Abstract
Isoniazid (INH) is a highly effective drug used in the treatment and prophylaxis of Mycobacterium tuberculosis infections. Resistance to INH in clinical isolates has been correlated with mutations in the inhA, katG, and ahpC genes. In this report, we describe a new mechanism for INH resistance in Mycobacterium smegmatis. Mutations that reduce NADH dehydrogenase activity (Ndh; type II) cause multiple phenotypes, including (i) coresistance to INH and a related drug, ethionamide; (ii) thermosensitive lethality; and (iii) auxotrophy. These phenotypes are corrected by expression of one of two enzymes: NADH dehydrogenase and the NADH-dependent malate dehydrogenase of the M. tuberculosis complex. The genetic data presented here indicate that defects in NADH oxidation cause all of the mutant traits and that an increase in the NADH/NAD+ ratio confers INH resistance.
Mycobacterium tuberculosis is exquisitely sensitive to the drug isoniazid (INH), and this sensitivity has made INH the core of tuberculosis chemotherapy and prophylaxis since the early 1950s (1, 37). However, mutant strains that are INH resistant (INHr) frequently arise, such that as many as 30% of tuberculosis isolates are INH resistant in many large cities (10, 25, 46). In our effort to develop better tuberculosis drugs, we are trying to understand how INH kills sensitive mycobacteria and how mycobacteria become resistant.
INH inhibits the synthesis of mycolic acids, which are long-chain fatty acids that constitute a major structural component of the mycobacterial cell envelope (48, 52). Inhibition of mycolic acid biosynthesis most likely causes the bactericidal activity of the drug (47). The InhA enzyme catalyzes an essential step in fatty acid elongation and mycolic acid synthesis and is a target for INH action in M. tuberculosis and the soil saprophyte Mycobacterium smegmatis (3, 36). Structural studies have shown that a reactive form of INH attacks the NAD(H) cofactor that is bound to the InhA active site and generates a covalent INH-NAD adduct (39). Mutations that lead to overproduction of InhA are found in roughly 15 to 30% of resistant tuberculosis isolates (15, 29, 31, 49); several inhA missense mutations have also been identified (30). Mutations in inhA also confer resistance to a second drug, ethionamide (ETH), that is a closely related chemical analog of INH (3, 15).
INH is a prodrug which is converted to an active form by the KatG catalase-peroxidase (51). Mutation of the katG gene is the most common mechanism of INH resistance in M. tuberculosis (15, 27, 31, 49, 55). KatG is thought to catalyze the oxidation of INH, generating an electrophilic species that reacts with targets such as InhA (18). INH activation may occur by the peroxidase activity of the KatG enzyme (20, 41).
Many clinical INHr isolates of M. tuberculosis (16%) have mutations that increase expression of the ahpC homolog, which is thought to encode one subunit of the NAD(P)H-dependent alkyl hydroperoxidase AhpCF (49). In some clinical strains, the increased expression of alkyl hydroperoxidase may compensate for a katG defect which causes peroxide sensitivity and reduced virulence (16, 40). A large fraction of the clinical isolates (13%) have ahpC promoter mutations and do not have katG mutations (49). The mechanism for INH resistance in these clinical isolates is unknown. Perhaps the increased expression of AhpC lowers the intracellular peroxide concentration and thereby prevents KatG-mediated peroxidation of INH (50, 56). A role for peroxides in INH sensitivity has been proposed (38).
This report describes a new mechanism for INH resistance in M. smegmatis. A survey of spontaneous INHr mutants shows that at least half are defective for NADH dehydrogenase (Ndh type II), which oxidizes NADH and transfers electrons to quinones of the respiratory chain. Many of the ndh mutants have multiple phenotypes, including thermosensitivity (Ts), auxotrophy, and resistance to ETH. Some of the Ndh mutants have a Ts lethal phenotype, which indicates that Ndh is essential for the viability of M. smegmatis. Our genetic data indicate that INHr and all of the other mutant phenotypes are due to defects in NADH oxidation which increase the intracellular NADH/NAD+ ratio. We propose that the increased NADH concentration causes INH resistance by two mechanisms: it may interfere with KatG-mediated peroxidation of the drug (like the ahpC promoter mutations), and/or it may displace the INH-NAD adduct from the InhA active site.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are listed in Table 1, and the plasmids employed are presented in Table 2. All M. smegmatis strains were derived from the laboratory strain mc2155, which is proficient for transformation (42). All plasmids were maintained in Escherichia coli STBL2 (Gibco BRL), which allows stable propagation of plasmids containing mycobacterial genes.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| E. coli STBL2 | mcrA Δ(mcrBC-hsdRMS-mrr) recA1 endA1 gyrA96 thi supE44 relA1 | Gibco BRL |
| M. smegmatis | ||
| mc2155 | ept-1 (laboratory wild-type strain) | 42 |
| mc2651 | ept-1 inhA1 | 3 |
| mc21390 | ept-1 ndh-4 | This study |
| mc21406 | ept-1 ndh-14 | This study |
| mc21407 | ept-1 ndh-15 | This study |
| mc21412 | ept-1 ndh-21 | This study |
| mc21416 | ept-1 ndh-29 | This study |
| mc21419 | ept-1 ndh-4/pYUB801 | This study |
| mc21420 | ept-1 ndh-4/pYUB806 | This study |
| mc21421 | ept-1/pMV261 | This study |
| mc21422 | ept-1/pYUB805 | This study |
| mc21423 | ept-1/pYUB807 | This study |
| mc21424 | ept-1 ndh-4/pMV261 | This study |
| mc21442 | ept-1 ndh-4/pYUB805 | This study |
| mc21443 | ept-1 ndh-4/pYUB808 | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pMV261 | KmrE. coli-mycobacterial shuttle plasmid | 40 |
| pYUB325 | pYUB328::mc2155 (45-kb cosmid; contains inhA, Tn5-seq1) | 9 |
| pYUB328 | Apr cosmid with PacI sites flanking the polylinker | 37 |
| pYUB412 | Apr HygrE. coli-mycobacterial shuttle cosmid (derivative of pYUB328); ColE1 origin; phage L5 int attP integrates into the attB site of the M. smegmatis chromosome | 36 |
| pYUB801 | pYUB412::mc2155 (45-kb cosmid; contains ndh) | This study |
| pYUB802 | pMV261::mc2155 (2.5-kb StuI subclone from pYUB801; contains ndh) | This study |
| pYUB803 | pMV261::ndh (1.6-kb PstI-HindIII subclone of pYUB802) | This study |
| CY359 | pYUB328::M. tuberculosis H37Rv (45-kb cosmid; contains ndh) | 6 |
| pYUB805 | pMV261::ndh (1.5-kb NheI-PstI subclone of CY359) | This study |
| pYUB806 | pYUB412::M. bovis BCG (45-kb cosmid; contains mdh) | This study |
| pYUB807 | pMV261::M. bovis BCG (2.5-kb EcoRV subclone of pYUB806; contains mdh) | This study |
| pYUB808 | pMV261::mdh (1.1-kb PCR product of pYUB807; contains mdh) | This study |
Media.
