituting the 3′-site of the aryl fragment with a bulky lipophilic group like the TBS group (1z) is highly favorable for antiproliferative activity. Additionally, combining substitution at the 3′-site with that in other sites, such as 2′,3′-dioxymethylene (1o) and 3′,5′-dimethoxy (1p), also enhances antiproliferative activity; (iii) the amine fragment, particularly that with a less flexible N–CH2–CH2–N moiety like N-methylpiperazine, exhibits the highest activity.
Fig. 4. The structure–activity relationships of hybrids 1.
Biological evaluation of 1z
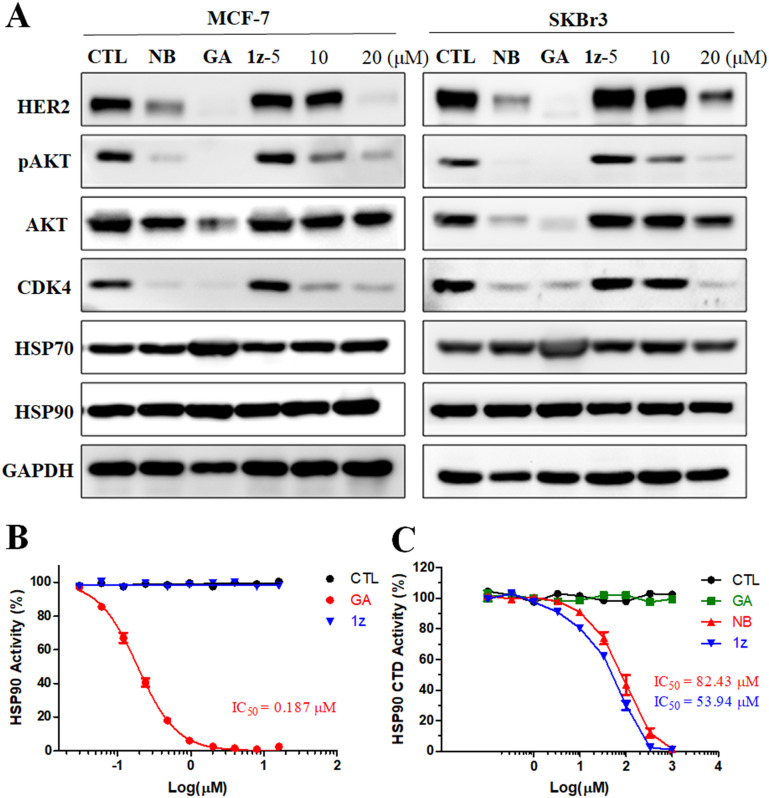
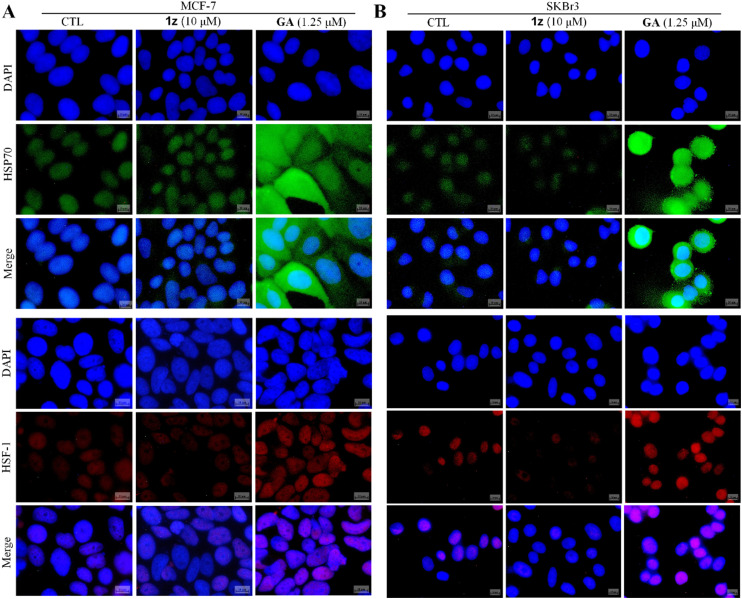
To ascertain whether the designed synthetic derivatives function as HSP90 inhibitors, the most potent compound 1z was chosen among them and its impact on HSP90, HSP70, and several crucial HSP90 client proteins in MCF-7 and SKBr3 cells was assessed. The results demonstrated that 1z downregulated HSP90 client proteins HER2, pAKT, and CDK4, but did not affect the expression level of AKT in MCF-7 cells (Fig. 5A), while these biomarkers could be significantly downregulated by 1z in SKBr3 cells (Fig. 5A). The expression of HSP90 and HSP70 was less impacted by 1z compared to GA, which exhibited similarities to NB in both cells. Subsequent enzyme-based experiments revealed that 1z lacked binding capacity to the HSP90 N-terminus (Fig. 5B), whereas it could disturb the binding of Cyp40 with the recombined HSP90 C-terminus with an IC50 value of 53.94 μM (Fig. 5C), surpassing NB in terms of inhibitory potency (IC50 = 82.43 μM), further corroborating our docking results of 1z interacting with the HSP90 C-terminus (Fig. 3). Furthermore, considering that the majority of HSP90 C-terminal inhibitors are not associated with heat shock response (HSR) due to their incapacity to promote the prosurvival factor HSP70,49,50 our investigation revealed that 1z does not elevate the expression levels of HSP70 in both MCF-7 and SKBr3 cells (Fig. 5A), thus suggesting its unlikelihood to induce HSR effects. Consequently, a cellular immunofluorescence experiment was performed and confirmed that 1z almost did not induce the alterations in HSP70 levels in MCF-7 and SKBr3 cells compared to GA (Fig. 6). To further address whether the unchanged HSP70 levels resulted from the inactivated upstream HSF-1/HSP90 axis, cellular immunofluorescence of HSF-1 in MCF-7 and SKBR3 cells was also performed. We found that 1z apparently precluded the nuclear accumulation of HSF-1 in both MCF-7 and SKBR3 cells (Fig. 6). Taken together, it can be inferred that 1z functions as a prototypical HSP90 C-terminal inhibitor without HSR induction.
Fig. 5. Effects of 1z on HSP90 and its client proteins in MCF-7 and SKBr3 cells. (A) Western blotting analysis of HER2, pAKT, AKT, CDK4, HSP70, and HSP90 protein expression in MCF-7 and SKBr3 cells after treatments with CTL (DMSO), GA (1.25 μM), NB (700 μM), and compound 1z (5, 10, and 20 μM) for 48 h. GAPDH was used as a loading control. (B) Unlike GA, 1z cannot compete the binding of FITC-GA (5 nM) with HSP90 in fluorescence polarization. (C) AlphaScreen-based HSP90 C-terminal inhibitory activities of 1z, GA, and NB (the results were determined by two dependent experiments).
Fig. 6. Hybrid 1z does not induce HSR. (A) MCF-7 and (B) SKBr3 cells were treated with CTL (DMSO), 1z (10 μM), and GA (1.25 μM) for 36 h and then immunostained with HSP70 (green), HSF-1 (red), and DAPI (blue). Scale bar: 10 μm.
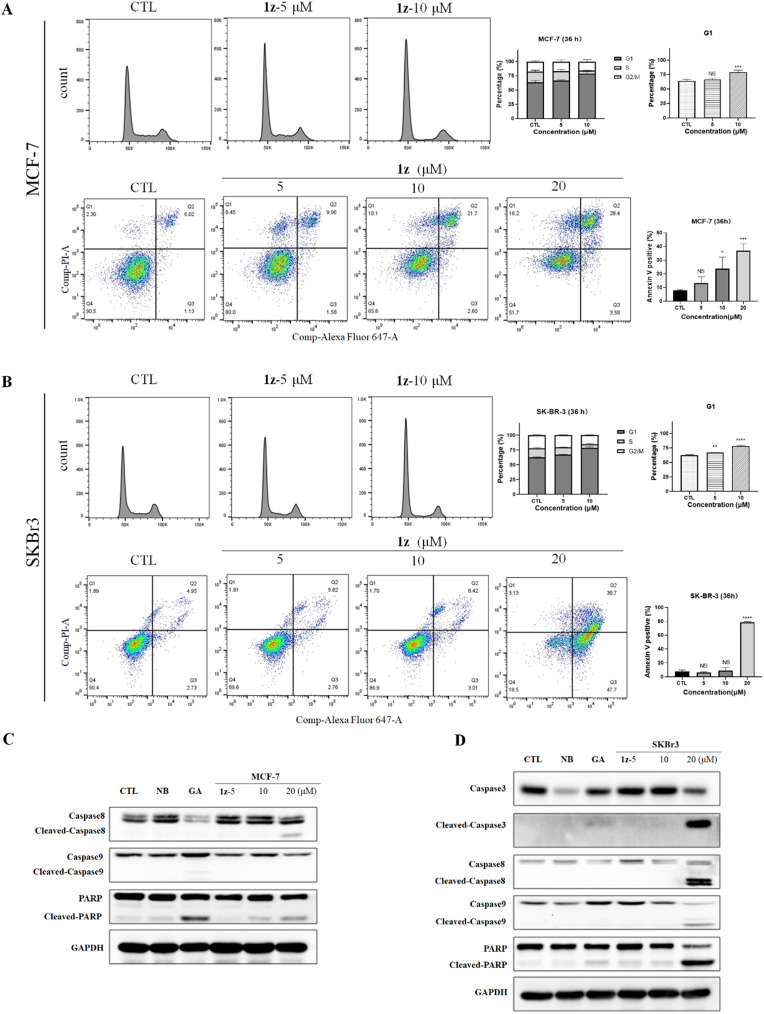
Subsequently, we investigated the distribution of the cell population in different stages of the cell cycle. Flow cytometric analyses were performed with MCF-7 and SKBr3 cells. As a result, 1z elevated the population of MCF-7 and SKBr3 cells in the G1 phase (***p < 0.001, Fig. 7A and B). Interestingly, 1z could induce obvious apoptosis of both cancer cells but in different manners, of which a late-stage apoptosis of MCF-7 cells and an early-stage apoptosis of SKBr3 cells were observed after treatments with 1z (0–20 μM) for 36 h (Fig. 7A and B). Furthermore, proteolytic activation of caspase-3, -8, and -9 was detected in 1z-teated MCF-7 and SKBr3 cells, respectively (Fig. 7C and D). The cleavage of pro-caspase-8 in MCF-7 cells and pro-caspase-3, -8, and -9 in SKBr3 cells as well as the cleavage of PARP in both cells was increased after 1z treatment. These observations suggested that 1z could induce cell cycle arrest and apoptosis, thereby resulting in its antiproliferative effects.
