Abstract
Factor XIII (FXIII) catalyzes formation of γ-glutamyl-ε-lysyl crosslinks between reactive glutamines (Q) and lysines (K). In plasma, FXIII is activated proteolytically (FXIII-A*) by the concerted action of thrombin and Ca2+. Cellular FXIII is activated non-proteolytically (FXIII-A°) by elevation of physiological Ca2+ concentrations. FXIII-A targets plasmatic and cellular substrates, but questions remain on correlating FXIII activation, resultant conformational changes, and crosslinking function to different physiological substrates. To address these issues, the characteristics of FXIII-A* versus FXIII-A° that contribute to transglutaminase activity and substrate specificities were investigated. Crosslinking of lysine mimics into a series of Q-containing substrates were measured using in-gel fluorescence, mass spectrometry, and UV-Vis spectroscopy. Covalent incorporation of fluorescent monodansylcadaverine revealed that FXIII-A* exhibits greater activity than FXIII-A° toward Q residues within Fbg αC (233–425 WT, Q328P Seoul II, and Q328PQ366N) and actin. FXIII-A* and FXIII-A° displayed similar activities toward α2-antiplasmin, fibronectin, and Fbg αC (233–388, missing FXIII binding site αC 389–402). Furthermore, the N-terminal α2-AP peptide (1–15) exhibited similar kinetic properties for FXIII-A* and FXIII-A°. MALDI-TOF mass spectrometry assays with glycine ethyl ester and Fbg αC (233–425 WT, αC E396A, and truncated αC (233–388) further documented that FXIII-A* exerts greater benefit from the αC 389–402 binding site than FXIII-A°. Conformational properties of FXIII-A* versus A° are proposed to help promote transglutaminase function toward different substrates. A combination of protein substrate disorder and secondary FXIII binding site exposure are utilized to control activity and specificity. From these studies, greater understandings of how FXIII-A targets different substrates are being achieved.
Keywords: antiplasmin, Factor XIII, fibrinogen, fibronectin, actin
Graphical Abstract
INTRODUCTION
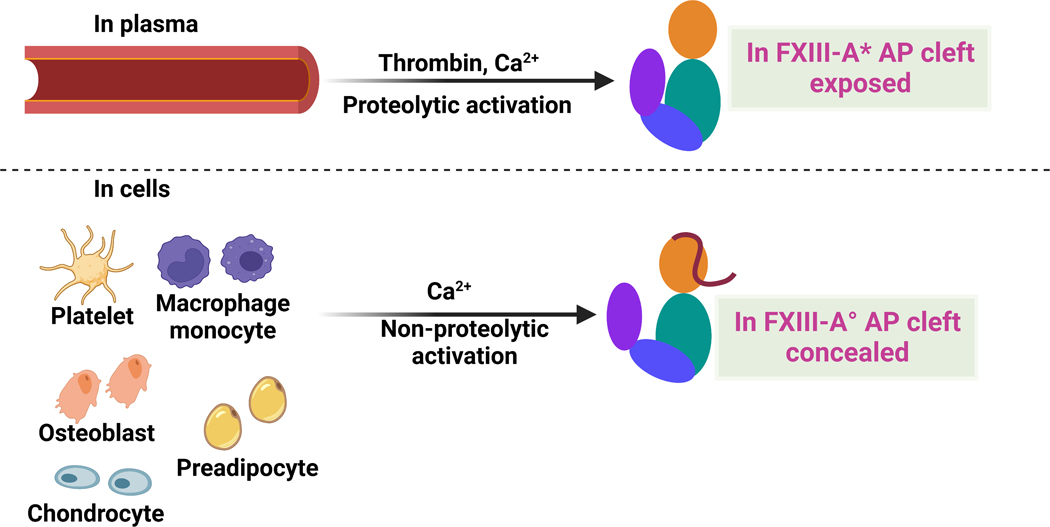
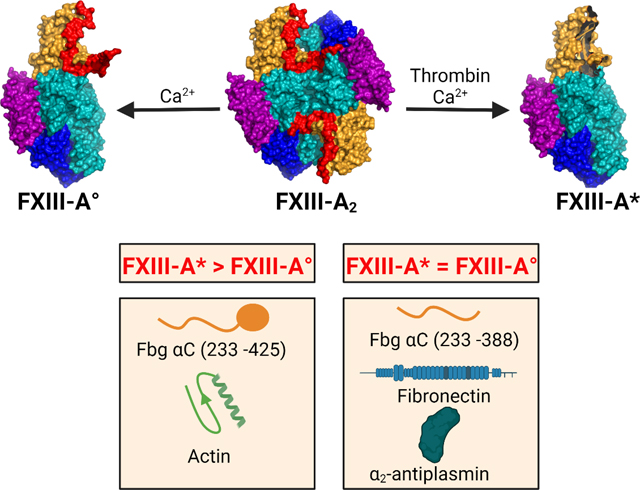
Factor XIII (FXIII), a member of the transglutaminase family, catalyzes the formation of γ-glutamyl-ε-lysyl peptide bonds between protein substrates.1–3 FXIII has been associated with blood coagulation, deep vein thrombosis, venous thromboembolism, wound healing, bone formation, arthritis, etc.2, 4–6 FXIII is found in the extracellular plasma and a series of intracellular environments (Fig. 1).
Fig. 1. Cartoon models highlighting FXIII locations and activation strategies.
pFXIII undergoes proteolytic activation by thrombin to form active FXIII-A* in which AP is removed and AP cleft is exposed. cFXIII is expressed in a variety of cells including platelets, monocytes, macrophages, chondrocytes, osteoblasts and preadipocytes. cFXIII undergoes nonproteolytic activation to form FXIII-A° in which AP is still associated with A subunit. In resting platelets and monocytes, FXIII is of cytoplasmic localization, however upon activation of these cells, FXIII-A is exposed on the surface of these cells. Cartoon model is created with BioRender.com.
Plasma FXIII (pFXIII) circulates as a FXIII-A2B2 zymogen composed of two protransglutaminase A subunits and two carrier B subunits.3, 7 pFXIII is activated proteolytically to generate FXIII-A* by thrombin cleavage and release of an activation peptide (AP) followed by Ca2+-induced conformational rearrangements to dissociate the B subunits.3, 7 Cellular FXIII (cFXIII) is a dimer of A subunits, FXIII-A2, and is found in platelets, monocytes, macrophages, chondrocytes, preadipocytes, bone osteoblasts, etc.5 cFXIII is non-proteolytically activated by elevated intracellular Ca2+ concentrations in a NaCl/KCl environment to form FXIII-A° (Fig. 1).8 In vitro, FXIII-A2 can also be activated by thrombin in the presence of Ca2+.9 Regardless of the FXIII source, the FXIII-A2 is converted to the active transglutaminase.
FXIII has a spectrum of plasmatic and cellular substrates and is involved in multiple physiological functions.1, 2, 5 FXIII-A* crosslinks fibrin γ and α chains making fibrin clots mechanically stable.10, 11 The fibrin α-α crosslinks are instrumental in increasing clot size by mediating red blood cell retention.12 FXIII-A* also crosslinks proteins such as α2-antiplasmin (α2-AP), and fibronectin (FN) to the fibrin αC region, providing anti-fibrinolytic and wound healing properties to the clot.13–15 cFXIII is of cytoplasmic localization; however, agonist stimulation of platelets and monocytes leads to translocation of cytoplasmic active FXIII-A° to the membrane.16–18 This FXIII-A° has the potential to target the thrombus by crosslinking fibrin and α2-AP proteins.16, 17 Additionally, FXIII-A° is involved in crosslinking cytoskeletal proteins like actin, filamin, and vinculin upon platelet activation.1, 2 Furthermore, FXIII- A in osteoblasts and osteoclasts was identified to regulate resorption, adipogenesis, and fibronectin homeostasis in bone and bone marrow.19
The fibrinogen (Fbg) αC region is composed of a disordered αC connector (Aα 221 – 391) and a globular, more structured αC domain (392 – 610).20 Crosslinks within the αC region lead to stiffer clots that are resistant to fibrinolysis.21 In vitro, Fbg variants with truncations at the αC 390 and αC 220 positions led to clots with reduced mechanical and fibrinolytic stability.22, 23 Fbg270/270 mice with truncation at Aα271 exhibited hypofibrinogenemia with protection from thrombosis.23 Reactive Fbg αC glutamines Q237, Q328, and Q366 are located within the predominantly unstructured αC connector (221–391). Mass spectrometric crosslinking studies of recombinant Fbg αC (233–425) and insoluble fibrin clots identified and ranked αC glutamine reactivities as Q237 >> Q328 ≈ Q366.24, 25 FXIII-A* in the presence of Ca2+ has been reported to bind to Fbg αC (389–402), with Fbg αC E396 serving as an important anchoring site.26–28 Thus, Fbg αC (233–425) provides an effective substrate model for FXIII-A.
