Abstract
Gas vesicles are intracellular, protein-coated, and hollow organelles found in cyanobacteria and halophilic archaea. They are permeable to ambient gases by diffusion and provide buoyancy, enabling cells to move upwards in liquid to access oxygen and/or light. In halobacteria, gas vesicle production is encoded in a 9-kb cluster of 14 genes (4 of known function). In cyanobacteria, the number of genes involved has not been determined. We now report the cloning and sequence analysis of an 8,142-bp cluster of 15 putative gas vesicle genes (gvp) from Bacillus megaterium VT1660 and their functional expression in Escherichia coli. Evidence includes homologies by sequence analysis to known gas vesicle genes, the buoyancy phenotype of E. coli strains that carry this gvp gene cluster, the presence of pressure-sensitive, refractile bodies in phase-contrast microscopy, structural details in phase-constrast microscopy, structural details in direct interference-contrast microscopy, and shape and size revealed by transmission electron microscopy. In B. megaterium, the gvp region carries a cluster of 15 putative genes arranged in one orientation; they are open reading frame 1 and gvpA, -P, -Q, -B, -R, -N, -F, -G, -L, -S, -K, -J, -T, and -U, of which the last 11 genes, in a 5.7-kb gene cluster, are the maximum required for gas vesicle synthesis and function in E. coli. To our knowledge, this is the first example of a functional gas vesicle gene cluster in nonaquatic bacteria and the first example of the interspecies transfer of genes resulting in the synthesis of a functional organelle.
Gas vesicles are intracellular hollow organelles found in many bacteria from aqueous environments and are most studied in cyanobacteria (8) and halophilic archaea (10, 13, 20). Gas vesicles are permeable to ambient gases by diffusion. The physiological role demonstrated for gas vesicles is that they provide buoyancy, enabling cells to move upwards in liquid to access oxygen and/or light (41). Gas vesicles range from 30 to 250 nm in width and from 50 nm to 1 μm in length (9, 41), but have a relatively constant size within each species. The morphology and main structural protein of gas vesicles are conserved among species. Gas vesicles are found both individually in cells and clustered together to form gas vacuoles that can be seen as refractile bodies in phase-contrast microscopy.
In the halophilic archaea Haloferax mediterranei and Halobacterium salinarium PHH1, a 9-kb cluster (vac) of 14 genes (gvp) is all that is required for gas vesicle production and regulation (15, 21). H. mediterranei has one chromosomal vac region (14, 15), while H. salinarium PHH1 has a chromosomal and a plasmid vac cluster of genes (15, 21, 31). The chromosomal vac is expressed only in the absence of the plasmid-borne vac region (22). Halobacterium halobium also has a chromosome- and a plasmid-borne vac cluster of genes (33, 43). These plasmid and chromosome-borne gvp genes are in the same order as each other and are arranged divergently in two groups, gvpDEFGHIJKLM, followed upstream by gvpACNO (11, 18, 21, 23). Another halophilic archaeon, Natronobacterium vacuolatum, that forms gas vesicles has a different vac arrangement; seven of its gvp genes are clustered as gvpACNOFGH and are cotranscribed (27, 32). In cyanobacteria, the total number of genes involved in gas vesicle formation has not been determined. There are two copies of gvpA and one copy of gvpC identified in Calothrix sp. strain 7601 (6, 8). In Anabaena flos-aquae, there are at least five copies of gvpA and one copy each of gvpC, -N, -J, -K, -F, and -L (19, 24).
The main structural protein of gas vesicles is GvpA. The 70-amino-acid, extremely hydrophobic protein was first cloned from Calothrix sp. strain PCC7601 (with an oligonucleotide based on the amino acid sequence) (37), and a probe based on this gvpA gene was subsequently used to clone gvpA from other cyanobacteria (7, 42) and halobacteria (13, 20, 36). All of the other cloned gvp genes were identified by being either homologous to or contiguous with gvpA. The highly conserved GvpA protein forms a linear crystalline array of ribs that make up the cylindrical shell and conical ends of the gas vesicle, while GvpC is located on the outer surfaces and adds strength and shape (3, 18, 30, 41). In H. halobium and H. salinarium, a nonessential homolog of GvpA named GvpB is located outside the well-studied gvp cluster (12, 20). GvpD and GvpE were shown to have a regulatory role in gvp gene expression (25, 29, 34). The functions of the other 10 gas vesicle gene products are still unknown. Apart from GvpA, GvpB, and, to a lesser extent, GvpN, sequence conservation of gvp gene products between genera is low.
Bacillus megaterium is generally considered to be a soil bacterium, although it has been found in diverse environments (38). Since gas vesicles have been described exclusively in bacteria from aqueous habitats and have not been found in the bacilli, the discovery of a functional gas vesicle gene cluster in B. megaterium is novel. Open reading frames (ORFs) with homologies to gvp genes were identified in B. megaterium VT1660 in the course of a screening of Tn917-LTV1 (4) transposon banks for polyhydroxyalkanoic acid mutants. One polyhydroxyalkanoic acid-overproducing mutant had the transposon inserted at a site contiguous with the gvp genes. In this paper, we report the discovery of a cluster of gvp genes in B. megaterium. We describe the cloning and sequence analysis of an 8,142-bp DNA fragment encoding a cluster of gvp genes that when transferred to Escherichia coli, conferred a buoyancy phenotype on its host resulting from the synthesis of gas vesicles.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | deoR endA1 gyrA96 hsdR17 (rK− mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80lacZΔM15 F−λ−; used for cloning purposes and for expression of gas vesicle genes | Clontech |
| B. megaterium 19213 | Part of the culture collection of the University of Texas, J. W. Foster wild-type strain | ATCCb |
| B. megaterium VT1660 | Arginine auxotroph, plasmidless derivative of B. megaterium 19213 | 40 |
| B. megaterium B001S | Tn917-LTV1 transposant of B. megaterium VT1660, Emr Lmr | This study |
| B. megaterium 11561 | Wild type | ATCC |
| Plasmids | ||
| pBluescriptIISK | Cloning vector, Ampr | Stratagene |
| pBluescriptIIKS | Cloning vector, Ampr | Stratagene |
| pHPS9 | Bacillus-E. coli shuttle vector, Emr Cmr | 17 |
| pNL4 | IR-L end of Tn917-LTV1 and 6.25-kb contiguous B. megaterium B001S sequences | This study |
| pNL21 | 6.4-kb SalI-BamHI fragment of pNL4, cloned in SalI-BamHI of pBluescriptIIKS | This study |
| pNL22 | 2.6-kb PstI fragment of B. megaterium VT1660 genomic DNA, cloned in PstI site of pBluescriptIISK | This study |
| pNL24 | 8,522-bp composite of pNL21 and pNL22, cloned in pBluescriptIIKS, all putative gvp genes present | This study |
| pNL25 | EcoRI in ORF1 to PstI in araC, cloned into EcoRI-PstI of pBluescriptIIKS | This study |
| pNL25-SK | Same as pNL25, except cloned in pBluescriptIISK | This study |
| pNL26 | HindIII in ORF1-gvpA intergenic region to PstI in araC, cloned into HindIII-PstI of pBluescriptIIKS | This study |
| pNL26-SK | Same as pNL26, except cloned in pBluescriptIISK | This study |
| pNL27 | EcoRI in gvpA to PstI in araC, cloned into EcoRI-PstI of pBluescriptIIKS | This study |
| pNL28 | EcoRV in gvpP to PstI in araC, cloned into EcoRV-PstI of pBluescriptIIKS | This study |
| pNL29 | HindIII in gvpQ to PstI in araC, cloned into HindIII-PstI of pBluescriptIIKS | This study |
| pNL30 | BglII in gvpB-gvpR intergenic region to PstI in araC, cloned into BamHI-PstI of pBluescriptIIKS | This study |
| pNL32 | HindIII in ORF1-gvpA intergenic region to HindIII in gvpU, cloned into HindIII of pBluescriptIIKS, orientation as in pNL24 | This study |
| pNL20 | 6.4-kb SalI-BamHI fragment of pNL4, cloned in SalI-BamHI of pHPS9 | This study |
| pNL40 | 8.5-kb SalI-BamHI in vector, cloned in SalI-BamHI of pHPS9, all putative gvp genes present | This study |
Emr, erythromycin resistant; Lmr, lincomycin resistant; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant.
