Abstract
Biological dinitrogen (N2) fixation supplies nitrogen to the oceans, supporting primary productivity, and is carried out by some bacteria and archaea referred to as diazotrophs. Cyanobacteria are conventionally considered to be the major contributors to marine N2 fixation, but non-cyanobacterial diazotrophs (NCDs) have been shown to be distributed throughout ocean ecosystems. However, the biogeochemical significance of marine NCDs has not been demonstrated. This review synthesizes multiple datasets, drawing from cultivation-independent molecular techniques and data from extensive oceanic expeditions, to provide a comprehensive view into the diversity, biogeography, ecophysiology, and activity of marine NCDs. A NCD nifH gene catalog was compiled containing sequences from both PCR-based and PCR-free methods, identifying taxa for future studies. NCD abundances from a novel database of NCD nifH-based abundances were colocalized with environmental data, unveiling distinct distributions and environmental drivers of individual taxa. Mechanisms that NCDs may use to fuel and regulate N2 fixation in response to oxygen and fixed nitrogen availability are discussed, based on a metabolic analysis of recently available Tara Oceans expedition data. The integration of multiple datasets provides a new perspective that enhances understanding of the biology, ecology, and biogeography of marine NCDs and provides tools and directions for future research.
Keywords: diazotrophs, marine nitrogen cycle, nitrogen fixation, non-cyanobacterial diazotrophs
The authors discuss recent advances that highlight marine non-cyanobacterial diazotrophs in marine ecosystems, with a focus on diversity, drivers of their global biogeography and potential ecophysiologies, and their significance to the nitrogen cycle in well-lit oxygenated euphotic waters.
Introduction
Biological dinitrogen (N2) fixation is the microbial process, whereby otherwise inert N2 gas is reduced to biologically available nitrogen (N) in the form of ammonia (NH3). In the oceans, the N supplied through this process can support primary productivity, especially in N-limited surface waters and, consequently, the vertical transport (export) of carbon (C) to the deep ocean (Karl et al. 1997, Knapp et al. 2018, Zehr and Capone 2021). More broadly, the balance between N inputs from N2 fixation and losses from denitrification and anaerobic ammonium (NH4+) oxidation set the oceanic inventory of reactive N, impacting the global C cycle and Earth’s climate system (Gruber and Galloway 2008). Rates of N2 fixation in the oceans have strong biogeographical patterns, with the highest rates measured in surface open-ocean and coastal waters of the tropics, subtropics, and temperate zones (e.g. Capone et al. 2005, Berthelot et al. 2017, Tang et al. 2019b), and lower rates in polar regions (Blais et al. 2012, Sipler et al. 2017, Shiozaki et al. 2020) and the deep sea (reviewed by Moisander et al. 2017, Benavides et al. 2018a). Marine N2 fixation has been best described in epipelagic environments, where rates are generally limited by temperature, nutrient [phosphorus (P) and iron (Fe)] concentrations, and N:P and/or N:Fe ratios (Mills et al. 2004, Moore et al. 2009, Letelier et al. 2019, Tang et al. 2019a).
While the importance of N2 fixation to marine biogeochemical cycling is indisputable, there is still uncertainty concerning which N2-fixing microorganisms (diazotrophs) contribute to this process in marine euphotic waters. Early microscopy-based analyses established the importance of filamentous cyanobacterial diazotrophs (Trichodesmium and heterocyst-forming symbionts of diatoms) in tropical and subtropical surface waters (e.g. Dugdale et al. 1961, Mague et al. 1974, Venrick 1974, Carpenter and Price 1977, Saino and Hatori 1980). More recently, marine diazotrophic diversity has been investigated using molecular approaches targeting the nifH gene, which encodes the two identical subunits of the Fe protein of the nitrogenase enzyme complex and serves as a phylogenetic marker (Zehr and McReynolds 1989, Gaby and Buckley 2011). These approaches led to the discovery of diverse cyanobacterial diazotrophs in the open ocean, as well as diazotrophs from many lineages not within the phylum Cyanobacteria, which we refer to as non-cyanobacterial diazotrophs (NCDs; see Box 1; Zehr et al. 1998). Subsequent ocean surveys showed that nifH genes from NCDs are ubiquitous in marine waters and can reach higher relative abundances than their cyanobacterial counterparts (Riemann et al. 2010, Farnelid et al. 2011). However, very few marine NCDs have been cultivated and the few isolates that exist have not provided unequivocal evidence of N2 fixation in situ (Bostrom et al. 2007, Farnelid et al. 2014, Bentzon-Tilia et al. 2015a, Martinez-Perez et al. 2018). To date, direct demonstrations of NCD N2 fixation in situ using cultivation-independent approaches are extremely limited. Thus, the biogeochemical significance of NCDs remains unknown.
Box 1.
The case for using the term ‘non-cyanobacterial diazotroph’
This review focuses on microorganisms referred to collectively as non-cyanobacterial diazotrophs, or NCDs. This term was previously defined by Moisander et al. (2017) and includes all diazotrophic bacteria and archaea not part of the phylum Cyanobacteria. We argue that the term NCD is more accurate than other terms used in the marine literature, such as ‘heterotrophic diazotrophs’ and ‘heterotrophic bacterial diazotrophs.’
Diazotrophic diversity is typically characterized by sequencing the nifH gene, which largely has congruence with 16S rRNA gene-based phylogeny (Zehr et al. 2003). This congruence is strong for cyanobacteria, such that if unknown sequences group within nifH Cluster 1B, they are phylogenetically distinguished as cyanobacteria. Most of these organisms are photoautotrophs, capable of using light as a source of energy and carbon dioxide as a C source, though there are some notable exceptions found among unicellular cyanobacterial symbionts of haptophytes and diatoms (Tripp et al. 2010, Nakayama and Inagaki 2017).
Unfortunately, nifH gene sequences are not easily used to predict the metabolic strategy a particular NCD may use to acquire energy and C. Although there is some congruence between nifH- and 16S rRNA gene-based phylogenies for non-cyanobacteria, there are exceptions for important marine phyla, including the proteobacteria, which are spread across multiple nifH clusters (Table S1, Supporting Information). More broadly, it can be challenging to infer metabolic potential from a single gene. Even well-established phylogenetic markers like the 16S rRNA gene are difficult to use for resolving species- and strain-level identities (Johnson et al. 2019), which are needed to infer metabolic strategies from genera containing high metabolic diversity, such as the γ-proteobacterial genus Pseudomonas (Jun et al. 2016).
Since nifH gene sequences cannot be used to accurately infer the metabolic potential of all NCDs, the terms ‘heterotrophic bacterial diazotrophs (HBDs)’ (Delmont et al. 2018) or ‘heterotrophic diazotrophs’ (Gradoville et al. 2017a) could potentially be inaccurate. For example, γ-proteobacterial diazotrophs within Cluster 1G include cultivated microorganisms with divergent metabolic strategies, e.g. M. purpuratum, which uses light as a source of energy and organic C as the source of cell C (photoheterotrophy), some strains of P. stutzeri, which are facultative anaerobes that oxidize inorganic compounds as a source of energy for growth (chemolithotrophy), and A. vinelandii which are aerobes that use organic C as their major source of energy and C (chemoheterotrophy). Furthermore, NCDs include archaea, and are thus not all bacterial.
Ideally, NCDs should be subclassified according to their potential metabolic traits. This will become more feasible with advances the isolation and genome sequencing of marine NCDs (see ‘Diversity and ecophysiological features inferred from MAGs’). Here, we decided to use the term NCD as an accurate and inclusive way to refer to this group without implying a particular metabolic strategy.
Much of the ambiguity concerning the significance of marine NCDs stems from uncertainties about their ecophysiology, particularly in well-lit, oxygenated marine habitats. There are several major challenges that marine NCDs must overcome (reviewed by Bombar et al. 2016). N2 fixation is energetically expensive, with high ATP and reductant requirements (Postgate 1982). Cyanobacterial diazotrophs can produce organic C and acquire energy through oxygenic photosynthesis, although there are some exceptions such as the photoheterotrophic symbiont UCYN-A (Tripp et al. 2010). In contrast, marine NCDs are thought to utilize organic substrates and/or alternative energy sources (including possibly light) to meet energy and C requirements. Similar to model NCDs from terrestrial systems (Dixon and Kahn 2004), various energy acquisition pathways have been observed in marine NCD genomes, which are speculated to be important strategies to fuel growth and N2 fixation activity (Bentzon-Tilia et al. 2015a, Martinez-Perez et al. 2018, Acinas et al. 2021). Many marine NCDs are presumed to be chemoheterotrophic, thus may have difficulty acquiring sufficient energy for N2 fixation in euphotic open-ocean waters, which are often depleted in labile organic C. Indeed, amendment experiments suggest that the abundances and/or N2 fixation activity of NCDs are limited by the availability of dissolved organic C (DOC) in many pelagic marine environments (Moisander et al. 2012, Dekaezemacker et al. 2013, Rahav et al. 2016).
A major challenge facing NCDs (and all diazotrophs) is the irreversible inactivation of the nitrogenase enzyme by oxygen (O2; Gallon 1992). The ecological advantage of N2 fixation is hindered by the costs of cellular strategies needed to protect nitrogenase from O2. Cyanobacterial diazotrophs have evolved numerous mechanisms to separate N2 fixation from O2 either in space or in time (or both), including cellular differentiation, symbiosis, membranes to restrict O2 diffusion, and increased rates of cellular respiration (reviewed by Zehr and Capone 2020). These mechanisms have energetic costs that would further exacerbate the C requirements for NCDs. Particles have been proposed as habitats for NCD N2 fixation, since they can provide both a C source and the potential for low O2 microzones (Paerl and Prufert 1987, Riemann et al. 2010, 2022, Bombar et al. 2016). However, active transcription of NCD nifH genes is also found in free-living size fractions (Salazar et al. 2019) and on particles too small to theoretically support low O2 conditions (Farnelid et al. 2019). In the terrestrial environment, free-living Azotobacter vinelandii fixes N2 under aerobic conditions through increased respiration to consume intracellular O2 and by investing in chemical barriers (like exopolysaccharides) and larger cell sizes to reduce O2 intrusion into the cell (Post et al. 1982, Poole and Hill 1997, Sabra et al. 2000). It is likely that marine NCDs inhabiting oxic environments may use similar strategies.
The presence of nitrate (NO3−) and NH4+ is generally thought to inhibit N2 fixation, either by supporting the growth of fast-growing phytoplankton that outcompete diazotrophs for other limiting nutrients (Ward et al. 2013), or by providing fixed N that some diazotrophs can utilize instead of investing in N2 fixation (although inhibition is variable, see Knapp 2012). However, fixed inorganic N is abundant in subeuphotic zone waters and coastal or upwelling ecosystems where N2 fixation by NCDs has been implicated (reviewed by Moisander et al. 2017), as well as in sediment systems where N2 fixation occurs (Capone 1983), underscoring that the sensitivity of NCDs to fixed N is not well-understood. Furthermore, in low O2 environments, N2 fixation in the presence of high NH4+ has been speculated to drive the balance of internal cellular redox states in Rhodopseudomonas palustris (isolate BAL398) a model NCD from the Baltic Sea (reviewed by Bombar et al. 2016). However, it is unknown whether some marine NCDs may use N2 fixation as a redox balancing strategy in low O2 environments (such as on particles).
Despite these challenges, NCDs appear to be widely distributed and occasionally transcriptionally active in marine euphotic waters. Measuring the activity of NCDs and quantifying their importance to marine N2 fixation is an active area of research. Fortunately, the past decade has seen a significant improvement in the tools and data available to study marine NCDs. High throughput sequencing (HTS) technologies and increased sampling efforts have exponentially increased the amount of nifH amplicon sequence data available for marine diazotrophs (e.g. Farnelid et al. 2011, Turk-Kubo et al. 2015, Shiozaki et al. 2017, Raes et al. 2020). Dozens of individual NCD nifH gene sequence types have been enumerated using quantitative PCR (qPCR), resulting in large datasets showing regional and seasonal abundance patterns (e.g. Langlois et al. 2015, Shiozaki et al. 2017, Cheung et al. 2021, Shao and Luo 2022). Furthermore, advances in sequencing technologies have enabled the detection of nifH genes from global ocean metagenomes and metatranscriptomes (Delmont et al. 2018, 2022, Salazar et al. 2019, Acinas et al. 2021, Pierella Karlusich et al. 2021, Poff et al. 2021), allowing for the validation of NCD distribution patterns without relying on the use of primers for the PCR-based amplification of NCD nifH gene sequences, and for the construction of metagenome-assembled genomes (MAGs) to evaluate the metabolic potential of uncultivated NCDs. Finally, advances in single-cell techniques are beginning to enable visualization and in situ single-cell N2 fixation rate measurements of NCDs (e.g. Martinez-Perez et al. 2018, Geisler et al. 2019).
This review synthesizes these new data to better understand the role of NCDs in marine waters. In ‘NCD diversity: nifH gene catalog,’ we introduce a novel catalog of marine NCD gene diversity that compiles available nifH sequence data from the NCBI Genbank non-redundant (nr) database and selected nifH HTS studies, qPCR targets, and global ocean metagenomes. ‘Habitats and environments of NCDs in marine systems’ reviews the known environments and habitats for NCDs in marine systems. In ‘Environmental drivers of NCD biogeography, activity, and presumed N2 fixation,’ we discuss known environmental controls on their abundances and activity, introduce a novel database of available NCD nifH gene abundances, and discuss the global distributions and environmental drivers of individual phylotypes. ‘NCD biogeography and ecophysiology from metagenomes and metatranscriptomes’ explores the potential ecophysiological features of marine NCDs using newly available MAGs from the global ocean (Delmont et al. 2022). In ‘Moving from genes to rates: are NCDs fixing N2 in the pelagic oceans?,’ we consider the current state of knowledge on NCD N2 fixation activity and in ‘Future perspectives’ we discuss remaining knowledge gaps and future perspectives for assessing the contribution of NCDs to N2 fixation in marine waters.