Analytical ultracentrifugation (AUC) experiments demonstrated that FXIII-A* is more conformationally flexible than FXIII-A°.29, 30 Moreover, crosslinking assays suggested that FXIII-A* could exhibit greater transglutaminase activity than FXIII-A°.30 Although FXIII-A*/ A° substrates and their reactive Q and K are known, questions remain on correlating FXIII activation, resultant conformational changes, and crosslinking function to different physiological substrates. The aim of the current study was to critically evaluate the abilities of FXIII-A* versus FXIII-A° to crosslink a series of substrates having diverse functions. Herein, we used in-gel fluorescence, UV-Vis spectroscopic, and mass spectrometric techniques to determine the crosslinking of lysine mimics to Fbg αC (233 – 425) and its variants, α2-AP, FN, and actin. Collectively, our data suggest that transglutaminase activities of FXIII-A* and A° may be regulated by FXIII conformational features, individual substrates, and presence of binding sites.
MATERIALS AND METHODS
Materials
Recombinant human FXIII-A2 was obtained from the late Dr. Paul Bishop (Zymogenetics, Seattle, WA), and recombinant human thrombin from E. Di Cera and L. Pelc (St. Louis University, MO). The activity of the recombinant human thrombin was determined using titration against hirudin.31 Human plasma fibronectin and Phe-Pro-Arg-chloromethyl ketone (PPACK) were supplied by Haematologic Technologies (Essex Junction, VT, USA). Bovine thrombin, human α2-antiplasmin, actin, ferulic acid, α-cyano-4-hydroxycinnamic acid (α-CHCA), N,N-Dimethyl-p-phenylenediamine (DMPDA), monodansylcadaverine (MDC), and Glycine ethyl ester (GEE) hydrochloride were obtained from Millipore Sigma (St. Louis, MO, USA). Peptides α2AP WT (1 – 15) and α2AP Q4S (1 – 15) were synthesized by New England Peptide (Gardner, MA, USA). Peptide stock concentrations were determined by quantitative amino acid analysis (AAA Service Laboratory, Damascus, OR, USA). Sequencing grade chymotrypsin and GluC endoproteinase, both from Roche, were supplied by Millipore Sigma (St. Louis, MO, USA). 30% Acrylamide/Bis solution, 4 – 20 % precast tris-glycine gels, and dual-color molecular weight standards were purchased from Bio-Rad Laboratories (Richmond, CA, USA).
Site directed mutagenesis, expression, and purification of fibrinogen αC (233 – 425) and variants
A series of site-directed mutagenesis projects was carried out using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA) with GST tagged Fbg αC (233 – 425) DNA serving as the template. See Supplementary Table S1 for the primer design sequences (IDT, Coralville, IA, USA). All Fbg αC (233 – 425) mutations were verified by DNA sequencing at the CGeMM DNA facility core (University of Louisville, KY, USA). The mutant pGEX-6P based plasmids were transformed into BL21 Gold DE3 (Agilent Technologies, Inc., Cedar Creek, TX, USA) or Acella (EdgeBio, San Jose, CA, USA) competent cells. Fbg αC (233 – 425) and variants were expressed in E. coli and purified as described previously.28, 30
Monodansylcadaverine incorporation assay
2 μM rFXIII-A2 was activated proteolytically with 50 nM recombinant human thrombin and 4 mM CaCl2 and non-proteolytically with 100 mM CaCl2 by incubating at 37 oC for 30 min in TBS buffer (50 mM tris acetate, 150 mM NaCl, pH 7.4). These conditions ensure full activation of FXIII in 30 minutes in both proteolytic and non-proteolytic assays.29 After 30 min of proteolytic activation, 95 μM PPACK was added to quench the activity of thrombin. Incorporation of 1 mM MDC to the glutamine donor proteins [5 μM Fbg αC (233 – 425) and variants or 2 μM α2-AP or 2 μM actin or 1μM FN] were triggered by the addition of activated FXIII-A (100 nM final concentration for all reactions except 200 nM for FN) in TBS buffer at 37 ºC. The final CaCl2 concentration in the reaction mixture for the proteolytic condition was 4 mM and 25 mM for non-proteolytic. At different time points, reaction aliquots were removed and quenched by the addition of 5 μl of 5x reducing sample loading buffer followed by heating at 100 ºC for 3 minutes. For each experiment, a reaction was prepared without the active FXIII-A that served as the zero-time point. As a positive control for the gel quantification process, MDC (1 mM) was crosslinked to 5μM N,N-dimethylated β-casein for 6 min under proteolytic conditions. The samples and the control were subjected to reducing condition SDS-PAGE on 15%T (for Fbg αC (233 – 425) and variants) or 10% T (for α2-antiplasmin and actin) or 4 – 20% gradient gel (for FN). After electrophoresis, the gels were washed in dI H2O and imaged on the Biorad Gel Doc XR Imager (used UV light), followed by total protein staining in 0.1% (w/v) coomassie blue stain solution. The coomassie-stained gel image was also imaged on a Biorad Gel Doc XR using white light.
MDC crosslinking data quantification and statistics
Fluorescent band intensities and total protein amounts loaded in each lane were quantified using ImageJ 1.53e (NIH).32 The in-gel fluorescent intensity was first normalized to its total protein signal from the coomassie blue-stained band for each substrate time point. Next, the data were normalized to the positive control β-casein band and reported as a fraction of the control. Data from three replicates were quantified, and the fully normalized data were plotted versus time. The resultant curves were fitted with the one phase exponential association model in GraphPad Prism 9.0 software. The equation for the model is given as Y = Y0 + (Plateau - Y0) * (1 – exp(−Kx)) where Y is the fluorescent band intensity as a fraction of the β-casein control, Y0 is the initial zero time-point, K is the rate constant, and Plateau (or Emax) is the Y value at infinite time, and t is the time in minutes. All data are reported as the mean ± SD from three replicates. In addition, t-tests were used to compare the activity values between proteolytic and non-proteolytic FXIII-A, and P < 0.05 was considered statistically significant.
UV-Vis spectrophotometric assay with Q-containing peptides for monitoring crosslinking reaction
Crosslinking of the chromogenic lysine mimic DMPDA (700 μM) to the α2-antiplasmin 1 – 15 (15 – 400 μM, WT, and Q4S)) was initiated by the addition of proteolytically or non-proteolytically activated FXIII-A (1 μM) to the reaction mixture incubated at 37 °C in borate buffer (40 mM boric acid, 300 mM NaCl, pH 7.8) and CaCl2 (4 mM for proteolytic and 25 mM for non-proteolytic). Activation of rFXIII-A2 was performed by following the same conditions as the MDC incorporation assay. An increase in the absorbance at 278 nm indicated the reaction’s progress, and the reaction velocity was determined over the absorbance curve’s initial part. The data were fitted to the Michaelis-Menten equation using GraphPad Prism 9.0, and velocities in μM min−1 were plotted as a function of the peptide concentrations. All the experiments were performed in triplicates and the kinetic values Km and Vmax are reported as mean ± SEM. One-way ANOVA calculations followed by Tukey multiple comparison tests were used for the statistical analysis of the kinetics data (GraphPad Prism 9.0.).
MALDI-TOF mass spectrometric kinetic assay
A previously optimized MALDI-TOF MS kinetic assay was used to compare the reactivities of FXIII-A* and FXIII-A° toward crosslinking GEE to glutamines within Fbg αC (233 – 425).24 FXIII-A2 was activated proteolytically by incubation in MALDI kinetic buffer (50 mM Tris-Acetate, 150 mM NaCl, 0.1 % PEG-8000, pH 7.4) with 8.4 NIH units/mL bovine thrombin and 4 mM CaCl2 at 37 °C for 10 minutes, followed by the addition of 190 μM PPACK. To activate non-proteolytically, 500 nM FXIII-A2 was incubated in MALDI kinetic buffer with 250 mM CaC12 for 30 minutes. To initiate crosslinking, 17 mM GEE and 13.6 μM of Fbg αC were added to the assay mixture. The final concentration of active FXIII-A was 50 nM while CaCl2 final concentrations were 4 mM (proteolytic) and 25 mM (non-proteolytic), respectively. The reaction was allowed to proceed at 37°C with 25 μL aliquots of the reaction mixture removed at different time points and quenched with 10 mM EDTA. Quenched sample aliquots were digested separately by either chymotrypsin or GluC endoproteinase, followed by zip-tipping, and MS analysis. Reaction rates were characterized by applying the peak height ratio method between reactive glutamines and their corresponding GEE-crosslinked products.33 With these values, the loss of reactant over time could be tracked. See Supplementary Fig. S1 for representative GEE-crosslinking related MS peaks and their identities.