The liquid medium used for M. smegmatis cultures was Mueller-Hinton broth (Difco); Tween 80 was added to 0.2% (vol/vol). The rich medium employed was Mueller-Hinton medium (Difco). The minimal medium used was Middlebrook 7H9 broth (Difco) supplemented with 0.2% (vol/vol) glycerol or 0.2% glucose. Anitimicrobial agents (Sigma) were used at the following concentrations: INH, 25 μg/ml; kanamycin A monosulfate, 10 μg/ml; hygromycin B (Boehringer Mannheim), 150 μg/ml; and streptomycin sulfate, 100 μg/ml. E. coli strains were grown in Luria-Bertani broth containing the appropriate antibiotics—ampicillin, hygromycin B, and kanamycin (50 μg/ml)—to maintain selection for plasmids.
Isolation of spontaneous INHr mutants.
Spontaneous INHr mutants were isolated from 25 independent cultures (30°C incubation; nonmutagenized) and plated onto selective medium containing INH at 25 μg/ml. The numbers of CFU in 10 of these cultures were determined by plating 10-fold serial dilutions onto nonselective medium. The plates were incubated at 30°C for 7 days and scored for the number of CFU. Plates used for isolation of INHr mutants were incubated at 37°C for 5 days.
Selection for INHr following EMS mutagenesis.
Ethyl methanesulfonate (EMS) mutagenesis was used to isolate one INHr Ts mutant, the ndh-4 mutant. The protocol of Miller, involving a 45-min exposure to the mutagen, was followed (28). Mutagenesis increased the frequency of INHr and Smr mutants ∼10-fold and caused no detectable decrease in culture viability.
Measurement of INH sensitivity.
Three independent cultures were grown to an optical density at 600 nm of 0.7, and 10-fold serial dilutions (0.1 ml each) were plated onto nonselective medium as well as medium containing INH; concentrations of the drug varied 2-fold, from 0.5 to 100 μg/ml. The MIC was scored as the concentration of drug that caused at least a 100-fold reduction in the number of CFU.
Tests of sensitivity to ETH, ethambutol, and streptomycin.
Sensitivity was tested with a zone of inhibition assay using a top agar plating method. ETH was dissolved in dimethyl sulfoxide (50 mg/ml), and 10 μl was spotted onto the lawn of cells. (Application of dimethyl sulfoxide alone did not inhibit cell growth.) BBL antimicrobial Sensi-Disks were used to test ethambutol (50 μg) and streptomycin (50 μg) sensitivity. Sensitivity was determined by measuring the diameter of the zone in which the drug inhibited growth. These assays were repeated three times for each strain.
Tests for Ts lethal phenotypes.
Tenfold serial dilutions of exponentially growing cultures were each spread onto two plates; one was incubated at 30°C to determine the number of CFU for that culture, while the other plate was incubated at 42°C for 2.5 days, scored for the presence or absence of colonies, and then transferred to 30°C. The data reported below are the fractions of cells that survive the 42°C incubation and form colonies upon transfer to 30°C and are the average values for data from three different experiments.
Tests of linkage to inhA.
Linkage analysis was performed with cosmid pYUB325 as described previously (3). Cosmid pYUB325 contains a 45-kb segment of the M. smegmatis chromosome that includes the inhA+ allele and a nearby Tn5-seq1 insertion (encoding kanamycin resistance). Recombination substrates were prepared by linearizing cosmid pYUB325 with PacI (New England Biolabs), which releases the chromosomal sequences as a single fragment. (The mycobacterial chromosome has no PacI sites.) This substrate (∼0.5 μg) was introduced into the mutant strains by electrotransformation (9). The entire transformation mixture was plated to kanamycin-containing medium, and the plates were incubated for 7 days at 30°C. Kmr recombinants (∼200) were tested for INH sensitivity by patching them onto kanamycin-containing medium with and without INH and were tested for thermosensitivity by patching them to nonselective medium and incubating the plates at 42°C.
Identification of complementing genes by using cosmid pYUB412 genomic libraries.
Genes which complement the mutant Ts phenotype were found by using large-fragment (45-kb) libraries of M. smegmatis and Mycobacterium bovis BCG and the integrative cosmid pYUB412 (Apr Hygr) (4). The M. smegmatis library was a gift from Franz C. Bange, and the M. bovis BCG library was a gift from Jordan Kriakov. The cosmid pYUB412 and the use of double cos-PacI cosmids have been described previously (2, 17, 33). The libraries were introduced into one Ts mutant, strain mc21390, by electrotransformation (9); complementation of the Ts phenotype was selected at 42°C. Temperature-resistant (Tr) transformants were tested for Hygr (vector marker), for sensitivity to INH, and for the ability to grow on minimal medium. Genomic DNA was purified from those transformants which met these criteria (17). The complementing fragments were recovered from the chromosome by PacI digestion, ligation into PacI arms of pYUB412, and in vitro packaging into phage lambda (Stratagene Gigapac II). The recovered cosmids were then tested for complementation of the original mutant, mc21390; all recovered cosmids complemented the original mutant (six tested). Two of these cosmids, pYUB801 (M. smegmatis) and pYUB806 (M. bovis BCG), were used for shotgun subcloning into the E. coli-mycobacterial shuttle plasmid pMV261 (44).
The insert of cosmid pYUB801 (M. smegmatis) was subcloned to a 2.5-kb fragment that retained the ability to complement all the ndh-4 mutant phenotypes (plasmid pYUB802). Sequence analysis identified one 1.4-kb open reading frame with significant homology to an M. tuberculosis gene on a 1.5-kb NheI-PstI fragment of cosmid MTCY359 (CDS19) (6). This M. tuberculosis gene, when subcloned into pMV261 (pYUB805), complemented all of the mutant phenotypes.
A 2.2-kb fragment of M. bovis BCG DNA that complemented all of the mutant phenotypes was obtained from shotgun subcloning of cosmid pYUB806 to generate pYUB807. Sequence analysis identified an open reading frame that encodes a protein 60% identical to the Thermus aquaticus Mdh enzyme (32). The M. bovis BCG mdh gene (1.1 kb) cloned into pMV261 (pYUB808) complemented all of the mutant phenotypes.
DNA sequencing methods and PCR amplification.
The inserts of plasmids pYUB802 (M. smegmatis; 2.5 kb) and pYUB806 (M. bovis BCG; 2.5 kb) were sequenced with the Applied Biosystems Prism Dye Terminator Cycle Sequencing Core kit with AmpliTaq DNA polymerase (Perkin-Elmer), using the Applied Biosystems model 377 automated DNA sequencer. The sequences of both strands were determined by primer walking.
The complementing gene of pYUB802 (M. smegmatis) was PCR amplified from chromosomal DNA of representative INHr mutants and the INH-sensitive (INHs) parent, mc2155; the nucleotide sequence was determined, and the presence of a mutation was verified by repeating the sequencing with different primers.
Preparation of cell extracts and membrane fractions.
