Abstract
Classical laboratory strains of Escherichia coli do not spontaneously colonize inert surfaces. However, when maintained in continuous culture for evolution studies or industrial processes, these strains usually generate adherent mutants which form a thick biofilm, visible with the naked eye, on the wall of the culture apparatus. Such a mutant was isolated to identify the genes and morphological structures involved in biofilm formation in the very well characterized E. coli K-12 context. This mutant acquired the ability to colonize hydrophilic (glass) and hydrophobic (polystyrene) surfaces and to form aggregation clumps. A single point mutation, resulting in the replacement of a leucine by an arginine residue at position 43 in the regulatory protein OmpR, was responsible for this phenotype. Observations by electron microscopy revealed the presence at the surfaces of the mutant bacteria of fibrillar structures looking like the particular fimbriae described by the Olsén group and designated curli (A. Olsén, A. Jonsson, and S. Normark, Nature 338:652–655, 1989). The production of curli (visualized by Congo red binding) and the expression of the csgA gene encoding curlin synthesis (monitored by coupling a reporter gene to its promoter) were significantly increased in the presence of the ompR allele described in this work. Transduction of knockout mutations in either csgA or ompR caused the loss of the adherence properties of several biofilm-forming E. coli strains, including all those which were isolated in this work from the wall of a continuous culture apparatus and two clinical strains isolated from patients with catheter-related infections. These results indicate that curli are morphological structures of major importance for inert surface colonization and biofilm formation and demonstrate that their synthesis is under the control of the EnvZ-OmpR two-component regulatory system.
Many bacteria can attach to solid surfaces. The first stage of adhesion seems to be reversible: the bacteria can be removed from the surface by washing. In a second phase, bacterial multiplication and production of extracellular polymers result in the formation of a slimy layer on the colonized surface, referred to as a biofilm (15, 16). Sophisticated methods recently revealed the structural and functional organization of biofilms. Bacteria are embedded in the polymer matrix and organized in mushroom-shaped microcolonies interspersed among less dense channels in which liquid flows have been measured (14, 30; reviewed in references 5 and 6). Bacterial life in a biofilm probably involves particular gene expression. Specific patterns of expression of the laf genes of Vibrio parahaemolyticus (17) and of the algC gene of Pseudomonas aeruginosa (7, 8) have been correlated with contact of the bacteria with solid surfaces.
Since microbial adhesion to solid surfaces is a very common phenomenon, biofilms develop on virtually every material that comes in contact with naturally occurring fluids, such as blood and seawater. Up to now, it has not been possible to design a nontoxic coating method able to prevent biofilm formation. Given the important medical and economical consequences of this situation, there is a strong need to understand the colonization process in order to discover a means of interfering with it. Increasing attention is being paid to the initial stages of adhesion. It is generally accepted that immersion of a clean substratum in a natural fluid is immediately followed by fast and efficient adsorption of organic molecules to the surface (33), forming a so-called “conditioning film.” Two types of bacterial interaction are then possible: weak chemical bonding between the bacterial envelope and the solid surface or the conditioning film and bridging mediated by specialized bacterial structures of adhesion. As pointed out by Marshall (16), because of strong repulsion forces, it seems unlikely that a large part of a bacterial surface could make direct contact with a solid surface. However, the contact could be consolidated by extracellular polymeric substances produced by the bacteria; these substances are subject to different colloidal interactions and could therefore form a link between the bacteria and the surface by various combinations of weak chemical bonds, dipole interactions, and hydrophobic interactions (16).
The present work was undertaken to gather information on the surface colonization processes in the very well characterized Escherichia coli K-12 context. Although E. coli is the most common bacterium found in biofilms that have developed on catheters introduced into the urinary tract (12), classical laboratory strains of this species do not spontaneously stick to surfaces. However, when maintained in continuous culture for long-term experiments or industrial processes, these laboratory strains usually generate adherent mutant cells which form a thick biofilm, visible with the naked eye, on the wall of the culture vessel (9). In this paper, we report the isolation of such E. coli K-12 adherent mutants and we show that, for one of them, a point mutation affecting the regulatory properties of the OmpR protein is responsible for the biofilm-forming phenotype. The surface-binding properties of this mutant are the result of the overproduction of curli, a particular class of envelope organelles. We generalized these observations by checking the role of curli and OmpR in other adherent mutants and in clinical isolates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All of the E. coli K-12 strains and plasmids used in this work are listed in Table 1. The E. coli clinical strains HH97496195 and HH97531012 have been isolated by G. Lina, F. Vandenesch, and J. Etienne at the Edouard Herriot Hospital of Lyon. HH97496195 was isolated from the percutaneous transhepatic catheter of a patient with cholecystitis, and HH97531012 was isolated from the urine of a patient with a urethral catheter-related infection. The bacteria were grown in complete Luria-Bertani (LB) medium (18) or in minimal M63 medium (18) supplemented with glucose (0.2%) as a carbon source. MOPS (morpholinepropanesulfonic acid) medium (20) was prepared as 10× stock and supplemented with K2HPO4 (1.32 mM), glycerol (0.4%), and thiamine (0.1 mg/ml). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 10 μg/ml. Congo red indicator plates (11) were made up of CFA agar containing 20 μg of Congo red (Sigma)/ml and 10 μg of Coomassie brilliant blue (Sigma)/ml. Curli-producing bacteria form red colonies, whereas nonproducing cells form white colonies.