ATCC, American Type Culture Collection.
Media and growth conditions.
Cultures were grown at 37°C in liquid media, aerated by rotation at 200 rpm in either Luria-Bertani (LB) broth (28) or M9 minimal salts (Life Technologies) with 1% (wt/vol) glucose. Cultures were also grown on LB broth and M9 minimal salts–1% glucose plates containing 1.2% agar (A4550 [Sigma]) at 37°C. To increase sporulation, B. megaterium was grown on sporulation medium, which contained 0.8% nutrient broth (Difco), 0.3% yeast extract (Difco), and 25 μg of MnSO4 · H2O per ml. For growth of B. megaterium VT1660 and derivatives, minimal medium was supplemented with arginine HCl at 50 μg/ml (wt/vol). For plasmid selections, the appropriate antibiotics were included in the media: ampicillin (100 μg/ml [AMP100]) or erythromycin (400 μg/ml [EM400]) for plasmid selection in E. coli and chloramphenicol (12 μg/ml [CM12]) or erythromycin (1 μg/ml [EM1]) and lincomycin (25 μg/ml [LM25]) for plasmid selection in Bacillus strains.
Transformations.
E. coli was transformed by electroporation of competent cells with the E. coli pulser (Bio-Rad Laboratories) and according to the manufacturer’s instructions. B. megaterium was transformed by a polyethylene glycol-mediated transformation method (16) with protoplasting and hypertonic media as previously described (5, 39).
Cloning of the gvp region.
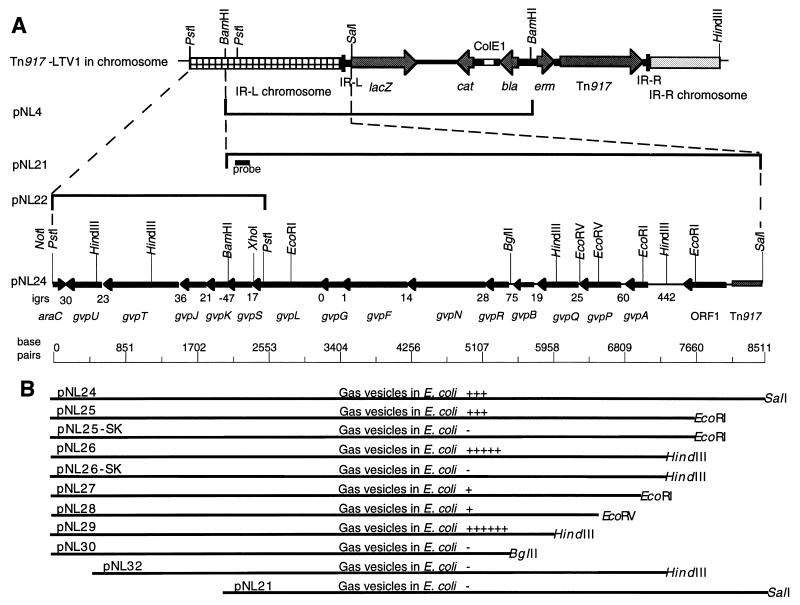
Purification of genomic and plasmid DNA, Southern blotting, colony hybridization, and dephosphorylation of DNA vectors were performed by standard procedures (35). To clone DNA sequences contiguous with the transposon in the transposant, B. megaterium B001S genomic DNA was cut with BamHI, self-ligated, and transformed into E. coli. Following selection on LB-AMP100 plates, colonies were screened. The plasmid pNL4 carried the left inverted repeat (IR-L) end of Tn917-LTV1 and flanking chromosomal DNA (Fig. 1). The SalI-BamHI fragment of pNL4 was ligated into the SalI-BamHI sites of pBluescriptIIKS. Following transformation of E. coli and selection on LB-AMP100, transformants were screened, thus yielding the plasmid pNL21. Chromosomal DNA sequences, overlapping and contiguous with pNL21 and distal to IR-L, were cloned from B. megaterium B001S genomic DNA. The PstI fragment size was identified in a Southern blot by using a 32P-, 5′-end-labeled synthetic oligonucleotide probe (5′-TCGGTTGAAACGCTTGTGC-3′) homologous to gvpS. The approximate-size fragments were excised from an agarose gel, extracted with Geneclean (Bio 101) and ligated into PstI-cut, dephosphorylated pBluescriptIISK. White colonies on LB-AMP100 plates, supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and ispropyl-β-d-thiogalactopyranoside (IPTG), were screened by colony hybridization and a plasmid, pNL22, carrying the 2.5-kb PstI fragment was identified in a Southern blot with the same 32P-, 5′-end-labeled probe homologous to gvpS. A plasmid with native DNA sequences of gvp genes was constructed as follows. pNL22 was completely cut with NotI and partially cut with BamHI; 2.1-kb NotI-BamHI fragments were then excised from an agarose gel, extracted with Geneclean, and ligated into the NotI-BamHI-linearized plasmid pNL21. The plasmid thus constructed, pNL24, was confirmed for accuracy across the BamHI junction by sequencing with the oligonucleotide probe (gvpS) described above used as a primer. Plasmids pNL25, pNL26, pNL27, pNL28, pNL29, and pNL30, are subclones of pNL24 in pBluescriptIIKS, unless otherwise noted (Table 1).