Fig. 7. Hybrid 1z induced cell cycle arrest and apoptosis of MCF-7 and SKBr3 cells. (A) MCF-7 and (B) SKBr3 cells were treated with 1z (0, 5, and 10 μM) for 36 h and stained with PI; cell cycle distribution was tested by flow cytometry. G1 population changes in cell cycle distribution (**p < 0.01, ***p < 0.001 vs. CTL, n = 3). Annexin V/PI staining was used to determine the early and late apoptotic cells after treating with 1z (0, 5, 10, and 20 μM) for 36 h (*p < 0.05, ***p < 0.001 vs. CTL, n = 3). (C) Effects of CTL (DMSO), NB (700 μM), GA (1.25 μM), and 1z (5–20 μM, 48 h) on expression of caspase-8, caspase-9, cleaved-caspase-8, cleaved-caspase-9, PARP, and cleaved-PARP in MCF-7 cells, and on expression of caspase-3, cleaved-caspase-3, caspase-8, caspase-9, cleaved-caspase-8, cleaved-caspase-9, PARP, and cleaved-PARP in SKBr3 cells (D).
Thermostability, in vitro metabolic stability, and preliminary toxicity study
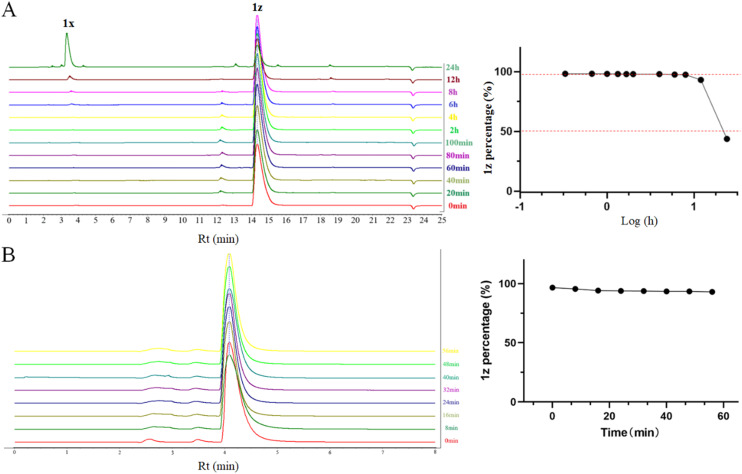
Previous work has indicated the thermal instability of vibsanin B derivatives. Despite some improvement in the thermal stability of VB4157 compared to vibsanin B,42 it fails to meet the prerequisites for comprehensive development. In this study, we eliminated the oxy-Cope rearrangement system in vibsanin B,43 which is responsible for its thermo-instability. Consequently, we developed and synthesized aryl/penta-1,4-dien-3-one/amine hybrids 1, aiming to fundamentally address the issue of thermo-instability. Subsequently, we examined the thermal stability of 1z in boiling toluene and found it was stable for approximately 12 hours at 120 °C. After 12 hours, the TBS moiety began to be removed, resulting in the formation of 1x (Fig. 8A). Nonetheless, the thermal stability of 1z (time for 50% decomposition > 12 hours) has shown a qualitative improvement compared to VB4157 (time for 50% decomposition <5 min). Moreover, the in vitro metabolic stability of 1z in simulated intestinal fluid was also tested (Fig. 8B). It was shown that 1z was stable at least for 1 hour in simulated intestinal fluid but unstable in simulated gastric fluid (data not shown). In addition, the preliminary toxicity of 1z was also tested using human embryonic kidney cells (293T).51 As a result, 1z [IC50 = 32.65 μM, selectivity index (SI) towards MCF-7: ∼4.2] was less toxic than both DDP (IC50 = 10.81 μM) and GA (IC50 = 0.53 μM), but more toxic than NB (IC50 > 100 μM, data not shown) (Fig. S1†), thereby establishing a foundation for subsequent optimization of this category of HSP90 inhibitors.
Fig. 8. The percentage of 1z after heating at 120 °C in toluene for 0–24 h was determined by HPLC (A). The percentage of 1z after incubating with simulated intestinal fluid at 37 °C for 1 h was determined by HPLC (B).
Conclusions
In conclusion, we have successfully designed and synthesized a novel class of aryl/penta-1,4-dien-3-one/amine hybrids as inhibitors targeting the HSP90 C-terminus. Among these hybrids, compound 1z showed significant antiproliferative effects on various human breast cancer cell lines, including MCF-7, HCC1806, MDA-MB-231, MDA-MB-468, SKBr3, and BT474. Furthermore, our findings revealed that 1z effectively suppressed the C-terminal activity of HSP90, resulting in the degradation of HSP90 client proteins HER2, pAKT, AKT, and CDK4, without inducing HSR. Consequently, by HSP90 inhibition, 1z induced cell cycle arrest and ultimately led to cell apoptosis through the activation of caspase-3, caspase-8, and caspase-9. Subsequent SARs analysis, complemented by molecular simulation, and cellular and enzyme-based experiments, highlighted the critical contributions of the penta-1,4-dien-3-one linker in the hybrid, the presence of a large bulky lipophilic silyl substitution at the 3′-site of the aryl fragment, and the inclusion of N-methylpiperazine in the amine fragment. These factors significantly influenced the observed antiproliferative activity. Additionally, 1z exhibited enhanced thermostability compared to vibsanin B derivatives and good in vitro metabolic stability in simulated intestinal fluid, representing one of the few silicon-containing HSP90 C-terminal inhibitors reported to date. These comprehensive findings provide a scientific foundation for the future development of HSP90C-terminal inhibitors.
Experimental section
Chemistry
General
All reactions were carried out under an atmosphere of argon in an oven-dried flask, and were monitored by analytical thin-layer chromatography (TLC), which was visualized by both ultraviolet light (254 nm) and improved Dragendorff's reagent. THF was distilled from Na, other reagents were obtained commercially. Purification of products was accomplished by flash column chromatography using silica gel (200–300 mesh). All NMR spectra were recorded using a Bruker AVANCE III 500 MHz or AVANCE III 600 MHz (1H NMR) spectrometer and 125 MHz or 150 MHz (13C NMR) in CDCl3: chemical shifts (δ) are given in ppm, coupling constants (J) in Hz, the solvent signals were used as references (CDCl3: δC = 77.16 ppm; residual CHCl3 in CDCl3: δH = 7.26 ppm).
General procedure for synthesis of enamines 5a–5f
To a solution of corresponding amines 3 (5 mmol) in THF (15 mL) was added NaOH (1.27 mL, 6 N in H2O, 7.5 mmol) dropwise at 0 °C; the resulting mixture was stirred for 1 h at 0 °C before 4-methoxy-3-butene-2-ketone (2) (5 mmol) was added. The resulting mixture was warmed to room temperature and stirred for 12–16 h. After consumption of the starting materials, the reaction was quenched by addition of water (2 mL) and extracted with DCM (6 × 10 mL). The combined organic layer was dried over Na2SO4, filtered, and concentrated in vacuo. The crude product was purified by silica gel column chromatography (DCM/MeOH = 20 : 1, v/v) to afford enamines 5a–5f.
(E)-4-(4-Methylpiperazin-1-yl)but-3-en-2-one (5a) (80% yield). 1H NMR (500 MHz, CDCl3) δ 7.36 (d, J = 13.0 Hz, 1H), 5.27 (d, J = 1.9 Hz, 1H), 5.13 (dd, J = 13.0, 1.9 Hz, 1H), 3.26 (brs, 4H), 2.39 (dd, J = 6.7, 3.4 Hz, 4H), 2.28 (d, J = 2.2 Hz, 3H), 2.06 (d, J = 2.2 Hz, 3H).