TABLE 1.
E. coli K-12 strains and plasmids used
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| β63 | MG1655 malA-kan mutT | P. Marlière |
| CT110 | CSH57 thiA(pULB110) | A. Toussaint |
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacYI proA2 rpsL20 xyl-5 mtl-1 recA13 Δ(mrcC-mrr) | Laboratory collection |
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | Laboratory collection |
| MG1655 | λ− F− prototroph | Laboratory collection |
| MH225 | MC4100 Φ(ompC-lacZ+)10–25 | 10 |
| MH513 | MC4100 Φ(ompF-lacZ+)16–13 | 10 |
| PHL628 | MG1655 malA-kan ompR234a | This study |
| PHL644 | MC4100 malA-kan ompR234 | This study |
| PHL645 | MC4100 malA-kan | This study |
| PHL690 | MG1655 malA-kan | This study |
| PHL691 | HB101(pR′7) | This study |
| PHL744 | MC4100 malT54::Tn10 ompR234 | This study |
| PHL745 | MC4100 malT54::Tn10 ompR234 csgA::kan | This study |
| PHL804 | MC4100 csgA::kan | This study |
| PHL806 | PHL804(pCSG4) | This study |
| PHL839 | MH513 malA-kan ompR234 | This study |
| PHL840 | MH225 malA-kan ompR234 | This study |
| PHL841 | MH513 malA-kan | This study |
| PHL842 | MH225 malA-kan | This study |
| PHL874 | PHL804(pOV874) | This study |
| PHL875 | PHL745(pCSG4) | This study |
| PHL876 | PHL745(pOV874) | This study |
| PHL893 | TK821 csgA::kan(pCSG4) | This study |
| PHL894 | TK821 csgA::kan(pOV874) | This study |
| RL101 | β63 ompR234 | This study |
| RL102 to -111 | β63 adr-102 to adr-111 | This study |
| TK821 | MC4100 ompR331::Tn10 | 10 |
| TST3 | F−araD139 Δ(argF-lac)169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR malT54::Tn10 | CGSC 6139 |
| YMel-1 | YMel csgA::kan | 21 |
| Plasmids | ||
| pAT003 | pBR322 with a 7.8-kb BamHI fragment harboring the ompB operon | 19 |
| pBR325 | Apr Tcr Cmr | Laboratory collection |
| pCSG4 | pUC19 with a 3.5-kb HindIII fragment containing the csgBA operon | 21 |
| pOV874 | pCSG4 carrying a uidA-kan fusion gene inserted at the unique ClaI site of the csgA gene | This study |
| pN496 | pBluescript Cmr containing a uidA-kan gene fusion | 13 |
| pOV711 | pBR325 with a 7.8-kb BamHI fragment harboring the ompR234 mutation | This study |
| pOV737 | pBR325 with a 1.285-kb EcoRI-SmaI fragment harboring the ompR234 mutation | This study |
| pR′7 | pULB110 derivative harboring the malA::kan marker and the ompR234 mutation of PHL628 | This study |
| pULB110 | RP4::Mu3A, Apr Tcr Kms | A. Toussaint |
ompR234 is equivalent to adr-101 (see text).
Genetic methods.
Random mutagenesis with phage Mu dX was performed as described by Baker et al. (1). Phage P1 vir was used for transductions, which were carried out following the procedure described by Miller (18). In vivo cloning was carried out with the RP4::mini-Mu plasmid pULB110 as described by Van Gijsegem and Toussaint (31).
Confocal laser microscopy.
Confocal laser microscopy was used to quantify biofilm development on microscope coverslips (4, 14). Glass coverslips (15 by 15 mm) were individually incubated in test tubes (diameter, 18 mm) containing 3 ml of bacterial culture. After an appropriate incubation time, the coverslips were removed and washed by immersion and agitation in 10 mM MgSO4. The coverslips were immediately stained by immersion for 30 min in a solution of acridine orange (10 μg/ml in 10 mM MgSO4). They were then rinsed for 30 s in two different baths of 10 mM MgSO4. Confocal laser microscopy was conducted with an LSM310 microscope (Zeiss) equipped with a 40×, 1.3 numerical aperture oil immersion, phase-contrast lens (P1-Neofl). An argon laser with a maximum-emission line at 488 nm was used as the excitation source. Horizontal optical thin sections were collected and digitized by the Zeiss interactive software. These images were collected at 1.0-μm intervals from the outer surface of the biofilm to the glass and serially arranged to create 3-D reconstitutions. Pseudocolors attributed to each point of these 3-D reconstitutions (according to its distance from the glass surface) by the image-processing system of the Zeiss microscope allowed easy evaluation of the thickness of the biofilm.
Quantification of the first stages of polystyrene surface colonization by biofilm retrieval.
During the first stages of biofilm formation, the bacteria do not firmly attach and can be removed from the surface by a moderate shear force (16). It is therefore possible to collect the liquid phase containing the free-living bacteria separately from the biofilm. For each point on the biofilm development curves, a polystyrene petri plate was filled with M63 liquid medium and inoculated. After a given incubation time, the liquid phase was removed and the biofilm which had developed on the bottom of the plate was washed twice with 10 mM MgSO4. Two milliliters of 10 mM MgSO4 was then introduced into the plate, and the biofilm was suspended by pipetting the solution up and down vigorously and repeatedly. The number of bacteria was estimated by measuring the optical density at 600 nm.