NCD diversity: nifH gene catalog
Knowledge of NCD diversity is based on the nucleic acid sequences of the genes that encode the nitrogenase enzyme. Nitrogenase is composed of dinitrogenase reductase and dinitrogenase, which are the Fe and molybdenum (Mo)-Fe (MoFe) containing metalloproteins, respectively, in the most common form. The MoFe protein-containing nitrogenase is encoded by the nifHDK operon; nifDK encodes the dinitrogenase alpha and beta subunits (containing MoFe) and nifH encodes dinitrogenase reductase (containing Fe). Less common nitrogenases substitute Fe or vanadium for Mo and are encoded by the vnfHDGK and anfHDGK genes, respectively (Seefeldt et al. 2009, Newton 2015). N2 fixation requires the involvement of many proteins and factors beyond nitrogenase, thus is a highly regulated and complex process ultimately set by intracellular N status and O2, Mo, and Fe concentrations, as well as available energy sources (Dixon and Kahn 2004, Leigh and Dodsworth 2007, Masepohl 2017).
The nifH gene is the most widely used proxy for N2 fixation potential in the marine environment. While lateral gene transfer has been observed in some taxa (Bolhuis et al. 2010), nifH gene-based phylogeny is broadly congruent with 16S rRNA gene-based phylogeny and four major nifH gene clusters have been defined (Zehr et al. 2003; Table S1, Supporting Information). Early application of ‘universal’ nifH PCR assays using pelagic marine samples by Zehr et al. (1998) established that there were multiple lineages of marine unicellular cyanobacteria along with ɑ-, β-, and γ-proteobacteria, sulfate reducers (δ-proteobacteria), and Clostridia among the picoplankton population. Over the past quarter-century, additional diazotrophic lineages have been identified, including marine representatives from each of the four major nifH clusters. Applications of new molecular techniques, including HTS of nifH amplicons and metagenomic-/transcriptomic-based approaches, have greatly increased the known nifH-based diversity of marine diazotrophs; however, most individual studies have focused on cyanobacterial diazotrophs, and no comprehensive compilations of the currently known NCD diversity exist.
We compiled a catalog of marine NCD nifH gene sequences (Table S2, Supporting Information; dataset doi: 10.5281/zenodo.6537451). This catalog is a new community resource that enables analysis of nifH datasets in the context of prior studies and identifies NCD sequences that have been recovered using different approaches. The NCD nifH gene catalog includes sequences recovered from: (1) studies using nifH PCR and cloning-based approaches (archived in the NCBI nr database); (2) select studies employing HTS of nifH PCR amplicons; (3) metagenomic/transcriptomic datasets from recent large-scale ocean surveys; and (4) targets for qPCR and digital droplet PCR (ddPCR) quantitative methods.
Briefly, marine-derived sequences from the NCBI nr database were selected from a curated nifH database (Heller et al. 2014 updated in June 2017) using available metadata. All marine nr-derived sequences (n = 23 848) were clustered at 97% amino acid identity using CD-HIT (Huang et al. 2010), and NCD operational taxonomic units (OTUs) with <100 sequences were removed. This resulted in 34 OTUs (representing 9360/23 848 of the total marine-derived sequences). Additionally, sequences representing the top three most abundant NCD OTUs (or amplicon sequence variants, ASVs) from nifH HTS gene studies that sampled the North Pacific (Shiozaki et al. 2017, Farnelid et al. 2019, Cabello et al. 2020, Gradoville et al. 2020, Sato et al. 2021, Turk-Kubo et al. 2021), South Pacific (Turk-Kubo et al. 2015), South Atlantic (Ribeiro et al. 2018), Indian Ocean (Wu et al. 2021), and polar regions (Shiozaki et al. 2018b, 2020) were also included. These studies were selected based on the accessibility of reference sequences for reported NCDs; sequences have been renamed according to the region, author, and specified name from the original publication (e.g. ‘Npac-Shio-otu00004’ for otu00004 from the North Pacific in Shiozaki et al. 2017; Table S1, Supporting Information). The catalog also includes NCD nifH sequences obtained using PCR-free approaches from Tara Oceans metagenomes, metatranscriptomes, and MAGs (Delmont et al. 2018, 2022, Salazar et al. 2019, Cornejo-Castillo and Zehr 2021, Pierella Karlusich et al. 2021) and the Polar Microbe R Gene Catalog (PM-RGC; Cao et al. 2020). PM-RGC nifH sequences were identified via blastp against a curated database containing genome-derived nifH sequences (jzehrlab.com/nifh). Finally, the catalog includes 55 reference sequences from NCDs targeted by published nifH qPCR/ddPCR assays, since these were identified as recurrently present diazotrophs from independent studies.
Together, the NCD nifH gene catalog contains 204 total sequences (Table S2, Supporting Information; dataset doi: 10.5281/zenodo.6537451). OTUs comprised of sequences with >99% amino acid identity were identified using CD-HIT (Huang et al. 2010) to explore which sequence types have been recovered using different methodological approaches. OTUs containing sequence types detected using both nifH PCR-based and PCR-free (derived from metagenomics/metatranscriptomics) methods are of particular interest, some of which are discussed below.
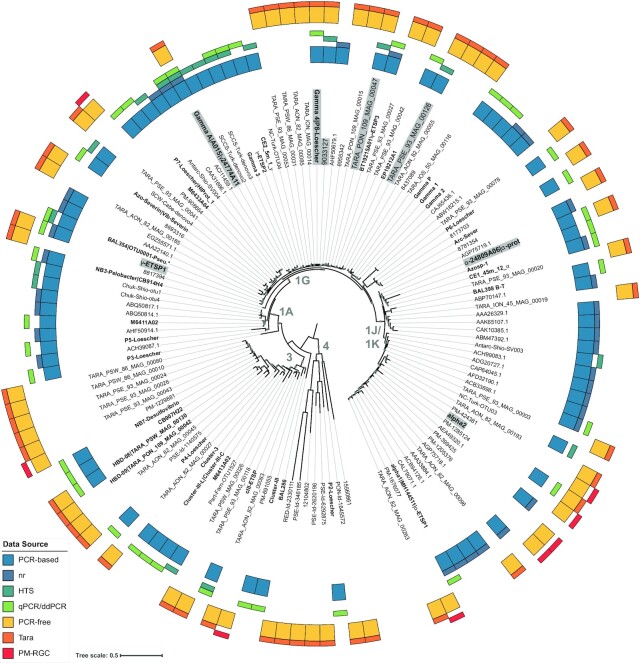
The Gamma A and Gamma 4 OTUs (1G) contain sequences from both PCR and non-PCR-based approaches (Fig. 1) and are arguably the best-studied marine NCD groups. The widespread distribution of Gamma A throughout tropical and subtropical surface waters has been demonstrated in qPCR-based studies (see ‘Environmental drivers of NCD biogeography, activity, and presumed N2 fixation’), nifH gene HTS studies (e.g. Npac-Shio-otu00004 from Shiozaki et al. 2017, SAtl_Ribe_otu006 from Ribeiro et al. 2018), and in a metagenome-based study (Cornejo-Castillo and Zehr 2021). Gamma A nifH transcripts have been detected in the environment (e.g. Bird et al. 2005, Moisander et al. 2014, Cornejo-Castillo and Zehr 2021) and this organism is hypothesized to be associated with small particles or picophytoplankton (Benavides et al. 2016b, Cornejo-Castillo and Zehr 2021; Fig. 2a). Gamma 4 is another γ-proteobacterium emerging as a potentially important marine NCD. Originally described as a qPCR target by Halm et al. ( 2012), and later observed in the Eastern Tropical South Pacific (P8; Löscher et al. 2014), this group now includes a MAG assembled from North Pacific samples collected during the Tara Oceans expedition (TARA_PON_109_MAG_00010, also referred to as HBD-06; Delmont et al. 2018, 2022) and can reach high abundances (5.8 × 106nifH gene copies l−1) in the North Pacific (Cheung et al. 2021).
Figure 1.
NCD diversity includes taxa found using PCR-based and PCR-free approaches. Phylogenetic tree represents the NCD nifH gene catalog based on amino acid sequences from OTU representatives. Sequences were aligned to the NifH/frxC family (Fer4_NifH; PF00142) using HMMalign in HMMER software v2.4 (Finn et al. 2011). Tree topology was calculated using FastTree 2.1.11 (Price et al. 2010) using maximum-likelihood rearrangements and the JTT model for nucleotide evolution. Branch support was determined using the Shimodaira–Hasegawa test (>50% support indicated with small gray squares on branches). iTOL 6.5.2 (Letunic and Bork 2021) was used to visualize the tree and display the source(s) of sequences in each cluster. nifH clusters are defined according to the convention established in Zehr et al. (2003) and are indicated in gray text in the center of the tree. Representative sequences affiliated with NCD qPCR/ddPCR assays are in bold. OTUs that contain sequences derived from both PCR-based and PCR-free approaches are in shaded boxes. Branches with multiple names indicate OTUs that contain sequences targeted by more than one qPCR assay. Thick outer color bars show if the sequences were acquired through PCR-based (teal) or PCR-free (yellow) methods, while the thinner color bars correspond to the specific method (PCR-based) or sampling campaign (PCR-free). Interactive tree publicly available at https://itol.embl.de/shared/1HrZrblPr7p4s. See Table S2 (Supporting Information) for supporting data.
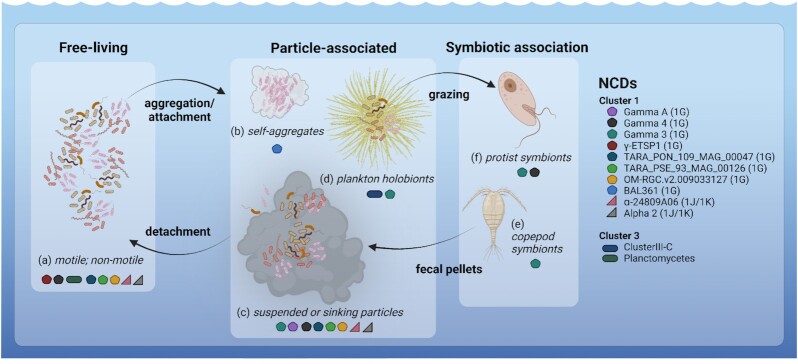
Figure 2.
NCDs occupy diverse marine habitats. NCDs in marine waters may be free-living and motile or nonmotile (a), associated with various particles including self-aggregates (b), suspended or sinking particles (c), plankton holobionts (d), or live in symbiosis with copepods (e) or other protists (f). Presumed habitats of some NCD taxa which are discussed throughout this review are indicated using symbols described in the legend. Created with BioRender.com.
Other potentially important γ-proteobacterial OTUs emerged from this meta-analysis. γ-ETSP1 was first described as a qPCR target in the South Pacific (Turk-Kubo et al. 2014), but also includes a MAG (TARA_AOS_82_MAG_00008), which was detected mainly in the Atlantic Ocean in <5µm size fractions (Delmont et al. 2022). In total, three additional γ-proteobacterial OTUs contained both MAGs and abundant sequence types from nifH gene HTS studies: TARA_PON_109_MAG_00047 included a sequence from Shiozaki et al. (2017), and TARA_PSE_93_MAG_00126 and OM-RGC.v2.009033127 included sequences from Gradoville et al. (2020; Table S2, Supporting Information). The recovery of numerous γ-proteobacterial sequences from PCR-based studies is notable, given recent assertions that primer mismatches could result in lack of amplification of this group (Delmont et al. 2018, 2022). Our analysis reinforces that γ-proteobacteria are recovered by both approaches and are among the most diverse NCD groups in the marine pelagic environment.
Several OTUs affiliated with nifH cluster 1J/1K were also found in both PCR-based and PCR-free studies (Fig. 1). The α-24809A06 OTU contains sequences related to putative ɑ-proteobacteria reported in the South China Sea (SCS; ɑ-24809A06; Moisander et al. 2007) and mesopelagic waters (Azosp_1; Hewson et al. 2007), and also includes a MAG (TARA_PSW_86_MAG_00238; Delmont et al. 2022) detected predominantly in the Pacific. A second putative α-proteobacterial OTU, Alpha 2, contained a sequence type targeted by qPCR in the Northern SCS (Alpha 2; Chen et al. 2019) and a MAG (TARA_AON_82_MAG_00070; Delmont et al. 2022) observed in the <20µm size fraction in several ocean basins including the Atlantic, Indian, and Pacific.
Unlike the proteobacteria, no putative δ-proteobacterial OTUs (cluster 3 or 1A) were found using both PCR-based and PCR-free approaches. Cluster 3 contains sequences from PCR-based studies and from MAGs including Planctomycetes lineages speculated to be important NCDs (Delmont et al. 2018, 2022). However, sequences affiliated with cluster 1A were recovered only by PCR-based techniques. In this dataset, cluster 1A sequences are predominant in coastal ecosystems (e.g. Short and Zehr 2007, Shiozaki et al. 2018b) that were not heavily sampled in the Tara Oceans expedition, which may partially explain this discrepancy.
Caveats and considerations for nifH sequence analysis
Although PCR-based approaches have been foundational in the study of marine diazotrophs, there are several challenges and caveats associated with using nifH sequences to explore the diversity of diazotrophs. Numerous sets of primers have been developed to amplify nifH genes, many of which have a high degree of degeneracy and are biased towards certain taxa (Gaby and Buckley 2011). The most widely applied assay in the marine environment (nifH1-4; Zehr and McReynolds 1989, Zani et al. 2000) has known biases, especially within the cyanobacteria (Caputo et al. 2018). However, recent assertions that nifH1-4 primers may not amplify some of the NCDs recovered using ‘omics-based approaches, e.g. nifH genes from Planctomycetes (Delmont et al. 2018, 2022), are primarily based on a single mismatch against the nifH4 primer located at the 5′ end of the priming site (Table S2, Supporting Information), which does not impact PCR amplification (Bru et al. 2008). Our NCD gene catalog shows that several MAG-derived sequences asserted to have incompatibilities with the nifH1-4 primers have been recovered from PCR-based surveys, underscoring that the 5′ mismatch does not prevent PCR amplification (Table S3, Supporting Information). Therefore, the nifH1-4 primers do not appear to be broadly incompatible with pelagic marine NCD taxa.