(E)-4-(Methyl(2-(4-methylpiperazin-1-yl)ethyl)amino)but-3-en-2-one (5b) (40% yield). 1H NMR (500 MHz, CD3OD) δ 7.68 (d, J = 12.5 Hz, 1H), 5.12 (brs, 1H), 3.46 (brs, 2H), 3.18 (brs, 1H), 2.89 (s, 3H), 2.58 (t, J = 6.3 Hz, 4H), 2.50 (brs, 5H), 2.28 (s, 3H), 2.10 (s, 3H).
(E)-4-(Methylamino)but-3-en-2-one (5c) (quantitative yield). 1H NMR (500 MHz, CDCl3) δ 9.65 (s, 1H), 6.61 (dd, J = 12.8, 7.3 Hz, 1H), 4.97 (d, J = 7.3 Hz, 1H), 2.98 (d, J = 5.0 Hz, 3H), 2.03 (s, 3H).
(E)-4-(Diethylamino)but-3-en-2-one (5d) (quantitative yield). 1H NMR (500 MHz, CDCl3) δ 7.45 (d, J = 11.5 Hz, 1H), 5.08 (d, J = 12.9 Hz, 1H), 3.20 (brs, 4H), 2.06 (s, 3H), 1.15 (t, J = 6.3 Hz, 6H).
(E)-4-((2-Methoxyethyl)(methyl)amino)but-3-en-2-one (5e) (85% yield). 1H NMR (500 MHz, CDCl3) δ 7.45 (d, J = 12.6 Hz, 1H), 5.04 (d, J = 12.9 Hz, 1H), 3.53–3.42 (m, 2H), 3.33 (brs, 2H), 3.31 (s, 3H), 2.82 (brs, 3H), 2.06 (s, 3H).
(E)-4-((2-(Dimethylamino)ethyl)(methyl)amino)but-3-en-2-one (5f) (74% yield). 1H NMR (500 MHz, CDCl3) δ 7.45 (d, J = 12.5 Hz, 1H), 5.06 (d, J = 12.8 Hz, 1H), 3.44 (t, J = 6.5 Hz, 2H), 2.88 (brs, 3H), 2.69 (t, J = 7.0 Hz, 2H), 2.42 (s, 6H), 2.06 (s, 3H).
General procedure for synthesis of aryl/penta-1,4-dien-3-one/amine hybrids 1a–1af
To a solution of corresponding enamines 5 (0.3 mmol) in THF (2 mL) was added dropwise LiHMDS (0.36 mmol) at −78 °C and stirred for 30 min before aldehydes 4 (0.3 mmol in 1 mL THF) was added; then the resulting mixture was warmed to room temperature and stirred for 12–20 h. The reaction was quenched by addition of water (2 mL) and extracted with DCM (10 mL × 5). The combined organic layer was dried over Na2SO4, filtered, and concentrated in vacuo. The residue was purified by silica gel column chromatography (DCM/MeOH/diethylamine = 100 : 5 : 1, v/v) to afford hybrids 1a–1af.
(1E,4E)-1-(3,4-Dimethoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1a) (51% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.66 (d, J = 12.7 Hz, 1H), 7.49 (d, J = 15.8 Hz, 1H), 7.10 (dd, J = 8.3, 1.8 Hz, 1H), 7.07 (d, J = 1.8 Hz, 1H), 6.83 (d, J = 8.3 Hz, 1H), 6.64 (d, J = 15.8 Hz, 1H), 5.39 (d, J = 12.7 Hz, 1H), 3.89 (s, 3H), 3.88 (s, 3H), 3.37 (brs, 4H), 2.46–2.42 (m, 4H), 2.31 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.0, 151.8, 150.4, 149.1, 138.9, 128.7, 126.4, 122.2, 111.1, 109.7, 96.1, 56.0, 55.9, 54.4 (×4), 46.2. HRESI-MS calculated for [M + H]+ C18H25N2O3+ (m/z): 317.1860; found: 317.1868. HPLC purity: 99.0% (tR = 11.03 min).
(1E,4E)-1-(6-Methoxy-2,3-dihydro-1H-inden-5-yl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1b) (28% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.87 (d, J = 16.0 Hz, 1H), 7.65 (d, J = 12.7 Hz, 1H), 7.41 (s, 1H), 6.79 (s, 1H), 6.78 (d, J = 16.0 Hz, 1H), 5.43 (d, J = 12.7 Hz, 1H), 3.84 (s, 3H), 3.38–3.35 (m, 3H), 2.89 (t, J = 7.4 Hz, 2H), 2.84 (t, J = 7.3 Hz, 2H), 2.47–2.41 (m, 4H), 2.32 (s, 3H), 2.09–2.04 (m, 2H). 13C NMR (126 MHz, CDCl3) δ 188.0, 157.5, 151.7, 147.7, 136.1, 134.8, 127.8, 123.6, 122.7, 107.6, 96.2, 55.8, 54.4 (×4), 46.2, 33.7, 32.1, 25.8. HRESI-MS calculated for [M + H]+ C20H27N2O2+ (m/z): 327.2067; found: 327.2075. HPLC purity: 99.0% (tR = 8.83 min).
(1E,4E)-1-(3-Methoxynaphthalen-2-yl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1c) (21% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 8.23 (d, J = 8.6 Hz, 1H), 8.17 (d, J = 16.0 Hz, 1H), 7.78 (d, J = 9.0 Hz, 1H), 7.75 (d, J = 8.1 Hz, 1H), 7.69 (d, J = 12.7 Hz, 1H), 7.46 (ddd, J = 8.4, 6.8, 1.3 Hz, 1H), 7.33 (t, J = 7.9 Hz, 1H), 7.24 (d, J = 1.0 Hz, 1H), 7.00 (d, J = 16.0 Hz, 1H), 5.42 (d, J = 12.7 Hz, 1H), 3.95 (s, 3H), 3.36 (brs, 4H), 2.47–2.40 (m, 4H), 2.30 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 187.7, 156.2, 151.9, 133.8, 132.9, 132.3, 130.6, 129.1, 128.5, 127.0, 124.0, 123.8, 118.5, 113.0, 96.7, 56.4, 54.4 (×4), 46.2. HRESI-MS calculated for [M + H]+ C21H25N2O2+ (m/z): 337.1911; found: 337.1912. HPLC purity: 97.8% (tR = 7.27 min).
(1E,4E)-1-(3-Fluoro-2-methoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1d) (55% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.76 (d, J = 16.0 Hz, 1H), 7.67 (d, J = 12.6 Hz, 1H), 7.31 (d, J = 7.7 Hz, 1H), 7.06–7.00 (m, 1H), 7.00–6.94 (m, 1H), 6.80 (d, J = 16.0 Hz, 1H), 5.39 (d, J = 12.6 Hz, 1H), 3.92 (d, J = 1.6 Hz, 3H), 3.38 (brs, 4H), 2.46–2.42 (m, 4H), 2.31 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.0, 156.9, 154.9, 152.2, 146.7, 146.6, 132.6, 132.5, 130.88, 130.9, 130.5, 123.7, 123.6, 123.0 (×2), 117.5, 117.3, 96.1, 61.8 (×2), 54.4 (×2), 54.2 (×2), 46.1. HRESI-MS calculated for [M + H]+ C17H22FN2O2+ (m/z): 305.1660; found: 305.1670. HPLC purity: 99.3% (tR = 5.20 min).
(1E,4E)-1-(4-Methylpiperazin-1-yl)-5-phenylpenta-1,4-dien-3-one (1e) (51% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.68 (d, J = 12.6 Hz, 1H), 7.56 (d, J = 15.8 Hz, 1H), 7.53 (d, J = 7.0 Hz, 2H), 7.37–7.30 (m, 3H), 6.77 (d, J = 15.8 Hz, 1H), 5.39 (d, J = 12.7 Hz, 1H), 3.38 (brs, 4H), 2.48–2.42 (m, 4H), 2.32 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.9, 152.1, 138.9, 135.8, 129.3, 128.8 (×2), 128.3, 128.0 (×2), 96.2, 54.5 (×4), 46.2. HRESI-MS calculated for [M + H]+ C16H21N2O+ (m/z): 257.1648; found: 257.1650. HPLC purity: 99.3% (tR = 12.84 min).
(1E,4E)-1-(4-Chlorophenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1f) (45% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.69 (d, J = 12.6 Hz, 1H), 7.50 (d, J = 15.8 Hz, 1H), 7.46 (d, J = 8.6 Hz, 2H), 7.32 (d, J = 8.5 Hz, 2H), 6.73 (d, J = 15.8 Hz, 1H), 5.37 (d, J = 12.6 Hz, 1H), 3.39 (brs, 4H), 2.48–2.43 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.5, 152.2, 137.5, 135.2, 134.4, 129.2 (×2), 129.1 (×2), 128.8, 96.2, 54.5 (×4), 46.2. HRESI-MS calculated for [M + H]+ C16H20ClN2O+ (m/z): 291.1259; found: 291.1260. HPLC purity: 99.3% (tR = 6.49 min).