Cultures (700 ml) of each strain were grown to an optical density at 600 nm of 0.6 to 0.7; the Ts mutants were grown at 30°C, and the Tr mutants were grown at 37°C. Cells were washed twice in cold buffer (50 mM KPO4 [pH 7.5], 5 mM MgSO4) and then resuspended in 1 ml of buffer per g of cell paste. DNase (20 μg) was added, and the cells were broken in a French pressure cell (16,000 lb/in2; 10 pressure applications). Cell debris was removed by centrifugation (12,000 × g, 30 min), and membrane particles from the clarified extracts were enriched by centrifugation (100,000 × g, 2 h) on a sucrose step gradient (20% and 60% [wt/vol]). Membrane fractions were recovered at the interface of the 20% and 60% sucrose layers. Membrane fractions of E. coli LE392 were prepared to ensure that this method did not destroy the unstable type I NADH dehydrogenase activity; large amounts of this activity were detected. The methods used for the preparation of cell extracts and membrane fractions were modified from previously reported techniques (13, 22).
Enzyme assays.
All assays were performed at room temperature in 50 mM KPO4 (pH 7.5)—5 mM MgSO4 in a total volume of 1 ml. Substrates were used at 100 μM (final concentration) unless otherwise stated. Reagents were purchased from Sigma Chemical Co. unless otherwise noted. NADH dehydrogenase activity in membrane fractions was measured spectrophotometrically by following the rate of NADH oxidation at 340 nm (ɛ340 = 6.2 mM−1 cm−1) in the presence of menadione (5 mM stock solution in acetonitrile) (22). NADH dehydrogenase activity was also determined by measuring the rate of DCIP (2,6-dichlorophenol-indophenol, sodium salt) reduction at 610 nm (ɛ610 = 16.5 mM−1 cm−1) in the presence of NADH (22). Type I NADH dehydrogenase activity (Nuo) was determined by measuring the rate of NHDH (reduced nicotinamide hypoxanthine dinucleotide) oxidation at 340 nm (22). Assays for succinate dehydrogenase activity were performed with disodium succinate (30 mM final concentration), measuring DCIP reduction. Since NADH dehydrogenase and succinate dehydrogenase are membrane-associated enzymes, assays were performed with membrane fractions in order to enrich these activities. Succinate dehydrogenase activity was measured to test for variation among the extracts prepared from the different strains. All strains showed similar succinate dehydrogenase activities. Malate dehydrogenase activity was measured in clarified cell extracts by following NADH oxidation in the presence and absence of oxaloacetic acid; activity was calculated as the oxaloacetate-dependent oxidation of NADH. KatG peroxidase activity was determined in clarified cell extracts by measuring the rate of o-dianisidine oxidation at 460 nm (ɛ460 = 11.3 mM−1 cm−1) in the presence of H2O2 (0.06%) (21). Protein concentrations were determined with the Pierce BCA protein assay. Extract preparation and assays were performed on the same day. Assays for each mutant strain were always performed in parallel with those for an INHs ndh+ control strain; the data presented are the averages for three repetitions of each assay.
Nucleotide sequence accession numbers.
The accession number for the ndh sequence of M. smegmatis is AF038423; the accession number for the mdh sequence of M. bovis is AF038422.
RESULTS
Isolation and phenotypic characterization of INHr Ts mutants.
We have taken a genetic approach that specifically identifies essential enzymes involved in INH action. Our initial hope was to identify Ts mutations in inhA in order to define the biochemical role of this enzyme in the growth of M. smegmatis. This problem was approached by isolating INHr mutants at 30°C (using INH at 25 μg/ml) and testing for those that were thermosensitive and hence unable to grow at 42°C (no INH added). These traits were expected to be caused by a mutation that confers resistance and also causes thermolability of the InhA enzyme. Since INHr Ts mutants were expected to be rare, resistant mutants were initially isolated from EMS-mutagenized cultures. Surprisingly, of 25 INHr mutants tested, 12 were temperature sensitive for growth. One of these Ts INHr mutants, the ndh-4 mutant, was further characterized (Table 3).
TABLE 3.
Growth phenotypes of the different INHr Ts mutants
| Allele | Mutant classa | INH susceptibility (MIC, μg/ml)b | Thermosensitivityc | % Viability after 42°C incubationc | Nutrient supplement requiredd | ETH susceptibility (diam of inhibition zone ± SD)e | Relative Ndh activityf |
|---|---|---|---|---|---|---|---|
| ndh+ | 5 | Tr | 100 | None | 4.3 ± 0.3 | 1 | |
| ndh-4 | I | >100 | Ts lethal | 4 | CAA | 0 | 0.04 |
| ndh-14 | I (0.4) | >100 | Ts lethal | 8 | CAA | 0.1 ± 0.3 | 0.04 |
| ndh-15 | II (0.4) | >100 | Ts | 40 | Ser or Gly | 0.4 | 0.18 |
| ndh-21 | III (0.2) | >100 | Ts | 40 | None | 2.7 ± 0.3 | 0.39 |
Mutants were isolated from the laboratory wild-type strain mc2155 by INHr selection at 30°C. The mutants were grouped according to their ability to grow on minimal medium. The number in parentheses is the fraction of spontaneous INHr Ts mutants that are of this class. Data for two class I mutants are presented to show that the phenotypes of the ndh-4 mutant are not an artifact of mutagenesis.
INH susceptibility was determined as the minimum concentration that reduced the number of CFU 100-fold (see Materials and Methods). Susceptibility was tested at the permissive temperature (30°C).
Thermosensitivity (Ts) was scored as the inability to grow at 42°C. The Ts lethal mutants could not survive 42°C incubation for 2.5 days, as indicated by a >90% reduction in viability after 42°C incubation. This viability, measured as the number of CFU that formed after transfer to the permissive temperature divided by the CFU of that culture, is the average of values from three different experiments.
Many of the mutants failed to grow on minimal medium plus glycerol or glucose at the permissive temperature. Some mutants required supplementation of Casamino Acids (CAA); others grew on minimal medium supplemented with serine or glycine.
ETH susceptibility was determined as the diameter of the zone (in millimeters) in which growth was inhibited by the drug; 500 μg was applied in a 10-μl volume.
Ndh activity is the rate of NADH oxidation with menadione as an electron acceptor (see the Fig. 3 legend and Materials and Methods).
The selections and screenings for INHr Ts mutants were repeated without mutagenesis. Spontaneous INHr (25 μg/ml) mutants arose at frequencies expected of single point mutations (10−6 to 10−7 per CFU). Of 66 different INHr mutants tested, 50% were temperature sensitive for growth at 42°C. Most of the Ts mutants did not have Ts lethal defects; they tolerated incubation at 42°C (2.5 days) and continued to grow after transfer to the permissive temperature (30°C) (Table 3). Four of the Ts mutants had lethal defects and could not survive the 42°C incubation (e.g., ndh-4 and ndh-14) (Table 3). All of the INHr Ts mutants were at least 20-fold more INH resistant than the parent strain (Table 3).
Most of the INHr Ts mutants (26 of 33) grew poorly at 30°C on minimal medium supplemented with glycerol or glucose. The INHr Ts mutants could be grouped into three classes on the basis of their nutrient requirements (Table 3). The first class grew poorly on all media and required Casamino Acid supplements for growth on minimal medium (40% of the Ts mutants; e.g., the ndh-4 and ndh-14 mutants). The second class grew well on rich medium and required serine or glycine for growth on minimal medium (40% of the Ts mutants; e.g., the ndh-15 mutant). The third class grew well on both minimal and rich media (e.g., the ndh-21 mutant). We noted that many of the Tr INHr mutants were also auxotrophs (Ser/Gly auxotrophs and general auxotrophs), but these mutants were not further characterized.