FIG. 1.
The gvp gene cluster and contiguous sequences in B. megaterium B001S. (A [from top to bottom]) Tn917-LTV1 and contiguous sequences in the chromosome of strain B001S and relevant restriction enzyme sites; cloned fragments of DNA in pNL4, pNL21, and pNL22 contiguous with the IR-L sequences; cloned DNA fragment in pNL24, map of putative genes, intergenic regions in base pairs (igrs), and relevant restriction enzyme sites; and ruler of sequence in base pairs. (B) Subclones of pNL24 and their ability to produce gas vesicles in E. coli. Gas vesicles were observed as refractile bodies by phase-contrast microscopy. ++++++, gas vesicles throughout all cells in culture; +++++, slightly reduced quantity of gas vesicles in most cells; +++, gas vesicles in approximately 50% of cells and in reduced quantity; +, very few gas vesicles in about 10% of cells; −, no gas vesicles observed. IR-L and IR-R, inverted repeats at lacZ end (left) and Tn917 end (right), respectively; ColE1, E. coli origin of replication; lacZ, β-galactosidase gene; cat, chloramphenicol resistance gene; bla, ampicillin resistance gene; erm, erythromycin resistance gene.
Cloned genomic DNA was subcloned into pHPS9 for expression in B. megaterium. The 6.4-kb SalI-BamHI fragment of pNL4 was subcloned from pNL4 into the SalI-BamHI sites of pHPS9, generating pNL20. This plasmid carries part of the gvp cluster. The complete cluster of cloned gas vesicle genes were cloned in pHPS9 by the digestion of pNL24 completely with SalI and partially with BamHI, followed by extraction of 8.5-kb SalI-BamHI fragments from agarose with Geneclean and ligation into the SalI-BamHI sites of pHPS9. The plasmid construct was confirmed by restriction mapping and was named pNL40.
Sequencing of the gvp region.
DNA fragments were subcloned into pBluescriptIISK, and sets of nested deletion clones from both sides were generated with the Erase-a-Base system (Promega). DNA was sequenced with USB Sequenase, version 2 (Amersham Life Science). Sequence assembly and analysis were performed with Lasergene (DNAStar, Inc.). Homologies to known genes were determined by Gapped BLAST and PSI BLAST sequence analysis programs (1) of the National Center for Biotechnology Information at the National Library of Medicine.
Buoyancy test.
Cells were grown on 100-mm-diameter plates in LB broth with ampicillin and IPTG for 24 h at 37°C. The cells from each plate were resuspended in 10 ml of saline in 13-mm-diameter test tubes. The tubes were left stationary and undisturbed at room temperature for at least 16 h, at which time the cell buoyancy was determined by the visual degree of turbidity of the culture medium.
Phase-contrast microscopy.
Wet mounts of cultures were visualized at ×1,000 magnification in a light microscope with phase-contrast attachments (Labophot-2; Nikon, Inc.).
DIC microscopy.
Wet mounts of cultures were visualized at ×1,000 magnification in a light microscope with Nomarski attachments for direct interference contrast (DIC) microscopy (Labophot-2).
Electron microscopy.
E. coli cells grown on LB broth with ampicillin and IPTG were resuspended in 10 mM Tris HCl (pH 7) and incubated at 25°C for 30 min with lysozyme (2 mg per ml) to generate protoplasts. To lyse these protoplasts, sodium dodecyl sulfate was added to a final concentration of 0.2%. Samples taken from the top of the supernatants of cell lysates and samples of protoplasts of E. coli were analyzed for gas vesicles with a Philips transmission electron microscope (CM10) at an 80-kV acceleration voltage. Drops of samples were adsorbed onto Formvar- and carbon-coated 400-mesh copper grids, negatively stained with 1% (wt/vol) uranyl acetate in water for 30 to 60 s, and blotted dry.
Pressure sensitivity test of refractile bodies.
Samples of overnight cultures grown in LB broth with ampicillin and IPTG were centrifuged at 16,000 × g for 5 to 40 min in a microcentrifuge. Pellets were gently resuspended in saline, and cells were visualized by phase-contrast microscopy for the presence of refractile bodies or left at room temperature for 16 h to test for effects on cell buoyancy.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBI, EMBL, and GenBank nucleotide databases under accession no. AF053765.
RESULTS
B. megaterium gvp gene cluster analysis.
E. coli synthesized functional gas vesicles when carrying an 8,142-bp region of DNA cloned from B. megaterium B001S (data below). Nucleotide and predicted amino acid sequence analyses of this region revealed a cluster of 1 partial and 15 complete, putative genes, as depicted on plasmid pNL24 in Fig. 1A. Construction of a map of this gvp region was based on a sequence analysis that revealed ORFs, putative ribosome binding sites, and homologies to known and putative genes in data banks (Table 2). These 15 putative genes read in one direction. Nine of the 15 have amino acid sequence similarity to known and putative gvp gene products, as determined by BLAST searches, while the remaining six have no significant homology to known genes. To comply with standard nomenclature, a four-letter designation was assigned to each putative gene according to the greatest homologies. Where paralogs to known genes exist, the gene of lesser homology to known genes was assigned a new letter, as were the six putative genes with no homology to known genes.
TABLE 2.