4-((1E,4E)-5-(4-methylpiperazin-1-yl)-3-oxopenta-1,4-dien-1-yl)benzonitrile (1g) (29% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.71 (d, J = 12.6 Hz, 1H), 7.60 (q, J = 8.3 Hz, 4H), 7.51 (d, J = 15.8 Hz, 1H), 6.81 (d, J = 15.8 Hz, 1H), 5.36 (d, J = 12.6 Hz, 1H), 3.39 (brs, 4H), 2.51–2.41 (m, 4H), 2.31 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 185.7, 152.6, 140.3, 136.3, 132.6 (×2), 131.5, 128.3 (×2), 118.8, 112.2, 96.2, 55.0, 53.7, 53.3, 46.1, 45.2. HRESI-MS calculated for [M + H]+ C17H20N3O+ (m/z): 282.1601; found: 282.1605. HPLC purity: >99.9% (tR = 11.05 min).
(1E,4E)-1-(4-Methoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1h) (38% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.66 (d, J = 12.7 Hz, 1H), 7.52 (d, J = 15.8 Hz, 1H), 7.48 (d, J = 8.7 Hz, 2H), 6.88 (d, J = 8.7 Hz, 2H), 6.65 (d, J = 15.8 Hz, 1H), 5.38 (d, J = 12.7 Hz, 1H), 3.82 (s, 3H), 3.42–3.32 (m, 4H), 2.50–2.41 (m, 4H), 2.32 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.2, 160.9, 151.8, 138.7, 129.6 (×2), 128.6, 126.2, 114.4 (×2), 96.4, 55.5, 54.5 (×4), 46.2. HRESI-MS calculated for [M + H]+ C17H23N2O2+ (m/z): 287.1754; found: 287.1756. HPLC purity: 98.7% (tR = 13.11 min).
(1E,4E)-1-(2-Methoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1i) (65% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.86 (d, J = 16.1 Hz, 1H), 7.66 (d, J = 12.7 Hz, 1H), 7.54 (dd, J = 7.6, 1.3 Hz, 1H), 7.33–7.27 (m, 1H), 6.94 (t, J = 7.5 Hz, 1H), 6.89 (d, J = 8.3 Hz, 1H), 6.83 (d, J = 16.0 Hz, 1H), 5.44 (d, J = 12.7 Hz, 1H), 3.87 (s, 3H), 3.43–3.32 (m, 4H), 2.51–2.43 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.7, 158.4, 151.8, 134.3, 130.6, 129.2, 128.5, 125.0, 120.8, 111.3, 96.2, 55.6, 54.5 (×4), 46.2. HRESI-MS calculated for [M + H]+ C17H23N2O2+ (m/z): 287.1754; found: 287.1752. HPLC purity: 97.5% (tR = 13.51 min).
(1E,4E)-1-(2-Methoxy-4-methylphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1j) (73% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.82 (d, J = 16.0 Hz, 1H), 7.65 (d, J = 12.7 Hz, 1H), 7.43 (d, J = 7.8 Hz, 1H), 6.80 (d, J = 16.0 Hz, 1H), 6.75 (d, J = 7.8 Hz, 1H), 6.70 (s, 1H), 5.43 (d, J = 12.7 Hz, 1H), 3.85 (s, 3H), 3.40–3.33 (m, 4H), 2.47–2.42 (m, 4H), 2.35 (s, 3H), 2.32 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.9, 158.3, 151.7, 141.3, 134.4, 128.5, 128.1, 122.0, 121.5, 112.0, 96.1, 55.5, 55.4 (×4), 46.2, 22.0. HRESI-MS calculated for [M + H]+ C18H25N2O2+ (m/z): 301.1911; found: 301.1912. HPLC purity: 98.8% (tR = 6.22 min).
3-Methoxy-4-((1E,4E)-5-(4-methylpiperazin-1-yl)-3-oxopenta-1,4-dien-1-yl)benzonitrile (1k) (51% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.79 (d, J = 16.0 Hz, 1H), 7.69 (d, J = 12.6 Hz, 1H), 7.58 (d, J = 8.0 Hz, 1H), 7.22 (dd, J = 7.9, 1.1 Hz, 1H), 7.10 (d, J = 1.0 Hz, 1H), 6.86 (d, J = 16.0 Hz, 1H), 5.39 (d, J = 12.6 Hz, 1H), 3.89 (s, 3H), 3.39 (brs, 4H), 2.50–2.43 (m, 4H), 2.32 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.5, 157.9, 152.4, 132.1, 131.9, 129.9, 128.8, 124.6, 118.9, 114.1, 113.0, 96.2, 55.9, 54.5 (×4), 46.2. HRESI-MS calculated for [M + H]+ C18H22N3O2+ (m/z): 312.1707; found: 312.1708. HPLC purity: 95.5% (tR = 11.44 min).
(1E,4E)-1-(2-Methoxy-5-nitrophenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1l) (44% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 8.43 (d, J = 2.7 Hz, 1H), 8.18 (dd, J = 9.1, 2.7 Hz, 1H), 7.82 (d, J = 15.9 Hz, 1H), 7.70 (d, J = 12.6 Hz, 1H), 6.94 (d, J = 9.1 Hz, 1H), 6.88 (d, J = 15.9 Hz, 1H), 5.39 (d, J = 12.6 Hz, 1H), 3.97 (s, 3H), 3.40 (s, 4H), 2.50–2.44 (m, 4H), 2.32 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.3, 162.6, 152.3, 141.5, 131.3, 131.2, 125.9 (×2), 123.3, 110.9, 96.6, 56.4, 54.4 (×4), 46.1. HRESI-MS calculated for [M + H]+ C17H22N3O4+ (m/z): 322.1605; found: 332.1608. HPLC purity: 97.8% (tR = 12.45 min).
(1E,4E)-1-(5-(Dimethylamino)-2-methoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1m) (38% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.81 (d, J = 15.9 Hz, 1H), 7.60 (d, J = 12.7 Hz, 1H), 7.42 (d, J = 8.7 Hz, 1H), 6.69 (d, J = 15.9 Hz, 1H), 6.28 (dd, J = 8.7, 1.8 Hz, 1H), 6.14 (d, J = 1.7 Hz, 1H), 5.42 (d, J = 12.7 Hz, 1H), 3.86 (s, 3H), 3.38–3.31 (m, 4H), 3.00 (s, 6H), 2.46–2.39 (m, 4H), 2.31 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 188.3, 160.0, 152.8, 151.1, 135.2, 130.0, 124.3, 113.2, 104.9, 96.4, 95.0, 55.4, 54.5 (×4), 46.2, 40.4 (×2). HRESI-MS calculated for [M + H]+ C19H28N3O2+ (m/z): 330.2176; found: 330.2170. HPLC purity: 96.4% (tR = 14.66 min).
(1E,4E)-1-(5-Isopropyl-2-methoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1n) (61% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.84 (d, J = 16.1 Hz, 1H), 7.66 (d, J = 12.7 Hz, 1H), 7.40 (d, J = 2.2 Hz, 1H), 7.15 (dd, J = 8.5, 2.2 Hz, 1H), 6.84 (d, J = 16.1 Hz, 1H), 6.82 (d, J = 8.5 Hz, 1H), 5.46 (d, J = 12.7 Hz, 1H), 3.85 (s, 3H), 3.38 (brs, 4H), 2.84 (dq, J = 13.7, 6.9 Hz, 1H), 2.47–2.43 (m, 4H), 2.32 (s, 3H), 1.22 (d, J = 6.9 Hz, 6H). 13C NMR (125 MHz, CDCl3) δ 187.9, 156.6, 151.8, 141.0, 134.6, 129.0, 128.6, 126.5, 124.4, 111.1, 96.0, 55.7, 54.5 (×4), 46.2, 33.4, 24.3 (×2). HRESI-MS calculated for [M + H]+ C20H29N2O2+ (m/z): 329.2224; found: 329.2226. HPLC purity: 98.1% (tR = 9.32 min).
(1E,4E)-1-(Benzo[d][1,3]dioxol-4-yl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1o) (55% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.67 (d, J = 12.7 Hz, 1H), 7.48 (d, J = 15.9 Hz, 1H), 6.96 (d, J = 15.8 Hz, 1H), 6.93 (dd, J = 7.6, 1.3 Hz, 1H), 6.83–6.75 (m, 2H), 6.03 (s, 2H), 5.38 (d, J = 12.7 Hz, 1H), 3.38 (s, 4H), 2.51–2.40 (m, 4H), 2.31 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.1, 152.1, 147.9, 146.2, 133.4, 131.0, 122.9, 121.8, 118.8, 109.0, 101.3, 96.6, 54.6 (×4), 46.2. HRESI-MS calculated for [M + H]+ C17H21N2O3+ (m/z): 301.1547; found: 301.1546. HPLC purity: 99.6% (tR = 12.96 min).
(1E,4E)-1-(3,5-Dimethoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1p) (23% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.69 (d, J = 12.6 Hz, 1H), 7.47 (d, J = 15.8 Hz, 1H), 6.73 (d, J = 15.8 Hz, 1H), 6.69 (s, 1H), 6.69 (s, 1H), 6.45 (t, J = 2.1 Hz, 1H), 5.40 (d, J = 12.6 Hz, 1H), 3.80 (s, 6H), 3.39 (brs, 4H), 2.50–2.41 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.9, 161.0 (×2), 152.1, 138.9, 137.8, 128.9, 105.9 (×2), 101.9, 96.1, 55.5, 54.3 (×4), 46.2. HRESI-MS calculated for [M + H]+ C18H25N2O3+ (m/z): 317.1860; found: 317.1866. HPLC purity: 99.3% (tR = 13.06 min).