One possible explanation for the observed INH resistance is that the mutants have a general metabolic defect that interferes with active-transport systems. This type of defect should confer resistance to other drugs as well. The possibility of a general transport defect was excluded by the finding that four representative mutants were equally sensitive to both streptomycin and ethambutol compared to the INHs parent strain (data not shown).
Mutations in the inhA gene are correlated with coresistance to INH and ETH in M. tuberculosis and M. smegmatis, whereas mutations in katG are correlated only with INH resistance (3, 15). The INHr Ts mutants also exhibited low-level resistance to ETH, which was indicated by the inhibitory zone being relatively smaller than that of the INHs strain (Table 3).
The mutations conferring INHr and Ts are not in the inhA gene.
Linkage analysis was performed to determine whether the newly acquired INH resistance was due to mutations in or near the inhA gene (3). This study used a recombination substrate, cosmid pYUB325, which contains a 45-kb segment of the M. smegmatis chromosome that includes the inhA+ allele and a nearby Kmr-encoding insertion. When linearized pYUB325 is introduced into M. smegmatis, Kmr recombinants can form by exchange of the homologous sequences flanking both sides of the Kmr element. This recombination can occur with allelic exchange at the inhA gene to incorporate the inhA+ allele. A recipient strain with an inhA resistance mutation (strain mc2651) forms Kmr transformants with coinheritance of the inhA+ allele in 30 to 70% of the transformants; these recombinants are INHs. Two of the INHr Ts mutants (mc21390 and mc21407) were tested for linkage to inhA by linear transformation with pYUB325. None of the Kmr transformants acquired an INHs or Tr phenotype. (One hundred Kmr recombinants from each mutant strain were tested.) We infer that the mutations causing INHr and thermosensitivity are not located in or near the inhA gene.
The ndh genes of M. smegmatis and M. tuberculosis complement the phenotypes of the INHr Ts mutants.
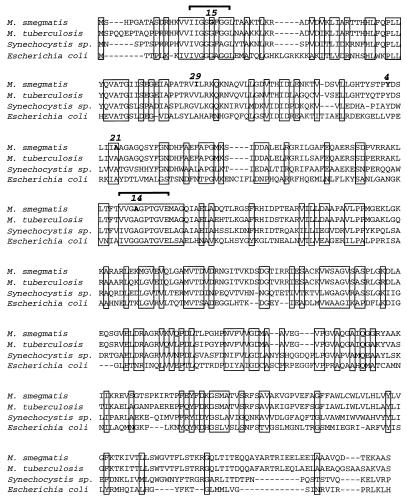
A 45-kb chromosomal fragment from the M. smegmatis genome that complements the Ts phenotype of one INHr mutant (ndh-4; mc21390) was identified from a large-insert genomic library (see Materials and Methods). The cosmids that conferred Tr growth also conferred INH and ETH sensitivity and prototrophic growth to the ndh-4 mutant (e.g., cosmid pYUB801). This complementation was not an artifact of gene overexpression, since the cosmid vector (pYUB412) integrates into the mycobacterial chromosome and the complementing gene is expressed from its own promoter. A 1.6-kb fragment that complements the ndh-4 mutant was isolated from cosmid pYUB801 (plasmid pYUB803). DNA sequence and BLAST analyses (1a) showed that the minimal plasmid pYUB803 has a 1.4-kb gene predicted to encode a 51-kDa protein similar to the type II NADH dehydrogenase of E. coli (Ndh; 13% identity and 56% similarity) and an Ndh homolog in a Synechocystis sp. (40% identity) (Fig. 1) (54).
FIG. 1.
Protein sequence alignment of the Ndh enzyme of E. coli and the putative homologs of M. smegmatis, M. tuberculosis, and a Synechocystis sp. The positions of amino acid substitutions of four different INHr ETHr Ts mutants are indicated with boldface print, and the corresponding mutant allele numbers are indicated above the substitution sites. Allele ndh-15 has a G→S substitution, ndh-4 has a Y→H substitution, ndh-21 has an A→T substitution, and ndh-14 has an A→S substitution. The ndh-29 mutation (I→F substitution) confers INHr but does not cause thermosensitivity; this mutant is described in the text. The first bracket indicates the putative FAD binding motif; the second bracket indicates the putative NAD binding motif (23). Conserved residues are boxed. The Ndh sequence of M. smegmatis is 81% identical to that of M. tuberculosis, 13% identical and 51% similar to the E. coli sequence, and 42% identical to the Synechocystis sp. sequence.
The ndh homolog from M. tuberculosis complements all of the ndh-4 mutant phenotypes (Fig. 2; Table 4). The M. tuberculosis ndh gene was identified on a 1.5-kb NheI-PstI fragment of cosmid MTCY356 and was subcloned into plasmid pMV261 (pYUB805) (6). The Ndh enzyme of M. tuberculosis is 81% identical to the M. smegmatis homolog. Eight Ts mutants representing the three different Ts mutant classes (the ndh-4, ndh-14, ndh-15, and ndh-21 mutants and four other mutants) were tested for complementation by the M. tuberculosis ndh gene; all of the phenotypes of each mutant were complemented.
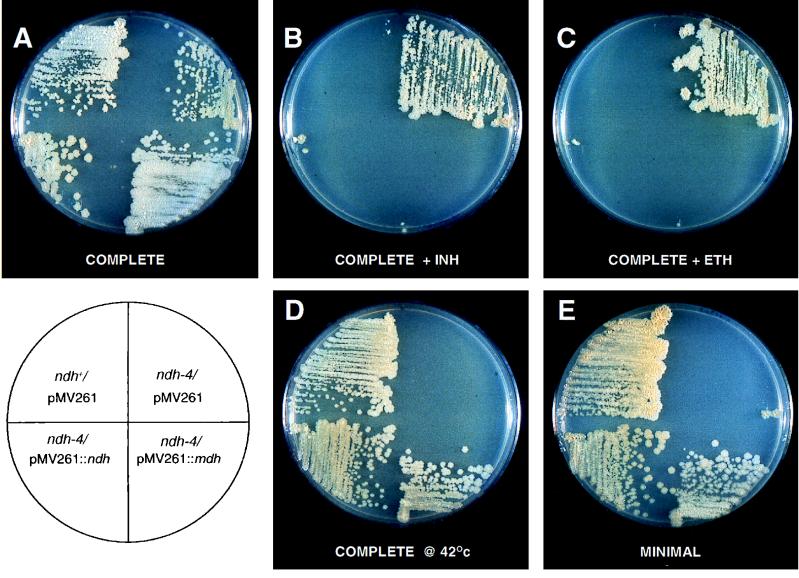
FIG. 2.
Phenotypes of one representative M. smegmatis INHr Ts mutant, the ndh-4 mutant, and complementation with the ndh and mdh genes. The ndh+ parent strain and the ndh-4 mutant shown here carry the E. coli-mycobacterial shuttle plasmid pMV261, which confers KmR. The lower half of each plate shows the ndh-4 mutant complemented by the ndh gene of M. tuberculosis (left; pYUB805) and the mdh gene of M. bovis BCG (right; pYUB808); these genes are expressed from the groEL promoter of plasmid pMV261. The plate in panel D was incubated at 42°C for 4 days, all other plates were incubated 6 days at 30°C. The concentration of both INH and ETH was 50 μg/ml, and all plates contained kanamycin.