There are several additional technical limitations of PCR-based approaches. Due to the generally low abundances of diazotrophs, amplification typically requires a nested PCR approach and many rounds of amplification, which introduces bias in relative abundances (Turk et al. 2011). Recovery of nifH gene fragments can also be influenced by contamination from numerous sources; differentiating between marine- and contaminant-sourced nifH genes can be challenging, particularly for NCDs (Zehr et al. 2003). This has occasionally been addressed in individual studies by processing reagent blanks or comparing data to known contaminant sequences (Bostrom et al. 2007, Farnelid et al. 2011, Blais et al. 2012, Moisander et al. 2014, Langlois et al. 2015, Fernandez-Mendez et al. 2016, Cheung et al. 2021), so that putative contaminant sequences can be removed during analysis.
Detection of nifH transcripts in environmental samples is sometimes used as a proxy for active N2 fixation, and in early studies was the standard for choosing targets for qPCR assays. However, detection of nifH transcripts may be heavily dependent on the time of sampling due to the diel pattern in nifH gene expression (observed in diazotrophs including marine cyanobacteria and terrestrial NCDs; Wyman et al. 1996, You et al. 2005), size fraction (which can reflect lifestyle, e.g. symbiont, particle-attached), and environmental controls (e.g. O2 concentration, presence of reduced N, and availability of organic C and Fe). Furthermore, while the detection of nifH transcripts may reflect active transcription of the nif operon, active N2 fixation is also under post-transcriptional control. Therefore, nifH transcript detection (or lack thereof) as evidence of active/inactive N2 fixation should be interpreted with caution.
Recovering nifH gene sequences from the environment, regardless of the approach, does not guarantee that they are sourced from organisms capable of fixing N2 as some taxa contain nifH genes but lack other genes required for a functional nitrogenase (Dos Santos et al. 2012, Mise et al. 2021). Furthermore, alternative nitrogenases in marine diazotrophs are not well-understood, but also signify the potential to fix N2 (McRose et al. 2017, Reeder and Löscher 2022), which is not captured in environmental nifH gene surveys. Nevertheless, approaches targeting partial nifH gene sequences have generated the majority of available marine NCD data and have enabled numerous insights into their diversity, distribution, and environmental drivers.
Habitats and environments of NCDs in marine systems
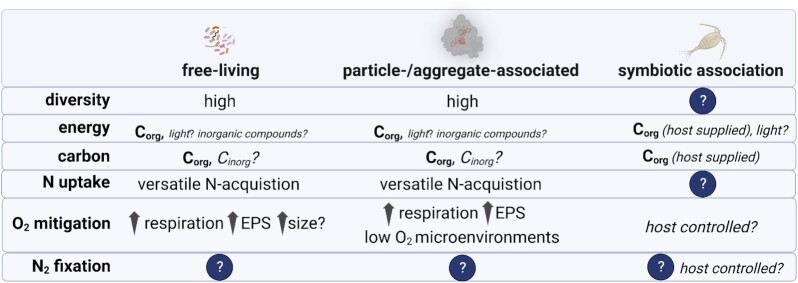
Marine euphotic waters are heterogeneous environments fostering free-living NCDs, NCDs attached to living or detrital particulate material, and NCDs likely living in symbiosis with protists (Fig. 2). These contrasting habitats can be found in close proximity, thus the scale of standard oceanographic sampling (typically milliliters–liters) can conceal complex life strategies and interactions. For example, some NCD taxa likely cycle through free-living (Fig. 2a) and particle-associated lifestyles (Fig. 2b-d), evidenced by their occurrence across size fractions (Pierella Karlusich et al. 2021) as well as thier potential for motility (Hallstrøm et al. 2022c) and the formation of self-aggregates (Bentzon-Tilia et al. 2015a). Other taxa may form facultative or obligate symbioses with other planktonic taxa (Fig. 2f and e). Rates and magnitudes of NCD N2 fixation in oxygenated euphotic waters are likely influenced by these different lifestyles. For example, N2 fixation by free-living or particle-associated NCDs would be dependent on a given taxa’s metabolic potential and ability to respond to a changing environment, while N2 fixation by NCDs living in symbiosis may be under host control like in other marine and terrestrial symbioses (Smercina et al. 2019, Landa et al. 2021, Mohr et al. 2021). Teasing apart the habitats and lifestyles of NCDs in specific environments remains a challenge but is key to linking diversity to active N2 fixation.
NCDs are also present in many marine environments beyond the euphotic zone (Table 1). Since the first reports of NCDs in the surface ocean (Paulsen et al. 1991, Coyer et al. 1996, Zehr et al. 1998), technological advances and ambitious sampling campaigns have highlighted a diversity of water column environments harboring NCDs, including coastal waters and estuaries, polar seas, aphotic environments, and O2 minimum zones (OMZs). Diverse benthic ecosystems also harbor NCDs and may seed them into the water column, where they become part of the rare biosphere (Pedros-Alio 2012, Troussellier et al. 2017). Here, we review known water column NCD habitats and environments (‘Habitats and environments of NCDs in marine systems’) before discussing global NCD distributions and environmental drivers (‘Environmental drivers of NCD biogeography, activity, and presumed N2 fixation’).
Table 1.
Marine NCD environments
| Environment | Known or theorized NCD habitat/lifestylea | Representative references | |
|---|---|---|---|
| Water column | Open ocean: euphotic | Free-living; suspended and sinking particles; plankton holobionts | Proctor (1997), Zehr et al. (1998), Langlois et al. (2005, 2008, 2015), Church et al. (2005a, b), Fong et al. (2008), Moisander et al. (2008, 2014), Farnelid et al. (2010,2011) Bombar et al. (2011), Kong et al. (2011), Zhang et al. (2011, 2016), Halm et al. (2012), Shiozaki et al. (2014,2017) Sunagawa et al. (2015), Azimuddin et al. (2016), Benavides et al. (2016b), Gradoville et al. (2017a), Delmont et al. (2018), Chen et al. (2019), Wu et al. (2019), Yang et al. (2019), Cheung et al. (2020, 2021), Raes et al. (2020), Pierella Karlusich et al. (2021), Hallstrøm et al. (2022a) |
| Open ocean: aphotic | Free-living; suspended and sinking particles | Hewson et al. (2007), Bonnet et al. (2013), Benavides et al. (2015, 2018c), Selden et al. (2019), Acinas et al. (2021) | |
| Oxygen minimum zones | Free-living; suspended and sinking particles | Fernandez et al. (2011), Hamersley et al. (2011), Löscher et al. (2014), Jayakumar et al. (2017), Chang et al. (2019), Reeder and Löscher (2022) | |
| Temperate coastal ecosystems | Free-living; suspended and sinking particles; plankton holobionts; resuspended sediments; terrestrial particles | Short et al. (2004), Moisander et al. (2007), Rees et al. (2009), Mulholland et al. (2012), Messer et al. (2015, 2021), Scavotto et al. (2015), Shiozaki et al. (2015), Bentzon-Tilia et al. (2015b), Gradoville et al. (2017a), Pedersen et al. (2018), Cabello et al. (2020), Turk-Kubo et al. (2021), Hallstrøm et al. (2022b) | |
| Inland seas | Free-living; suspended and sinking particles | Bostrom et al. (2007), Man-Aharonovich et al. (2007), Farnelid et al. (2013), Rahav et al. (2013, 2016), Benavides et al. (2016a), Kirkpatrick et al. (2018), Geisler et al. (2020), Ridame et al. (2022) | |
| Polar seas | Free-living; suspended and sinking particles; sediment resuspension; terrestrial input | Blais et al. (2012), Fernandez-Mendez et al. (2016), Shiozaki et al. (2018b, 2020) | |
| Benthic | Coral reefs and sponges | Associated with released mucus, coral tissues, coral skeletons, associated macroalgae, and microbial mats, surrounding seawater | Mohamed et al. (2008), Olson et al. (2009), Lema et al. (2012, 2014), Ribes et al. (2015), Yang et al. (2016), Liang et al. (2020), Moynihan et al. (2022) |
| Macroalgae | Living and decomposing tissue, surrounding seawater | Hamersley et al. (2015), Zhang et al. (2015), Raut et al. (2018), Raut and Capone (2021) | |
| Coastal and continental shelf sediments | Sediment-associated | Fulweiler et al. (2013), Brown and Jenkins (2014), Newell et al. (2016), Jabir et al. (2018) | |
| Oxygen-deficient zone sediments | Sediment-associated | Gier et al. (2016, 2017) | |
| Abyssal pelagic | Associated with methane seeps, hydrothermal vents, whale falls | Mehta et al. (2003), Mehta and Baross (2006), Pernthaler et al. (2008), Dang et al. (2009), Dekas et al. (2009, 2014, 2016, 2018), Miyazaki et al. (2009), Kapili et al. (2020) | |
| Microbial mats | Microbial mat associated, surrounding water | Zehr et al. (1995), Steppe et al. (1996), Olson et al. (1999) | |
| Seagrass rhizosphere | Sediments, roots | McGlathery et al. (1998), Mohr et al. (2021) |
Allocation of NCDs between free-living and particle-association is not well-resolved.
Marine particles
Although little is known about the physiology of NCDs and how they fix N2 in oxygenated surface waters, for over two decades it has been theorized that NCDs residing in sunlit surface waters have a particle- or aggregate-associated lifestyle (Fig. 2b-d; Paerl and Prufert 1987, Church et al. 2005a, Riemann et al. 2010, Bombar et al. 2013, Rahav et al. 2016, Pedersen et al. 2018). Marine particles are an amalgamation of living and detrital material formed throughout the water column, with highest concentrations in the euphotic zone, especially in areas of high productivity, such as coastal and upwelling regions (Simon et al. 2002, Azam and Malfatti 2007). Particles provide substrates for attachment as well as access to organic C that could fuel chemoheterotrophic N2 fixation, while promoting the formation of low O2 microenvironments that provide protection for the O2 sensitive nitrogenase enzyme (Paerl 1985, Ploug et al. 1997, Ploug 2001, Ploug and Bergkvist 2015). Cyanobacterial aggregates (Klawonn et al. 2015) or self-produced NCD aggregates bound together by O2-impermeable exopolysaccharides may likewise form anoxic microniches for NCDs as exemplified by a Pseudomonas stutzeri strain (BAL361) isolated from the Baltic Sea (Fig. 2b; Bentzon-Tilia et al. 2015a, Paerl et al. 2018). A recent cellular model of A. vinelandii showed that the energetic costs to maintain low intracellular O2 for a free-living chemoheterotrophic diazotroph through increased respiration, synthesis of thicker cell membranes, or polysaccharide production surpassed the cost of N2 fixation (Inomura et al. 2017), underscoring the potential importance of particle- or aggregate-associated lifestyles.
To better understand the potential for particles as N2 fixation hot spots, Chakraborty et al. (2021) designed a mathematical model to investigate N2 fixation by NCDs bound to large (ca. 250 mm) sinking marine particles (reviewed by Riemann et al. 2022). Cells were modeled as facultative diazotrophs that acquire C and N from particle-supplied polysaccharides and polypeptides and only supplement cellular N requirements with N2 fixation once other N sources are exhausted, producing high C:N ratios favorable for heterotrophic N2-fixers. Microbial respiration depletes O2, however, the anoxic or microanoxic conditions created in the particle interior is temporary. Cells eventually become C-limited and O2 diffusion into the particle again exceeds microbial respiration, which in turn inhibits N2 fixation. NCD N2 fixation in the modeled particle interior thus depends on the surrounding water O2 concentration, initial composition and ratio of C to N in the particle, particle size (minimum of 0.6 mm diameter), and sinking speed. Based on this simulation, the amount of time a particle may harbor an anoxic or low O2 microzone and support N2 fixation can be short-lived (<1 day), which may make this process challenging to validate with experimental data.
A few recent studies show the most direct evidence to date of N2 fixation by NCDs bound to particles. In eutrophic waters of the Qishon estuary (Israel), which flow into the Mediterranean Sea, immunolabeling of the nitrogenase protein was used to demonstrate that NCDs colonized aggregates (Geisler et al. 2020). Furthermore, active N2 fixation by NCDs was implicated based on the detection of N2 fixation in particle-enrichment incubations where cyanobacterial photosynthesis was inhibited. This immunolabeling approach requires intact nitrogenase proteins but provides no phylogenetic information making interpretation challenging. Another recent study surveyed well-lit oxygenated pelagic waters of the North Pacific and measured the incorporation of 15N2 into putative NCDs (defined as cells with 15N enrichment, but not 13C enrichment after 24h 15N2/13C-bicarbonate incubations) residing on small particles (<210 μm, smaller than those predictied to host anaerobic microzones by Chakraborty et al. 2021) using particle-targeted nanoSIMS (Harding et al. 2022). Single cell N2 fixation rates ranged from low to quite high (0.02 to 8.61 fmol N cell-1 d-1) and in some cells could entirely fulfill cellular N requirements. Complementary nifH gene HTS indicated γ-proteobacterial NCDs, including Gamma A, were prevalent in the water column. However, particle-targeted nanoSIMS does not provide phylogenetic information needed to identify taxonomic affiliations of the active NCDs.