(1E,4E)-1-(Benzo[d][1,3]dioxol-4-yl)-5-(methylamino)penta-1,4-dien-3-one (1q) (20% yield); yellow oil. 1H NMR (600 MHz, CDCl3) δ 10.24 (s, 1H), 7.42 (d, J = 15.9 Hz, 1H), 6.94 (dd, J = 7.9, 0.9 Hz, 1H), 6.88 (d, J = 15.9 Hz, 1H), 6.84 (dd, J = 12.7, 7.2 Hz, 1H), 6.80 (d, J = 7.8 Hz, 1H), 6.77 (dd, J = 7.7, 1.1 Hz, 1H), 6.04 (s, 2H), 5.20 (d, J = 7.1 Hz, 1H), 3.06 (d, J = 4.9 Hz, 3H). 13C NMR (150 MHz, CDCl3) δ 188.2, 155.5, 147.9, 146.1, 132.5, 130.9, 122.8, 121.8, 118.9, 108.9, 101.3, 95.1, 35.6. HRESI-MS calculated for [M + H]+ C18H25N2O3+ (m/z): 317.1860; found: 317.1866. HPLC purity: 98.7% (tR = 12.50 min).
(1E,4E)-1-(3,5-Dimethoxyphenyl)-5-(methylamino)penta-1,4-dien-3-one (1r) (34% yield); yellow oil. 1H NMR (600 MHz, CDCl3) δ 10.22 (s, 1H), 7.39 (d, J = 15.8 Hz, 1H), 6.85 (dd, J = 12.7, 7.1 Hz, 1H), 6.68 (s, 1H), 6.68 (s, 1H), 6.65 (d, J = 15.8 Hz, 1H), 6.44 (t, J = 2.2 Hz, 1H), 5.21 (d, J = 7.1 Hz, 1H), 3.81 (s, 6H), 3.07 (d, J = 5.0 Hz, 3H). 13C NMR (150 MHz, CDCl3) δ 187.9, 161.0 (×2), 155.6, 137.9, 128.7, 105.8 (×2), 101.7, 94.8, 94.8, 55.5 (×2), 35.7. HRESI-MS calculated for [M + H]+ C14H18NO3+ (m/z): 248.1281; found: 248.1286. HPLC purity: 99.0% (tR = 12.71 min).
(1E,4E)-1-(benzo[d][1,3]dioxol-4-yl)-5-(diethylamino)penta-1,4-dien-3-one (1s) (28% yield); yellow oil. 1H NMR (600 MHz, CDCl3) δ 7.77 (d, J = 12.6 Hz, 1H), 7.48 (d, J = 15.4 Hz, 1H), 7.01–6.91 (m, 2H), 6.84–6.74 (m, 2H), 6.05 (s, 2H), 5.31 (d, J = 12.6 Hz, 1H), 3.38–3.21 (m, 4H), 1.24–1.17 (m, 6H). 13C NMR (150 MHz, CDCl3) δ 186.6, 151.9, 151.9, 147.9, 147.9, 146.1, 133.0, 123.0, 121.8, 119.0, 108.9, 101.3, 50.7, 42.9, 14.9, 11.7. HRESI-MS calculated for [M + H]+ C16H20NO3+ (m/z): 274.1438; found: 274.1436. HPLC purity: 97.3% (tR = 6.09 min).
(1E,4E)-1-(Diethylamino)-5-(3,5-dimethoxyphenyl)penta-1,4-dien-3-one (1t) (34% yield); yellow oil. 1H NMR (600 MHz, CDCl3) δ 7.77 (d, J = 12.6 Hz, 1H), 7.48 (d, J = 15.7 Hz, 1H), 6.75 (d, J = 15.7 Hz, 1H), 6.70 (s, 1H), 6.70 (s, 1H), 6.44 (t, J = 2.2 Hz, 1H), 5.32 (d, J = 12.6 Hz, 1H), 3.81 (s, 6H), 3.31 (dd, J = 35.4, 5.6 Hz, 4H), 1.26–1.21 (m, 6H). 13C NMR (150 MHz, CDCl3) δ 186.3, 161.0 (×2), 151.9, 138.4, 138.0, 129.2, 105.8 (×2), 95.9, 101.7, 101.7, 55.5 (×2), 50.8, 42.9, 14.9, 11.7. HRESI-MS calculated for [M + H]+ C17H24NO3+ (m/z): 290.1751; found: 290.1755. HPLC purity: 97.0% (tR = 6.31 min).
(1E,4E)-1-(Benzo[d][1,3]dioxol-4-yl)-5-(methyl(2-(4-methylpiperazin-1-yl)ethyl)amino)penta-1,4-dien-3-one (1u) (25% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.75 (d, J = 12.0 Hz, 1H), 7.49 (d, J = 15.9 Hz, 1H), 6.97 (d, J = 15.8 Hz, 1H), 6.94 (d, J = 7.5 Hz, 1H), 6.79 (dt, J = 14.2, 7.3 Hz, 2H), 6.04 (s, 2H), 5.27 (d, J = 12.7 Hz, 1H), 3.37 (s, 3H), 2.89 (brs, 2H), 2.69–2.49 (m, 10H), 2.37 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.7, 153.4, 147.9, 146.2, 133.2, 131.1, 127.9, 122.9, 121.9, 118.9, 114.0, 109.0, 101.3, 97.0, 56.3, 55.5, 54.9, 52.5, 45.4, 36.2. HRESI-MS calculated for [M + H]+ C20H28N3O3+ (m/z): 358.2125; found: 358.2120. HPLC purity: 99.6% (tR = 9.08 min).
(1E,4E)-1-(3,5-Dimethoxyphenyl)-5-(methyl(2-(4-methylpiperazin-1-yl)ethyl)amino)penta-1,4-dien-3-one (1v) (22% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.75 (d, J = 11.8 Hz, 1H), 7.45 (d, J = 15.8 Hz, 1H), 6.73 (d, J = 15.8 Hz, 1H), 6.68 (d, J = 2.2 Hz, 2H), 6.43 (t, J = 2.1 Hz, 1H), 5.27 (d, J = 12.2 Hz, 1H), 3.79 (s, 6H), 3.37 (s, 3H), 2.89 (brs, 2H), 2.70–2.42 (m, 11H), 2.35 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.4, 161.0 (×2), 153.4, 138.7, 137.8, 128.9, 105.9 (×2), 101.8, 96.6, 56.5, 55.5 (×2), 54.9 (×4), 52.7, 45.6, 36.3. HRESI-MS calculated for [M + H]+ C21H32N3O3+ (m/z): 374.2438; found: 374.2440. HPLC purity: 99.8% (tR = 13.92 min).
(1E,4E)-1-(3,5-Dimethoxyphenyl)-5-((2-methoxyethyl)(methyl)amino)penta-1,4-dien-3-one (1w) (39% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.76 (d, J = 12.5 Hz, 1H), 7.47 (d, J = 15.8 Hz, 1H), 6.75 (d, J = 15.8 Hz, 1H), 6.70 (d, J = 2.0 Hz, 2H), 6.44 (t, J = 2.1 Hz, 1H), 5.30 (d, J = 12.5 Hz, 1H), 3.81 (s, 6H), 3.54 (brs, 2H), 3.43 (brs, 2H), 3.35 (s, 3H), 2.94 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.6, 161.1 (×2), 153.3, 138.7, 138.0, 129.0, 106.0 (×2), 101.9, 96.8, 71.3, 59.2, 57.7, 55.5 (×2), 36.7. HRESI-MS calculated for [M + H]+ C17H24NO4+ (m/z): 306.1700; found: 306.1704. HPLC purity: >99.9% (tR = 6.05 min).
(1E,4E)-1-(3-Hydroxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1x) (45% yield); yellow oil. Compound 1x was prepared from 1z by removal of the TBS group under the condition of tetrabutylammonium fluoride (TBAF) (2.0 eq.) in a mixed solvent of THF/AcOH (20 : 1, v/v).421H NMR (500 MHz, CDCl3) δ 7.72 (d, J = 12.6 Hz, 1H), 7.61 (d, J = 15.7 Hz, 1H), 7.22 (dd, J = 9.1, 6.6 Hz, 2H), 7.09 (d, J = 7.7 Hz, 1H), 6.89 (dd, J = 8.0, 1.8 Hz, 1H), 6.78 (d, J = 15.7 Hz, 1H), 5.39 (d, J = 12.6 Hz, 1H), 3.38 (s, 4H), 2.52–2.39 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.3, 157.3, 152.7, 139.7, 136.8, 129.8, 127.9, 119.3, 117.2, 115.4, 96.3, 54.2 (×4), 45.9. HRESI-MS calculated for [M + H]+ C16H21N2O2+ (m/z): 273.1598; found: 273.1597. HPLC purity: >99.9% (tR = 9.46 min).