TABLE 4.
Complementation of the ndh-4 mutant by minimal plasmids carrying the ndh or mdh genea
| Strain | Relevant genotype | Thermosensitivity | INH susceptibility (MIC, μg/ml) | ETH susceptibility (inhibitory zone diam ± SD) | Supplement required |
|---|---|---|---|---|---|
| mc21421 | ndh+/pMV261 | Tr | 5 | 3.5 ± 0.3 | None |
| mc21422 | ndh+/pMV261::ndh (M. tuberculosis) | Tr | 5 | 3.4 ± 0.1 | None |
| mc21423 | ndh+/pMV261::mdh (M. bovis BCG) | Tr | 1 | 4.6 ± 0.1 | None |
| mc21424 | ndh-4/pMV261 | Ts lethal | >100 | 0 | CAA |
| mc21425 | ndh-4/pMV261::ndh (M. tuberculosis) | Tr | 2.5 | 3.3 ± 0.3 | None |
| mc21426 | ndh-4/pMV261::mdh (M. tuberculosis complex) | Tr | 1 | 5.8 ± 0.1 | None |
All strains were derived from the ndh+ parent strain mc2155. Plasmid pYUB805 is pMV261::ndh (M. tuberculosis); plasmid pYUB808 is pMV261::mdh (M. tuberculosis complex). See footnotes to Table 3 for explanations of data categories.
Identification of mutations in the ndh gene.
The ndh gene was PCR amplified from the chromosomes of four INHr ETHr Ts mutants (the ndh-4, ndh-14, ndh-15, and ndh-21 mutants). Sequence analysis identified a different missense mutation in each mutant, and two of the mutants, the ndh-14 and ndh-15 mutants, had substitutions in the highly conserved NAD and flavin adenine dinucleotide (FAD) binding domains (Fig. 1) (23).
A second gene, mdh, complements the ndh mutant phenotypes.
An M. bovis BCG genomic library was used to identify genes of the M. tuberculosis complex that complement the ndh-4 mutant. (The M. tuberculosis complex includes M. tuberculosis, M. bovis, other closely related virulent species, and the avirulent M. bovis BCG vaccine strain.) Surprisingly, a different complementing gene was identified. Subcloning and sequence analyses identified a gene encoding a 36-kDa protein that is 60% identical to the T. aquaticus malate dehydrogenase enzyme (Mdh; EC 1.1.1.37) (32). Mdh catalyzes the NADH-dependent interconversion of oxaloacetate and malate, a reaction of the tricarboxylic acid (TCA) cycle (35).
A minimal 1.1-kb fragment containing mdh fully complements all of the ndh-4 mutant phenotypes when expressed from the pMV261 vector (pYUB808) (Fig. 2 and Table 4). Furthermore, in both the ndh-4 mutant and the parent strain, mc2155, mdh expression causes hypersensitivity to INH and ETH (Table 4). Eight Ts mutants representing the three different Ts mutant classes (the ndh-4, ndh-14, ndh-15, and ndh-21 mutants and four other mutants) were tested for complementation by mdh; all of the phenotypes of each mutant were complemented. The mdh gene was PCR amplified from M. tuberculosis H37Rv genomic DNA, and the nucleotide sequence was found to be 100% identical to the M. bovis BCG sequence. (PCR amplification and sequencing were performed twice to verify this identity.)
The INHr Ts mutants are defective for NADH dehydrogenase activity.
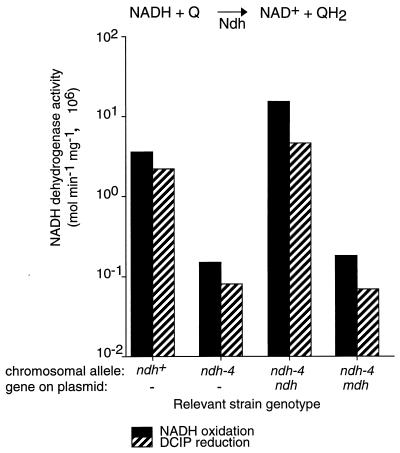
The Ndh enzyme of E. coli catalyzes the first step in the respiratory chain: NADH oxidation and quinone reduction (54). Based on sequence similarity, we predicted that the mycobacterial homolog would have the same activity and that the mutants would be defective. Two different assays were used to measure NADH dehydrogenase activity: oxidation of NADH in the presence of a quinone (menadione), and the NADH-dependent reduction of an electron acceptor (DCIP) (Fig. 3). These assays were performed with membrane fractions because the Ndh enzyme of E. coli is a peripheral membrane protein.
FIG. 3.
Comparison of NADH dehydrogenase activities in the INHs parent strain, the INHr ndh-4 mutant, and the ndh-4 mutant complemented by the ndh and mdh genes. For each strain, the chromosomal allele and the plasmid-borne gene are indicated below the horizontal axis. The rate of NADH oxidation was measured with menadione as an electron acceptor (solid black bars). The rate of NADH-dependent DCIP reduction is represented by bars with diagonal stripes. Cultures were grown at 30°C, and assays were performed at room temperature. Membrane fractions were used for these assays; reaction conditions are described in Materials and Methods. The error (standard deviation) is less than 10% for all data points and is not shown.
NADH dehydrogenase activity was measured in the ndh+ control, the ndh-4 mutant, and strains with the ndh-4 mutation and the complementing plasmids (Fig. 3). The ndh-4 mutant exhibited reduced NADH dehydrogenase activity relative to the ndh+ control in both assays (25-fold reduction), and the complementing M. tuberculosis ndh gene restored activity to the ndh-4 mutant (plasmid pYUB805). NADH dehydrogenase activity is specific for NADH; no oxidation of deamino-NADH (reduced nicotinamide hypoxanthine dinucleotide) or NADPH was detected in our membrane fractions. The complementing mdh gene did not restore NADH dehydrogenase activity to the ndh-4 mutant (pYUB808).
Representatives of the three INHr Ts mutant classes were tested for Ndh defects by measuring NADH oxidation (Table 3). The defects in Ndh activity correlated with the mutants’ growth defects. The largest Ndh defect (25-fold reduction) was observed among mutants (the ndh-4 and ndh-14 mutants) with the most severe growth defects: Ts lethality, general auxotrophy, and poor growth under permissive conditions. A Ser-Gly auxotroph (the ndh-15 mutant) exhibited a sixfold reduction in Ndh activity relative to the ndh+ control, and the prototroph (the ndh-21 mutant) showed a threefold reduction. Data for five representative mutants are presented; however, the tests were performed on at least two different mutants of each phenotypic class, and the results were consistent with those shown in Table 3. Ndh activity was also measured by the NADH-dependent DCIP reduction assay, and the results were similar to those of the NADH oxidation assay.