Assays for each experimental condition were performed in triplicate with a maximal reaction time of 30 minutes. Data were reported as mean ± SD, and individual data points were analyzed by the Student’s t-test and by P-values (GraphPad Prism 9.0.1). In addition, the data from each GEE-crosslinking assay were fit using a one phase exponential decay model [Q]remaining,t = ([Q]0 – [Q]plateau)(e-Kt) + [Q]plateau. [Q]remaining,t is the concentration of uncrosslinked reactive glutamine in μM for time point t in minutes, [Q]0 is the theoretical initial concentration of uncrosslinked reactive glutamine in μM, [Q]plateau is the theoretical concentration of uncrosslinked reactive glutamine in μM when t = ∞ minutes, and K is the rate constant in min.−1.
RESULTS
Crosslinking of MDC to glutamine containing proteins reveals substrate specificity differences between FXIII-A* and FXIII-A°
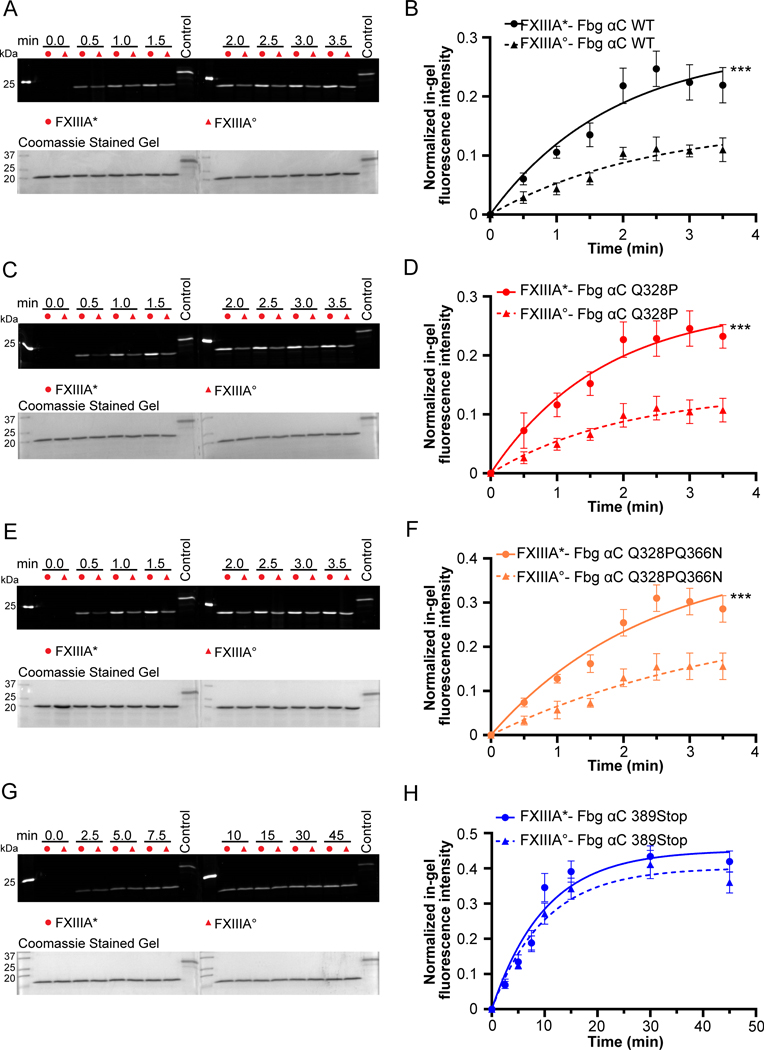
The fluorescent MDC incorporation assay was performed to globally compare the cross-linking rates of FXIII-A* and FXIII-A° enzymes. For crosslinking of MDC into Fbg αC (233–425), a higher incorporation rate (K, 1.3-fold) and higher fluorescent intensity plateau (Emax, 1.9-fold) occurred for FXIII-A* than for A° (P < 0.001, Fig 2A–B, Supplementary Table S2). Fbg αC Q328P is a natural variant (Seoul II) that causes fibrin α crosslinking impairment and can lead to myocardial infarction.34 Fig 2C–D show that both activated forms of FXIII could catalyze crosslinking reactions with the Fbg αC Q328P variant. FXIII-A* exhibited higher K (1.3-fold) and Emax (1.9-fold) than FXIII-A° (Supplementary Table S2). Fig. 2E–F displays reactions with Fbg αC (233–425, Q328P Q366N), where Q237 is the only reactive Q responsible for crosslinking. This Fbg αC variant revealed that FXIII-A* is still more active than FXIII-A° indicated by 1.6-fold increase in K and Emax (P < 0.001). FXIII-A* catalyzed MDC crosslinking to Fbg αC WT and variants exhibited a sudden increase in activity from 2 min and onwards, unlike FXIII-A° which showed a gradual increase in activity. These effects suggest that FXIII-A* approached maximal activity faster for these substrates than FXIII-A° and thus the overall activity was higher for FXIII-A*. To assess the consequences of losing the Fbg αC binding site on FXIII-A activity, Fbg αC (233–388, 389Stop), a truncated variant, was expressed. Interestingly, K and Emax values with Fbg αC (233–388) were similar for FXIII-A* and A° (Fig 2G–H, Supplementary Table S2). FXIII-A* reactivity had weakened and was approaching the level of FXIII-A°. Moreover, both reactions started to plateau beyond 20 minutes of crosslinking time. These MDC results clearly demonstrate that transglutaminase activities of FXIII-A* and A° were affected by removal of the αC binding sequence.
Fig. 2. Incorporation of monodansyl cadaverine (MDC) into Fbg αC (233 – 425) WT and variants using FXIII-A* and FXIII-A°.
The crosslinking reaction between Fbg αC (233 – 425) WT and variants (Fbg αC Q328P, Fbg αC Q328PQ366N, and Fbg αC 389Stop) and MDC was initiated by adding 100 nM proteolytically or non-proteolytically activated FXIII-A (FXIII-A*/FXIII-A°). The positive control used was MDC crosslinked into N, N-dimethylated β-casein. The quantification of the gels was performed as described in the methods section. The curves represent one phase exponential association fits of the data as a function of time. Data were reported as mean ± SD (N=3). The gels for the MDC incorporation and the quantification curves include Fbg αC (233 – 425) WT (A and B), Fbg αC (233 – 425) Q328P (C and D), Fbg αC (233 – 425) Q328PQ366N (E and F), and Fbg αC (233 – 388) 389Stop (G and H). t-tests analysis showed that activity differences between FXIII-A* and FXIII-A° are significant (***P < 0.001) for all substrates except Fbg αC (233 – 388) 389Stop.