Despite this recent progress, most current knowledge of particle-associated NCDs is based on molecular surveys. Accounts of NCDs from large size fractions or marine particles, based on both PCR-based and PCR-free approaches (Turk-Kubo et al. 2014, Benavides et al. 2016b, Gradoville et al. 2020, Pierella Karlusich et al. 2021), collectively suggest diverse NCD communities attached to suspended particles, sinking particles (marine snow), and fecal pellets (Riemann et al. 2010), as well as forming self-produced aggregates (Bentzon-Tilia et al. 2015a). Notably, NCDs hypothesized to be associated with larger organisms either as symbionts or as prey items (Farnelid et al. 2009, Scavotto et al. 2015, Pierella Karlusich et al. 2021) could also be recovered from large size fractions. Techniques linking specific taxa to active N2 fixation are needed to assess the biogeochemical importance of these diverse particle-associated NCDs in the euphotic zone.
NCDs are also found on particles sinking out of the euphotic zone (Farnelid et al. 2019) and in the deep ocean (Rahav et al. 2013, Acinas et al. 2021, Poff et al. 2021). Particles (20–500 µm) sampled from floating net traps deployed at a depth of 150 m in the North Pacific often contained NCD sequences from Cluster 1 (including Part-Farn-OTU1012 and Part-Farn-OTU1107, which are Gamma 3 and Gamma A, respectively; Fig. 2c) and Cluster 3 (Farnelid et al. 2019; Table S2, Supporting Information). Notably, some sequences from individual particles (>20µm) were not well-represented in the whole water column diazotrophic community in this study, suggesting that preference for particle attachment may be taxa-specific. Additionally, three novel NCD MAGs have recently been described from mesopelagic particles including a member of the Micavibrionaceae family (Poff et al. 2021), a putative sulfur-oxidizing lithotroph (α-proteobacteria MAG0509), and a Gamma 4 MAG (MAG0081; Acinas et al. 2021). It remains unclear whether these sinking particle-associated NCDs are active N2-fixers, but collectively these studies suggest that particles delivered to the deep ocean harbor both distinct NCD lineages and those known to also reside in the surface ocean.
There remain large knowledge gaps about the role of marine particles in N2 fixation by NCDs. Most critically, although NCDs are clearly present on marine particles and there is some evidence of NCDs fixing N2, no studies have demonstrated N2 fixation by a taxonomically defined, particle-bound NCD. Further work is needed to determine the magnitude of potential fixed N inputs from particle-bound NCDs given the high concentration of particles in marine systems (Riemann et al. 2022).
Plankton holobionts
NCDs are often found associated with planktonic organisms, although the exact nature of these interactions is not well-described. NCDs affiliated with ɑ-, β-, and γ-proteobacteria, Clusters 2 and 3 have been found in association with heterotrophic dinoflagellate–cyanobacteria consortia (Farnelid et al. 2010). Cheung et al. (2021) noted that one of the prevalent γ-proteobacterial sequences found with several dinoflagellate genera described in Farnelid et al. (2010) was Gamma 4 (Fig. 2f); the lack of host specificity suggests a facultative symbiotic interaction, or possibly grazing on Gamma 4. Trichodesmium colonies also contain diverse assemblages of Cluster 3 and ɑ-, β-, and γ-proteobacteria, notably including ClusterIII-C (Fig. 2d; Gradoville et al. 2014, 2017b). More generally, δ-proteobacterial (1A) and γ-proteobacterial taxa (1G) have been found actively transcribing nifH in net tow samples (>100µm) (defined as ‘plankton associated diazotrophs’; Yang et al. 2019), including a putative Marichromatium-like sequence type closely related to Gamma 3 (KY774962.1, 98.4% nucleotide identity). NCDs are also associated with small photosynthetic picoeukaryotes, but these observations are sparse (Bombar et al. 2013).
Pelagic copepods have long been suspected to form symbiotic associations with N2-fixing bacteria, originally based on the visible growth of purple sulfur bacteria from copepod enrichment cultures allowed to develop anoxia, after which N2 fixation could be detected (Proctor 1997). Subsequent studies have described diverse NCD taxa presumed to be affiliated with copepods (Braun et al. 1999, Scavotto et al. 2015, Azimuddin et al. 2016). In addition, Scavotto et al. (2015) reported distinct NCD community compositions between full-gut and starved copepods that suggest δ-proteobacteria diazotrophs may be grazed from particles, while γ-proteobacteria may form more permanent symbioses with copepods.
Beyond the presence of NCD nifH genes, very little is known about these associations. Unique challenges exist in sampling plankton holobionts. For example, plankton nets collect (and concentrate) various types of aggregates in addition to plankton, thus do not allow for confident differentiation between plankton- and particle-associated lifestyles. Nonetheless, holobionts are understudied habitats for marine NCDs and more research is needed to better characterize these associations, and to elucidate the role a host or partner may have in regulating N2 fixation in plankton-NCD symbioses.
Coastal ecosystems
The availability of fixed inorganic N in coastal waters has been argued to select against N2 fixation, as diazotrophs are generally considered to be poor competitors for acquiring P and Fe compared to faster growing photoautotrophs such as diatoms (Ward et al. 2013, Landolfi et al. 2015). However, some cyanobacterial diazotrophs (most notably the UCYN-A symbiosis, e.g. Short and Zehr 2007, Mulholland et al. 2012, Turk-Kubo et al. 2021) and NCDs are now recognized as components of coastal microbial communities (Table 1). NCDs are often found in nutrient-replete coastal waters and can reach high relative abundances, particularly in regions with terrestrial input, e.g. from riverine or estuarine sources (Moisander et al. 2008, Bombar et al. 2011, Kong et al. 2011, Hashimoto et al. 2012, Shiozaki et al. 2015, Rahav et al. 2016, Geisler et al. 2020, Selden et al. 2021, Hallstrøm et al. 2022b). It is important to note, however, that very little is known about the physiology or activity of NCDs in coastal areas; their presence suggests that coastal NCDs may be insensitive to dissolved inorganic N availability and/or ‘facultative’ diazotrophs capable of utilizing alternate N sources (discussed in ‘Diversity and ecophysiological features inferred from MAGs’).
NCDs reported in euphotic coastal waters often reflect the typical diversity of marine sediments, which suggests that sediment resuspension may be an important mechanism for introducing NCDs into the water column in coastal ecosystems. Periodic resuspension events are prevalent in shallow waters and areas with high wind or wave activity (Zouch et al. 2018). In particular, Cluster 3 nifH sequences have been widely reported in coastal regions, occasionally transcribing nifH (Short et al. 2004, Bentzon-Tilia et al. 2015b, Shiozaki et al. 2020, Hallstrøm et al. 2022b), suggesting NCD N2 fixation in the oxygenated water column. In Western North Atlantic coastal waters, NCD nifH genes and transcripts dominate over those from cyanobacteria where salinity decreases due to freshwater input and turbidity increases through the resuspension of terrestrial and coastal sediments (Mulholland et al. 2019, Geisler et al. 2020, Selden et al. 2021, Hallstrøm et al. 2022b).
In the eutrophic Roskilde Fjord (Denmark), N2 fixation rates were higher in sediment-amended seawater than in the surrounding seawater (217 nmol N l−1 d−1 vs. 1.7 nmol N l−1 d−1, respectively), supporting the idea that NCDs resuspended from sediment fix N2 in the water column (Pedersen et al. 2018). This same study demonstrated that NCDs were secondary colonizers of artificial particles in natural seawater, indicating their capability for motility and particle attachment. This successional attachment to particles may be indicative of a preference for sufficiently large particles that support low O2 zones due to high activity of microbial respiration (see ‘Marine particles’).
Surface pelagic ocean waters
Open ocean gyres are typically N-limited and are known habitats for diazotrophs. Although the biogeography and environmental determinants are better understood for cyanobacterial diazotroph taxa, NCDs have been commonly recovered in nifH gene surveys dating back to early clone library-based studies in the North and South Pacific Oceans and the Mediterranean Sea (Zehr et al. 1998, 2003, Church et al. 2005b, Man-Aharonovich et al. 2007, Fong et al. 2008). Farnelid et al. (2011) were the first to apply nifH gene HTS in the global surface ocean, establishing that NCDs were broadly distributed and co-occur with cyanobacterial diazotrophs in tropical and subtropical waters. Farnelid et al. (2011) also revealed global taxa-specific NCD distribution patterns and confirmed that many NCDs were actively transcribing nifH, including those in small (<10µm) size fractions. Since this foundational work, studies leveraging nifH gene HTS have significantly expanded the known habitats for NCDs in the ocean, most notably in pelagic ecosystems (Table 1).
Several studies have demonstrated that distinct NCD communities may be found in different regions of the global ocean (Farnelid et al. 2011, Shiozaki et al. 2017, Raes et al. 2020), but there are also several NCDs that appear to have more cosmopolitan distributions. This includes arguably the best-studied NCD Gamma A (Langlois et al. 2005), also referred to as AO15 (Zehr et al. 1998), UMB (Bird et al. 2005), and γ-24774A11 (Moisander et al. 2008).
Gamma A is broadly distributed throughout the tropical and subtropical North Atlantic and Pacific (reviewed by Langlois et al. 2015) and is theorized to be an oligotrophic specialist. Evidence of Gamma A as a potential contributor to N2 fixation in oligotrophic gyres has been demonstrated in multiple surveys of nifH transcripts showing that Gamma A can account for a majority of the total transcript pool (Bird et al. 2005, Langlois et al. 2005, Church et al. 2005b, Bombar et al. 2011, Farnelid et al. 2011, Moisander et al. 2014, Messer et al. 2015, Shiozaki et al. 2017, Gradoville et al. 2020, Selden et al. 2021). Additionally, in the Western South Pacific, Gamma A nifH genes and transcripts positively correlate with cyanobacterial diazotroph abundances, temperature and DOC, and negatively with depth, chlorophyll a, and nutrients, suggesting similar physiological constraints as Trichodesmium and Crocosphaera (Moisander et al. 2014). The presence and nifH expression of Gamma A in well-lit surface waters suggests it benefits directly or indirectly from light. It has been speculated that Gamma A may have a photoheterotrophic lifestyle and utilize rhodopsin or bacteriochlorophyll-based energy generation or may rely on photosynthate from a photoautotroph (Langlois et al. 2015, Benavides et al. 2016b, Cornejo-Castillo and Zehr 2021). It is also found in association with larger size fractions (>3µm), which has raised speculation of a symbiotic- or particle-bound lifestyle (Benavides et al. 2016b, Cornejo-Castillo and Zehr 2021; Fig. 2a).
Many other NCD taxa are routinely recovered in nifH gene surveys and, more recently, in metagenomic surveys (see ‘Global ocean surveys using ‘omics provide insights into NCD abundance and diversity’) of the pelagic oceans. Several recent studies have demonstrated latitudinal shifts in NCD communities across the Pacific (Shiozaki et al. 2017, Gradoville et al. 2020, Raes et al. 2020). For example, cold, nutrient-rich sub-Antarctic waters support NCDs affiliated with α-, δ-, and ε-proteobacteria, along with Actinobacteria, while diverse γ-proteobacteria were seen in warmer, low latitude Pacific waters (Raes et al. 2020). Currently, the biological (e.g. nutrient limitation, grazing) or physical (e.g. advection on currents) mechanisms behind these observed latitudinal shifts in NCD community composition are not well-understood.
Aphotic waters and O2-deficient zones
Active N2 fixation attributed to NCDs has been reported in aphotic waters in the Pacific (Fernandez et al. 2011, Hamersley et al. 2011, Bonnet et al. 2013, Löscher et al. 2014, Benavides et al. 2015, 2018c, Jayakumar et al. 2017, Selden et al. 2019), Mediterranean Sea (Rahav et al. 2013, Benavides et al. 2016a), Baltic Sea (Farnelid et al. 2013), and Black Sea (Kirkpatrick et al. 2018). Aphotic N2 fixation rates are typically low (< 1 nmol N l−1 d−1) making them difficult to measure with accuracy and precision, especially given methodological challenges associated with the low biomass often recovered and the different 15N2 techniques (see ‘Moving from genes to rates: are NCDs fixing N2 in the pelagic oceans?’). However, considering the vastness of the marine aphotic zone, low but persistent rates of N2 fixation could significantly affect the global marine N budget (Bonnet et al. 2013, Benavides et al. 2016a). Several early studies reported aphotic N2 fixation rates in OMZs and suboxic waters (e.g. Fernandez et al. 2011, Hamersley et al. 2011, Farnelid et al. 2013), where NCDs are thought to be favored by O2-deplete and Fe-replete conditions where denitrification creates low N:P ratios (Deutsch et al. 2007, Löscher et al. 2014). However, in more recent studies, N2 fixation was largely undetectable in the OMZs and suboxic waters in the Pacific and Indian Oceans (Chang et al. 2019, Selden et al. 2019, Löscher et al. 2020), implying spatial or temporal variability of aphotic N2 fixation and/or changes to the calculation and reporting of detection limits (White et al. 2020).
NCDs present in aphotic waters are mostly Cluster 1 α-proteobacteria and γ-proteobacteria and Cluster 3 anaerobes affiliated with δ-proteobacteria and Clostridia (Fernandez et al. 2011, Hamersley et al. 2011, Bonnet et al. 2013, Farnelid et al. 2013, Löscher et al. 2014, Benavides et al. 2015). Clusters 2 and 4 nifH sequences have also been associated with OMZs (Löscher et al. 2014, Jayakumar et al. 2017). Although NCDs are well-established as the dominant diazotrophs in aphotic waters and low rates of N2 fixation have been measured, little is known about which NCDs are active and the factors controlling their activity (see discussion in ‘Environmental drivers of NCD biogeography, activity, and presumed N2 fixation’).