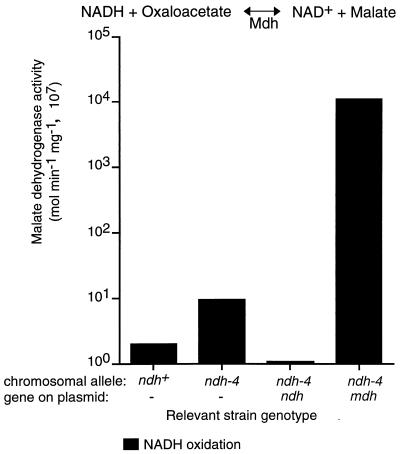
Mdh is highly active in oxidation of NADH.
Expression of the M. tuberculosis complex Mdh enzyme corrected the phenotypes of the ndh mutants and conferred hypersensitivity to INH and ETH. The only common function shared by Mdh and Ndh is oxidation of NADH. We therefore reasoned that the Mdh enzyme might complement the mutant defects by oxidizing NADH with oxaloacetate as an electron acceptor. Mdh activity was determined by measuring oxaloacetate-dependent NADH oxidation with clarified whole-cell extracts of the ndh-4 mutant and strains with the complementing plasmids. The ndh+ strains and the ndh-4 mutant exhibited extremely low Mdh activities (Fig. 4); the Mdh activity of the ndh-4 mutant was about fivefold higher than those of the ndh+ strains. Low Mdh activity was expected since previous studies did not detect this activity in M. smegmatis; this species possesses a different malate oxidation system, one that uses the FAD-dependent malate-vitamin K reductase (35). Expression of the M. tuberculosis complex mdh gene greatly increased NADH oxidation only when oxaloacetate was used as an electron acceptor (1,000-fold increase).
FIG. 4.
Comparison of Mdh activity of the INHs parent strain, the INHr ndh-4 mutant, and the ndh-4 mutant complemented by the ndh and mdh genes. For each strain, the chromosomal allele and the plasmid-borne gene are indicated below the horizontal axis. These assays measure oxaloacetate-dependent NADH oxidation in clarified cell extracts. Oxaloacetate-independent NADH oxidation (background) is subtracted from these data. Assay conditions are described in the legend to Fig. 3 and in Materials and Methods.
ndh mutants are frequently recovered from INHr selections at 37°C.
It is surprising that Ndh defects are found in such a large fraction of INHr mutants isolated at 30°C. Perhaps the reduced growth temperature favors the recovery of ndh mutants or limits the recovery of inhA and katG mutants. The selection for INH resistance was repeated at 37°C to determine whether the selection temperature biases the recovery of mutants. Twenty INHr mutants were isolated from 10 different cultures of strain mc2155. A plasmid with the M. smegmatis ndh gene (pYUB802) was introduced into each of the INHr mutants, and the transformants were tested for INH sensitivity. Expression of the ndh gene restored sensitivity to 50% of the INHr mutants isolated at 37°C. Six of these were tested for complementation by mdh of the M. tuberculosis complex; all were complemented (pYUB808). Unlike the mutants isolated at 30°C, most (70%) of these putative ndh mutants isolated at 37°C grew well on minimal medium and were thermoresistant. Assays of NADH dehydrogenase activity of one Tr prototrophic mutant, ndh-29, showed a small Ndh defect (3.7-fold reduction). Sequence analysis identified a missense mutation in the ndh-29 mutant (Fig. 1). We infer that Ndh defects confer INHr in about 50% of mutants isolated at 37°C and that this resistance is not typically associated with growth defects.
Resistance of the ndh mutants is not due to lower-level KatG expression.
One possible explanation for the mutants’ resistance is that the Ndh defect reduces katG expression by decreasing the amount of H2O2 produced from the respiratory chain. Lower katG expression has been observed in ndh mutants of E. coli (14). KatG expression in different ndh mutant strains was measured by determining the amount of peroxidase activity.
KatG expression was lower in some ndh mutants; however, KatG activity did not correlate with INH susceptibility. Mutants that have small Ndh defects had normal peroxidase activity (e.g., the ndh-21 and ndh-29 mutants) (Table 5). These mutants were resistant to INH (MICs, >100 μg/ml). Reduced peroxidase activity was observed in mutants with larger Ndh defects (the ndh-4, ndh-14, and ndh-15 mutants), and complementation with the ndh+ gene restored peroxidase activity (Table 5). However, Mdh expression did not restore peroxidase activity to the ndh-4 mutant but conferred hypersensitivity to INH and ETH.
TABLE 5.
KatG peroxidase activity of the ndh mutant strains
| Relevant genotypea | INH resistance (MIC, μg/ml)b | KatG peroxidase activityc |
|---|---|---|
| ndh+ | 5 | 1 |
| ndh-14 | >100 | 0.5 |
| ndh-15 | >100 | 0.7 |
| ndh-21 | >100 | 0.9 |
| ndh-29 | >100 | 1.4 |
| ndh-4/pMV261 | >100 | 0.14 |
| ndh-4/pMV261::ndh (M. tuberculosis) | 2.5 | 1.1 |
| ndh-4/pMV261::mdh (M. bovis BCG) | 1 | 0.4 |
All strains are derived from the INHs parent strain mc2155 (Table 1).
KatG activity was measured as the rate of o-dianisidine oxidation in the presence of H2O2 as described in Materials and Methods. All assays were repeated three times with the same extracts; in all cases, error was less than 10%. All activities are expressed relative to that of the ndh+ control, strain mc2155 or mc21421; the assays were performed in parallel.
DISCUSSION
Mutants that are defective for NADH dehydrogenase (Ndh; type II) are frequently found among spontaneous INH-resistant mutants of M. smegmatis. These mutations often confer low-level resistance to the structurally related drug ETH. Many ndh mutants have multiple growth phenotypes, including temperature sensitivity and the inability to grow on minimal medium supplemented with glycerol or glucose. Expression of the malate dehydrogenase enzyme (Mdh) of the M. tuberculosis complex restored thermoresistance and prototrophic growth to the ndh mutants and also caused increased sensitivity to INH and ETH (Table 4).
We propose that all of the ndh mutant phenotypes, INHr, ETHr, Ts, and auxotrophy, are due to defects in NADH oxidation which result in NADH accumulation and NAD+ depletion. The increased NADH concentration may inhibit reactions that are sensitive to this nucleotide; a lower NAD+ concentration may reduce the rates of NAD+-dependent reactions. These two factors may impair operation of the TCA cycle and/or inhibit biosynthetic pathways; they also could explain why the most defective Ndh mutants cannot grow on minimal medium. Biochemical studies of E. coli have shown that NADH is a potent inhibitor of the first committed reaction in the biosynthetic pathway for serine and glycine, which would explain the specific serine or glycine requirement of many ndh mutants (45). Expression of the Mdh enzyme would correct the mutant phenotypes by oxidizing NADH and reducing oxaloacetate—the energetically favorable reaction. This Mdh activity is robust in strains expressing Mdh; these strains show increased sensitivity to INH and ETH (Fig. 4; Table 4).