Sequence analysis results
| Gene name | No. of amino acids | Mol mass (Da) | Isoelectric point | Homologies to known and putative genes (accession no.)a | BLAST score |
|---|---|---|---|---|---|
| ORF1 | 161 | 16,147 | 5.39 | None | |
| gvpA | 86 | 9,291 | 4.37 | GVPA_MICBC (P08412) | 95.3 |
| GVPA_APHFL (P10397) | 93.5 | ||||
| gvpP | 161 | 17,945 | 4.89 | None | |
| gvpQ | 157 | 17,583 | 9.40 | None | |
| gvpB | 88 | 9,618 | 4.44 | GVPA_MICBC (P08412) | 94.6 |
| GVPA_APHFL (P10397) | 92.8 | ||||
| gvpR | 88 | 10,344 | 5.18 | None | |
| gvpN | 308 | 34,700 | 5.19 | GVPN_APHFL (P55150) | 228.0 |
| GVPN_HALME (P02239) | 178.0 | ||||
| GVPN_HALHA (P33952) | 178.0 | ||||
| GVPN_HALSA (P33965) | 178.0 | ||||
| gvpF | 255 | 28,864 | 5.15 | GVPL_APHFL (P55149) | 67.0 |
| GVPF_HALME (P02231) | 65.5 | ||||
| GVPF_HALHA (P24370) | 63.0 | ||||
| gvpG | 88 | 10,540 | 4.60 | GVPG_HALHA (P24371) | 33.9 |
| GVPG_HALSA (P33960) | 32.8 | ||||
| gvpL | 269 | 31,253 | 5.02 | GVPL_HALHA (P24376) | 68.5 |
| GVPL_HALME (P02237) | 61.5 | ||||
| GVPF_HALHA (P24370) | 50.1 | ||||
| GVPL_APHFL (P55149) | 48.2 | ||||
| gvpS | 95 | 10,422 | 4.53 | GVPJ_APHFL (P55147) | 37.6 |
| GVPA_PSEAN (P22453) | 31.7 | ||||
| GVPA_APHFL (P10397) | 31.3 | ||||
| gvpK | 94 | 10,448 | 4.18 | GVPK_APHFL (P55148) | 65.2 |
| GVPK_HALHA (P24375) | 58.9 | ||||
| GVPK_HALME (P02236) | 55.6 | ||||
| GVPK_HALSA (P33963) | 53.8 | ||||
| gvpJ | 100 | 11,261 | 4.95 | GVPJ_APHFL (P55147) | 107.0 |
| GVPA_APHFL (P10397) | 49.7 | ||||
| GVPJ_HALME (P02235) | 49.7 | ||||
| GVPA_FREDI (P07060) | 49.4 | ||||
| GVPA_PSEAN (P22453) | 48.6 | ||||
| GVPJ_HALSA (P33956) | 48.6 | ||||
| GVPJ_HALHA (P24374) | 48.2 | ||||
| GVPA_MICBC (P08412) | 47.9 | ||||
| GVPK_APHFL (P55148) | 39.4 | ||||
| gvpT | 292 | 32,739 | 4.26 | None | |
| gvpU | 138 | 14,960 | 5.72 | None | |
| araC | 49b | 5,831 | Not done | HTH_ARAC (AB001488)c | 60.4 |
Loci and accession numbers are SWISS-PROT unless otherwise stated. None, no discernible similarity to known sequences.
Partial protein.
DDBJ.
GvpA and GvpB have high homology to each other and to other known GvpA proteins (Fig. 2A). GvpS and GvpJ have 26% identity to each other and less sequence similarity to other known GvpJ proteins. B. megaterium GvpA and GvpB show sequence similarity to GvpS and GvpJ and less homology to the low-molecular-weight protein GvpK (Fig. 2B). As well as sequence similarity, GvpA, -B, -S, and -J have low molecular weights and hydropathy profiles that show the presence of a center hydrophobic domain, bordered at the C and N termini by hydrophilic amino acid sequences. GvpF and GvpL have 24% identity to each other, and both show low sequence similarities to GvpF and GvpL from both the cyanobacteria and halophilic archaea. GvpN and GvpG have no homology to other genes within this cluster. GvpP, GvpQ, and GvpR are extremely hydrophilic putative proteins, but they show no obvious amino acid identity to each other or to the other gvp gene products in this region. However, an alignment of the amino acid sequence of GvpQ with that of GvpI of H. halobium demonstrates sequence similarities in intervals (Fig. 2C). While this pattern of amino acid identity is insufficient for homology to be recognized in database searches, it could point to a common function for these two proteins. ORF1, although it reads in the same direction as the other 14 putative genes, is separated from them by 442 bp and may not be part of this gvp gene cluster (see below). AraC reads in the opposite orientation to the other 15 putative gene products and extends beyond the sequenced fragment of the gvp cluster. The 49 amino acids at the C terminus of this ORF have sequence similarity to the putative AraC transcriptional activator of Bacillus subtilis (26).
FIG. 2.
Homologies of predicted amino acid sequences. (A) Multiple sequence alignment of B. megaterium GvpA and GvpB with representative homologs from cyanobacteria and the halophilic archaea, with the majority consensus sequences shown in black. (B) Multiple sequence alignment of B. megaterium GvpA, GvpB, GvpJ, GvpS, and GvpK, with multiple consensus sequences shown in black. (C) Pairwise alignment of H. halobium GvpI with B. megaterium GvpQ, with amino acid identities shown in black. The Clustal method with the PAM250 residue weight table was used. GVPA_APHFL, GvpA of A. aquae-flos; GvpA1_HALHA, GvpA (plasmid borne) of H. halobium; GVPI_HALHA, GvpI of H. halobium; Bmeg, B. megaterium.
There are very few or no intergenic sequences between 14 of the putative gene products, from GvpA to GvpU, while GvpS and -K overlap by 47 bp (Fig. 1A). Upstream from GvpA is a 442-bp region with no discernible coding function. This region has a G+C ratio of 25%, compared to 34 to 45% for the putative gvp coding sequences, and possibly carries a promoter for gvpA and downstream genes. Upstream of this intergenic region is ORF1, which is preceded by 63 bp on this cloned gvp region of B. megaterium DNA. A study of the regulation of gvp gene expression is in progress, but culture conditions that result in a buoyancy phenotype of B. megaterium have not been determined.