DNA manipulations.
Standard techniques were used for chromosomal DNA preparation, plasmid extraction, gel electrophoresis, and DNA sequencing (28). Restriction endonucleases, DNA polymerase I, and DNA T4 ligase were used as recommended by the manufacturers.
SDS-PAGE of outer membrane proteins.
The outer membrane proteins were prepared from overnight cultures grown in MOPS medium at 30°C. Cells were harvested, washed, and resuspended in 25 mM Tris HCl (pH 7.5) and then sonicated. Membrane fractions were pelleted by ultracentrifugation at 350,000 × g for 2 h. The pellets were resuspended in 25 mM Tris HCl, pH 7.5. Separating sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with 6 M urea and an acrylamide concentration of 12%.
Scanning electron microscopy.
The bacteria were grown for 24 h in M63 medium at 30°C. The cells were harvested, resuspended in 10 mM MgSO4, and allowed to adhere to carbon-coated 200-mesh grids. After being stained with 1% phosphotungstic acid (Sigma), the grids were examined with a Philips CM120 electron microscope.
Assays of enzymatic activities encoded by reporter genes.
β-Galactosidase activity was assayed as described by Miller (18). β-Glucuronidase activity was measured by following the degradation of p-nitrophenyl-β-d-glucuronide into p-nitrophenol that absorbs at 405 nm (2). Specific activities of these two enzymes are expressed as nanomoles of products liberated per minute per milligram (dry weight) of bacteria.
RESULTS
Isolation of adherent mutants.
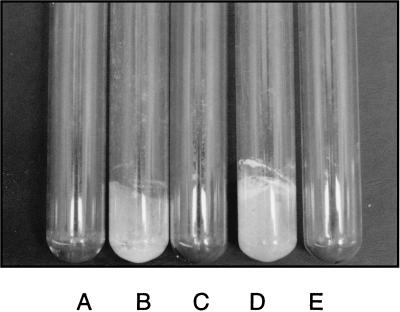
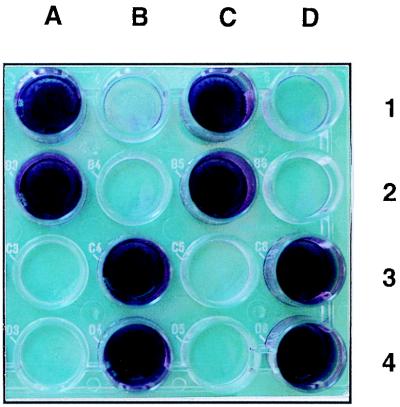
When maintained in continuous culture for long-term experiments (for instance, to address fundamental issues concerning population genetics and evolutionary processes), E. coli K-12 strains usually generate adherent mutant cells which form a thick biofilm on the wall of the culture vessel (14a). During such an experiment with the mutT strain β63 (cultivated in cyclic-flow conditions in a synthetic medium [14b]), a sample of the biofilm was taken and clones were isolated by streaking on LB plates. About 25% of these clones were able to develop a visible biofilm on the walls of glass test tubes (a hydrophilic substratum) in less than 24 h (Fig. 1). All of these adherent mutants were also able to colonize the hydrophobic surfaces of polystyrene microtiter plates (Fig. 2) or petri plates. One of these clones was named RL101 and retained for subsequent experiments.
FIG. 1.
Biofilm development on the walls of glass test tubes containing β63 (A), RL101 (B), MG1655 (C), PHL628 (D), and no bacteria (control) (E). After inoculation, the tubes were gently shaken at 30°C. After 24 h, the medium (M63) was removed and the tubes were dried for 1 h at room temperature.
FIG. 2.
Biofilm development in the wells of a polystyrene microtiter plate. The wells contained RL101 (A1), β63 (B1), PHL628 (MG1655 adr-101) (C1), MG1655 (D1), PHL644 (MC4100 adr-101) (A2), PHL645 (MC4100 adr+) (B2), MG1655(pR′7) (C2), MG1655(pULB110) (D2), TK821 (MC4100 ompR331::Tn10) (A3), MC4100(pR′7) (B3), MC4100(pAT003) (C3), MC4100(pOV711) (D3), PHL745 (MC4100 adr-101 csgA::kan) (A4), PHL745(pCSG4) (B4), MC4100 csgA::kan(pCSG4) (C4), and MC4100(pOV737) (D4). The wells were filled with M63 medium. After inoculation, the plate was incubated for 48 h at 30°C. The liquid was removed from each well, and a drop of crystal violet was added to intensify the contrast. The wells were then washed twice (with 10 mM MgSO4), and the plate was dried.
Genetic transfer of the adr-101 mutation.
RL101, like its β63 parent, contains a mutT allele which confers a high frequency of spontaneous mutagenesis. In order to avoid interference by secondary mutations, we had to transfer the adr-101 mutation, assumed to be responsible for the adherent phenotype of RL101, to a mutT+ strain. To reach this objective, random insertions of Mu dX (a transposable element conferring resistance to ampicillin) were performed in RL101. Several thousand transposed clones were selected (on LB plates containing ampicillin) and pooled. This set of clones was infected by phage P1 vir. A lysate was obtained and used to transduce the mutT+ strain MG1655. Transduction of the ampicillin resistance determinant was selected, and 192 recombinant clones were screened for cotransduction of the adherent phenotype. Only one clone showed this phenotype. Unexpectedly, this clone also acquired resistance to kanamycin, indicating a genetic linkage between adr-101 and the malA-kan mutation originally present in RL101 and its parental strain, β63. This linkage was confirmed by P1 vir transduction of malA-kan from RL101 to MG1655: 54% of the recombinant clones also received the adr-101 mutation.