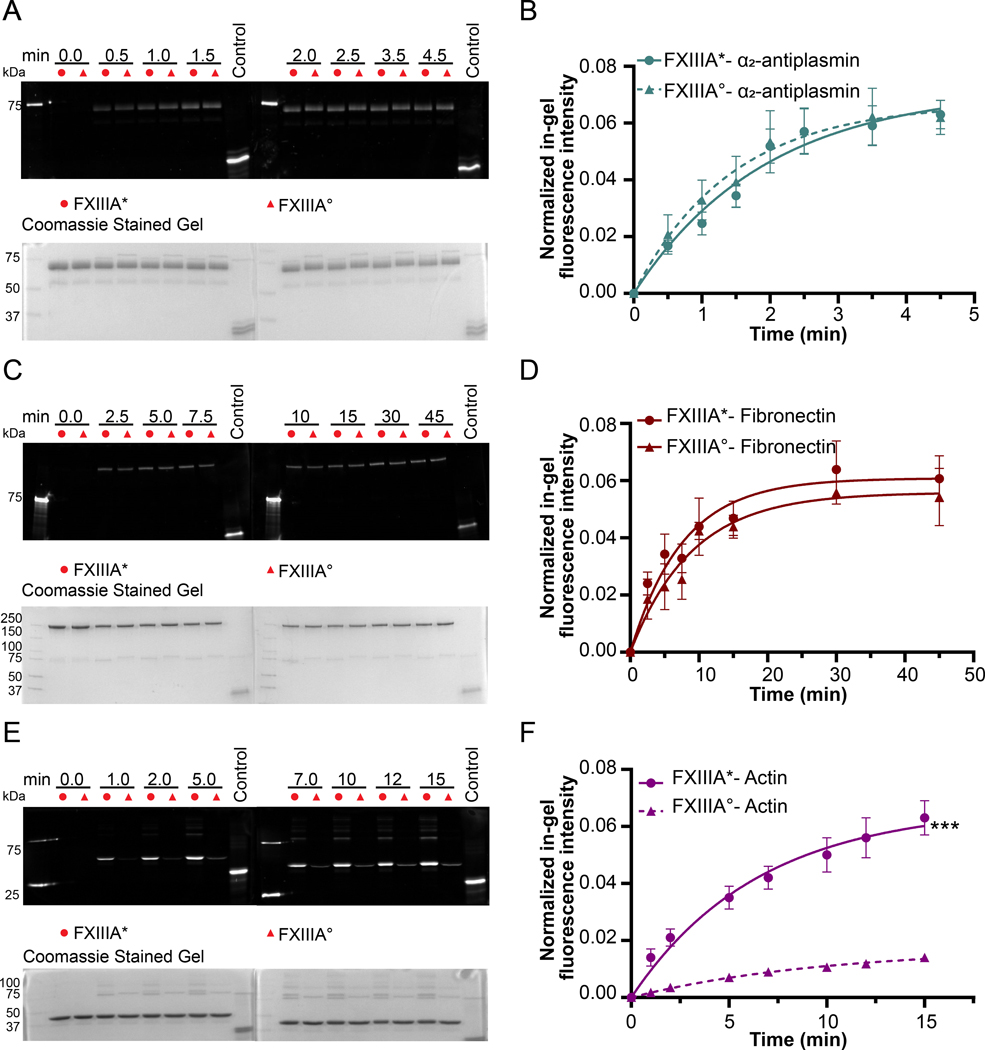
The intriguing results with Fbg αC (233–425) and Fbg αC (233– 388) led to interest in probing other Q-containing FXIII substrates. Studies with anti-fibrinolytic α2 – AP followed a trend resembling that of Fbg αC (233–388), where K and Emax values were similar for FXIII-A* and FXIII-A° (Fig. 3A–B, Supplementary Table S2). FN is part of the extracellular matrix and exists in plasmatic and cellular forms.35 As observed with Fbg αC (233–388) and α2-AP, the crosslinking profiles of FXIII-A* and FXIII-A° toward plasmatic FN were similar (Fig 3C–D, Supplementary Table S2). Comparable to Fbg αC (233–388), longer time points were needed before FN crosslinking approached the plateau region. Actin filaments are major building blocks of the platelet cytoskeleton, and they control the platelet cell shape.36 Moreover, actin was identified as a cellular substrate of FXIII.37 As shown in Fig 3E–F, FXIII-A* exhibited greater reactivity toward actin than FXIII-A° (3.5-fold increase in Emax; P < 0.001). Actin also formed inter-molecular high molecular weight (HMW) crosslinked polymers in the presence of FXIII-A*. MDC was also crosslinked to these polymers suggesting multiple reactive Q and K residues. However, these polymers were not observed with FXIII-A°.
Fig. 3. Incorporation of monodansyl cadaverine (MDC) into α2 -antiplasmin, fibronectin and actin using FXIII-A* and FXIII-A°.
The addition of 100 nM FXIII-A* or FXIII-A° initiated the incorporation of MDC (1 mM) into α2 -antiplasmin and actin (2 μM). For fibronectin (1 μM), 200 nM FXIII-A*/FXIII-A° were used. The positive control used was MDC crosslinked into N, N-dimethylated β-casein. The quantification of the gels was performed as described in the methods section. The curves represent one phase exponential association fits of the data as a function of time. Data were reported as mean ± SD (N=3). The gels for the MDC incorporation and the quantification curves include α2 -antiplasmin (A and B), fibronectin (C and D), and actin (E and F). t-tests analysis showed that activity differences between FXIII-A* and FXIII-A° are significant (***P < 0.001) for actin and not for (P > 0.05) α2 -antiplasmin and fibronectin.
Comparing kinetic parameters for FXIII-A* and FXIII-A° catalyzed reactions involving α2-antiplasmin (1–15) peptides and the lysine mimic DMPDA
A continuous UV/Vis assay was used to determine individual kinetic parameters for FXIII-A* and FXIII-A°. The two Q-reactive peptides examined were α2-AP WT (1–15, 1NQEQVSPLTLLKLGN15) and α2-AP Q4S (1–15, 1NQESVSPLTLLKLGN15) and DMPDA was the chromogenic lysine mimic. For both peptides, Q2 is the sole reactive glutamine, and K12 is not reactive. For these α2-AP peptides, there were no statistical differences among the Km and Vmax values for each FXIII-A* versus FXIII-A° set (Table 1, P > 0.05 and Supplementary Fig. 3). These peptide kinetic results mirrored what was observed in the MDC assays of α2-AP protein. Both FXIII-A* and FXIII-A° predominantly recognize the N-terminal segment of the α2-AP protein.
Table 1.
Curve fit data of DMPDA incorporation into α2-AP based peptide substrates of FXIII-A
| Substrate | FXIIIA Species | Km (μM) | Vmax (μM/min) |
|---|---|---|---|
| α2-AP (1–15) | FXIIIA* FXIIIA° |
26 ± 5 21 ± 4 |
100 ± 8 84 ± 4 |
| α2-AP (1–15, Q4S) | FXIIIA* FXIIIA° |
29 ± 2 34 ± 4 |
39 ± 1 40 ± 1 |
Data from each DMPDA incorporation assay were fit Michaelis-Menten equation V0 = (Vmax*[S]) / (Km+[s]) in GraphPad Prism 9.0 software. In this equation, V0 is the initial velocity in μM/min, Vmax is the maximum velocity in μM/min, [S] is the substrate concentration in μM, and Km is the Michaelis-Menten constant in μM.
A mass spectrometric assay reveals that FXIII-A° catalyzed GEE-crosslinking to the Q237 of Fbg αC WT (233 – 425) is slower than with FXIII-A*
Our mass spectrometry based assay complements and extends beyond the global fluorescent MDC assay by permitting reactive Qs within the FXIII substrate Fbg αC (233–425) to be individually probed. At a physiological concentration of 50 nM FXIII, Q237-GEE production dominates the mass spectral results (Supplementary Fig S2).24 With a focus on highly reactive Fbg αC Q237, the current studies probed the influences of αC E396 and surrounding residues on the transglutaminase activities of 50 nM FXIII-A* versus FXIII-A°.