The thermosensitive lethality of many ndh mutations indicates that Ndh is the primary enzyme responsible for NADH oxidation in M. smegmatis and is essential for viability. The thermosensitive lethality of some ndh mutations may be due to a severe imbalance in the NADH/NAD+ ratio at the nonpermissive temperature that is caused by temperature-sensitive Ndh activity. Oxidation of NADH produced via glycolysis and the TCA cycle is essential for maintenance of metabolic flux. However, the essentiality of Ndh is surprising because most organisms have multiple enzymes and pathways for oxidizing NADH. E. coli has two different NADH dehydrogenase enzymes (Nuo and Ndh) and can oxidize NADH by fermentation (8, 12). An E. coli ndh nuo double mutant has an impaired ability to oxidize NADH and grows poorly on minimal medium supplemented with glucose, presumably because of an increased NADH/NAD+ ratio (53). From sequence analysis, it appears that M. tuberculosis has genes encoding the Nuo system (Sanger database; cosmid SCY03A2). We believe that the Nuo system is not active in M. smegmatis, since some ndh mutants have very low NADH dehydrogenase activity (∼25-fold reduction). These mutants arise spontaneously at a high frequency (10−6 to 10−7 per CFU), so the defect is not likely to be caused by multiple mutations. Furthermore, assays for Nuo activity (measuring oxidation of deamino-NADH, a Nuo-specific substrate) in our wild-type strain were negative. It is possible that M. smegmatis has genes encoding the Nuo system and that nuo expression is repressed under the conditions of high aeration and rich medium used in our experiments. Oxygen tension influences nuo and ndh expression in E. coli (7, 43).
Defects in Ndh activity cause resistance to INH and ETH in M. smegmatis. Since the ndh mutations that confer resistance are always recessive and occur in conjunction with reduced Ndh activity, it seems unlikely that Ndh is an INH target (as proposed in reference 11). Our genetic data indicate that Ndh potentiates the action of INH and ETH by oxidizing NADH and that an increase in the NADH/NAD+ ratio confers resistance. Recent structural studies showed that INH reacts with the NADH located within the active site of the InhA enzyme (39). Enzymatic studies show that NADH is required for INH to inhibit the InhA enzyme and that mutations which reduce the affinity for NADH confer resistance (5, 19, 36). Therefore, it is surprising that mutations which increase the NADH/NAD+ ratio confer resistance. We propose that an increase in the NADH concentration prevents the action of INH and ETH by two different mechanisms which may act in conjunction to confer high-level resistance. First, NADH may displace the active drug from the InhA enzyme, such that an increased NADH concentration may prevent target inhibition. An increased NADH concentration may also prevent peroxidation reactions that are required to activate the drugs. NADH is a substrate for peroxidases such as AhpCF and the KatG catalase peroxidase (21, 34). Increased amounts of NADH may drive peroxide reduction, thereby preventing KatG-mediated peroxidation of INH. NADH may also competitively inhibit peroxidation of INH by KatG.
Mutants with thermosensitive defects in the Ndh enzyme were the most frequent class of INHr mutants recovered from selections performed at 30°C. One expects that katG mutations would be recovered more frequently than ndh mutations because any katG deletion or point mutation that eliminates or reduces activity should confer INH resistance. In contrast, Ndh is essential, so ndh mutations which eliminate activity would be forbidden. The paucity of katG deletion mutants recovered in our selections is enigmatic and may indicate that INH can be activated by multiple enzymes in M. smegmatis. Another unexpected finding is that a large proportion of the ndh mutants isolated at 30°C have a thermosensitive phenotype. This may be due to properties of Ndh, which is a peripheral membrane enzyme. The high frequency of ndh Ts mutants has complicated the isolation of the inhA Ts mutants, which are expected to be more rare.
The genetic data presented here indicate that NADH oxidation systems influence INH sensitivity. While others have suggested that the exquisite INH sensitivity of M. tuberculosis is due to a species-specific target (24), we believe that metabolic functions such as differences in the KatG enzyme, alkyl hydroperoxide reductase expression, peroxide concentrations, and the NADH oxidation systems could fully explain the different sensitivities of M. tuberculosis and M. smegmatis. Results reported in the early literature of Middlebrook and Cohn suggest that INH resistance in M. tuberculosis can occur via metabolic defects that may be similar to the ndh defects found in M. smegmatis (26). These investigators attempted to culture INH-resistant tubercle bacilli from patients treated with INH monotherapy and found that 8 of 21 patients produced tubercle bacilli exhibiting growth defects. Of these eight isolates, five were auxotrophs and three were not culturable in rich or minimal medium. Perhaps these resistant tubercle bacilli had NADH oxidation defects that prevented their growth on certain types of media.
The isolation of drug-resistant mutants that are thermosensitive for growth provides a means to identify essential enzymes involved in drug action. These types of enzymes may be useful for the development of new antimicrobial agents. This approach should work with any microorganism that is amenable to genetic analysis and should apply to any antimicrobial agent. Our studies unexpectedly found that the ndh-encoded NADH dehydrogenase is essential for the viability of M. smegmatis and that this enzyme normally promotes INH action by oxidizing NADH.
ACKNOWLEDGMENTS
We thank Paras Doshi for technical assistance and Franz Bange, Jordan Kriakov, Stewart Cole, and Stoyan Bardorov for providing cosmids and cosmid libraries. L.M. is very grateful to Stoyan Bardorov, John Blanchard, Miriam Braunstein, Jeffrey Cox, Marty Pavelka, James C. Sacchettini, and Howard Steinman for intellectual contributions to this work and for critically reviewing the manuscript.
R.B. was supported by a presidential award from the Queens College of the City University of New York. This work was supported by NIH grant AI36849.
REFERENCES
- 1.Ad Hoc Committee of the Scientific Assembly on Microbiology, Tuberculosis, and Pulmonary Infections. Treatment of tuberculosis and tuberculosis infection in adults and children. Clin Infect Dis. 1995;21:9–27. [Google Scholar]
- 1a.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A, Dubnau E, Quémard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R., Jr inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 4.Bardorov, S. Unpublished data.