Phenotype conferred by the adr-101 mutation. (i) Glass colonization.
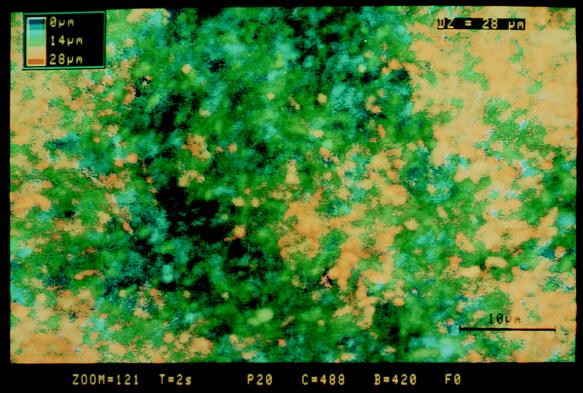
To visualize the glass colonization properties conferred by the adr-101 mutation, test tubes containing 3 ml of M63 medium were inoculated either with MG1655 malA-kan adr-101 (PHL628) or with MG1655 malA-kan (PHL690) and a glass microscope coverslip was introduced into each tube. After a given culture time, each slide was removed and observed by confocal laser microscopy in order to evaluate the thickness of the biofilm (see Materials and Methods). Coverslips incubated with the adr+ strain PHL690 were never colonized, and only individual bacteria could be visualized on the glass surface by confocal microscopy (data not shown). On the other hand, 24 h after inoculation, the adr-101 strain PHL628 generated visible biofilms (about 20 μm deep) (Fig. 3). Over longer incubation periods, the thicknesses of the biofilms were stable and were maintained at between 20 and 30 μm (maximum incubation time, 7 days).
FIG. 3.
3-D reconstitution of the biofilm developed by the adherent strain PHL628 on a glass coverslip. A test tube containing 3 ml of M63 medium was inoculated with the bacteria, and a glass coverslip was introduced into the tube. After being cultured for 24 h at 30°C, the slide was removed, stained with acridine orange, and observed by confocal laser microscopy (see Materials and Methods). Horizontal optical sections were collected at 1.0-μm intervals, digitized, and serially arranged to create 3-D reconstitutions. Pseudocolors were attributed to each point according to its distance from the glass surface. The color-to-distance conversion scale is shown at the upper left corner of the micrograph. Bar, 10 μm.
(ii) Polystyrene colonization.
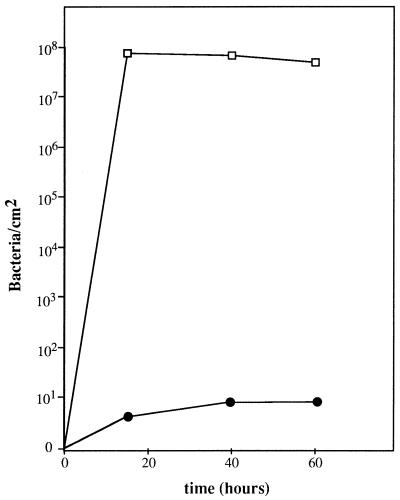
The adr-101 mutation also confers the ability to adhere to hydrophobic surfaces, such as polystyrene (Fig. 2). The colonization properties of strain PHL644 (MC4100 adr-101) were quantified on the surfaces of petri plates by a method of biofilm retrieval (see Materials and Methods). As shown in Fig. 4, this strain established a biofilm containing about 108 bacteria per cm2 in 15 h. Under the same conditions, the adr+ strain PHL645 was unable to colonize the polystyrene surface, even after 60 h.
FIG. 4.
Quantification of biofilm development on the surface of polystyrene petri plates. Colonization by the adr-101 strain PHL644 (open squares) and the wild-type strain PHL645 (solid circles) is shown. The plates contained liquid M63 medium and were incubated at 30°C. The number of bacteria in the biofilm was measured as described in Materials and Methods.
(iii) Autoaggregation.
In addition to the wall-growing properties, another phenotype could be correlated with the adr-101 mutation. When cultivated in gently shaken containers, cells of the PHL628 strain spontaneously interacted to form visible aggregates which settled to the bottom of the tube. In strongly shaken Erlenmeyer flasks, this autoaggregation process did not occur. However, when PHL628 was cultivated under these conditions to an optical density at 660 nm of 0.8 and then left without shaking at room temperature, cell clusters formed within minutes. In the transduction experiments reported above, the wall-growing and autoaggregation phenotypes were always cotransferred at 100%.
Mapping of adr-101.
malA-kan and adr-101 are cotransduced at 54%, and malA is located at 76.5 min (3). Different well-characterized insertions of the transposon Tn10 were used to map more precisely the adr-101 mutation. One hundred-percent cotransduction was observed when RL101 was transduced with a lysate obtained on the ompR331::Tn10 strain TK821. A total of 350 tetracycline-resistant transductants were tested in microtiter plates for their colonization properties: all of them lost the adherent phenotype. As a control for this linkage frequency, a lysate obtained on RL101 was used to transduce TK821; clones resistant to kanamycin (transfer of malA-kan) were selected, and adherent clones (cotransfer of adr-101) were screened on microtiter plates. The same cotransduction frequency between adr-101 and ompR was observed: 100% of the 96 adherent clones tested were sensitive to tetracycline (transfer of the ompR allele of RL101). The adr-101 mutation was therefore localized in the very near vicinity of the ompR-envZ operon (76.1 min).