Polar seas
Polar waters have long been assumed to be devoid of N2 fixation. However, recent efforts to measure N2 fixation in polar waters suggest it is an active process in both the Arctic and Antarctic Oceans, particularly in coastal and continental shelf regions (Blais et al. 2012, Sipler et al. 2017, Harding et al. 2018, Shiozaki et al. 2018b, 2020). While N2 fixation by the UCYN-A/haptophyte symbiosis has been confirmed in polar waters (Harding et al. 2018), NCDs are also prevalent (Farnelid et al. 2011, Blais et al. 2012, Fernandez-Mendez et al. 2016, Shiozaki et al. 2018b) and are suspected to account for some portion of the N2 fixation (Blais et al. 2012, Harding et al. 2018). In the Arctic, nifH gene surveys suggest that the NCD community in polar waters is primarily composed of δ-proteobacterial sequence types affiliating with Clusters 1A and 3 that are not closely related to sequence types recovered from other regions of the ocean (Blais et al. 2012, Fernandez-Mendez et al. 2016, Shiozaki et al. 2018b). Cluster 3 sequence types that appear endemic to the coastal Antarctic Ocean have also been described by Shiozaki et al. (2020). Collectively, the prevalence of putative anaerobes suggest that sediment resuspension plays an important role in shaping the pelagic diazotrophic community in these regions. However, several γ-proteobacterial NCDs have also been reported in the Antarctic with high nifH sequence similarity to oligotrophic taxa [including Gamma A, identified as SV009 in Shiozaki et al. (2020)].
Interestingly, a recent study reported the presence of ‘ultrasmall’ (<0.22µm) NCDs (Pierella Karlusich et al. 2021) comprising up to 10% of the ultrasmall bacterioplankton in Arctic Ocean waters based on metagenome-derived abundances (described in ‘Global ocean surveys using ‘omics provide insights into NCD abundance and diversity’). An ε-proteobacterium, Arcobacter nitrofigilis, dominated bacterioplankton populations at the surface and deep chlorophyll maximum, while a γ-proteobacterium dominated in the mesopelagic. Both ultrasmall diazotrophs were also present in larger size fractions, suggesting they may have particle- or symbiont-associated lifestyles. At present, the ecological and biogeochemical importance of ultrasmall NCDs is unknown in the global oceans.
Environmental drivers of NCD biogeography, activity, and presumed N2 fixation
Nutrient perturbation experiments
We are still in the early stages of understanding the environmental controls on marine NCD biogeography and activity. Since few marine NCDs are available in culture, environmental controls must be inferred from biogeographical surveys coupled to environmental data (discussed in ‘Meta-analysis of nifH gene abundance data’) and from in situ experiments involving nutrient and/or environmental manipulations. Such experiments have yielded important insights but can be challenging to interpret given the regular co-occurrence of cyanobacterial diazotrophs and NCDs, the complexity of microbial interactions among the broader community, and the lack of standardized approaches across experiments. For example, some studies employ nifH amplicon libraries, while others quantitatively target specific NCDs with qPCR/ddPCR. Additionally, some studies note enhanced N2 fixation rates without observed changes in NCD abundances/nifH transcription, while other studies characterize the diazotroph community composition in the region, but not in the experiments themselves making interpretation challenging. Table 2 summarizes some environmental drivers of NCD abundance, nifH transcription or putative N2 fixation based on representative studies using a variety of approaches.
Table 2.
Changes in NCD abundances, activity, and putative N2 fixation in response to nutrient perturbations provide insights into their environmental drivers. NPSG—North Pacific Subtropical Gyre; WSP—Western South Pacific; ETSP—Eastern Tropical South Pacific; SCS—South China Sea; MedSea—Mediterranean Sea; RT-qPCR—reverse transcription qPCR; N—nitrogen; Fe—iron; P—phosphate; DOC—dissolved organic C; DON—dissolved organic N; DOP—dissolved organic P; DCMU—photosynthesis inhibitor (3-(3,4-dichlorophenyl)-1,1-dimethylurea); and GX—xanthan gum.
| NCD(s)a | Region (depth) | Type of analysesc | Environmental perturbation(s) | Synthesis of findings | Reference |
|---|---|---|---|---|---|
| Gamma A (AO15) | NPSG (0 – 25 m) | RT-qPCR; N2 fixation | P | No significant stimulation of N2 fixation or nifH transcripts | Zehr et al. (2007) |
| Cluster-3 | NPSG (40 m) | qPCR | DOC | Cluster-3 increased in +DOC | Bombar et al. (2013) |
| Gamma A (γ24774A11) | WSP (5 m) | qPCR | N, P, Fe, DOC, Fe/P, N/P, N/Fe, N/P/Fe/DOC | Abundances increased in response to Fe and Fe/P in westernmost stations; decreased in +N | Moisander et al. (2012) |
| Gamma A (γ24774A11) | WSP (3 m) | qPCR & RT-qPCR; N2 fixation | DOC, DON, DOP, inhibition of photosynthesis using DCMU | No nifH expression across all treatment implying they were not actively fixing N2; DCMU additions suppressed nearly all N2 fixation implying that the most active N2-fixers were phototrophs | Benavides et al. (2018b) |
| P1, P4, P7 | ETSP (95 m) | qPCR, N2 fixation | Glucose, O2 | N2 fixation rates increased in +glucose, +O2; P7 abundances increased in +glucose, +O2 | Löscher et al. (2014) |
| Unknownb | ESTP (15 m) | N2 fixation rates | Fe, N or N/Fe, P, glucose | N2 fixation rates increased in +Fe and +glucose and occasionally in +N/Fe | Dekaezemacker et al. (2013) |
| P2, P4, P6, P7 | ESTP (0 m) | nifH transcript sequencing & RT-qPCR; N2 fixation | Glucose | N2 fixation stimulated by +glucose in eddy cores; nifH transcription from P2, P4, P6, P7 in eddy samples (not measured in experiments) | Löscher et al. (2016) |
| Unknownb | WNA (coastal) | N2 fixation (ARA) | DOC, organic detritus, light and dark incubations | Water column N2 fixation stimulated in +DOC, + organic detritus treatments | Paerl and Prufert (1987) |
| Gamma A, gamma P, Clll-Church | ENA (1 – 3 m) | qPCR, N2 fixation | N, P, Fe, dust | Gamma A abundances increased most in +NFe and +dust treatments; Gamma P and CIII were undetected or not quantifiable | Langlois et al. (2012) |
| Mainly 1G | MedSea (5 m) | nifH sequencing; N2 fixation | Dust under contemporary and future temp. and pH scenarios | Increased N2 fixation rates in response to dust additions in stations dominated by NCDs (cyanobacteria also present) | Ridame et al. (2022) |
| Diverse cluster I and III NCDs | MedSea (5 m) | nifH transcript sequencing; N2 fixation | GX, N, P, DOC, NP, DOC/P, DOC/N, DOC/N/P; light and dark incubations | N2 fixation stimulated by DOC in both light and dark incubations; increased relative abundances of NCD nifH transcripts in +DOC/N/P; increased N2 fixation and NCD transcript relative abundances in +GX (cyanobacteria also present) | Rahav et al. (2016) |
| M6411A02, M6413A02, M6433A04 | WSP (aphotic) | qPCR (environmental samples, not nutrient exp.); N2 fixation | DOC, DON | N2 fixation stimulated by amino acid (+DON) additions; assumed to be NCDs, but recently Trichodesmium has been shown to fix N2 in mesopelagic waters (Benavides et al. 2022) | Benavides et al. (2015) |
| αETSP-2, cIII-ETSP | ETSP (aphotic) | nifH sequencing & qPCR, N2 fixation | Amino acids, simple sugars | Aphotic N2 fixation rates increased in +amino acids and +DOC treatments; identified NCDs did not change in abundance. | Bonnet et al. (2013) |
| Unknown (Gulf of Aqaba), 1 G (Med Sea) | MedSea,Gulf of Aqaba (aphotic) | nifH sequencing (environmental samples, not nutrient exp.; N2 fixation | GX, amino acids | Aphotic N2 fixation rates increased in +amino acids (Gulf of Aqaba) and +GX (Med Sea) | Rahav et al. (2013) |
NCD nifH catalog name referenced when possible.
NCDs suspected to be dominant N2-fixers.
N2 fixation measured on the whole community.
Together these experiments indicate that NCD abundances and nifH transcription are sometimes limited by the availability of Fe, P (or both), and/or DOC in oligotrophic euphotic waters (Table 2). Gamma A abundances appear to be influenced (at least in part) by the availability of Fe in the Western South Pacific (Moisander et al. 2012). Regional differences are seen in the response of Gamma A to N availability; N additions resulted in decreased abundances of Gamma A in the Western South Pacific (Moisander et al. 2012), whereas in the North Atlantic, abundances increased in experiments with fixed N additions (Langlois et al. 2012). Aeolian dust input is an important source of Fe to surface oceans and has been shown to influence NCD abundances in the North Atlantic Ocean (Langlois et al. 2012) and the Mediterranean Sea (Ridame et al. 2022). The availability of labile DOC is also a control on N2 fixation in regions where NCDs are thought to be the most prevalent diazotrophs, like in the Eastern Tropical South Pacific (Dekaezemacker et al. 2013, Turk-Kubo et al. 2014, Knapp et al. 2016).
DOC availability may also be a particularly important control on aphotic N2 fixation (Table 2). For example, aphotic N2 fixation was positively correlated with transparent exopolymeric particles (TEPs) in the oxygenated waters of the South Pacific Ocean and the Mediterranean Sea (Rahav et al. 2013, Benavides et al. 2015), suggesting that TEPs could provide C-rich and/or O2-depleted microenvironments that favor NCD N2 fixation. Moreover, additions of glucose and amino acids occasionally enhance aphotic N2 fixation (Bonnet et al. 2013, Rahav et al. 2013, Löscher et al. 2014, Benavides et al. 2015, Gradoville et al. 2017a).
Although these studies illustrate that perturbations in nutrient availability (e.g. nutrient, trace metal, and labile organic C) or environmental conditions (e.g. availability of light) can lead to changes in the abundance and nifH transcription of NCDs, data from these experiments are relatively sparse and spatially heterogeneous. Fortunately, valuable insights into the environmental drivers behind diazotroph biogeography can also be inferred from biogeographical surveys coupled to environmental data (‘Meta-analysis of nifH gene abundance data’).
Meta-analysis of nifH gene abundance data
In the absence of direct cell counts, quantifying nifH gene abundances (via qPCR or ddPCR) is arguably the best method for enumerating NCDs and determining the biogeographical patterns and environmental drivers of specific taxa. Unfortunately, oceanographic sample collection, DNA extraction, and nifH gene quantification are laborious compared to more high-throughput methods (e.g. automated flow cytometry) and the coverage of qPCR/ddPCR nifH gene abundance datasets are often sparse in space, time, and NCD targets. Thus, it can be difficult to discern the global distribution patterns and environmental predictors of diverse NCD taxa from a single dataset.
A global nifH qPCR database has been compiled for marine cyanobacterial diazotrophs (Luo et al. 2012, Tang et al. 2019a), revealing taxon-specific biogeographical patterns. Linking these qPCR abundances to environmental data has revealed both common and distinct environmental drivers among taxa. For example, abundances of UCYN-A, Crocosphaera, Trichodesmium, and Richelia all correlate positively with temperature and negatively with depth, but UCYN-A is distributed across a larger temperature range, specifically being present in lower temperature waters (Tang et al. 2019a).
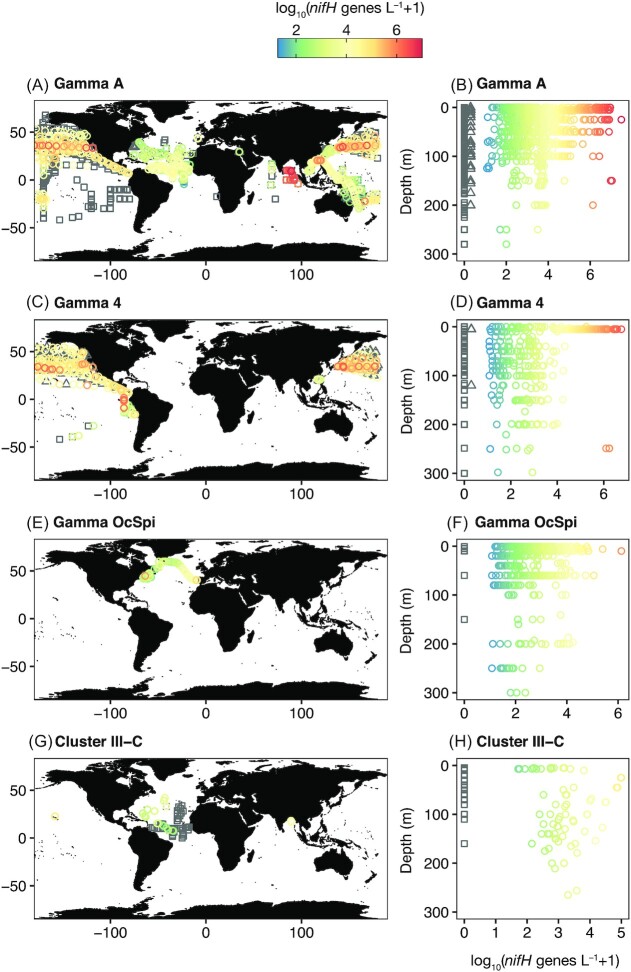
To our knowledge, previous compilations of global NCD nifH gene abundance data have only targeted Gamma A (Langlois et al. 2015, Shao and Luo 2022). To increase understanding of the distribution of NCDs beyond this phylotype, we compiled qPCR/ddPCR data from 59 published studies of the 55 targets included in our NCD nifH catalog (Fig. 1; Table S2, Supporting Information), yielding a total of 7385 water column observations (dataset doi: 10.5281/zenodo.6537451). This database shows that while NCD taxa have been quantified from many ocean regions, there has been a strong sampling bias, with the most samples collected from the North Pacific and few samples collected from the southern hemisphere (Figure S1, Supporting Information). Data is sparse for some NCD taxa (e.g. n = 34 observations of β-proteobacteria) but is particularly rich for γ-proteobacteria (n = 4138 observations). Here, we discuss the global distributions and environmental predictors of the three phylotypes for which the most data are available: Gamma A, Gamma 4, and a γ-proteobacterium of the order Oceanospirilalles isolated by Ratten (2017) (‘Gamma OcSpi’), as well as the most data-rich Cluster 3 phylotype (‘ClusterIII-C’; Church et al. 2005a, Langlois et al. 2008). nifH gene transcripts of these groups have all been observed, except for Gamma OcSpi, for which no nifH RT-qPCR data (either presence or absence) have been reported (Figure S2, Supporting Information). There were eight additional NCD phylotypes with >100 nifH gene abundance observations; however, in most cases data for these phylotypes are only available from a single ocean region (Figure S3, Supporting Information).