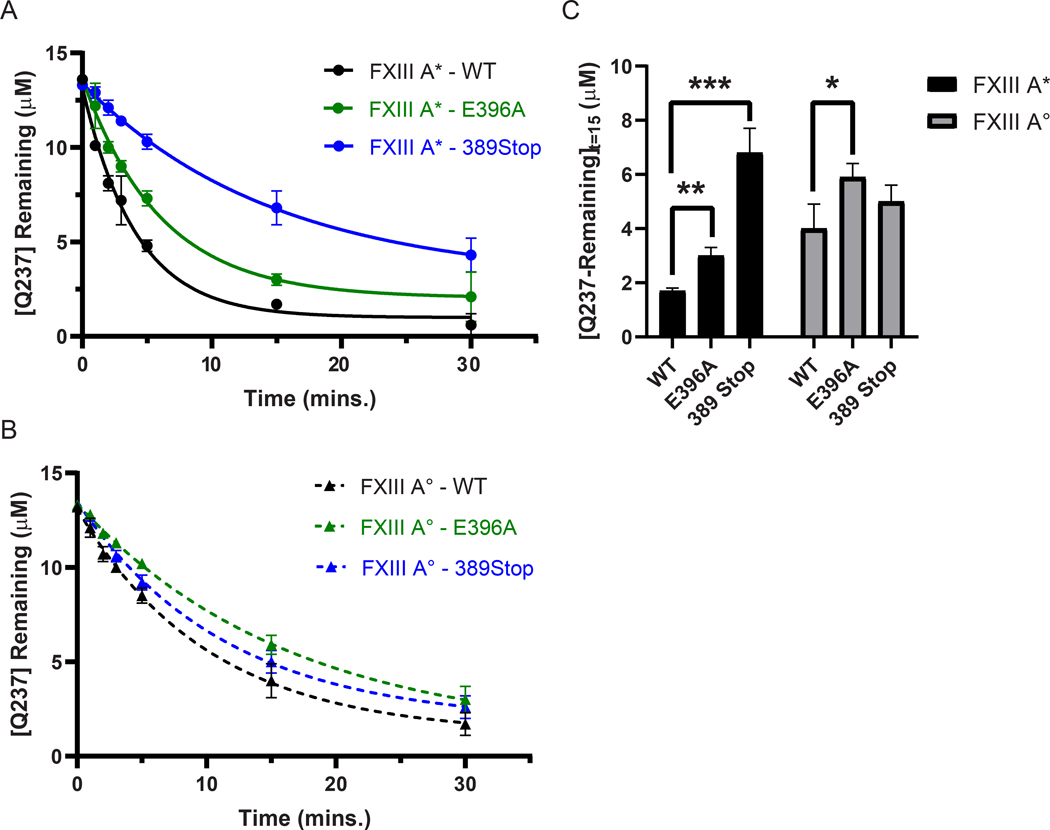
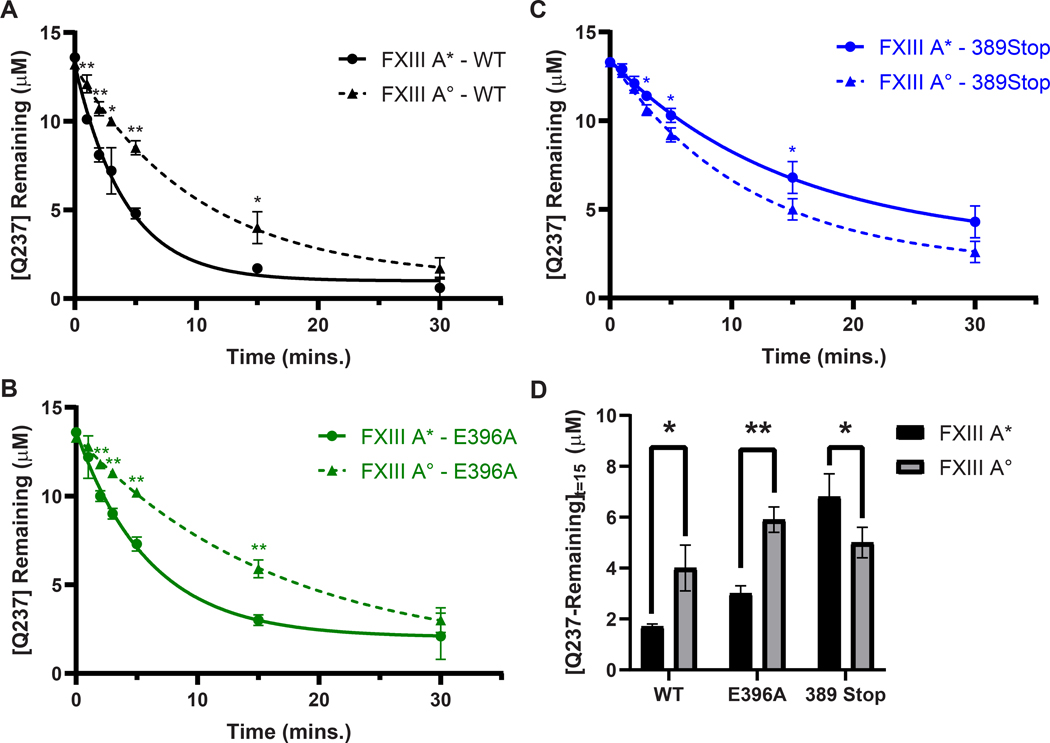
The ability of FXIII-A* to catalyze GEE-crosslinking varied between Fbg αC (233–425) WT, Fbg αC (233–425) E396A, and Fbg αC (233–388) 389Stop (Fig 4A, Supplementary Table S3). The rates of Q237 consumption by FXIII-A* ranked as Fbg αC WT > E396A > 389 stop (Supplementary Table S3). The disparity in reactivities is highlighted at t = 15 min, where the concentrations of unreacted Q237 in αC E396A and αC 389Stop were ~2-fold higher (P ≤ 0.01) and ~4-fold higher (P ≤ 0.001) relative to WT, respectively (Fig 4A, 4C). For FXIII-A*, the full FXIII binding region αC (389–402) played a more prominent role in promoting Q237-GEE crosslinking than just the key anchoring residue αC E396. In contrast, FXIII-A° reactivity toward αC Q237 was not as affected by the binding site mutations as FXIII-A* (Fig. 4B and 4C). Rates of substrate consumption were comparable for the FXIII-A° catalyzed reactions with Fbg αC WT > E396A ≈ 389 stop (Supplementary Table S3).
Fig. 4. Transglutaminase activity of FXIII-A* is more affected by Fbg αC binding site mutations than FXIII-A°.
FXIII-catalyzed Q237-GEE crosslinking assays between WT Fbg αC (233 – 425, black) and the FXIII binding site variants E396A (green) and 389 Stop (blue) were performed using 50 nM of either (A) FXIII-A* (circles, solid line), or (B) FXIII-A° (triangles, dashed line). Reactions were monitored by measuring the concentration of uncrosslinked Q237 remaining at each time point. Plots represent one phase exponential decay fits of uncrosslinked Q237 concentrations as a function of time. Data were reported as mean ± SD (N = 3). (C) FXIII GEE-crosslinking activity after 15 minutes of reaction time was compared between Fbg. αC WT, E396A, and 389 Stop. Shown here is the mean uncrosslinked Q237 concentration ± SD in μM after 15 minutes of reaction time (N = 3). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001).
Evaluating the ability of FXIII-A* versus FXIII-A° to catalyze GEE crosslinking reactions with Fbg αC (233–425) WT, Fbg αC (233–425) E396A, and Fbg αC (233–388) 389Stop were also critical to examine. A comparison of αC Q237-GEE crosslinking plots revealed that FXIII-A* exhibited faster rate of transglutaminase activity toward Fbg αC (233–425) WT than FXIII-A° (Fig. 5A, 5D and Supplementary Table S3). Similarly, experiments with Fbg αC (233–425) E396A indicated that the rate of the FXIII-A* catalyzed crosslinking reaction was greater than that with FXIII-A° (Fig 5B, 5D and Supplementary Table S3). Interestingly, the full removal of the αC 389 – 402 binding region modestly increased the crosslinking rate for FXIII-A° with respect to FXIII-A* (Fig. 5C, 5D and Supplementary Table S3).
Fig. 5. Transglutaminase activities between FXIII-A* and FXIII-A° on Fbg. αC Q237 are significantly different.
FXIII-catalyzed Q237-GEE crosslinking assays between (A) WT Fbg αC (233 – 425, black) and the FXIII binding site variants (B) E396A (green) and (C) 389 Stop (blue) were performed using 50 nM of either FXIII-A* (circles, solid lines), or FXIII-A° (triangles, dashed lines). Reactions were monitored by measuring the concentration of uncrosslinked Q237 remaining at each time point. Plots represent one phase exponential decay fits of uncrosslinked Q237 concentrations as a function of time. Data were reported as mean ± SD (N = 3). (D) FXIII GEE-crosslinking activity after 15 minutes of reaction time was compared between Fbg. αC WT, E396A, and 389 Stop. Shown here is the mean uncrosslinked Q237 concentration ± SD in μM after 15 minutes of reaction time (N = 3). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.01).
This second set of crosslinking comparisons revealed FXIII-A* exhibits greater reactivity toward Fbg αC (233–425) WT and αC E396A than FXIII-A°. By contrast, rates of Q-substrate consumption are more comparable between FXIII-A* and FXIII-A° when utilizing the truncated Fbg αC (233–338). The FXIII binding region within αC (389–402) is proposed to be more beneficial for promoting the transglutaminase activities of FXIII-A* over that of FXIII-A°.