Cloning and sequencing of the adr-101 mutation.
In vivo cloning of the chromosomal region surrounding the malA-kan insertion was carried out with the RP4::mini-Mu plasmid pULB110, a kanamycin-sensitive derivative of pULB113 (31). The malA-kan adr-101 strain PHL628 (pULB110) was mated with HB101. Kanamycin-resistant transconjugants were selected (on LB plates containing kanamycin and streptomycin), and 15 R-prime plasmids were retained. One of them (pR′7) was able to confer the adherence phenotype when it was transferred to MG1655 or MC4100. Considering the dominant character of the adr-101 mutation revealed by this result, we successively subcloned in pBR325 a 7.8-kb BamHI fragment and a 1.285-kb SmaI-EcoRI fragment (Fig. 5) able to confer the adherence phenotype. As a control, we compared the adherence properties conferred by pOV711 (containing the 7.8-kb fragment) and pAT003 (containing the equivalent fragment previously cloned [19] from a wild-type strain). As shown in Fig. 2 (wells C3 and D3), only pOV711 conferred adherence.
FIG. 5.
Restriction maps of the chromosomal fragments cloned in this work and sequence of the ompR234 mutation. The ompR region of PHL628 was cloned in vivo on pR′7 (see text). A 7.8-kb BamHI fragment and a 1.285-kb SmaI-EcoRI fragment conferring the adherent phenotype were successively subcloned into pBR325 (to generate plasmids pOV711 and pOV737). Part of the nucleotide sequence of ompR (32) is also shown. The single-base pair mutation is indicated by a black square, with the corresponding modification of the amino acid sequence given in the solid box. The mutation created a NaeI restriction site (see text).
The nucleotide sequence of the 1.285-kb fragment was determined (Fig. 5). This fragment contains only one complete open reading frame, corresponding to the ompR gene. Comparison with the published sequence of ompR (32) revealed only one point mutation: a transversion of the T residue at position 234 (counting from the +1 transcription start site) into a G residue. This mutation produces a new NaeI site (Fig. 5), which was confirmed by the presence of an additional restriction fragment after digestion of plasmid pOV711 (adr-101) with NaeI, followed by comparison with the adr+ plasmid pAT003 (data not shown). At the amino acid sequence level, the adr-101 mutation results in replacement of the leucine at position 43 by an arginine. The adr-101 mutation was therefore renamed ompR234 (instead of ompR101, to avoid confusion with the ompR101 allele already described by Garrett et al. [10]).
Identification of the morphological structure responsible for the phenotype.
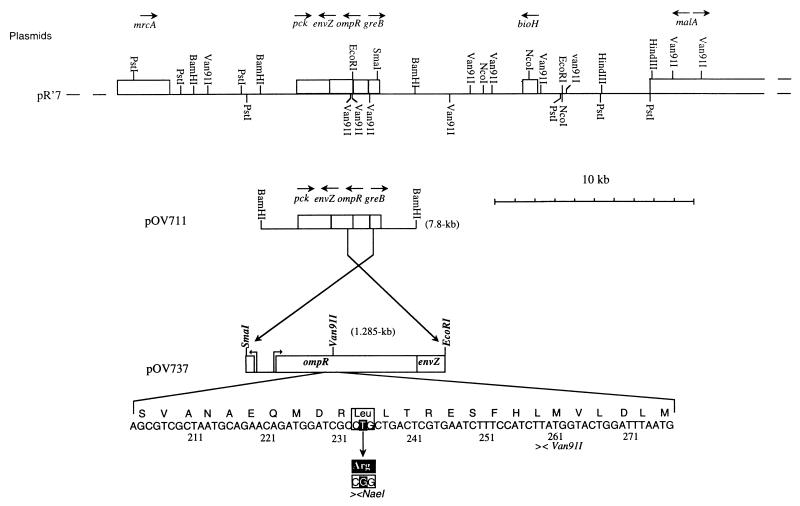
To visualize the adhesive structure induced in an ompR234 background, electron microscopic observations of negatively stained bacteria of the strain PHL628 were carried out. These observations revealed that the ompR234 cells were usually arranged in clumps, which often had copious numbers of fibrillar structures closely associated with the cells (Fig. 6). On the other hand, ompR+ strains, such as MG1655, did not present this organization (data not shown).
FIG. 6.
Electron micrographs of negatively stained bacteria of the biofilm-forming mutant PHL628. Bars, 1 μm (top) and 0.5 μm (bottom). The cells were cultivated in M63 medium at 30°C for 24 h.
These fibrillar structures looked like the thin and flexible fimbriae described as fibronectin-binding curli by Olsén et al. (22). To monitor curli expression, the bacteria were grown at 26°C on the CFA-Congo red indicator plates described by Hammar et al. (11). Although the ompR234 strain PHL644 and the ompR+ strain PHL645 developed red colonies on this medium, the colonies of PHL644 were more intensely stained, indicating curli overproduction in the ompR234 strain. In contrast, the curli-negative mutant Ymel-1 (21) produced white colonies when tested under the same conditions.