B. megaterium gvp gene cluster functions in E. coli.
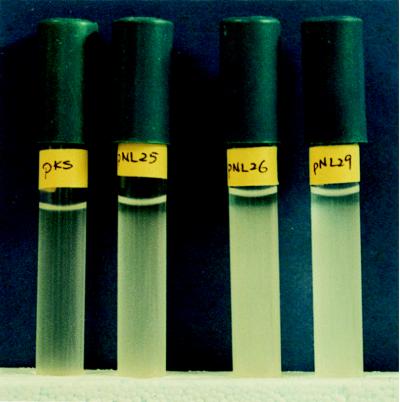
The 8,142-bp region of the B. megaterium chromosomal DNA that is contiguous with Tn917-LTV1 in the mutant B001S and that carries genes whose predicted amino acid sequences show homology to known gvp genes was tested for gas vesicle function in E. coli. This experiment was carried out by first cloning the 8,522-bp SalI-PstI fragment from B001S in pBluescriptIIKS to make pNL24 as described above. Following overnight growth at 37°C on LB broth with ampicillin and IPTG, E. coli(pNL24) and E. coli(pBluescriptIIKS) were tested for a buoyancy phenotype and for the presence of gas vesicles by DIC and phase-contrast microscopy. The results of this experiment showed that on standing, the majority of the E. coli(pNL24) cells remained suspended in the medium rather than sinking to the bottom of the tube, as was the case with the control E. coli(pBluescriptIIKS) cells. Following overnight growth at 37°C, cells carrying pNL24 also showed the presence of refractile bodies in phase-contrast microscopy and hollow-looking structures in DIC microscopy, while no such structures were present in the control. These results were consistent with the 8,124-bp region of B. megaterium DNA coding for the synthesis of functional gas vesicles. In order to determine the minimum length of DNA required for the putative gas vesicle synthesis, deletion derivatives of pNL24 were constructed, and E. coli strains carrying these plasmids were tested for buoyancy phenotype and for the presence of gas vesicles as was E. coli(pNL24). The plasmid constructs are described in Table 1. The results of phase-contrast and DIC microscopy gave an approximation of the quantities of presumptive gas vesicles in the cells and are summarized in Fig. 1B. The results of both types of microscopy were in agreement with each other and with the results of the buoyancy test. The turbidity (buoyancy) of the cultures on standing at room temperature for 16 h and longer, as observed by the naked eye, was directly related to the quantities of presumptive gas vesicles in the cells at time zero (following overnight growth), as determined by phase-contrast and DIC microscopy. The buoyancy phenotypes are shown in Fig. 3, and the DIC microscopy results are demonstrated in Fig. 4. Figure 3 shows the results for E. coli(pNL25), E. coli(pNL26), and E. coli(pNL29) compared to E. coli(pBluescriptIIKS) after 16 h of standing at room temperature. The cultures carrying the B. megaterium cloned fragments remained dispersed throughout the medium, whereas the culture carrying the cloning vector alone sank to the bottom of the tube. This buoyancy phenotype was maintained for at least 3 days and was not observed thereafter. When the cultures with buoyancy phenotypes were disturbed by shaking after 16 h of standing at room temperature and then allowed to stand for a further 16 h, the buoyancy phenotype was reestablished and maintained for at least 3 days.
FIG. 3.
Buoyancy of E. coli strains carrying plasmids with cloned B. megaterium gvp genes as labeled: pNL25, pNL26, pNL29, and the control, pKS (pBluescriptIIKS).
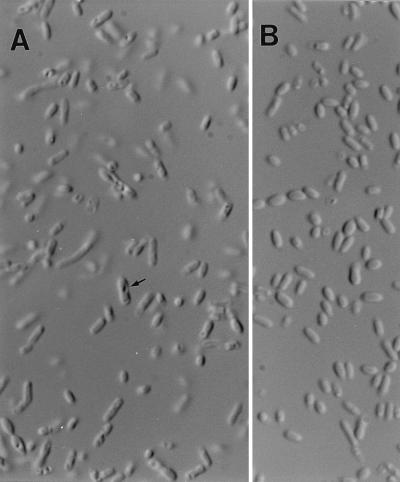
FIG. 4.
DIC microscopy of E. coli(pNL26) (A) and E. coli(pBluescriptIIKS) (B). In panel A, the cells are longer and have hollow-looking structures (arrow), and most of the cells tend towards a vertical position in the wet mount compared to those in panel B. Magnification, ×1,000.
The shortest fragment of DNA shown to produce the presumptive gas vesicles in E. coli was the 6,040-bp HindIII-PstI fragment in pNL29. This fragment carries GvpB, -R, -N, -F, -G, -L, -S, -K, -J, -T, and -U and parts of GvpQ and AraC. We hypothesize, therefore, that these 11 complete genes are the maximum number of B. megaterium genes required by E. coli to produce functional gas vesicles. Since each of the genes within this 6,040-bp fragment has not been deleted, it is not yet possible to say that all of these 11 genes are necessary. GvpU and GvpB are required to produce gas vesicles under the conditions tested, based on the fact that pNL32 and pNL30, respectively, gave negative results for synthesis of gas vesicles in E. coli. Whether or not GvpA can substitute for GvpB has not been tested. In E. coli, the plasmid pNL29, with its shorter fragment of cloned DNA, produced the most gas vesicles and the most pronounced buoyancy phenotype of all of the plasmids tested.
The plasmids that gave rise to gas vesicles had their DNA fragment inserted in the cloning vector in the same orientation as the lacZ promoter of pBluescriptIIKS. In pNL25-SK and pNL26-SK, in which the inserted fragment of DNA reads in the opposite orientation to the lacZ promoter, no gas vesicles and no buoyancy phenotypes were observed, indicating that expression of at least some, if not all, of the gvp genes on pNL24 were transcribed from the lacZ promoter. Given these results, it is not surprising that pNL24 and pNL25 produced fewer gas vesicles than pNL26, due to a possible reduction in the rate of transcription at the end of ORF1. If this were the only factor determining the quantity of gas vesicles produced by these plasmids, one would not expect pNL29 to produce more gas vesicles than pNL26, pNL27, and pNL28. The results show otherwise (Fig. 1B), indicating that GvpP and/or GvpQ may play a regulatory role. A comparison of the results for pNL27 and pNL28 indicates that GvpP has no regulatory function under the experimental conditions used, while a comparison of the results for pNL28 and pNL29 shows clearly that the absence of GvpQ makes a significant difference in the quantity of gas vesicles synthesized, suggesting that GvpQ could be a negative regulator of gas vesicle synthesis. However, these data give no indication of the mode or mechanism by which such regulation may occur.
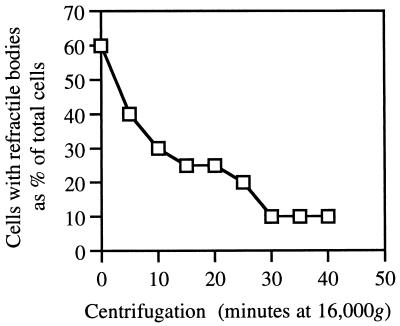
That the refractile bodies observed by phase-contrast microscopy are hollow vesicles is supported by the fact that the quantity of refractile bodies in E. coli(pNL26) decreased in proportion to centrifugation times (Fig. 5). Following 30 min of centrifugation at 16,000 × g, refractile bodies were present in approximately 10% of cells compared with those seen in 60% of cells prior to centrifugation. Loss of refractile bodies coincided with loss of buoyancy.