The four most data-rich NCD phylotypes appear to have different global and depth distributions (Fig. 3). Abundances of the three γ-proteobacterial phylotypes are all highest at the surface and decrease with depth. Gamma A has the best sampling coverage of any NCD phylotype (n = 2339 samples), with nifH genes detected in 63% of samples. The highest Gamma A abundances (>10 6nifH gene copies l–1) have been observed in the North Pacific Ocean (Cheung et al. 2020), Indian Ocean (Wu et al. 2019), SCS (Liu et al. 2020), and New Caledonia Lagoon (Benavides et al. 2018b); Gamma A has not been detected in any samples collected from the Eastern South Pacific (Halm et al. 2012, Turk-Kubo et al. 2014, Shiozaki et al. 2018a; Fig. 3A). Gamma 4 has only been quantified in the Pacific Ocean, where its nifH gene sequence has been detected in 81% of samples (n = 943). The highest abundances (>10 6nifH gene copies l–1) have been observed in the North Pacific and Eastern Tropical South Pacific. Gamma 4 values below detection limits have been observed in the North Pacific, Central South Pacific, Eastern Tropical South Pacific, and SCS (Halm et al. 2012, Löscher et al. 2014, Chen et al. 2019, Cheung et al. 2021). Gamma OcSpi has only been explored in the upper 300 m of the North Atlantic, where nifH genes were detected in 98% of samples (n = 440; Ratten 2017). Maximum abundances of Gamma OcSpi were generally lower (<10 5nifH gene copies l–1) compared to Gamma A and Gamma 4 and the only observations of Gamma OcSpi below detection limits occurred in the western North Atlantic. Finally, ClusterIII-C nifH genes have been detected in 51% of samples (n = 147) and though a smaller number of samples have been collected than for the γ-proteobacterial phylotypes, coverage has been distributed over many ocean basins (Fig. 3). The highest ClusterIII-C abundances (>10 5nifH gene copies l–1) have been observed in the North Pacific Subtropical Gyre (Church et al. 2005a), while relatively high abundances (>10 4nifH gene copies l–1) have been detected in the North Atlantic and the Bay of Bengal (Langlois et al. 2008, Löscher et al. 2020). Measurements of ClusterIII-C below detection limits have been reported in the Atlantic (Langlois et al. 2008). Notably, abundances of ClusterIII-C do not decrease with depth (Fig. 3), unlike the γ-proteobacterial phylotypes, and instead have a positive depth trend.
Figure 3.
NCD taxa have distinct global and depth distribution patterns. Maps (A), (C), (E), and (G) show the global nifH gene abundances of four NCD taxa at all sampling depths (0–4000 m) while depth plots (B), (D), (F), and (H) show the upper 300 m only. Observations of no detect are represented by gray squares; observations of detect but not quantifiable (DNQ) were given a nominal value of 1 nifH gene l–1 in the database and are represented by gray triangles. Dataset doi: 10.5281/zenodo.6537451.
Comparing the distribution of these four NCD phylotypes is complicated by differences in sampling efforts and coverage (Fig. 3). For instance, samples for Gamma 4 and Gamma OcSpi have only been collected in the Pacific and Atlantic Oceans, respectively, and the three γ-proteobacterial phylotypes are heavily biased toward surface samples. Sampling strategies designed to target NCDs in undersampled ocean regions may be particularly useful for helping to constrain NCD biogeography. Reporting non-detects is also useful for efforts to model diazotroph distributions (Meiler et al. 2022) and we encourage researchers to do so in future studies.
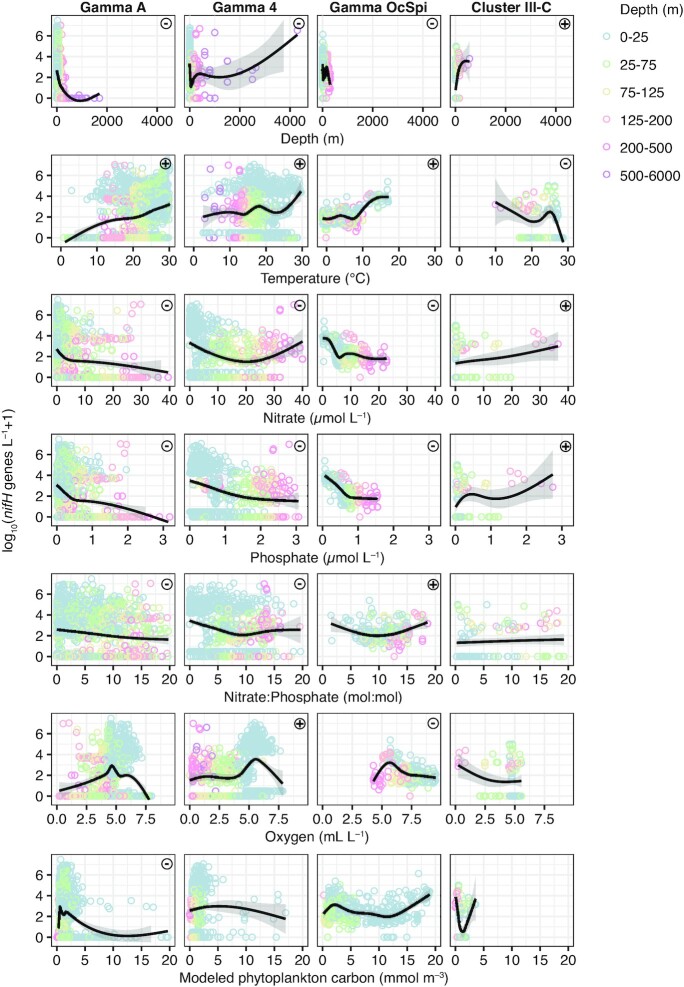
To investigate the potential environmental determinants of NCD phylotypes, we colocalized qPCR abundance data with available World Ocean Atlas climatology, satellite observations, Argo float data, and PICES model products using the Simons Collaborative Marine Atlas Project (Simons C-MAP, https://simonscmap.com, Ashkezari et al. 2021). Details on variables and space/time/depth tolerances used for colocalization are provided in Table S4 (Supporting Information). All sampling depths were included in this analysis, and while many environmental variables covaried with depth, their relationships with nifH gene abundances were generally consistent when restricting analyses to surface samples (Figure S4, Supporting Information).
Environmental drivers of NCDs appear to differ among the four phylotypes investigated and diverge from those described for cyanobacterial diazotrophs (Fig. 4). Abundances of the three γ-proteobacteria correlated positively with temperature and negatively with NO3− and phosphate (PO43−) concentrations; these same trends have been observed for cyanobacterial diazotrophs (Tang et al. 2019a). In contrast, abundances of ClusterIII-C correlated negatively with temperature and positively with NO3− and PO43− concentrations (Fig. 4), reflecting the positive depth trend for this phylotype (Figure S4, Supporting Information). Gamma A and Gamma 4 abundances correlated negatively with the NO3−:PO43− (N:P) ratio, a trend also observed for most cyanobacterial diazotrophs (Tang et al. 2019a), while Gamma OcSpi and ClusterIII-C had weakly positive but nonsignificant correlations to N:P. The highest abundances of the γ-proteobacterial phylotypes were associated with moderate O2 concentrations (∼4.5–6.5 ml l–1), while their abundances were lower at low O2 concentrations (deep samples) and at high O2 concentrations (supersaturated and/or cold surface ocean waters). Cluster III-C abundances appear negatively related to O2 (although this trend was not statistically significant), with some of the highest abundances observed in deep, low-O2 waters (<1ml l–1). This is consistent with a potentially anaerobic or facultative anaerobic lifestyle, i.e. a characteristic of many Cluster 3 diazotrophs. Relationships between NCD abundance and additional environmental variables are presented in Figure S4 (Supporting Information).
Figure 4.
Environmental predictors vary among NCD taxa. NCD nifH gene abundances are plotted against environmental metadata from World Ocean Atlas monthly climatology (temperature, nitrate, phosphate, N:P, and O2 concentration) and Pisces model output of phytoplankton biomass in units of carbon (modeled phytoplankton C). Tolerances for colocalization are presented in Table S4 (Supporting Information). Each point represents an individual nifH gene abundance sample, with sample depth shown in color. Note that a small fraction of data with outlying x-axis values were excluded from plots. Black lines and gray shading represent the smoothed conditional mean and 95% confidence intervals. Symbols (+/-) reflect positive and negative correlations (Spearman rank with Bonferroni correction for 28 comparisons).
Overall, our analysis suggests greater variability in the biogeography and environmental predictors among NCD taxa than among cyanobacterial diazotroph taxa. This likely reflects the taxonomic and metabolic diversity of NCDs, which are distributed over broad bacterial and archaeal lineages (Fig. 1) and have diverse energy generation and nutrient transport mechanisms (see ‘Diversity and ecophysiological features inferred from MAGs’). While genome comparisons among the four NCD taxa examined here are not yet possible, we presume that they have divergent ecophysiological features that may explain their different environmental drivers and distributions.
NCD biogeography and ecophysiology from metagenomes and metatranscriptomes
Global ocean surveys using ‘omics provide insights into NCD abundance and diversity
Investigations of the global diversity and distribution of NCDs from an ‘omics perspective are now possible due to recent improvements in HTS techniques, decreases in sequencing costs, and global ocean surveys focused on primer independent sequencing including Tara Oceans (Karsenti et al. 2011) and the Malaspina (Duarte 2015) expeditions. The Tara Oceans expedition has provided the most samples and deepest metagenomic sequencing effort to date for the open ocean (Sunagawa et al. 2015, Carradec et al. 2018, Salazar et al. 2019), with sampling coverage including all major ocean regions from the euphotic layer to the mesopelagic sea. The first version of the Ocean Microbiome Reference Gene Catalog (OM-RGC; Sunagawa et al. 2015) focused on the free-living size faction (<3µm) and provided a total of 28 nifH gene sequence variants, only two of which belonged to cyanobacteria (Cornejo-Castillo 2017). In recent years, increased metagenomic data available from undersampled environments, including polar regions and the deep ocean, has revealed new NCD nifH sequence variants, altogether showing that NCDs are globally distributed in surface and mesopelagic layers (Cornejo-Castillo 2017, Salazar et al. 2019, Acinas et al. 2021, Pierella Karlusich et al. 2021, Delmont et al. 2022). Some NCD nifH gene sequences are different from sequences identified with primer-based approaches, suggesting they may represent novel diazotrophic diversity not found in previous studies (Fig. 1; Cornejo-Castillo 2017, Delmont et al. 2018, 2022). In addition, the reconstruction of MAGs has provided vital insight into the genomic content of some marine NCDs, which is crucial to gain a better understanding of their physiology and ecology (see ‘Diversity and ecophysiological features inferred from MAGs’).
One advantage of metagenomic approaches is that normalization of nitrogenase genes to other genetic markers can provide an estimate of the relative abundance of NCDs to the total bacterioplankton community. Using this method, NCDs have been estimated to be more abundant than some previous reports (∼106 cells l−1; Delmont et al. 2018); NCDs were also among the top contributors to the nifH transcript pool in Tara Ocean samples (Salazar et al. 2019). The relative contributions of NCDs to total bacterioplankton were higher in large (5–20 µm) size fractions than in small (0.2–3 µm) size fractions (Pierella Karlusich et al. 2021) and relative abundances were significantly higher in the mesopelagic than in surface waters (Cornejo-Castillo 2017, Salazar et al. 2019, Pierella Karlusich et al. 2021). However, these estimations should be interpreted with caution since they have yet to be corroborated using other quantitative approaches.