In order to demonstrate that the curli are the morphological structures responsible for the adherence of the ompR234 strains, a null mutation in csgA, the structural gene for curlin synthesis (21), was transduced from Ymel-1 to PHL744 (MC4100 malT::Tn10 ompR234). The resulting strain (PHL745) was unable to colonize polystyrene surfaces (Fig. 2) and lost its aggregation properties (data not shown). Introduction of plasmid pCSG4 carrying the complete csgBA operon (21) restored biofilm formation (Fig. 2) and aggregation (data not shown) in MC4100 ompR234 csgA::kan (PHL875) but was unable to confer these phenotypes on an ompR+ strain, such as PHL804.
Transcriptional regulation of porin and curli synthesis by the ompR234 allele.
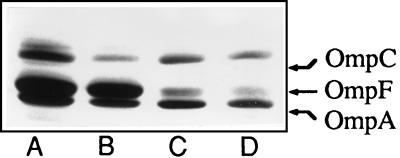
OmpR is a regulatory protein known to modulate the expression of the ompC and ompF genes coding for two major outer membrane proteins (for a review, see reference 24). In order to determine whether the ompR234 mutation had an effect on the synthesis of OmpC or OmpF, the outer membrane proteins of the wild-type strain PHL645 and the ompR234 strain PHL644 (grown at 30°C in MOPS medium) were extracted and separated by electrophoresis on a 6 M urea–SDS-polyacrylamide gel. As shown in Fig. 7, no significant differences in the porin patterns were observed.
FIG. 7.
Outer membrane proteins of the ompR234 strain PHL644 (lanes A and C) and the wild-type strain PHL645 (lanes B and D). Bacteria were grown to mid-log phase at 30°C in MOPS medium (lanes A and B) or in MOPS medium with 300 mM NaCl (lanes C and D). Equal amounts of each membrane fraction protein were separated by 6 M urea–SDS-PAGE.
The effects of the ompR234 mutation on the expression of ompC-lacZ and ompF-lacZ operon fusions were also examined (Table 2). The assays of the ompR+ strains reflected the normal osmotic regulation of porin gene expression: ompF is preferentially expressed at low osmolarity and ompC at high osmolarity. A similar pattern of expression was observed after introduction of the ompR234 mutation. However, the levels of both ompC and ompF expression were increased (about two times) at low osmolarity. At high osmolarity, the levels of expression were similar in the mutant and the wild-type strains. As discussed below, these results indicate that the OmpR-EnvZ system of the ompR234 mutants is still able to detect osmotic variations and to react appropriately. Nevertheless, the regulatory properties of the mutant OmpR protein seem sufficiently altered to explain the activation of curli synthesis.
TABLE 2.
Effect of ompR234 on expression of ompC and ompF at low and high osmolarities
| Strain | Relevant genotype | β-Galactosidase activitya
|
|
|---|---|---|---|
| −NaCl | +NaCl | ||
| PHL841 | ompF-lacZ | 264.0 | 11.5 |
| PHL839 | ompF-lacZ ompR234 | 454.0 | 16.7 |
| PHL842 | ompC-lacZ | 118.7 | 682.0 |
| PHL840 | ompC-lacZ ompR234 | 242.4 | 634.3 |
Bacteria were grown at 30°C in MOPS medium (−NaCl) or in MOPS medium containing 300 mM NaCl (+NaCl). β-Galactosidase activity was assayed at mid-log growth phase. The values are means from three independent cultures and are expressed as nanomoles of products liberated per minute per milligram (dry weight) of bacteria.
To monitor this activation at the transcriptional level, a reporter gene encoding β-glucuronidase was inserted into the csgBA operon carried by pCSG4. A 3.8-kb SmaI fragment containing a uidA-kan cassette (2) was extracted from plasmid pN496 (13) and ligated into the unique ClaI site (filled by DNA polymerase I) of pCSG4. This site is located in csgA (21). The resulting plasmid (pOV874) was introduced into MC4100 csgA::kan ompR+ (PHL874) and MC4100 csgA::kan ompR234 (PHL876). These strains were grown to stationary phase in M63 medium at 30°C, and β-glucuronidase assays were performed. A 3.5-fold-higher activity was measured in the ompR234 strain (503 U in PHL876 versus 144 U in PHL874). This result confirmed that curli are overexpressed in the mutant strains.
Effects of ompR and csgA null mutations on wall-growing and clinical isolates.
Transduction of the ompR331::Tn10 allele into RL101 suppresses adherence (see “Mapping of adr-101” above). To quantify this effect, the expression of the csgA-uidA gene fusion carried by pOV874 was measured in an ompR331::Tn10 context (PHL894). Under the same culture conditions as those for PHL874 and PHL876 (see preceding paragraph), we were unable to detect any β-glucuronidase activity in this strain. This finding was confirmed on CFA-Congo red indicator plates: the ompR+ strain PHL645 (yielding red colonies) and the ompR234 strain PHL644 (yielding dark-red colonies) produced white colonies when transduced by ompR331::Tn10 (data not shown). These results demonstrate the crucial role of OmpR in the activation of curli synthesis.