FIG. 5.
Pressure sensitivity of refractile bodies in E. coli(pNL26). See Materials and Methods for details. Triplicate samples of the same culture had a 5% margin of error.
Electron microscopy identifies gas vesicles in E. coli.
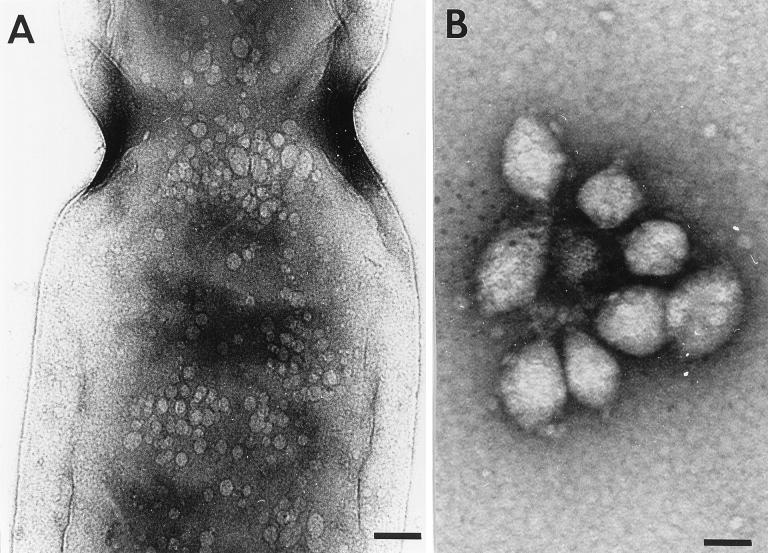
Transmission electron microscopy of E. coli(pNL26) showed structures within protoplasts that resembled the shape and size of gas vesicles found in cyanobacteria and halophilic archaea (Fig. 6). The gas vesicles in E. coli(pNL26) ranged from 25 to 70 nm in width and from 60 to 110 nm in length, but most of the gas vesicles observed were approximately 50 nm in width and 75 nm in length. A rib structure, as has been previously described for gas vesicles, can be observed around the width of the gas vesicles from E. coli. Also, the gas vesicles appeared to be cylindrical with cone-shaped ends; those gas vesicles that appeared round were possibly being viewed from one end. Gas vesicles, both released and in cells, were found mainly in clusters, known as gas vacuoles, and are consistent with the structures that were observed as refractile bodies in phase-contrast microscopy and as hollow structures in DIC microscopy. In keeping with this is our observation by phase-contrast microscopy that the gas vesicles (as gas vacuoles) clustered together over a 15-min period following lysis of the protoplasts and release of gas vesicles. This observation is consistent with the known hydrophobicity of the gas vesicle structural protein, GvpA.
FIG. 6.
Electron micrographs of E. coli(pNL26). (A) Protoplast of a dividing cell shows gas vesicles within the cell. Bar, 123 nm. (B) Gas vesicles from cell lysate. Bar, 44 nm.
Expression of a gvp cluster in B. megaterium.
Buoyancy phenotypes have not been observed in (i) B. megaterium B001S, the transposon mutant from which the gvp cluster was cloned; (ii) B. megaterium VT1660, the progenitor of the mutant B001S; (iii) B. megaterium 19213, the progenitor of VT1660; or (iv) B. megaterium 11561. These strains, with and without the plasmids pNL20 and pNL40, were cultured with a wide range of media and culture conditions, but gas vesicles could not be confirmed by the methods described above for E. coli, because they were observed in very small quantities and inconsistently. Since the plasmid pNL40 carries the 8,142-bp cluster of functional gvp genes, as demonstrated in E. coli, at a plasmid copy number of approximately five per cell, it is probable that the expression of gvp genes in B. megaterium is stringently regulated. The B. megaterium strains used in this study were motile. Motility was greatest in fresh cultures, while in late-stationary-phase cultures, the cells showed very little or no motility.
gvp genes in other Bacillus strains.
Sequences with high homology to gvpA and gvpS were identified in B. megaterium 11561 by Southern hybridization with gvpA and gvpS probes. This result indicated that at least one other strain of B. megaterium as well as strain 19213 carries gvp genes. A similar analysis of B. subtilis 168 showed no homology to the B. megaterium gvp gene probes. Genome sequence data for B. subtilis 168 have since confirmed the absence of gvp genes in this strain (26).
DISCUSSION
The only known physiological role for gas vesicles is that they provide buoyancy to cells for vertical movement in liquid, and thus cells can position themselves at a depth that allows other metabolic activities to function. With gas vesicles, cells are known to position themselves in liquid for optimal light and oxygen. Gas vesicles have been previously described in microorganisms from aqueous environments, but have not been previously identified in B. megaterium. However, it is not surprising that gas vesicles should occur in soil organisms. It is normal for soil to become wet or flooded, and under such conditions, bacteria with gas vesicles and/or other means of motility could avoid being washed down deep into the earth, becoming depleted in the upper layers of soil. Since B. megaterium is an obligate aerobe and motile, the survival advantage of gas vesicles over other motility methods could be due to the lack of energy requirements by gas vesicles for function. Since older cultures of the B. megaterium strains used in this study had reduced motility, it is reasonable to speculate that older cells, especially large cells with gas vesicles, could have a survival advantage. Since a functional cluster of gvp genes is present in B. megaterium, as demonstrated by expression in E. coli, it is reasonable to assume that this gene cluster functions to provide buoyancy to B. megaterium under certain environmental conditions not yet defined. In size and shape, the B. megaterium gas vesicles synthesized in E. coli are similar to those described in cyanobacteria and halophilic archaea. Gas vesicle synthesis has not been reported in E. coli with known gvp genes.