(1E,4E)-1-(3-Methoxyphenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1y) (67% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.69 (d, J = 12.7 Hz, 1H), 7.53 (d, J = 15.8 Hz, 1H), 7.28 (dd, J = 8.8, 7.0 Hz, 2H), 7.14 (d, J = 7.7 Hz, 1H), 6.89 (dd, J = 8.2, 2.3 Hz, 1H), 6.76 (d, J = 15.8 Hz, 1H), 5.41 (d, J = 12.7 Hz, 1H), 3.83 (s, 3H), 3.40 (brs, 4H), 2.52–2.43 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.9, 159.9, 152.1, 138.8, 137.2, 129.8, 128.6, 124.3, 120.7, 115.3, 112.9, 96.2, 55.4, 54.4 (×4), 46.1. HRESI-MS calculated for [M + H]+ C17H23N2O2+ (m/z): 287.1754; found: 287.1755. HPLC purity: 99.1% (tR = 12.74 min).
(1E,4E)-1-(3-((Tert-butyldimethylsilyl)oxy)phenyl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1z) (55% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.68 (d, J = 12.7 Hz, 1H), 7.48 (d, J = 15.8 Hz, 1H), 7.20 (t, J = 7.8 Hz, 1H), 7.13 (d, J = 7.7 Hz, 1H), 7.03–6.97 (m, 1H), 6.83–6.76 (m, 1H), 6.71 (d, J = 15.8 Hz, 1H), 5.39 (d, J = 12.7 Hz, 1H), 3.38 (brs, 4H), 2.48–2.42 (m, 4H), 2.32 (s, 3H), 0.98 (s, 9H), 0.19 (s, 6H). 13C NMR (125 MHz, CDCl3) δ 187.0, 156.1, 152.1, 138.8, 137.4, 129.8, 128.5, 121.3 (×2), 119.4, 96.2, 54.5 (×4), 46.2, 25.8 (×3), 18.4, −4.3 (×2). HRESI-MS calculated for [M + H]+ C22H35N2O2Si+ (m/z): 387.2462; found: 387.2466. HPLC purity: 99.2% (tR = 14.30 min).
(2Z,4E)-1-(Benzo[d][1,3]dioxol-4-yl)-3-hydroxy-5-((2-methoxyethyl)(methyl)amino)penta-2,4-dien-1-one (1aa) (24% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.68 (d, J = 12.6 Hz, 1H), 7.43 (dd, J = 6.6, 2.8 Hz, 1H), 6.91–6.85 (m, 2H), 6.17 (s, 1H), 6.06 (s, 2H), 4.93 (d, J = 12.7 Hz, 1H), 3.53 (s, 2H), 3.41 (s, 2H), 3.35 (s, 3H), 2.88 (d, J = 41.6 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ 188.3, 173.6, 151.4, 148.0, 145.9, 122.1, 121.6, 120.4, 119.1, 110.5, 101.3, 99.2, 94.1, 71.2, 59.2, 57.7, 36.5. HRESI-MS calculated for [M + H]+ C16H20NO5+ (m/z): 306.1336; found: 306.1338. HPLC purity: >99.9% (tR = 6.17 min).
(1E,4E)-1-(4-methylpiperazin-1-yl)-5-(3,4,5-trimethoxyphenyl)penta-1,4-dien-3-one (1ab) (42%); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.70 (d, J = 12.6 Hz, 1H), 7.47 (d, J = 15.7 Hz, 1H), 6.77 (s, 2H), 6.67 (d, J = 15.7 Hz, 1H), 5.41 (d, J = 12.6 Hz, 1H), 3.87 (s, 6H), 3.86 (s, 3H), 3.41 (brs, 4H), 2.54–2.46 (m, 4H), 2.35 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.9, 153.5 (×2), 152.1, 139.6, 139.1, 131.4, 127.8, 105.2 (×2), 96.2, 61.1, 56.2 (×2), 54.3 (×4), 46.0. HRESI-MS calculated for [M + H]+ C16H20NO5+ (m/z): 306.1336; found: 306.1338. HPLC purity: 99.7% (tR = 11.19 min).
(1E,4E)-1-(3,5-Dimethoxyphenyl)-5-((2-(dimethylamino)ethyl)(methyl)amino)penta-1,4-dien-3-one (1ac) (27%); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.76 (d, J = 11.0 Hz, 1H), 7.46 (d, J = 15.8 Hz, 1H), 6.74 (d, J = 15.8 Hz, 1H), 6.69 (d, J = 1.9 Hz, 2H), 6.44 (s, 1H), 5.29 (d, J = 12.4 Hz, 1H), 3.80 (s, 6H), 3.37 (brs, 2H), 2.91 (brs, 2H), 2.49 (t, J = 6.1 Hz, 2H), 2.26 (s, 6H). 13C NMR (126 MHz, CDCl3) δ 186.5, 161.0 (×2), 153.2, 138.7, 137.9, 129.0, 105.9 (×2), 101.9, 96.7, 58.1, 56.1, 55.5 (×2), 45.7 (×2), 36.2. HRESI-MS calculated for [M + H]+ C18H27N2O3+ (m/z): 319.2016; found: 319.2015. HPLC purity: 98.1% (tR = 9.41 min).
(1E,4E)-1-(furan-3-yl)-5-(4-methylpiperazin-1-yl)penta-1,4-dien-3-one (1ad) (63% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.66 (d, J = 12.7 Hz, 1H), 7.62 (s, 1H), 7.46 (d, J = 15.7 Hz, 1H), 7.41 (s, 1H), 6.60 (d, J = 1.5 Hz, 1H), 6.51 (d, J = 15.6 Hz, 1H), 5.35 (d, J = 12.7 Hz, 1H), 3.38 (brs, 4H), 2.50–2.41 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 186.8, 151.8, 144.0, 143.9, 128.7, 128.2, 123.4, 107.6, 95.9, 54.3 (×4), 46.0. HRESI-MS calculated for [M + H]+ C14H19N2O2+ (m/z): 247.1441; found: 247.1440. HPLC purity: 98.2% (tR = 9.85 min).
(1E,4E)-1-(4-Methylpiperazin-1-yl)-5-(thiophen-3-yl)penta-1,4-dien-3-one (1ae) (78% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 7.66 (d, J = 12.7 Hz, 1H), 7.54 (d, J = 15.7 Hz, 1H), 7.42 (s, 1H), 7.29 (d, J = 1.7 Hz, 2H), 6.59 (d, J = 15.7 Hz, 1H), 5.36 (d, J = 12.7 Hz, 1H), 3.37 (brs, 4H), 2.48–2.41 (m, 4H), 2.31 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 187.1, 152.0, 138.7, 132.6, 128.0, 127.1, 126.6, 125.2, 96.0, 54.9, 53.7, 53.0, 46.2, 45.0. HRESI-MS calculated for [M + H]+ C14H19N2OS+ (m/z): 263.1213; found: 263.1216. HPLC purity: 95.6% (tR = 11.78 min).
(1E,4E)-1-(4-Methylpiperazin-1-yl)-5-(pyridin-3-yl)penta-1,4-dien-3-one (1af) (64% yield); yellow oil. 1H NMR (500 MHz, CDCl3) δ 8.76 (s, 1H), 8.54 (s, 1H), 7.81 (d, J = 7.8 Hz, 1H), 7.70 (d, J = 12.5 Hz, 1H), 7.52 (d, J = 15.8 Hz, 1H), 7.28 (s, 1H), 6.81 (d, J = 15.8 Hz, 1H), 5.38 (d, J = 12.6 Hz, 1H), 3.41 (s, 4H), 2.54–2.43 (m, 4H), 2.34 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 186.0, 152.4, 150.0, 149.5, 135.1, 134.3, 131.7, 130.2, 123.9, 96.1, 54.4 (×4), 46.1. HRESI-MS calculated for [M + H]+ C14H19N2OS+ (m/z): 263.1213; found: 263.1216. HPLC purity: 96.4% (tR = 7.88 min).
Biology
MTT assay
Cell viability after treatments of tested compounds (20 μM) was evaluated by MTT assay according to our previous work.42 The IC50 values were determined by non-linear regression analysis using GraphPad Prism 5 software.
Fluorescence polarization (FP)
HSP90 NTD inhibitory activity was determined by a competitive binding assay against FITC-geldanamycin (GA) according to the literature.52 The tested compounds were dissolved in DMSO and then appropriately diluted to obtain a gradient concentration (0.03–16.0 μM) for addition to the final reaction system. The HSP90 reaction system consisted of HSP90α (10 nM), BSA (1%), FITC-GA (5 nM), and reaction buffer. After thoroughly mixing all components (DMSO <1%), the reaction system was incubated at room temperature for 3 h. Subsequently, the fluorescence was measured at 485 nm excitation wavelength and 530 nm emission wavelength using a SpectraMax M5 plate reader. HSP90 NTD activity% = (FPdrug − FPbackgroud)/(FPenzyme − FPbackgroud) × 100%. Curve fitting was performed using GraphPad Prism 5.