To generalize these observations, 10 adherent clones (RL102 to RL111) isolated from the wall of the culture vessel at the same time as RL101 were transduced by ompR331::Tn10. In each case, the adherence and autoaggregation properties were lost (data not shown). To make sure that the adherence structures activated in the mutants RL102 to RL111 were curli, the csgA::kan mutation of strain YMel-1 was transduced in these clones (we first had to eliminate their malA-kan allele by transduction of the malT54::Tn10 mutation of strain TST3 and to screen for recombinants which had conserved their adherent properties). In all cases, again, the biofilm-forming phenotype was lost.
Furthermore, the E. coli clinical isolates HH97496195 and HH97531012, which were isolated from patients with catheter-related bacteremia (see Materials and Methods), were also transduced. These two strains produced dark-red colonies on CFA-Congo red plates and were able to develop visible biofilms on glass and polystyrene surfaces in 24 h. When transduced by either ompR331::Tn10 or csgA::kan, they produced white colonies on the indicator plates and lost their adherence properties.
DISCUSSION
The mechanisms by which bacteria adhere to inert surfaces are not well understood. The aim of this work was to elucidate the structural and genetic changes induced in a classical E. coli K-12 strain by a mutation allowing surface colonization. Electron microscopy of negatively stained bacteria revealed the presence of thin fibrillar pili at the surfaces of the mutant cells (Fig. 6). These particular pili seemed to be identical to the Congo red-binding structures described by Olsén and coworkers (22) as curli. When grown on CFA-Congo red indicator plates, the mutant strains were more intensely stained, indicating curli overproduction. The curli subunits are encoded by the csgA gene (21). Insertion of a reporter cassette into this gene revealed a 3.5-fold-higher transcription in the presence of the mutation responsible for the adherent phenotype. Furthermore, the introduction of a csgA null mutation totally suppressed the biofilm-forming properties (Fig. 2), demonstrating that curli production is necessary for the adhesion of mutant bacteria. Ten additional adherent clones, obtained under the same conditions as those for the mutant strain characterized in this work, and two clinical strains isolated from patients with catheter-related infections were also transduced by the csgA null mutation. In all cases, the adherence properties were lost. Curli seem therefore to be envelope structures of major importance for surface colonization, not only in fermentation processes disrupted by wall growth but also in indwelling medical device-related bacteremia.
The mutation causing curli overexpression was localized in the ompR gene. In wild-type classical laboratory strains, curli are not synthesized at a level sufficient to allow adherence, even in the presence of csgA on a multicopy plasmid (see above). These strains need a regulatory mutation, such as ompR234, to activate curli expression and gain adherence. On the other hand, the two clinical isolates are naturally able to form biofilms. Interestingly, however, the introduction of an ompR null mutation into these strains suppresses the adherence properties and curli production. Furthermore, the transduction of the ompR null mutation in strains containing the csgA-uidA fusion decreased csgA transcription to an undetectable level. These results are suggestive of an important role for the EnvZ-OmpR two-component system in regulating curli expression in response to environmental fluctuations.
OmpR is a 239-amino-acid cytoplasmic protein known to transcriptionally regulate the ompC and ompF porin genes (for a review, see reference 24). The EnvZ-mediated phosphorylation of OmpR is assumed to change its binding affinity for different sites in the target promoters (25). Data presented by Silhavy and coworkers (23, 24, 26, 27) indicate that a low intracellular concentration of phosphorylated OmpR corresponds to the low-osmolarity state and a higher concentration of phosphorylated OmpR corresponds to the high-osmolarity state. These authors have proposed that, below a given threshold, increasing concentrations of OmpR-phosphate proportionally increase the levels of expression of both ompF and ompC; beyond this threshold, OmpR-phosphate binds to other operator sequences in the ompF promoter and represses its expression, whereas ompC expression continues to increase proportionally to the OmpR-phosphate concentration. This model integrates the results of Russo and coworkers (27), who isolated a collection of ompR mutations and defined three classes on the basis of porin gene regulation: those that prevent both the repression of ompF and the activation of ompC at high osmolarity, those that prevent the repression of ompF while still allowing the activation of ompC, and those that show reduced expression of both ompF and ompC. The ompR mutation isolated in this work clearly falls into a fourth class: neither the repression of ompF nor the activation of ompC was prevented at high osmolarity, and the levels of expression of both ompF and ompC at low osmolarity were higher in the mutant than in the wild-type strain (Table 2). According to the model presented above, we believe that the replacement of a leucine by an arginine at position 43 in the OmpR protein corresponding to the ompR234 allele results in an increased efficiency of interaction with the regulatory sites or with the RNA polymerase. This hypothesis, which could explain curli overexpression in the ompR234 strains, could also account for the higher expression of both ompF and ompC observed in the mutant strains at low osmolarity (Table 2). At high osmolarity, the levels of expression of ompC (high) and ompF (low) are similar in the wild-type and in the mutant strains (Table 2). This result probably reflects the saturation of the regulatory sites. Even with the wild-type OmpR protein, the levels of activation of ompC and repression of ompF reach a plateau at given levels of osmolarity: above these thresholds, increasing the intracellular OmpR-phosphate concentration does not enhance the regulatory effects any further (24).