It has been suggested that many of the gas vesicle proteins with possible structural function in the archaea may play a role in gas vesicle assembly, but the roles of these proteins have not been described in detail. GvpA, the main structural protein of gas vesicles, and its paralog, GvpB, have a highly conserved amino acid sequence, indicating that the physical structure of gas vesicles is conserved. GvpC, a minor structural protein involved in shaping and strengthening gas vesicles, has been identified in both cyanobacteria and archaea, but a homolog of GvpC has not been identified in the B. megaterium gvp gene cluster. GvpB, -F, -G, -H, -I, -J, -K, -L, -M, and -N are possibly minor structural proteins of gas vesicles in halobacteria (10, 41). Of these genes, only GvpJ, -K, -F/-L, and -N have been identified in cyanobacteria. B. megaterium homologs of GvpB, -F, -G, -J, -K, -L, and -N are described in this paper, but homologs of GvpH, -I, and -M, which are possible minor structural proteins, and GvpD, -E, and -O, which have possible regulatory functions, have not been identified. It is not surprising that none of the gvp gene products with regulatory functions has been identified, because regulation in B. megaterium may be very different from that in the archaea.
Nonorthologous functional equivalents of GvpH, -I, and -M may be among the B. megaterium GvpP, -Q, -R, -T, and -U proteins. The gvp cluster provides an interesting opportunity to examine the possibility of nonorthologous functional alternatives thus providing a functional identity to unclassified genes. In the B. megaterium gvp cluster, 38% of the ORFs (5 in 14, or 6 in 15 [including ORF1]) show no significant homology to known genes. The E. coli genome has 1,632 hypothetical and unclassified ORFs, based on sequence homology, which amounts to 38% of the total genome sequence (2). In B. subtilis, the function of 42% of the hypothetical genes cannot be predicted by homology to genes of known function (26). Nonorthologous gene alternatives could provide new insights not only into gene function but also into the evolutionary origins of organisms. In a comparison of the B. megaterium and H. halobium predicted gvp gene products, it is notable that GvpQ and GvpI have a number of features in common. GvpQ (17.6 kDa) has an isoelectric point of 9.4, while all of the other putative gvp gene products from B. megaterium have isoelectric points ranging from 4.2 to 5.7 (Table 2). The gvp gene products of halophilic archaea have isoelectric points ranging from 3.9 to 4.9, with the exception of GvpI (16 kDa) (21), which has an isoelectric point of 10.8. This fact, together with the similarity of their sizes, their sequence identity (at intervals), and similar hydropathy profiles, suggests that GvpQ of B. megaterium and GvpI of H. halobium could be nonorthologous functional equivalents.
We have identified a maximum of 11 genes in a 5.7-kb cluster from B. megaterium that are required for gas vesicle formation and function in E. coli. They are gvpB, -R, -N, -F, -G, -L, -S, -K, -J, -T, and -U. As well as these 11 genes, we have provided evidence that 3 and possibly 4 additional genes may be involved in gas vesicle formation; they are gvpA, -P, and -Q and, less likely, ORF1. The presence of paralogous genes in the gvp cluster of B. megaterium, which are also present in the gvp clusters in archaea and cyanobacteria, is interesting from both functional and evolutionary perspectives. This discovery of the B. megaterium gvp cluster and its functional expression in E. coli will enable the study of gas vesicle biogenesis, including assembly, gene product function, and regulation.
ACKNOWLEDGMENTS
We thank Frank Cannon for critically reading the manuscript and Curt Thorne for the Bacillus strains. We also thank Tracy Guillemette and Heather Baker for excellent technical assistance and Lucy Yin for contributions to electron microscopy and photography.
This research was supported in part by a grant from the National Science Foundation (MCB 97-28066). The University of Massachusetts, Amherst, Central Microscopy Facility is supported by a grant from the NSF (NSF BBS 8714235).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, et al. The complete genome sequence of E. coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz B E, Hayes P K, Walsby A E. The distribution of the outer gas vesicle protein, GvpC, on the Anabaena gas vesicle, and its ratio to GvpA. J Gen Microbiol. 1993;139:2353–2363. doi: 10.1099/00221287-139-10-2353. [DOI] [PubMed] [Google Scholar]
- 4.Camilli A, Portnoy D A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang S, Cohen S N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 6.Damerval T, Castets A-M, Guglielmi G, Houmard J, Tandeau de Marsac N. Occurrence and distribution of gas vesicle genes among cyanobacteria. J Bacteriol. 1997;171:1445–1452. doi: 10.1128/jb.171.3.1445-1452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damerval T, Castets A M, Houmard J, Tandeau de Marsac N. Gas vesicle synthesis in the cyanobacterium Pseudanabaena sp.: occurrence of a single photoregulated gene. Mol Microbiol. 1991;5:657–664. doi: 10.1111/j.1365-2958.1991.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 8.Damerval T, Houmard J, Guglielmi G, Csiszar K, Tandeau de Marsac N. A developmentally regulated gvpABC operon is involved in the formation of gas vesicles in Cyanobacterium calothrix 7601. Gene. 1987;54:83–92. doi: 10.1016/0378-1119(87)90350-7. [DOI] [PubMed] [Google Scholar]
- 9.DasSarma S, Arora P. Gas vesicle proteins and genes. Mol Biol. 1993;12:93–98. [Google Scholar]
- 10.DasSarma S, Arora P. Genetic analysis of the gas vesicle gene cluster in haloarchaea. FEMS Microbiol Lett. 1997;153:1–10. [Google Scholar]
- 11.DasSarma S, Arora P, Lin F, Molinari E, Ru-Siu Yin L. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J Bacteriol. 1994;176:7646–7652. doi: 10.1128/jb.176.24.7646-7652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DasSarma S, Arora P, Perkel J, Ng W-L, Hackett N. Abstracts of the 92nd General Meeting of the American Society for Microbiology 1992. Washington, D.C: American Society for Microbiology; 1992. The second copy of the gas vesicle gene cluster of Halobacterium halobium NRC-1 is located on a large plasmid, abstr. H-128; p. 204. [Google Scholar]