Intrinsic disorder propensities of Fbg αC (233–425), actin, α2-antiplasmin and fibronectin
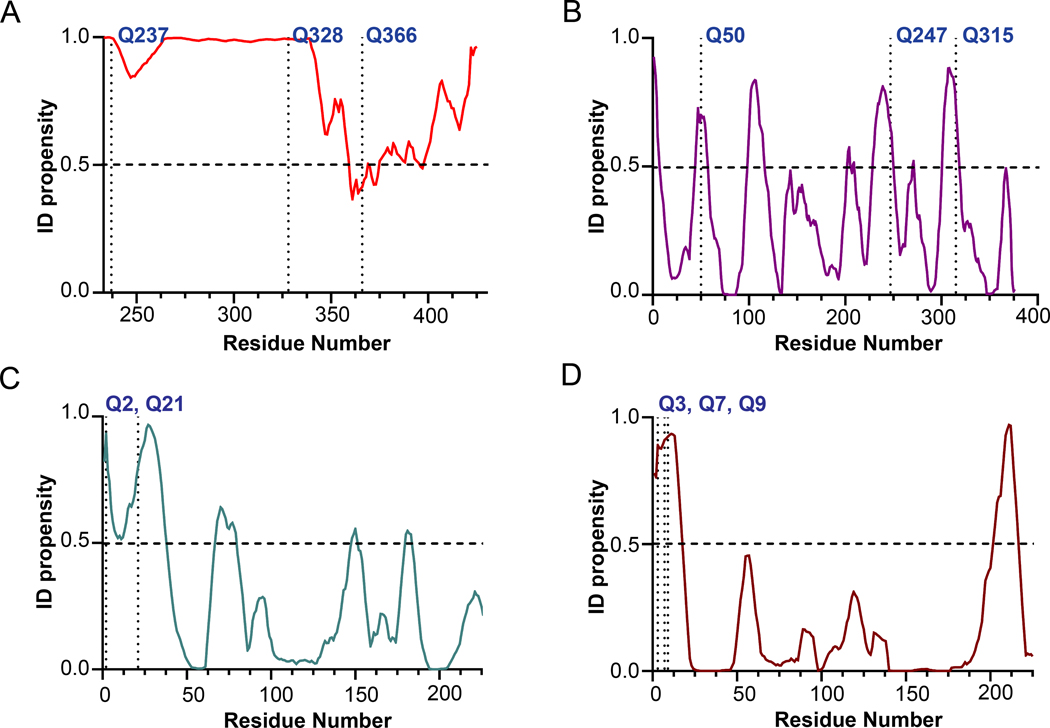
The amino acid sequences of the substrates Fbg αC (233 – 425), actin, α2-AP, and FN were analyzed using the PONDR-VLXT program to check for the regions of disorder. An ID propensity score ≥ 0.5 for a residue indicates it is in a disordered region, while a score < 0.5 indicates an ordered region. For the 233 – 340 segment of Fbg αC (233 – 425), a score > 0.9 was predicted, indicating a high degree of disorder (Fig 6A). The primary reactive Q237 is also present in this region 24. For actin, 23% of the total amino acids have a score > 0.5, and Q50, Q247, and Q315 are in the disordered segments of the protein (Fig 6B). Sequence analyses of α2-AP and FN predict that the N-terminal region is among the most disordered segments (Fig 6C and 6D). This segment contains the major reactive Q2 of α2-AP and Q3 of FN in addition to other reactive glutamines.14, 38
Fig. 6. PONDR Analysis of Fbg αC (233–425), actin, and α2-antiplasmin and fibronectin for order-disorder characteristics.
Intrinsic disorder propensity (ID propensity) of the amino acid sequences of (A) Fbg αC (233–425) (B) actin (C) α2-antiplasmin (1–225) and (D) Fibronectin (1–225) were calculated with the PONDR VLXT algorithm (www.pondr.com). The amino acid sequences of the proteins were accessed from the UniProt database (Uniprot ID for actin (P68135), α2-antiplasmin (P08697), and Fibronectin (P02751)). ID propensity value equal to or greater than 0.5 indicated disorder. The major and other potential reactive glutamines are labeled in the graph. For α2-antiplasmin and fibronectin only 225 amino acid residues from the N-terminus were used for plotting since the major reactive Qs are located in the said region.
DISCUSSION
The transglutaminase FXIII can be activated both proteolytically (FXIII-A*) and nonproteolytically (FXIII-A°), and following activation, the catalytic FXIII-A interacts with an array of substrates in different environments.1–3 There are numerous reports on the identification of FXIII substrates, and the potential reactive glutamine and lysine residues have been characterized using various biochemical methods.1, 2 However, most of the studies used FXIII-A* as the active enzyme form, and only a few studies also made comparisons to FXIII-A°.30 In this project, we characterized in greater depth the reactivities of FXIII-A* versus FXIII-A° toward glutamine-containing substrates based on Fbg αC (233–425), α2-AP, FN, and actin. The results obtained have been further correlated with protein substrate disorder and with the presence of a secondary substrate anchoring site involving the FXIII AP binding cleft.
MDC crosslinking studies revealed FXIII-A* exhibited higher activity than FXIII-A° toward Fbg αC (233–425), an effective Q-containing FXIII substrate system. Fbg αC Q328P (Seoul II) has been reported in a patient with increased susceptibility to myocardial infarction.34 However, the Q328P substitution did not hinder reactivity of Fbg αC (233–425) suggesting the two remaining glutamines play compensatory roles. The reduced clotting properties observed in Fbg Seoul II must result from additional impacts on fibrin α-α crosslinking. Additionally, PONDR-VLXT analysis shows that Q328P is located within a highly disordered component, and hence the P328 mutation is not expected to introduce a major structural change to this disordered segment.
Fbg αC (389–402) starts at the end of the disordered αC connector (Aα 221–391) and extends into the structured C-terminal portion (Aα 392–610).20 FXIII-A* is hypothesized to adopt a conformation that can take advantage of a FXIII binding site region within Fbg αC (389–402). PONDR analysis suggests this Fbg αC segment has the greatest potential for introducing some modest protein ordering. Electrostatic anchoring of Fbg αC E396 is proposed to play a contributing role in FXIII-A* reactivity.26, 28 A bigger hindrance to FXIII-A* occurred in this project when Fbg αC (233–425) residues beyond 388 were eliminated, thus removing the full FXIII-A* binding region (αC 389–402). Physiologically, Fbg Keokuk (Q328stop) contains an even greater truncation leading to hypofibrinogenemia and postsurgical thrombosis and miscarriages.39 Unlike FXIII-A*, the current results indicated that the FXIII-A° conformation is less reliant on αC (389–402), generating activities comparable to that observed with αC (233–425).
Using peptides derived from Fbg αC (389–402), Smith et al. reported binding to the FXIII-A* AP cleft that includes key electrostatic residue R158.27 This cleft region is located within the β-sandwich domain and becomes exposed following thrombin-catalyzed release of FXIII AP. Thomas et al. performed docking studies with the entire Fbg α chain, and active site inhibited FXIII-A°. Docking contacts were reported within the FXIII-A β-sandwich and catalytic core, including amino acids in the 160s, 170s, and 200s regions.40
Conformational differences between FXIII-A* and FXIII-A° are important to consider for elucidating possible sources of substrate specificities. AUC results demonstrated that FXIII-A* exhibits a more flexible conformation, whereas FXIII-A° has a tighter, more homogeneous conformation stabilized by higher mM Ca2+ binding.30 With FXIII-A*, the cleaved FXIII AP segment may be partially or fully displaced from the FXIII surface.41 Both X-ray crystallography and HDX-MS studies have documented that the unhydrolyzed AP can remain associated with FXIII-A° in the presence of Ca2+.42, 43 A well-defined crystal structure for the less soluble, active site inhibited FXIII-A* is not yet available. However, a more exposed AP cleft of FXIII-A* is predicted to contribute to Q-substrate binding and have the potential to promote higher transglutaminase activity. In addition, the more flexible FXIII-A* may contribute further conformational states for effective substrate binding.
It is important to point out that the zymogen FXIII-A2 does not bind to Fbg αC (233–425). The binding sites on FXIII are only exposed upon activation, such as for FXIII-A* in the presence of Ca2+.27 Current results demonstrate FXIII-A° is proposed to have less need for the FXIII binding site region (389–402), or the FXIII-A° conformation cannot take advantage of this binding region. In further support, Hornyak et al. reported that KD values for binding fibrin polymers ranked as FXIII-A2 > A2° > A2* with A2* having the strongest affinity for fibrin.44
FXIII-A V34L, a common polymorphism associated with venous thrombosis protection, is activated more readily via thrombin-catalyzed hydrolysis of the FXIII AP R37-G38 peptide bond than FXIII V34, and the resultant FXIII-A* leads to reduced fibrin clot weights.45, 46 FXIII P36S, a type II deficiency variant, has also been documented in patient plasma. This variant cannot be cleaved by thrombin, and crosslinking ability must be derived from FXIII-A° activated slowly by low physiological Ca2+ levels.47 These two variants reveal both FXIII-A* and A° can be encountered in plasma. Further examples of wild-type FXIII-A° are found in cellular environments.5 In our experimental conditions, we used preactivated FXIII-A*/A° to analyze the crosslinking reactions, to avoid complications arising from different activation rates, and thus a focus on substrate specificities could be achieved.