AlphaScreen-based HSP90 C-terminal inhibition
The HSP90 C-terminal inhibitory activity was determined by a modified AlphaScreen assay system (PerkinElmer, Waltham, MA) according to our previous work.42
Western blot
Cells were digested with trypsin, centrifuged, washed with PBS once before recentrifugation, and lysed in RIPA lysate containing a cocktail of phosphatase and protease inhibitors. The supernatant was collected (14 000 rpm, 4 °C, 15 min) and the protein concentration determined using a BCA reagent kit (Beyotime Shanghai, China). Twenty micrograms protein was separated by SDS-PAGE and transferred to an Immobilon-P Transfer Membrane (Millipore Corporation, Billerica, MA, USA). After blocking in 5% non-fat milk for 1 h at room temperature, the membrane was incubated with primary antibodies overnight at 4 °C; primary antibody dilutions were as follows: HER2 (1 : 1000), AKT (1 : 1000), pAKT (1 : 1000), CDK4 (1 : 1000), HSP70 (1 : 1000), HSP90 (1 : 1000), caspase-3 (1 : 1000), CL-caspase-3 (1 : 1000), PARP (1 : 1000), caspase-8 (1 : 1000), caspase-9 (1 : 1000), and GAPDH (1 : 10000), followed by incubation with HRP-conjugated rabbit or rat secondary antibodies (1 : 1000–1 : 5000). An ECL Chemiluminescence Substrate Kit (Biosharp, Anhui, China) was used for detection.
Cell cycle analysis and annexin V/PI assay
Pancreatic enzyme digestion of cells, centrifugation collection, 70% anhydrous ethanol fixation overnight or longer were performed. Absolute ethanol was washed off and cells were incubated with RNAase (0.2 mg ml−1) and propidium iodide (PI, 0.1 mg mL−1) for 20 min. For the annexin V/PI assay, a FITC-conjugated annexin V apoptosis detection kit (Solarbio, Beijing, China) was used according to the manufacturer's protocol. Stained cells were analyzed by flow cytometry using BD FACS Celesta.
Immunofluorescence analysis
For immunofluorescence analysis, cells were seeded in a 24-well plate that was previously inserted into the cell ladder and then treated with the tested compounds. The cells were washed twice with PBS in a warm bath and fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 for 5 min, followed by blocking with BSA (3%, 15 min), and then incubated with primary antibody (1 h, room temperature) and fluorescein-conjugated secondary antibody (1 h, room temperature). Cells were mounted with antifade mounting medium with DAPI (Beyotime Shanghai, China) and images were obtained using a Zeiss upright fluorescence microscope.
Molecular modeling
The ligand (1z) and receptor (PDB 3Q6M)47 were prepared using Autodock Tools v1.56 (ref. 53) according to our previous work.42 A grid box that covered the whole HSP90 C-terminus in 3Q6M was chosen. Docking parameters were set as the default values. Docking conformations were classified into different clusters by binding energy, and the cluster with the lowest binding energy was selected. In the selected cluster, conformations with the lowest binding energy and RMSD (<2.0 Å) were finally chosen to analyze the receptor–ligand interaction.
Stability
Thermostability was tested using 1z (1 mg mL−1) in boiling toluene (distilled); the remaining percentage of 1z was detected by an HPLC instrument (Agilent 1260) equipped with an Eclipse XDB-C18 (5 μm, 4.6 mm × 250 mm) column after the appointed time. HPLC analysis method A: 0.00–2.00 min: 70% MeOH/30% H2O; 2.01–15.00 min: from 70% MeOH/30% H2O to 100% MeOH; 15.00–20.00 min: 100% MeOH; 20.01–25.00 min: 70% MeOH/30% H2O; method B: 0.00–2.00 min: 45% MeOH/55% H2O; 2.01–15.00 min: from 45% MeOH/55% H2O to 100% MeOH; 15.00–20.00 min: 100% MeOH; 20.01–25.00 min: 45% MeOH/55% H2O.
In vitro metabolic stability was tested in simulated intestinal fluid (Phygene) which was incubated with 1z (1 mg mL−1); the remaining percentage of 1z was detected by an HPLC instrument (Agilent 1260) equipped with an Eclipse XDB-C18 (5 μm, 4.6 mm × 250 mm) column after the appointed time. HPLC analysis method: 95% MeOH/5% H2O.
Author contributions
Yu-Ting Liao, Xin-Ye Du, and Mei Wang: investigation and writing – original draft. Chun-Xia Zheng: software. Dashan Li: data curation. Chuan-Huizi Chen: methodology, formal analysis. Rong-Tao Li: writing – review & editing, supervision, resources. Li-Dong Shao: conceptualization, project administration, resources, supervision, writing – original draft, writing – review & editing.
Conflicts of interest
The authors declare that they have no known conflicts of interest.
Supplementary Material
Acknowledgments
The authors are grateful for the financial support from the National Natural Science Foundation of China (81960631, 82260683, and 22067012), the Yunnan Fundamental Research Project (202001AS070038), the Top Young Talent of Ten Thousand Talents Program of Yunnan Province (D. L. and L.-D. S.), the Bioactive Ethnopharmacol Molecules Chemical Conversion and Application Innovation Team of the Department of Education of Yunnan Province (2022), and the Start-up Fund of Yunnan University of Chinese Medicine (2019YZG03).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3md00431g
References
- Sung H. Ferlay J. Siegel R. L. Laversanne M. Soerjomataram I. Jemal A. Bray F. Ca-Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Giaquinto A. N. Sung H. Miller K. D. Kramer J. L. Newman L. A. Minihan A. Jemal A. Siegel R. L. Ca-Cancer J. Clin. 2022;72:524–541. doi: 10.3322/caac.21754. [DOI] [PubMed] [Google Scholar]
- Zheng R. Zhang S. Zeng H. Wang S. Sun K. Chen R. Li L. Wei W. He J. J. Natl. Cancer Inst. 2022;2:1–9. doi: 10.1093/jnci/djab126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G.-T. Jiang Y.-Z. Shi J.-X. Yang F. Li X.-G. Pei Y.-C. Zhang C.-H. Ma D. Xiao Y. Hu P.-C. Wang H. Yang Y.-S. Guo L.-W. Lu X.-X. Xue M.-Z. Wang P. Cao A. Y. Ling H. Wang Z.-H. Yu K.-D. Di G.-H. Li D.-Q. Wang Y.-J. Yu Y. Shi L.-M. Hu X. Huang W. Shao Z.-M. Nat. Commun. 2020;11:5679. doi: 10.1038/s41467-020-19342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Kim Y.-J. Park J. M. Park M. Nam K. D. Farrand L. Nguyen C.-T. La M. T. Ann J. Lee J. Kim J. Y. Seo J. H. Cell Death Discovery. 2021;7:354. doi: 10.1038/s41420-021-00743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M. Kim Y.-J. Park S. Park M. Farrand L. Nguyen C.-T. Ann J. Nam G. Park H.-J. Lee J. Kim J. Y. Seo J. H. Mol. Cancer. 2020;19:161. doi: 10.1186/s12943-020-01283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L. B. Blagg B. S. Future Med. Chem. 2009;1:267–283. doi: 10.4155/fmc.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matts R. L. Dixit A. Peterson L. B. Sun L. Voruganti S. Kalyanaraman P. Hartson S. D. Verkhivker G. M. Blagg B. S. J. ACS Chem. Biol. 2011;6:800–807. doi: 10.1021/cb200052x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Wang L. You Q.-D. Xu X.-L. J. Med. Chem. 2020;63:1798–1822. doi: 10.1021/acs.jmedchem.9b00940. [DOI] [PubMed] [Google Scholar]
- Amatya E. Blagg B. S. J. Bioorg. Med. Chem. Lett. 2023;80:129111. doi: 10.1016/j.bmcl.2022.129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Zhang C. Song C. Eur. J. Med. Chem. 2022;238:114516. doi: 10.1016/j.ejmech.2022.114516. [DOI] [PubMed] [Google Scholar]
- Trepel J. Mollapour M. Giaccone G. Neckers L. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren J. and Blagg B. S. J., in HSF1 and Molecular Chaperones in Biology and Cancer, ed. M. L. Mendillo, D. Pincus and R. Scherz-Shouval, Springer International Publishing, Cham, 2020, pp. 135–146, 10.1007/978-3-030-40204-4_9 [DOI] [Google Scholar]
- Bickel D. Gohlke H. Bioorg. Med. Chem. 2019;27:115080. doi: 10.1016/j.bmc.2019.115080. [DOI] [PubMed] [Google Scholar]