- 13.DasSarma S, Damerval T, Jones G J, Tandeau de Marsac N. A plasmid-encoded gas vesicle protein gene in halophilic archaebacterium. Mol Microbiol. 1987;1:365–370. doi: 10.1111/j.1365-2958.1987.tb01943.x. [DOI] [PubMed] [Google Scholar]
- 14.Englert C, Horne M, Pfeifer F. Expression of the major gas vesicle protein gene in the halophilic archaebacterium Haloferax mediterranei. Mol Gen Genet. 1990;222:225–232. doi: 10.1007/BF00633822. [DOI] [PubMed] [Google Scholar]
- 15.Englert C, Kruger K, Offner S, Pfeifer F. Three different but related gene clusters encoding gas vesicles in halophilic archaea. J Mol Biol. 1992;227:586–592. doi: 10.1016/0022-2836(92)90914-6. [DOI] [PubMed] [Google Scholar]
- 16.Fodor K, Hadlaczky G, Alföldi L. Reversion of Bacillus megaterium protoplasts to the bacillary form. J Bacteriol. 1975;121:390–391. doi: 10.1128/jb.121.1.390-391.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haima P, Bron S, Venema G. Novel plasmid marker rescue transformation system for molecular cloning in Bacillus subtilis enabling direct selection of recombinants. Mol Gen Genet. 1990;223:185–191. doi: 10.1007/BF00265052. [DOI] [PubMed] [Google Scholar]
- 18.Halladay J T, Jones J G, Lin F, MacDonald A B, DasSarma S. The rightward gas vesicle operon in Halobacterium plasmid pNRC100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J Bacteriol. 1993;175:684–692. doi: 10.1128/jb.175.3.684-692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes P K, Powell R S. The gvpA/C cluster of Anabaena flos-aquae has multiple copies of a gene encoding GvpA. Arch Microbiol. 1995;164:50–57. doi: 10.1007/BF02568734. [DOI] [PubMed] [Google Scholar]
- 20.Horne M, Englert C, Pfeifer F. Two genes encoding gas vacuole proteins in Halobacterium halobium. Mol Gen Genet. 1988;213:459–464. doi: 10.1007/BF00339616. [DOI] [PubMed] [Google Scholar]
- 21.Horne M, Englert C, Wimmer C, Pfeifer F. A DNA region of 9 kbp contains all genes necessary for gas vesicle synthesis in halophilic archaebacteria. Mol Microbiol. 1991;5:1159–1174. doi: 10.1111/j.1365-2958.1991.tb01889.x. [DOI] [PubMed] [Google Scholar]
- 22.Horne M, Pfeifer F. Expression of two gas vacuole protein genes in Halobacterium halobium and other related species. Mol Gen Genet. 1989;218:437–444. doi: 10.1007/BF00332407. [DOI] [PubMed] [Google Scholar]
- 23.Jones J G, Young D C, DasSarma S. Structure and organization of the gas vesicle gene cluster on the Halobacterium halobium plasmid pNRC100. Gene. 1991;102:1017–1022. doi: 10.1016/0378-1119(91)90549-q. [DOI] [PubMed] [Google Scholar]
- 24.Kinsman R, Hayes P K. Genes encoding proteins homologous to halobacterial GvpN, J, K, F and L are located downstream of gvpC in the cyanobacterium Anabaena flos-aquae. DNA Seq. 1997;7:97–106. doi: 10.3109/10425179709020156. [DOI] [PubMed] [Google Scholar]
- 25.Krüger K, Pfeifer F. Transcript analysis of the c-vac region and differential synthesis of the two regulatory gas vesicle proteins GvpD and GvpE in Halobacterium salinarium PHH4. J Bacteriol. 1996;178:4012–4019. doi: 10.1128/jb.178.14.4012-4019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst N, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 27.Mayr A, Pfeifer F. The characterization of the nv-gvpACNOFGH gene cluster involved in gas vesicle formation in Natronobacterium vacuolatum. Arch Microbiol. 1997;168:24–32. doi: 10.1007/s002030050465. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Offner S, Pfeifer F. Complementation studies with the gas vesicle-encoding p-vac region of Halobacterium salinarium PHH1 reveal a regulatory role for the p-gvpDE gene. Mol Microbiol. 1995;16:9–19. doi: 10.1111/j.1365-2958.1995.tb02387.x. [DOI] [PubMed] [Google Scholar]
- 30.Offner S, Wanner G, Pfeifer F. Functional studies of the gvpACNO operon of Halobacterium salinarium reveal that the GvpC protein shapes gas vesicles. J Bacteriol. 1996;178:2071–2078. doi: 10.1128/jb.178.7.2071-2078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeifer F, Ghahraman P. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas-vesicle gene cluster and insertion elements. Mol Gen Genet. 1993;238:193–200. doi: 10.1007/BF00279547. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer F, Kruger K, Roder R, Mayr R, Ziesche A, Offner S. Gas vesicle formation in halophilic archaea. Arch Microbiol. 1997;167:259–268. doi: 10.1007/s002030050441. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer F, Weidinger G, Goebel W. Characterization of plasmids in halobacteria. J Bacteriol. 1981;145:369–374. doi: 10.1128/jb.145.1.369-374.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roder R, Pfeifer F. Influence of salt on the transcription of the gas-vesicle genes of Haloferax mediterranei and identification of the endogenous transcriptional activator gene. Microbiology. 1996;142:1715–1723. doi: 10.1099/13500872-142-7-1715. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Surek B, Pillay B, Rdest U, Beyreuther K, Goebel W. Evidence for two different gas vesicle proteins and genes in Halobacterium halobium. J Bacteriol. 1988;170:1746–1751. doi: 10.1128/jb.170.4.1746-1751.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tandeau de Marsac N, Mazel D, Bryant D A, Houmard J. Molecular cloning and nucleotide sequence of a developmentally regulated gene from the cyanobacterium Calothrix PCC 7601: a gas vesicle protein gene. Nucleic Acids Res. 1985;13:7223–7236. doi: 10.1093/nar/13.20.7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vary P S. Prime time for Bacillus megaterium. Microbiology. 1994;140:1001–1013. doi: 10.1099/13500872-140-5-1001. [DOI] [PubMed] [Google Scholar]
- 39.Von Tersch M A, Carlton B C. Megacinogenic plasmids of Bacillus megaterium. J Bacteriol. 1983;155:872–877. doi: 10.1128/jb.155.2.872-877.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Von Tersch M A, Carlton B C. Molecular cloning of structural and immunity genes for megacins A-216 and A-19213 in Bacillus megaterium. J Bacteriol. 1984;160:854–859. doi: 10.1128/jb.160.3.854-859.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsby A E. Gas vesicles. Microbiol Rev. 1994;58:94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsby A E, Hayes P K. Gas vesicle proteins. Biochem J. 1989;264:313–322. doi: 10.1042/bj2640313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weidinger G, Klotz G, Goebel W. A large plasmid from Halobacterium halobium carrying genetic information for gas vacuole formation. Plasmid. 1979;2:377–386. doi: 10.1016/0147-619x(79)90021-0. [DOI] [PubMed] [Google Scholar]