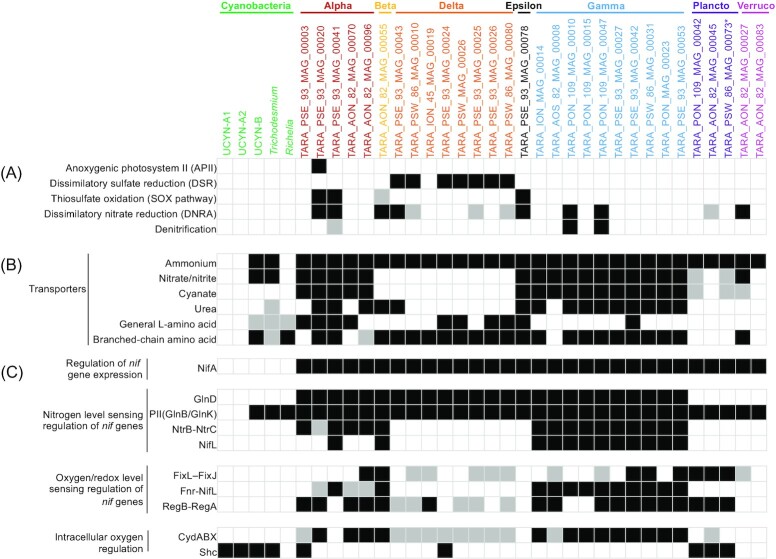
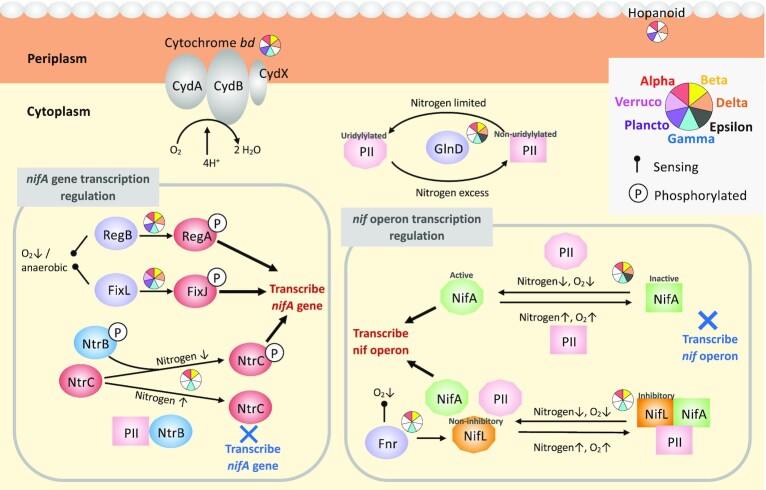
Diversity and ecophysiological features inferred from MAGs
Acquiring NCD genomes from cultivated isolates and the assembly and binning of MAGs from the metagenomic datasets described above enables the prediction of metabolic pathways, and hence ecophysiological features, of marine NCDs from the surface ocean, estuaries, OMZs, and the bathypelagic (Bentzon-Tilia et al. 2015b, Martinez-Perez et al. 2018, Acinas et al. 2021, Cheung et al. 2021, Poff et al. 2021, Delmont et al. 2022). Recently, 40 putative NCD MAGs with a high percent of completion were reconstructed from the Tara Oceans expedition (termed Heterotrophic Bacterial Diazotrophs, see Box 1) providing the largest dataset of the genomic potential of marine NCDs to date (Delmont et al. 2022). These MAGs represent diverse NCDs affiliated to Proteobacteria, Planctomycetes, and Verrucomicrobia (nifH Clusters 1 and 3). NCD MAGs contributed nearly half of the total nifH gene sequences detected in the Tara Oceans metagenomic dataset, although their nifH gene transcripts were greatly outnumbered by those of cyanobacterial diazotrophs (Figure S5A, Supporting Information). Among the NCD MAGs, γ-proteobacteria contributed the majority of the nifH sequences in both the metagenomic (55%) and metatranscriptomic (78%) datasets, followed by δ-proteobacteria (∼15% of both nifH genes and transcripts). In contrast, Planctomycetes and ɑ-proteobacteria contributed 2%–3% of the nifH transcripts, while their relative abundances at DNA level were similar to δ-proteobacteria. The remaining NCD MAGs were affiliated with β-proteobacteria, ε-proteobacteria, and Verrucomicrobiota, which contributed <3% to the total nifH genes and transcripts (Figure S5B, Supporting Information). These findings suggest that γ-proteobacteria and δ-proteobacteria may be the dominant NCDs in the pelagic oceans.
We examined the ecophysiological features of NCD MAGs, focusing specifically on pathways involved in energy generation other than aerobic respiration and in the transport of fixed N, which can inhibit cyanobacterial N2 fixation (Knapp 2012). In addition, we inferred potential regulatory mechanisms of N2 fixation based on our understanding of similar pathways from terrestrial model NCDs, described in detail below. All MAGs with >90% completeness (as estimated by Anvi’o; Eren et al. 2015) containing all the nitrogenase structural genes (nifHDK) were included in this analysis (30 MAGs) and potential pathways in these MAGs were reconstructed using KEGGMAPPER (Kanehisa et al. 2012).
Although there have been reports of photoheterotrophic and chemolithoautotrophic NCDs in estuarine and bathypelagic waters (Bentzon-Tilia et al. 2015a, Acinas et al. 2021), all Tara Ocean NCD MAGs were originally identified as chemoheterotrophs (Delmont et al. 2022). Nevertheless, several energy generating pathways other than aerobic respiration were detected in the NCD MAGs, including e.g. anoxygenic photosystem II (APII), dissimilatory sulfate reduction (DSR), the SOX pathway (i.e. sulfur oxidation), dissimilatory NO3− reduction to NH4+ (DNRA), and denitrification (Fig. 5A; Hu et al. 2002, Kraft et al. 2011, Santos et al. 2015, Grabarczyk and Berks 2017). APII was only detected in an α-proteobacterial MAG and DSR was constrained to δ-proteobacteria. In contrast, the SOX pathway and DNRA were detected across MAGs of various groups. Denitrification was found in an α-proteobacterial MAG and two γ-proteobacterial MAGs. Approximately, half of the NCD MAGs contained at least one of these energy generating pathways, and one-third of them contained multiple pathways.
Figure 5.
Ecophysiological features of NCDs include pathways involved in energy generation and transport of fixed N not present in cyanobacterial diazotrophs. Color represents presence/absence of metabolic pathways (black: all genes; gray: one missing gene; and white: no genes) related to energy acquisition (A), N uptake (B), and regulation of nitrogenase synthesis and activity (C) of NCDs and cyanobacterial diazotrophs as inferred from reconstructed Tara Oceans MAGs (Delmont et al. 2022). Alpha = ɑ-proteobacteria; Beta = β-proteobacteria; Delta = δ-proteobacteria; Epsilon = ε-proteobacteria; Gamma = γ-proteobacteria; Plancto = Planctomycetes; and Verruco = Verrucomicrobiota. See Fig. 6 for more details about these regulatory pathways and the proteins involved.
The NCD MAGs contained the genes for ABC transporters of various chemical forms of N, P, and Fe (Fig. 5B; Figure S6, Supporting Information). The genes encoding NH4+ transporters were detected in all NCD MAGs but only in some cyanobacterial diazotrophic MAGs. Additionally, the γ-proteobacterial and α-proteobacterial NCDs were genetically capable of transporting NO3−, NO2−, cyanate, urea, and amino acids, while the other NCDs and cyanobacterial diazotrophs generally lacked some or all genes in these pathways. The genetic capacity for uptake/transport of more forms of fixed N in NCDs than in cyanobacterial diazotrophs suggests that some NCDs may be facultative diazotrophs and switch to alternative fixed N forms when available under certain conditions. Most NCD MAGs also contained diverse P and Fe uptake pathways (Figure S6, Supporting Information).
N2 fixation is a highly regulated process, due both to the energetic demands (high ATP and reductant requirements) and O2 sensitivity of nitrogenase. As such, there are multiple complex pathways for the sensing of alternative N sources and intracellular O2 conditions. Many potential N/O2 sensing regulatory mechanisms of N2 fixation were found in the NCD MAGs (Figs 5C and 6). All NCD MAGs contained the nifA gene, which is the major regulator that interacts with the upstream environmental sensing pathways and controls the transcription of the nif operon in terrestrial NCDs (Dixon and Kahn 2004). In terms of known N-sensing regulatory mechanisms, only the proteobacterial MAGs contained the genes encoding the N sensing protein (GlnD) and PII proteins (GlnB or GlnK). Additionally, within that group, nearly all the α-proteobacterial and γ-proteobacterial MAGs contained the genes for the two-component system (NtrB-NtrC) that regulates the expression of nifA gene. Briefly, when alternative N forms are in excess, activation of nifA transcription is inhibited by the removal of uridylyl groups from PII by GlnD, which then interacts with NtrB and inhibits NtrC (Bueno Batista and Dixon 2019). When N is limiting, the PII proteins are uridylylated by GlnD, which cannot interact with NtrB-NtrC; uridylylated PII proteins can also directly activate the NifA protein, and thus allow the expression of the nif operon (Dixon and Kahn 2004). Another regulatory protein (NifL), known to interact with non-uridylylated PII and inhibit NifA under excess N conditions in the terrestrial γ-proteobacterial diazotroph A. vinelandii (Little et al. 2000) was also detected, but mostly in γ-proteobacterial MAGs (Fig 5C).
Figure 6.
NCDs use diverse regulatory pathways for N2 fixation. Circular diagrams next to each regulatory pathway represent the presence or absence of the corresponding genes in at least one MAG from each NCD group (Alpha = ɑ-proteobacteria; Beta = β-proteobacteria; Delta = δ-proteobacteria; Epsilon = ε-proteobacteria; Gamma = γ-proteobacteria; Plancto = Planctomycetes; and Verruco = Verrucomicrobiota) as indicated by color (presence) or white (absence).
Some NCD MAGs also contained genes for O2 (FixL-FixJ) and redox (RegB-RegA) sensing two-component regulatory systems (Figs 5C and 6). When the intracellular conditions favor the activity of the O2-sensitive nitrogenases, these systems can activate the transcription of the nifA gene and allow the synthesis of nitrogenase (Dixon and Kahn 2004). The Fnr protein, which was mostly detected in γ-proteobacterial MAGs, can also sense O2 levels and is important in maintaining the non-inhibitory form of NifL and the functioning of NifA in the terrestrial γ-proteobacterial diazotroph Klebsiella pneumoniae (Grabbe et al. 2001). Moreover, the NifA in some NifL-lacking terrestrial NCDs can also directly sense O2 level (Dixon and Kahn 2004), which could theoretically be the case for some of the marine NCDs that appear to lack known O2 sensing systems.
In addition to these regulatory processes, genes coding for cytochrome bd (CydABX) were found in most γ-proteobacterial and α-proteobacterial MAGs (Figs 5C and 6). The cytochrome bd consumes large amounts of O2 during ATP generation, which was found to be important in reducing intracellular O2 levels through respiration for aerobic N2 fixation in A. vinelandii (Poole and Hill 1997). Additionally, of relevance to intracellular O2 regulation, a recent study has proposed that hopanoids can act as an O2-diffusion barrier that protects the O2-sensitive nitrogenase in marine cyanobacteria (Cornejo-Castillo and Zehr 2019); however, the gene for hopanoid synthesis [squalene-hopene cyclase (shc)] was absent in most NCD MAGs, except Planctomycetes (Fig. 5C). Interestingly, many of these regulatory pathways were not detected in the MAGs of cyanobacterial diazotrophs (Fig. 5).
Acquisition and analysis of these MAGs provides unprecedented insight into the genomic potential of marine NCDs. The emerging pattern suggests that collectively this group may rely on alternative N sources in place of N2 fixation under oxygenated conditions, as evidenced by the diverse N uptake proteins detected, along with the presence of N/O2-sensing regulatory pathways. Future studies leveraging in situ whole genome transcription from NCDs are needed to link metabolic status (e.g. fixing N2 vs. relying on N-uptake) with environmental conditions.
Moving from genes to rates: are NCDs fixing N2 in the pelagic oceans?
The high diversity and wide distribution of NCDs and the occasional detection of nifH transcripts in PCR-based and metatranscriptomic datasets are often referenced as indicators of the significance of NCDs to marine N2 fixation. Numerous attempts have been made to link NCD presence and/or active transcription of nifH to whole community N2 fixation. This is challenging in the well-lit surface ocean given the generally low abundance of NCDs relative to co-occurring cyanobacterial diazotrophs (Moisander et al. 2017). However, there are some reports of N2 fixation rates where NCDs are abundant and cyanobacterial diazotrophs are absent (Yogev et al. 2011, Großkopf et al. 2012, Halm et al. 2012, Löscher et al. 2014, Shiozaki et al. 2014,2017, Bentzon-Tilia et al. 2015b, Rahav et al. 2016), occasionally at high rates (e.g. 24.8 ± 8.4 nmol N l-1 d-1; Löscher et al. 2014). But even when such direct comparisons are possible (e.g. NCD cell abundances and N2 fixation rates are both reported), discrepancies often exist between NCD abundances and potential N2 fixation rates (Turk-Kubo et al. 2014), suggesting underestimates of NCD abundances and/or cell-specific rates or overestimates of whole community N2 fixation rates.
Proving that NCDs fix N2 in aphotic waters is also challenging. The low rates reported for many aphotic samples (<1nmol N l−1 d−1) are near detection limits for the 15N2 uptake assay, which have not historically been reported throughout the literature (Gradoville et al. 2017a). Reports of commercial 15N2 gas stocks contaminated with 15NH4+ and/or 15NO3−/15N-nitrite (NO2−) raise a concern that some apparent rates could be artifacts driven by fixed N uptake (Dabundo et al. 2014). Additionally, measuring accurate N2 fixation rates from the mesopelagic, where particulate N concentrations are low, can be logistically challenging due to the large filtration volumes needed to produce sufficient N content for mass spectrometry (White et al. 2020). Fortunately, the scientific community is now coming to a consensus regarding best practices for measuring and reporting N2 fixation rates in the face of these challenges (White et al. 2020). However, more data validating active N2 fixation by NCDs in aphotic waters is needed and linking rates to a particular NCD taxon remains a challenge.
Measurements of taxonomically-resolved single-cell NCD N2 fixation rates in marine waters are extremely sparse. Farnelid et al. (2014) isolated more than 60 strains of putative NCDs from Baltic Sea surface waters, more than half of which fixed N2 under culture conditions. Single-cell N2 fixation rates (up to 0.17 fmol N cell−1 h−1) of Baltic Sea isolates indicated that NCDs could be a locally-important N source if they fix N2 at these rates in the environment given in situ abundances (104 cells L-1; Bentzon-Tilia et al. 2015a). Additionally, Martinez-Perez et al. (2018) isolated an α-proteobacterial NCD, Sagittula castanea (targeted by the qPCR assay CE1_45m_12_ɑ, Zhang et al. 2011), from sulfidic waters in the upwelling region off the coast of Peru and successfully measured single-cell rates of 0.0008–0.060 fmol N cell−1 d−1 under culture conditions. N2 fixation was only detected under anoxic conditions (despite being isolated from oxic waters) and was stimulated by NO2− and inhibited by NH4+. Importantly, in situ single-cell N2 fixation rates by S. castanea were undetectable, despite measurable bulk rates during the time of isolation, suggesting that in situ N2 fixation by S. castanea may be transient and dependent on the development of anoxia.