FXIII-A incorporates α2-AP and FN into the fibrin clot rendering anti-fibrinolytic and wound healing, properties respectively.13 FXIII-A* and FXIII-A° both crosslinked MDC into α2-AP to similar extents. The same property was observed with FN. With FN, an exosite that promotes FXIII transglutaminase activity is proposed to be located within the FN 4th-5th Type I module.38 For α2AP and FN, FXIII-A* conformations do not seem to enhance transglutaminase activity over that of FXIII-A°.
FXIII-A is highly selective for reactive glutamines in flexible regions of protein substrates. Physiologically, FXIII-A crosslinks reactive Qs from α2AP and FN to specific reactive Ks within the fibrin network.1 PONDR analysis shows that the major reactive glutamine Q2 of α2-AP and Q3 of FN are in highly disordered regions of these proteins. High affinity binding sites for α2-AP and FN are located within αC 502–610 and αC 221–391, respectively.48, 49 With both α2-AP and FN, FXIII-A is proposed to utilize the fibrin α chain to provide further anchoring regions that may enhance transglutaminase activity.
Actin is abundant in platelets and exhibits fundamental roles in the dynamics of platelet cell shape.36, 37 Previous studies showed that actin could bind to both FXIIIA* and FXIII-A°, but not to zymogen A2.50 Our current study demonstrated that FXIII-A* shows higher activity than FXIII-A° when crosslinking MDC to actin. HMW crosslinked polymers were observed in FXIII-A*, suggesting multiple reactive Q and K residues. According to the PONDR analysis, the potential reactive glutamines are in the flexible regions of the actin. The higher binding affinity and transglutaminase activity of FXIII-A* for actin could be arising from a binding site that is not exposed in FXIII-A°. Additionally, the presence of reactive glutamines in the flexible regions of actin could be accessed more by the dynamic conformation of FXIII-A*.
In conclusion, transglutaminase activity differences between FXIII-A* and A° have been elucidated for four physiological substrates and correlated with protein disorder and presence of binding sites. For Fbg αC (233–425) and actin, FXIII-A* had higher activity than FXIII-A°, whereas for α2-AP and FN, FXIII-A* and FXIII-A° had similar activities. However, the removal of the FXIII binding region αC (389–402) diminished the activity of FXIII-A* making the crosslinking ability approach that of FXIII-A°. Overall, the transglutaminase activities of FXIII-A* and A° may be regulated by FXIII conformational features, individual substrates, and presence of binding sites. The novel features of FXIII-A* will help differentiate this crosslinking enzyme from other transglutaminases. The knowledge gained provides critical explanations for what drives the substrate specificity of FXIII-A and provides vital tools for future control of its transglutaminase activity.
Supplementary Material
SUMMARY TABLE.
What is known on this topic?
Plasma FXIII is activated proteolytically (FXIII-A*) by thrombin and Ca2+ whereas cellular FXIII is activated non-proteolytically (FXIII-A°) by elevation of physiological Ca2+concentrations.
FXIII-A catalyzes covalent γ-glutamyl-ε-lysyl peptide bond formation among diverse substrates that include the fibrin α and γ chains, α2-antiplasmin, fibronectin, and actin.
In FXIII-A*, the activation peptide (AP) is cleaved exposing a substrate binding site region that accommodates fibrinogen αC (389–402) whereas FXIII-A° adopts a more rigid structure stabilized by excess Ca2+ binding in which AP remains intact.
What does this paper add?
Crosslinking activity of FXIII-A* > FXIII-A° for Fibrinogen (Fbg) αC (233–425) and actin substrates whereas FXIII-A* and FXIII-A° had similar activities for α2-antiplasmin (α2-AP), fibronectin (FN), and Fbg αC (233–388, missing FXIII binding site αC 389–402),
Protein substrate disorder and secondary FXIII binding site exposure are utilized to control activity and specificity.
The novel characteristics of FXIII-A* versus A° help explain what drives FXIII-A substrate specificity and may guide future strategies to control transglutaminase activity.
Acknowledgements
The authors acknowledge the late Paul Bishop (Zymogenetics, Seattle, WA, USA) for recombinant human FXIII-A2, expressed in Saccharomyces cerevisiae and E. Di Cera and L. Pelc (St. Louis University, MO, USA) for generously providing recombinant human thrombin. The authors thank R. Lumata for helpful discussions related to MDC assay applications, and we appreciate his reviews of the manuscript. We also want to note that H. Butala participated in early studies involving MDC assay development. This research was supported by a grant from the National Institutes of Health R15 HL120068.
Footnotes
Conflict of Interest
The authors declare no conflict of interest
REFERENCES
- 1.Richardson VR, Cordell P, Standeven KF, Carter AM. Substrates of Factor XIII-A: roles in thrombosis and wound healing. Clin Sci (Lond). 2013;124(3):123–37 [DOI] [PubMed] [Google Scholar]
- 2.Schroeder V, Kohler HP. Factor XIII: Structure and Function. Semin Thromb Hemost. 2016;42(4):422–8 [DOI] [PubMed] [Google Scholar]
- 3.Muszbek L, Bereczky Z, Bagoly Z, Komaromi I, Katona E. Factor XIII: a coagulation factor with multiple plasmatic and cellular functions. Physiol Rev. 2011;91(3):931–72 [DOI] [PubMed] [Google Scholar]
- 4.Byrnes JR, Wolberg AS. Newly-Recognized Roles of Factor XIII in Thrombosis. Semin Thromb Hemost. 2016;42(4):445–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell JL, Mutch NJ. Let’s cross-link: diverse functions of the promiscuous cellular transglutaminase factor XIII-A. J Thromb Haemost. 2019;17(1):19–30 [DOI] [PubMed] [Google Scholar]
- 6.Alshehri FSM, Whyte CS, Mutch NJ. Factor XIII-A: An Indispensable “Factor” in Haemostasis and Wound Healing. Int J Mol Sci. 2021;22(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorand L. Factor XIII: structure, activation, and interactions with fibrinogen and fibrin. Ann N Y Acad Sci. 2001;936:291–311 [DOI] [PubMed] [Google Scholar]
- 8.Polgar J, Hidasi V, Muszbek L. Non-proteolytic activation of cellular protransglutaminase (placenta macrophage factor XIII). Biochem J. 1990;267(2):557–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornyak TJ, Bishop PD, Shafer JA. Alpha-thrombin-catalyzed activation of human platelet factor XIII: relationship between proteolysis and factor XIIIa activity. Biochemistry. 1989;28(18):7326–32 [DOI] [PubMed] [Google Scholar]
- 10.Standeven KF, Carter AM, Grant PJ, et al. Functional analysis of fibrin {gamma}-chain cross-linking by activated factor XIII: determination of a cross-linking pattern that maximizes clot stiffness. Blood. 2007;110(3):902–7 [DOI] [PubMed] [Google Scholar]
- 11.Helms CC, Ariens RA, Uitte de Willige S, Standeven KF, Guthold M. alpha-alpha Cross-links increase fibrin fiber elasticity and stiffness. Biophys J. 2012;102(1):168–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrnes JR, Duval C, Wang Y, et al. Factor XIIIa-dependent retention of red blood cells in clots is mediated by fibrin alpha-chain crosslinking. Blood. 2015;126(16):1940–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakata Y, Aoki N. Cross-linking of alpha 2-plasmin inhibitor to fibrin by fibrin-stabilizing factor. J Clin Invest. 1980;65(2):290–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KN, Lee CS, Tae WC, Jackson KW, Christiansen VJ, McKee PA. Cross-linking of wild-type and mutant alpha 2-antiplasmins to fibrin by activated factor XIII and by a tissue transglutaminase. J Biol Chem. 2000;275(48):37382–9 [DOI] [PubMed] [Google Scholar]
- 15.Matsuka YV, Migliorini MM, Ingham KC. Cross-linking of fibronectin to C-terminal fragments of the fibrinogen alpha-chain by factor XIIIa. J Protein Chem. 1997;16(8):739–45 [DOI] [PubMed] [Google Scholar]