Based on an analogy to CheY, OmpR is presumed to be phosphorylated on an aspartic acid residue at position 55 (24). This residue is located in an acidic pocket at the N-terminal end of the protein (29). Thus, we can suggest that the replacement of a leucine residue by an arginine residue at position 43 could facilitate OmpR phosphorylation and consequently its DNA binding. Another hypothesis to explain the increased activation properties of the mutant OmpR protein comes from the results of Pratt and Silhavy (23). These authors identified five amino acids which are directly involved in the interaction between OmpR and the α subunit of the RNA polymerase. Four are located at the C-terminal end of OmpR, and one is located at the N-terminal end. This residue is an arginine at position 42. It is therefore possible to assume that the replacement of a leucine by another arginine at the next position could enhance the affinity of OmpR for the RNA polymerase.
Further work will focus on testing these hypotheses. Additionally, we hope to gain information on the mechanisms of curli overexpression and to identify other functions involved in biofilm formation.
ACKNOWLEDGMENTS
We acknowledge Catherine Souchier for helping us with confocal laser microscopy and M. L. Bernardini, P. Bertin, J. Etienne, M. Hammar, M. Heyde, P. Laloi, G. Lina, S. Normark, A. Olsén, P. Sansonetti, T. Silhavy, and F. Vandenesch for gifts of strains and helpful discussions. We also thank V. James for suggestions regarding the manuscript and the members of the Laboratoire de Génétique Moléculaire des Microorganismes for their kind interest in this study.
This work was partially supported by a grant from the French Defense Ministry (96-048/DRET).
REFERENCES
- 1.Baker T A, Howe M M, Gross C A. Mu dX, a derivative of Mu d1 (lac Apr) which makes stable lacZ fusions at high temperature. J Bacteriol. 1983;156:970–974. doi: 10.1128/jb.156.2.970-974.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardonnet N, Blanco C. uidA antibiotic resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol Lett. 1992;93:243–248. doi: 10.1016/0378-1097(92)90469-5. [DOI] [PubMed] [Google Scholar]
- 3.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 4.Caldwell D E, Korber D R, Lawrence J R. Confocal laser microscopy and digital image analysis in microbial ecology. Adv Microb Ecol. 1992;12:1–67. [Google Scholar]
- 5.Costerton J W, Lewandowski Z, De Beer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 7.Davies D G, Chakrabarty A M, Geesey G G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies D G, Geesey G G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61:860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykhuizen D E, Hartl D L. Selection in chemostats. Microbiol Rev. 1983;47:150–168. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett S, Taylor R K, Silhavy T J. Isolation and characterization of chain-terminating nonsense mutations in a porin regulator gene, envZ. J Bacteriol. 1983;156:62–69. doi: 10.1128/jb.156.1.62-69.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 12.Hessen M T, Kaye D. Infections associated with foreign bodies in the urinary tract. In: Bisno A L, Waldvogel F A, editors. Infections associated with indwelling medical devices. Washington, D.C: American Society for Microbiology; 1989. pp. 199–213. [Google Scholar]
- 13.Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. Environmental conditions affect transcription of the pectinase genes of Erwinia chrysanthemi 3937. J Bacteriol. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence J R, Korber D R, Hoyle B D, Costerton J W, Caldwell D E. Optical sectioning of microbial biofilms. J Bacteriol. 1991;173:6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Longin, R., and P. Marlière. Unpublished observations.
- 14b.Longin, R., et al. Unpublished data.
- 15.Marshall K C, Stout R, Mitchell R. Mechanisms of the initial events in the sorption of marine bacteria to surfaces. J Gen Microbiol. 1971;68:337–348. [Google Scholar]
- 16.Marshall K C. Biofilms: an overview of bacterial adhesion, activity, and control at surfaces. ASM News. 1992;58:202–207. [Google Scholar]
- 17.McCarter L L, Showalter R E, Silverman M R. Genetic analysis of surface sensing in Vibrio parahaemolyticus. Biofouling. 1992;5:163–175. [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Mizuno T, Wurtzel E T, Inouye M. Cloning of the regulatory genes (ompR and envZ) for the matrix proteins of the Escherichia coli outer membrane. J Bacteriol. 1982;150:1462–1466. doi: 10.1128/jb.150.3.1462-1466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 22.Olsén A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 23.Pratt L A, Silhavy T J. OmpR mutants specifically defective for transcriptional activation. J Mol Biol. 1994;243:579–594. doi: 10.1016/0022-2836(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 24.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 25.Rampersaud A, Harlockers S L, Inouye M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 26.Russo F D, Silhavy T J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 27.Russo F D, Slauch J M, Silhavy T J. Mutations that affect separate functions of OmpR the phosphorylated regulator of porin transcription in Escherichia coli. J Mol Biol. 1993;231:261–273. doi: 10.1006/jmbi.1993.1281. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Stock J B, Stock A M, Mottonen J M. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 30.Stoodley P, De Beer D, Lewandowski Z. Liquid flow in biofilm systems. Appl Environ Microbiol. 1994;60:2711–2716. doi: 10.1128/aem.60.8.2711-2716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Gijsegem F, Toussaint A. Chromosome transfer and R-prime formation by an RP4::mini-Mu derivative in Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, and Proteus mirabilis. Plasmid. 1982;7:30–44. doi: 10.1016/0147-619x(82)90024-5. [DOI] [PubMed] [Google Scholar]
- 32.Wurtzel E T, Chou M Y, Inouye M. Osmoregulation of gene expression. I. Dna sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982;257:13685–13691. [PubMed] [Google Scholar]
- 33.ZoBell C E. The effects of solid surfaces upon bacterial activity. J Bacteriol. 1943;46:39–56. doi: 10.1128/jb.46.1.39-56.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]