- 5.Basso L A, Zheng R, Blanchard J S. Kinetics of inactivation of WT and C243S mutant of Mycobacterium tuberculosis enoyl reductase by activated isoniazid. J Am Chem Soc. 1996;118:11301–11302. [Google Scholar]
- 6.Bergh S, Cole S T. MycDB: an integrated mycobacterial database. Mol Microbiol. 1994;12:517–534. doi: 10.1111/j.1365-2958.1994.tb01039.x. [DOI] [PubMed] [Google Scholar]
- 7.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-nuoN) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 8.Calhoun M W, Gennis R B. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol. 1993;175:3013–3019. doi: 10.1128/jb.175.10.3013-3019.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo J D, Weisbrod T R, Jacobs W R., Jr . Efficient electrotransformation of Mycobacterium smegmatis. Technical bulletin no. 1360. Richmond, Calif: Bio-Rad Laboratories; 1993. [Google Scholar]
- 10.Cohn, D. L., F. Bustreo, and M. C. Raviglione. 1997. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD global surveillance project. Clin. Infect. Dis. 24(Suppl. 1):S121–S130. [DOI] [PubMed]
- 11.Davis W B, Weber M M. Specificity of isoniazid on growth inhibition and competition for an oxidized nicotinamide adenine dinucleotide regulatory site on the electron transport pathway in Mycobacterium phlei. Antimicrob Agents Chemother. 1977;12:213–218. doi: 10.1128/aac.12.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gennis R B, Stewart V. Respiration. In: Niedhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 13.Gillespie J, Barton L L, Rypka E W. Influence of oxygen tension on the respiratory activity of Mycobacterium phlei. J Gen Microbiol. 1988;134:247–252. doi: 10.1099/00221287-134-1-247. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez B F, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 15.Heym B, Honoré N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D A, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 16.Heym B, Stavropoulos E, Honoré N, Domenech P, Saint-Joanis B, Wilson T M, Collins D M, Colston M J, Cole S T. Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect Immun. 1997;65:1395–1401. doi: 10.1128/iai.65.4.1395-1401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Udani R A, Jones W D, Jr, Barletta R, Bloom B R. Genetic systems for the mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 18.Johnsson K, Schultz P G. Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J Am Chem Soc. 1994;116:7425–7426. [Google Scholar]
- 19.Johnsson K, King D S, Shultz P G. Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J Am Chem Soc. 1995;117:5009–5010. [Google Scholar]
- 20.Magliozzo R S, Marcinkeviciene J A. Evidence for isoniazid oxidation by oxyferrous mycobacterial catalase-peroxidase. J Am Chem Soc. 1996;118:11303–11304. [Google Scholar]
- 21.Marcinkeviciene J A, Magliozzo R S, Blanchard J S. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J Biol Chem. 1995;270:22290–22295. doi: 10.1074/jbc.270.38.22290. [DOI] [PubMed] [Google Scholar]
- 22.Matsushita K, Ohnishi T, Kaback H R. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- 23.McKie J H, Douglas K T. Evidence for gene duplication forming similar binding folds for NAD(P)H and FAD in pyridine nucleotide-dependent flavoenzymes. FEBS Lett. 1991;279:5–8. doi: 10.1016/0014-5793(91)80236-v. [DOI] [PubMed] [Google Scholar]
- 24.Mdluli K, Sherman D, Hickey M J, Kreiswirth B N, Morris S, Stover C K, Barry C E., III Biochemical and genetic data suggest that InhA is not a primary target for activated isoniazid in Mycobacterium tuberculosis. J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 25.Middlebrook G. Sterilization of tubercle bacilli by isonicotinic acid hydrazide and the incidence of variants resistant to the drug in vitro. Am Rev Tuberc. 1952;65:765–767. [PubMed] [Google Scholar]
- 26.Middlebrook G, Cohn M L. Some observations on the pathogenicity of isoniazid-resistant variants of tubercle bacilli. Science. 1953;118:297–299. doi: 10.1126/science.118.3063.297. [DOI] [PubMed] [Google Scholar]
- 27.Middlebrook G. Isoniazid resistance and catalase activity of tubercle bacilli. Am Rev Tuberc. 1954;69:471–472. doi: 10.1164/art.1954.69.3.471. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 29.Morris S, Bai G H, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 30.Musser, J. Unpublished data.
- 31.Musser J M, Kapur V, Williams D L, Kreiswirth B N, van Soolingen D, van Embden J D A. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 32.Nicholls D J, Sundaram T K, Atkinson T, Minton N P. Cloning and nucleotide sequences of the mdh and sucD genes from Thermus aquaticus B. FEMS Microbiol Lett. 1990;70:7–14. doi: 10.1016/0378-1097(90)90093-6. [DOI] [PubMed] [Google Scholar]
- 33.Pavelka M S, Jr, Jacobs W R., Jr Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole L B, Ellis H R. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 35.Prasada Reddy T L, Murthy R S, Venkitasubramanian T A. Variations in the pathways of malate oxidation and phosphorylation in different species of mycobacteria. Biochim Biophys Acta. 1975;376:210–218. doi: 10.1016/0005-2728(75)90012-2. [DOI] [PubMed] [Google Scholar]
- 36.Quémard A, Sacchettini J C, Dessen A, Vilcheze C, Bittman R, Jacobs W R, Jr, Blanchard J S. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 37.Robitzek E H, Selikoff I J. Hydrazine derivative of isonicotinic acid (rimifon, marsilid) in the treatment of active progressive caseous-pneumonic tuberculosis. Am Rev Tuberc. 1952;65:402–428. doi: 10.1164/art.1952.65.4.402. [DOI] [PubMed] [Google Scholar]
- 38.Rosner J L, Storz G. Effects of peroxides on susceptibilities of Escherichia coli and Mycobacterium smegmatis to isoniazid. Antimicrob Agents Chemother. 1994;38:1829–1833. doi: 10.1128/aac.38.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozwarski D A, Grant G A, Barton D H R, Jacobs W R, Jr, Sacchettini J C. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 40.Sherman D R, Mdluli K, Hickey M J, Arain T M, Morris S L, Barry III C E, Stover C K. Compensatory ahpC gene expression in isoniazid resistant Mycobacterium tuberculosis. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 41.Shoeb H A, Bowman B U, Jr, Ottolenghi A C, Merola A J. Peroxidase-mediated oxidation of isoniazid. Antimicrob Agents Chemother. 1985;27:399–403. doi: 10.1128/aac.27.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 43.Spiro S, Roberts R E, Guest J R. FNR-dependent repression of the ndh gene of Escherichia coli and metal ion requirement for FNR-regulated gene expression. Mol Microbiol. 1989;3:601–608. doi: 10.1111/j.1365-2958.1989.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 44.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Bennett L A L T, Bansal G P, Young J F, Lee M H, Hatfull G F, Snapper S B, Barletta R G, Jacobs W R, Jr, Bloom B R. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 45.Sugimoto E, Pizer L I. The mechanism of end product inhibition of serine biosynthesis. J Biol Chem. 1968;243:2081–2089. [PubMed] [Google Scholar]
- 46.Szybalski W, Bryson V. Bacterial resistance studies with derivatives of isonicotinic acid. Am Rev Tuberc. 1952;65:768–770. [PubMed] [Google Scholar]
- 47.Takayama K, Wang L, David H L. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1972;2:29–35. doi: 10.1128/aac.2.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takayama K, Schnoes H K, Armstrong E L, Boyle R W. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J Lipid Res. 1975;16:308–317. [PubMed] [Google Scholar]
- 49.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson T M, Collins D M. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol Microbiol. 1996;19:1025–1034. doi: 10.1046/j.1365-2958.1996.449980.x. [DOI] [PubMed] [Google Scholar]
- 51.Winder F. Catalase and peroxidase in mycobacteria. Am Rev Respir Dis. 1960;81:68–78. doi: 10.1164/arrd.1960.81.1P1.68. [DOI] [PubMed] [Google Scholar]
- 52.Winder F G. Mode of action of the antimycobacterial agents and associated aspects of the molecular biology of the mycobacteria. In: Ratledge C, Stanford J, editors. The biology of the mycobacteria. New York, N.Y: Academic Press; 1982. pp. 353–438. [Google Scholar]
- 53.Young I G, Wallace B J. Mutations affecting the reduced nicotinamide adenine dinucleotide dehydrogenase complex of Escherichia coli. Biochim Biophys Acta. 1976;449:376–385. doi: 10.1016/0005-2728(76)90149-3. [DOI] [PubMed] [Google Scholar]
- 54.Young I G, Jaworowski A, Poulis M I. Amplification of the respiratory NADH dehydrogenase of Escherichia coli by gene cloning. Gene. 1978;4:25–36. doi: 10.1016/0378-1119(78)90012-4. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Dhandayuthapani S, Deretic V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]