- 16.Alshehri FSM, Whyte CS, Tuncay A, Williams ML, Wilson HM, Mutch NJ. Monocytes Expose Factor XIII-A and Stabilize Thrombi against Fibrinolytic Degradation. Int J Mol Sci. 2021;22(12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell JL, Lionikiene AS, Fraser SR, Whyte CS, Booth NA, Mutch NJ. Functional factor XIII-A is exposed on the stimulated platelet surface. Blood. 2014;124(26):3982–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somodi L, Beke Debreceni I, Kis G, et al. Activation mechanism dependent surface exposure of cellular factor XIII on activated platelets and platelet microparticles. J Thromb Haemost. 2022;20(5):1223–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mousa A, Cui C, Song A, et al. Transglutaminases factor XIII-A and TG2 regulate resorption, adipogenesis and plasma fibronectin homeostasis in bone and bone marrow. Cell Death Differ. 2017;24(5):844–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsurupa G, Tsonev L, Medved L. Structural organization of the fibrin(ogen) alpha C-domain. Biochemistry. 2002;41(20):6449–59 [DOI] [PubMed] [Google Scholar]
- 21.Duval C, Allan P, Connell SD, Ridger VC, Philippou H, Ariens RA. Roles of fibrin alpha- and gamma-chain specific cross-linking by FXIIIa in fibrin structure and function. Thromb Haemost. 2014;111(5):842–50 [DOI] [PubMed] [Google Scholar]
- 22.McPherson H, Duval C, Baker SR, et al. Fibrinogen alphaC-subregions critically contribute blood clot fibre growth, mechanical stability and resistance to fibrinolysis. Elife. 2021;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hur WS, Paul DS, Bouck EG, et al. Hypofibrinogenemia with preserved hemostasis and protection from thrombosis in mice with a Fga truncation mutation. Blood. 2022;139(9):1374–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouapi KN, Bell JD, Smith KA, Ariens RA, Philippou H, Maurer MC. Ranking reactive glutamines in the fibrinogen alphaC region that are targeted by blood coagulant factor XIII. Blood. 2016;127(18):2241–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt LR, Henderson R, Barrett A, et al. Mass spectrometry-based molecular mapping of native FXIIIa cross-links in insoluble fibrin clots. J Biol Chem. 2019;294(22):8773–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith KA, Adamson PJ, Pease RJ, et al. Interactions between factor XIII and the alphaC region of fibrinogen. Blood. 2011;117(12):3460–8 [DOI] [PubMed] [Google Scholar]
- 27.Smith KA, Pease RJ, Avery CA, et al. The activation peptide cleft exposed by thrombin cleavage of FXIII-A(2) contains a recognition site for the fibrinogen alpha chain. Blood. 2013;121(11):2117–26 [DOI] [PubMed] [Google Scholar]
- 28.Mouapi KN, Wagner LJ, Stephens CA, et al. Evaluating the Effects of Fibrinogen alphaC Mutations on the Ability of Factor XIII to Crosslink the Reactive alphaC Glutamines (Q237, Q328, Q366). Thromb Haemost. 2019;119(7):1048–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anokhin BA, Stribinskis V, Dean WL, Maurer MC. Activation of factor XIII is accompanied by a change in oligomerization state. FEBS J. 2017;284(22):3849–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anokhin BA, Dean WL, Smith KA, et al. Proteolytic and nonproteolytic activation mechanisms result in conformationally and functionally different forms of coagulation factor XIII A. FEBS J. 2020;287(3):452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayala YM, Cantwell AM, Rose T, Bush LA, Arosio D, Di Cera E. Molecular mapping of thrombin-receptor interactions. Proteins. 2001;45(2):107–16 [DOI] [PubMed] [Google Scholar]
- 32.Murrey HE, Judkins JC, Am Ende CW, et al. Systematic Evaluation of Bioorthogonal Reactions in Live Cells with Clickable HaloTag Ligands: Implications for Intracellular Imaging. J Am Chem Soc. 2015;137(35):11461–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doiphode PG, Malovichko MV, Mouapi KN, Maurer MC. Evaluating factor XIII specificity for glutamine-containing substrates using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay. Anal Biochem. 2014;457:74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park R, Doh HJ, An SS, Choi JR, Chung KH, Song KS. A novel fibrinogen variant (fibrinogen Seoul II; AalphaGln328Pro) characterized by impaired fibrin alpha-chain cross-linking. Blood. 2006;108(6):1919–24 [DOI] [PubMed] [Google Scholar]
- 35.To WS, Midwood KS. Plasma and cellular fibronectin: distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair. 2011;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bender M, Palankar R. Platelet Shape Changes during Thrombus Formation: Role of Actin-Based Protrusions. Hamostaseologie. 2021;41(1):14–21 [DOI] [PubMed] [Google Scholar]
- 37.Cohen I, Blankenberg TA, Borden D, Kahn DR, Veis A. Factor XIIIa-catalyzed cross-linking of platelet and muscle actin. Regulation by nucleotides. Biochim Biophys Acta. 1980;628(3):365–75 [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann BR, Annis DS, Mosher DF. Reactivity of the N-terminal region of fibronectin protein to transglutaminase 2 and factor XIIIA. J Biol Chem. 2011;286(37):32220–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefebvre P, Velasco PT, Dear A, et al. Severe hypodysfibrinogenemia in compound heterozygotes of the fibrinogen AalphaIVS4 + 1G>T mutation and an AalphaGln328 truncation (fibrinogen Keokuk). Blood. 2004;103(7):2571–6 [DOI] [PubMed] [Google Scholar]
- 40.Thomas A, Biswas A, Dodt J, et al. Coagulation Factor XIIIA Subunit Missense Mutations Affect Structure and Function at the Various Steps of Factor XIII Action. Hum Mutat. 2016;37(10):1030–41 [DOI] [PubMed] [Google Scholar]
- 41.Schroeder V, Vuissoz JM, Caflisch A, Kohler HP. Factor XIII activation peptide is released into plasma upon cleavage by thrombin and shows a different structure compared to its bound form. Thromb Haemost. 2007;97(6):890–8 [PubMed] [Google Scholar]
- 42.Stieler M, Weber J, Hils M, et al. Structure of active coagulation factor XIII triggered by calcium binding: basis for the design of next-generation anticoagulants. Angew Chem Int Ed Engl. 2013;52(45):11930–4 [DOI] [PubMed] [Google Scholar]
- 43.Woofter RT, Maurer MC. Role of calcium in the conformational dynamics of factor XIII activation examined by hydrogen-deuterium exchange coupled with MALDI-TOF MS. Arch Biochem Biophys. 2011;512(1):87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornyak TJ, Shafer JA. Interactions of factor XIII with fibrin as substrate and cofactor. Biochemistry. 1992;31(2):423–9 [DOI] [PubMed] [Google Scholar]
- 45.Kattula S, Bagoly Z, Toth NK, Muszbek L, Wolberg AS. The factor XIII-A Val34Leu polymorphism decreases whole blood clot mass at high fibrinogen concentrations. J Thromb Haemost. 2020;18(4):885–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trumbo TA, Maurer MC. Examining thrombin hydrolysis of the factor XIII activation peptide segment leads to a proposal for explaining the cardioprotective effects observed with the factor XIII V34L mutation. J Biol Chem. 2000;275(27):20627–31 [DOI] [PubMed] [Google Scholar]
- 47.Li B, Billur R, Maurer MC, et al. Proline 36 of the Factor XIII Activation Peptide Plays a Crucial Role in Substrate Recognition and Zymogen Activation. Thromb Haemost. 2018;118(12):2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsurupa G, Yakovlev S, McKee P, Medved L. Noncovalent interaction of alpha(2)-antiplasmin with fibrin(ogen): localization of alpha(2)-antiplasmin-binding sites. Biochemistry. 2010;49(35):7643–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makogonenko E, Tsurupa G, Ingham K, Medved L. Interaction of fibrin(ogen) with fibronectin: further characterization and localization of the fibronectin-binding site. Biochemistry. 2002;41(25):7907–13 [DOI] [PubMed] [Google Scholar]
- 50.Serrano K, Devine DV. Intracellular factor XIII crosslinks platelet cytoskeletal elements upon platelet activation. Thromb Haemost. 2002;88(2):315–20 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.