- Burlison J. A. Neckers L. Smith A. B. Maxwell A. Blagg B. S. J. J. Am. Chem. Soc. 2006;128:15529–15536. doi: 10.1021/ja065793p. [DOI] [PubMed] [Google Scholar]
- Shen G. Yu X. M. Blagg B. S. J. Bioorg. Med. Chem. Lett. 2004;14:5903–5906. doi: 10.1016/j.bmcl.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Yu X. M. Shen G. Neckers L. Blake H. Holzbeierlein J. Cronk B. Blagg B. S. J. J. Am. Chem. Soc. 2005;127:12778–12779. doi: 10.1021/ja0535864. [DOI] [PubMed] [Google Scholar]
- Donnelly A. C. Mays J. R. Burlison J. A. Nelson J. T. Vielhauer G. Holzbeierlein J. Blagg B. S. J. J. Org. Chem. 2008;73:8901–8920. doi: 10.1021/jo801312r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlison J. A. Avila C. Vielhauer G. Lubbers D. J. Holzbeierlein J. Blagg B. S. J. J. Org. Chem. 2008;73:2130–2137. doi: 10.1021/jo702191a. [DOI] [PubMed] [Google Scholar]
- Mays J. R. Hill S. A. Moyers J. T. Blagg B. S. J. Bioorg. Med. Chem. 2010;18:249–266. doi: 10.1016/j.bmc.2009.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L. B. Blagg B. S. J. Bioorg. Med. Chem. Lett. 2010;20:3957–3960. doi: 10.1016/j.bmcl.2010.04.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. Donnelly A. C. Kusuma B. R. Brandt G. E. L. Brown D. Rajewski R. A. Vielhauer G. Holzbeierlein J. Cohen M. S. Blagg B. S. J. J. Med. Chem. 2011;54:3839–3853. doi: 10.1021/jm200148p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma B. R. Duerfeldt A. S. Blagg B. S. J. Bioorg. Med. Chem. Lett. 2011;21:7170–7174. doi: 10.1016/j.bmcl.2011.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. Blagg B. S. J. Bioorg. Med. Chem. Lett. 2013;23:552–557. doi: 10.1016/j.bmcl.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma B. R. Khandelwal A. Gu W. Brown D. Liu W. Vielhauer G. Holzbeierlein J. Blagg B. S. J. Bioorg. Med. Chem. 2014;22:1441–1449. doi: 10.1016/j.bmc.2013.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K. M. Subramanian C. Sanchez J. Motiwala H. F. Liu W. Cohen M. S. Holzbeierlein J. Blagg B. S. J. Chem. – Eur. J. 2016;22:6921–6931. doi: 10.1002/chem.201504955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G. Forsberg L. K. Zhao H. Blagg B. S. J. Chem. – Eur. J. 2017;23:16574–16585. doi: 10.1002/chem.201703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg L. K. Davis R. E. Wimalasena V. K. Blagg B. S. J. Bioorg. Med. Chem. 2018;26:3096–3110. doi: 10.1016/j.bmc.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh K. W. Zhang Z. Wang J. Xu X. Munthali V. Zuo A. Blagg B. S. J. ACS Med. Chem. Lett. 2020;11:1535–1538. doi: 10.1021/acsmedchemlett.0c00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C.-T. Thanh La M. Ann J. Nam G. Park H.-J. Min Park J. Kim Y.-J. Young Kim J. Hong Seo J. Lee J. Bioorg. Med. Chem. Lett. 2021;45:128134. doi: 10.1016/j.bmcl.2021.128134. [DOI] [PubMed] [Google Scholar]
- Lee S.-C. Min H.-Y. Choi H. Kim H. S. Kim K.-C. Park S.-J. Seong M. A. Seo J. H. Park H.-J. Suh Y.-G. Kim K.-W. Hong H.-S. Kim H. Lee M.-Y. Lee J. Lee H.-Y. Mol. Pharmacol. 2015;88:245–255. doi: 10.1124/mol.114.096883. [DOI] [PubMed] [Google Scholar]
- Kim H. S. Hong M. Ann J. Yoon S. Nguyen C.-T. Lee S.-C. Lee H.-Y. Suh Y.-G. Seo J. H. Choi H. Kim J. Y. Kim K.-W. Kim J. Kim Y.-M. Park S.-J. Park H.-J. Lee J. Bioorg. Med. Chem. 2016;24:6082–6093. doi: 10.1016/j.bmc.2016.09.067. [DOI] [PubMed] [Google Scholar]
- Cho T.-M. Kim J. Y. Kim Y.-J. Sung D. Oh E. Jang S. Farrand L. Hoang V.-H. Nguyen C.-T. Ann J. Lee J. Seo J. H. Cancer Lett. 2019;447:141–153. doi: 10.1016/j.canlet.2019.01.029. [DOI] [PubMed] [Google Scholar]
- Kim H. S. Hoang V.-H. Hong M. Kim K. C. Ann J. Nguyen C.-T. Seo J. H. Choi H. Yong Kim J. Kim K.-W. Sub Byun W. Lee S. Lee S. Suh Y.-G. Chen J. Park H.-J. Cho T.-M. Kim J. Y. Seo J. H. Lee J. Bioorg. Med. Chem. 2019;27:1370–1381. doi: 10.1016/j.bmc.2019.02.040. [DOI] [PubMed] [Google Scholar]
- Nguyen C.-T. Ann J. Sahu R. Byun W. S. Lee S. Nam G. Park H.-J. Park S. Kim Y.-J. Kim J. Y. Seo J. H. Lee J. Bioorg. Med. Chem. Lett. 2020;30:127374. doi: 10.1016/j.bmcl.2020.127374. [DOI] [PubMed] [Google Scholar]
- Alexander L. D. Sellers R. P. Davis M. R. Ardi V. C. Johnson V. A. Vasko R. C. McAlpine S. R. J. Med. Chem. 2009;52:7927–7930. doi: 10.1021/jm901566c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasko R. C. Rodriguez R. A. Cunningham C. N. Ardi V. C. Agard D. A. McAlpine S. R. ACS Med. Chem. Lett. 2010;1:4–8. doi: 10.1021/ml900003t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardi V. C. Alexander L. D. Johnson V. A. McAlpine S. R. ACS Chem. Biol. 2011;6:1357–1366. doi: 10.1021/cb200203m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay Y. C. McConnell J. R. Wang Y. Kim S. J. Buckton L. K. Mansour F. McAlpine S. R. ACS Med. Chem. Lett. 2014;5:771–776. doi: 10.1021/ml500114p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi M. N. Buckton L. K. Zaiter S. S. Kho J. Chan V. Guo A. Konesan J. Kwon S. Lam L. K. O. Lawler M. F. Leong M. Moldovan G. D. Neale D. A. Thornton G. McAlpine S. R. ACS Med. Chem. Lett. 2018;9:73–77. doi: 10.1021/acsmedchemlett.7b00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Chen N. Xia D. Xu S. Dai W. Tong Y. Wang L. Jiang Z. You Q. Xu X. Cell Chem. Biol. 2021;28:1446–1459.e1446. doi: 10.1016/j.chembiol.2021.03.016. [DOI] [PubMed] [Google Scholar]
- Shao L.-D. Su J. Ye B. Liu J.-X. Zuo Z.-L. Li Y. Wang Y.-Y. Xia C. Zhao Q.-S. J. Med. Chem. 2017;60:9053–9066. doi: 10.1021/acs.jmedchem.7b01395. [DOI] [PubMed] [Google Scholar]
- Fukuyama Y. Kubo M. Esumi T. Harada K. Hioki H. Heterocycles. 2010;81:1571–1602. doi: 10.3987/REV-10-671. [DOI] [Google Scholar]
- Salehi B. Stojanović-Radić Z. Matejić J. Sharifi-Rad M. Kumar N. V. A. Martins N. Sharifi-Rad J. Eur. J. Med. Chem. 2019;163:527–545. doi: 10.1016/j.ejmech.2018.12.016. [DOI] [PubMed] [Google Scholar]
- Tran P. L. Kim S.-A. Choi H. S. Yoon J.-H. Ahn S.-G. BMC Cancer. 2010;10:276. doi: 10.1186/1471-2407-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak O. W. Sharma N. Reynisson J. Leung I. K. H. Bioorg. Med. Chem. Lett. 2021;38:127857. doi: 10.1016/j.bmcl.2021.127857. [DOI] [PubMed] [Google Scholar]
- Bhatia S. Spanier L. Bickel D. Dienstbier N. Woloschin V. Vogt M. Pols H. Lungerich B. Reiners J. Aghaallaei N. Diedrich D. Frieg B. Schliehe-Diecks J. Bopp B. Lang F. Gopalswamy M. Loschwitz J. Bajohgli B. Skokowa J. Borkhardt A. Hauer J. Hansen F. K. Smits S. H. J. Jose J. Gohlke H. Kurz T. ACS Cent. Sci. 2022;8:636–655. doi: 10.1021/acscentsci.2c00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareva N. F. Baryshok V. P. Lazarev I. M. Arch. Pharm. 2018;351:e1700297. doi: 10.1002/ardp.201700297. [DOI] [PubMed] [Google Scholar]
- Kurop M. K. Huyen C. M. Kelly J. H. Blagg B. S. J. Eur. J. Med. Chem. 2021;226:113846. doi: 10.1016/j.ejmech.2021.113846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J. R. Alexander L. A. McAlpine S. R. Bioorg. Med. Chem. Lett. 2014;24:661–666. doi: 10.1016/j.bmcl.2013.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L.-D. Chen Y. Wang M. Xiao N. Zhang Z.-J. Li D. Li R.-T. Org. Chem. Front. 2022;9:2308–2315. doi: 10.1039/D1QO01871J. [DOI] [Google Scholar]
- Khandelwal A. Kent C. N. Balch M. Peng S. Mishra S. J. Deng J. Day V. W. Liu W. Subramanian C. Cohen M. Holzbeierlein J. M. Matts R. Blagg B. S. J. Nat. Commun. 2018;9:425. doi: 10.1038/s41467-017-02013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M. Huey R. Lindstrom W. Sanner M. F. Belew R. K. Goodsell D. S. Olson A. J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.