Single-cell N2 fixation rates from marine NCDs, both in situ and from nutrient perturbation experiments, are critically needed to validate their role in marine N2 fixation and better understand environmental controls on their activity. Advances in single cell visualization techniques including CARD-gene-FISH (Moraru et al. 2010), mRNA-FISH (Pilhofer et al. 2009, McInnes et al. 2014), FISH-TAMB (Harris et al. 2021), and nitrogenase immunolabeling (Geisler et al. 2019, 2020), hold promise for measuring targeted NCD single-cell N2 fixation rates if they could be successfully coupled to 15N2 incubations and nanoscale secondary ion mass spectrometry (nanoSIMS). However, technical challenges arising from low cell concentrations in complex environmental samples and sample preparation steps that dilute the 15N signal (Meyer et al. 2021) require innovative solutions. Furthermore, given that NCD N2 fixation may be a transient process, especially in particle microenvironments (Riemann et al. 2022), capturing active NCD N2 fixation may currently be hampered by insufficient temporal and spatial sampling resolution (Benavides and Robidart 2020).
Future perspectives
The past two decades of research have greatly increased our understanding of marine NCDs and a framework for understanding these organisms is now emerging (Fig. 7). Diverse NCD sequences have been detected from many ocean environments and habitats using both primer-based and primer-free approaches, underscoring their prevalence in the marine system. Insights from cultivation-based studies and marine NCD MAGs have revealed potential physiological strategies that marine NCDs may use to counter the inhibition of nitrogenase by O2 and acquire enough energy to fuel N2 fixation, such as forming self-aggregates in microaerophilic conditions and the ability to use alternative energy sources (Bentzon-Tilia et al. 2015a, Martinez-Perez et al. 2018, Acinas et al. 2021; Fig. 5A). Furthermore, reconstructing the metabolic potential of NCDs from MAGs shows that the phylogenetic diversity of this group is mirrored by diverse ecophysiological strategies. Marine NCDs have complex genetic systems to sense O2 and N-availability and are able to use many alternative N sources, suggesting that some NCDs may be functioning as facultative diazotrophs. Collectively, these findings help explain why NCDs appear to inhabit such disparate habitats, from sunlit surface waters to the bathypelagic and to diverse benthic systems.
Figure 7.
Predicated features of euphotic marine NCD communities are likely influenced by habitats. Potential strategies used by NCDs to acquire energy and C needed to support N2 fixation with energetic constraints and O2 inactivation of nitrogenase. Question marks emphasize where many open questions remain. Notably, there is recent evidence that NCDs may be fixing N2 on particles, as discussed in this review (Geisler et al. 2019). EPS—extracellular polymeric substances. Created with BioRender.com.
Despite these advances, many open questions remain (emphasized in Fig. 7). In addition to the critical need to demonstrate active N2 fixation in marine NCDs (discussed in ‘Moving from genes to rates: are NCDs fixing N2 in the pelagic oceans?’), better characterizing habitats and lifestyles of NCDs will be key to understanding the factors promoting active N2 fixation. This includes determining whether NCD N2 fixation is restricted only to low O2 microhabitats (e.g. particle- or aggregate-associated), or if NCDs can fix N2 aerobically (like some terrestrial counterparts) and if so, how they mitigate O2 inactivation of nitrogenase. Another interesting question to resolve is whether some NCD taxa have formed obligate symbioses with other planktonic species, an evolutionary strategy that has led to many symbioses between cyanobacterial diazotrophs and algae (Villareal 1992, Thompson et al. 2012, Schvarcz et al. 2022).
At this time, the substantial knowledge gaps discussed throughout this review impede obtaining reasonable estimates of NCD contributions to the global marine N cycle, and the biogeochemical significance of NCDs remains uncertain. Ultimately, determining the importance of marine NCDs will require measurements of single cell N2 fixation rates as well as advances in techniques to collect, enrich, characterize, and manipulate aggregates and particles, cultivate rare marine microbes, and increase temporal and spatial resolution of N2 fixation rates coupled to PCR-based and PCR-free surveys.
Supplementary Material
Acknowledgments
We gratefully acknowledge Mohammad Ashkezari and the Simons CMAP technical support team (University of Washington) for help with metadata colocalization and Ellen Salomon (University of Copenhagen) for feedback on the manuscript. We also thank Mar Benavides (Mediterranean Institute of Oceanography), Hugo Berthelot (European Molecular Biology Laboratory), Deniz Bombar (FUJIFILM Diosynth Biotechnologies), Julie LaRoche (Dalhousie University), Carolin Löscher (University of Southern Denmark), Clara Martínez-Pérez (ETH Zürich), Takuhei Shiozaki (University of Tokyo), and Yao Zhang (Xiamen University) for providing depths, times and/or locations from their qPCR-based studies that enabled ingestion into CMAP.
Contributor Information
Kendra A Turk-Kubo, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States.
Mary R Gradoville, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States; Columbia River Inter-Tribal Fish Commission, Portland, OR, United States.
Shunyan Cheung, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States.
Francisco M Cornejo-Castillo, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States; Department of Marine Biology and Oceanography, Institute of Marine Sciences (ICM-CSIC), Pg. Marítim Barceloneta, 37-49 08003 Barcelona, Spain.
Katie J Harding, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States; Marine Biology Research Division, Scripps Institute of Oceanography, 9500 Gilman Drive, La Jolla, CA 92093, United States.
Michael Morando, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States.
Matthew Mills, Department of Earth System Science, Stanford University, 473 Via Ortega, Stanford, CA 94305, United States.
Jonathan P Zehr, Ocean Sciences Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, United States.
Conflict of interest statement
None declared.
Funding
This work was supported by the National Science Foundation (grant number OCE-2023498 to K.A.T.-K., and OCE-2023278 to K.R.A., Stanford); the Simons Foundation (award ID 824082 to J.P.Z.); Simons Collaboration on Ocean Processes and Ecology (SCOPE, award ID 724220 to J.P.Z.); 'La Caixa' Foundation (ID 100010434, grant 105090 to F.M.C.-C.); the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie (grant 847648 to F.M.C.-C.]; and the ‘Severo Ochoa Centre of Excellence’ accreditation through the Spanish Government (grant CEX2019-000928-S to F.M.C.-C.).
References
- Acinas SG, Sanchez P, Salazar Get al. Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Commun Biol. 2021;4:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkezari MD, Hagen NR, Denholtz Met al. Simons Collaborative Marine Atlas Project (Simons CMAP): an open-source portal to share, visualize, and analyze ocean data. Limnol Oceanogr Methods. 2021;19:488–96. [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. [DOI] [PubMed] [Google Scholar]
- Azimuddin KM, Hirai J, Suzuki Set al. Possible association of diazotrophs with marine zooplankton in the Pacific Ocean. MicrobiologyOpen. 2016;5:1016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides M, Bonnet S, Berman-Frank Iet al. Deep into oceanic N2 fixation. Front Mar Sci. 2018a;5:1–4.29552559 [Google Scholar]
- Benavides M, Bonnet S, Hernández Net al. Basin-wide N2 fixation in the deep waters of the Mediterranean Sea. Glob Biogeochem Cycl. 2016a;30:952–61. [Google Scholar]
- Benavides M, Bonnet S, Le Moigne FACet al. Sinking Trichodesmium fixes nitrogen in the dark ocean. ISME J. 2022;1:2398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides M, Martias C, Elifantz Het al. Dissolved organic matter influences N2 fixation in the New Caledonian lagoon (Western Tropical South Pacific). Front Mar Sci. 2018b;5:1–11.29552559 [Google Scholar]
- Benavides M, Moisander PH, Berthelot Het al. Mesopelagic N2 fixation related to organic matter composition in the Solomon and Bismarck Seas (Southwest Pacific). PLoS ONE. 2015;10:e0143775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides M, Moisander PH, Daley MCet al. Longitudinal variability of diazotroph abundances in the subtropical North Atlantic Ocean. J Plankton Res. 2016b;38:662–72. [Google Scholar]
- Benavides M, Robidart J. Bridging the spatiotemporal gap in diazotroph activity and diversity with high-resolution measurements. Front Mar Sci. 2020;7:568876. [Google Scholar]
- Benavides M, Shoemaker KM, Moisander PHet al. Aphotic N2 fixation along an oligotrophic to ultraoligotrophic transect in the western tropical South Pacific Ocean. Biogeosciences. 2018c;15:3107–19. [Google Scholar]
- Bentzon-Tilia M, Severin I, Hansen LHet al. Genomics and ecophysiology of heterotrophic nitrogen-fixing bacteria isolated from estuarine surface water. mBio. 2015a;6:e00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzon-Tilia M, Traving SJ, Mantikci Met al. Significant N2 fixation by heterotrophs, photoheterotrophs and heterocystous cyanobacteria in two temperate estuaries. ISME J. 2015b;9:273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot H, Benavides M, Moisander PHet al. High-nitrogen fixation rates in the particulate and dissolved pools in the Western Tropical Pacific (Solomon and Bismarck Seas). Geophys Res Lett. 2017;44:8414–23. [Google Scholar]
- Bird C, Martinez Martinez J, O'Donnell AGet al. Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic γ-Proteobacteria in the Arabian Sea. Appl Environ Microbiol. 2005;71:2079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais M, Tremblay JÉ, Jungblut ADet al. Nitrogen fixation and identification of potential diazotrophs in the Canadian Arctic. Glob Biogeochem Cycl. 2012;26:GB3022. [Google Scholar]
- Bolhuis H, Severin I, Confurius-Guns Vet al. Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISME J. 2010;4:121–30. [DOI] [PubMed] [Google Scholar]
- Bombar D, Moisander PH, Dippner JWet al. Distribution of diazotrophic microorganisms and nifH gene expression in the Mekong River plume during intermonsoon. Mar Ecol Progr Ser. 2011;424:39–52. [Google Scholar]
- Bombar D, Paerl RW, Riemann L. Marine non-cyanobacterial diazotrophs: moving beyond molecular detection. Trends Microbiol. 2016;24:916–27. [DOI] [PubMed] [Google Scholar]
- Bombar D, Turk-Kubo KA, Robidart Jet al. Non-cyanobacterial nifH phylotypes in the North Pacific Subtropical Gyre detected by flow-cytometry cell sorting. Environ Microbiol Rep. 2013;5:705–15. [DOI] [PubMed] [Google Scholar]
- Bonnet S, Dekaezemacker J, Turk-Kubo KAet al. Aphotic N2 fixation in the Eastern Tropical South Pacific Ocean. PLoS ONE. 2013;8:e81265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom KH, Riemann L, Kuhl Met al. Isolation and gene quantification of heterotrophic N2-fixing bacterioplankton in the Baltic Sea. Environ Microbiol. 2007;9:152–64. [DOI] [PubMed] [Google Scholar]
- Braun S, Proctor L, Zani Set al. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol Ecol. 1999;28:273–9. [Google Scholar]
- Brown SM, Jenkins BD. Profiling gene expression to distinguish the likely active diazotrophs from a sea of genetic potential in marine sediments. Environ Microbiol. 2014;16:3128–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru D, Martin-Laurent F, Philippot L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl Environ Microbiol. 2008;74:1660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno Batista M, Dixon R. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit. Biochem Soc Trans. 2019;47:603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello AM, Turk-Kubo KA, Hayashi Ket al. Unexpected presence of the nitrogen-fixing symbiotic cyanobacterium UCYN-A in Monterey Bay, California. J Phycol. 2020;56:1521–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Zhang W, Ding Wet al. Structure and function of the Arctic and Antarctic marine microbiota as revealed by metagenomics. Microbiome. 2020;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone DG, Burns JA, Montoya JPet al. Nitrogen fixation by Trichodesmium spp.: an important source of new nitrogen to the tropical and subtropical North Atlantic Ocean. Glob Biogeochem Cycl. 2005;19:G03002. [Google Scholar]
- Capone DG. 1983; Benthic Nitrogen Fixation. Nitrogen in the Marine Environment. Carpenter EJ, Capone DG (eds.), New York: Academic Press, 105–37. [Google Scholar]
- Caputo A, Stenegren M, Pernice MCet al. A short comparison of two marine planktonic diazotrophic symbioses highlights an un-quantified disparity. Front Mar Sci. 2018;5:1–8.29552559 [Google Scholar]
- Carpenter EJ, Price CC. Nitrogen fixation, distribution, and production of Oscillatoria (Trichodesmium) spp. in the western Sargasso and Caribbean Seas. Limnol Oceanogr. 1977;22:60–72. [Google Scholar]
- Carradec Q, Pelletier E, Da Silva Cet al. A global ocean atlas of eukaryotic genes. Nat Commun. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Andersen KH, Visser AWet al. Quantifying nitrogen fixation by heterotrophic bacteria in sinking marine particles. Nat Commun. 2021;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BX, Jayakumar A, Widner Bet al. Low rates of dinitrogen fixation in the eastern tropical South Pacific. Limnol Oceanogr. 2019;64:1913–23. [Google Scholar]
- Chen T-Y, Chen Y-lL, Sheu D-Set al. Community and abundance of heterotrophic diazotrophs in the northern South China Sea: revealing the potential importance of a new alphaproteobacterium in N2 fixation. Deep Sea Res Part I. 2019;143:104–14. [Google Scholar]
- Cheung S, Nitanai R, Tsurumoto Cet al. Physical forcing controls the basin-scale occurrence of nitrogen-fixing organisms in the North Pacific Ocean. Glob Biogeochem Cycl. 2020;34:p.e2019GB006452. [Google Scholar]
- Cheung S, Zehr JP, Xia Xet al. Gamma4: a genetically versatile Gammaproteobacterial nifH phylotype that is widely distributed in the North Pacific Ocean. Environ Microbiol. 2021;23:4246–59. [DOI] [PubMed] [Google Scholar]
- Church MJ, Jenkins BD, Karl DMet al. Vertical distributions of nitrogen-fixing phylotypes at Stn ALOHA in the oligotrophic North Pacific Ocean. Aquat Microb Ecol. 2005a;38:3–14. [Google Scholar]
- Church MJ, Short CM, Jenkins BDet al. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2005b;71:5362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo-Castillo FM, Zehr JP. Hopanoid lipids may facilitate aerobic nitrogen fixation in the ocean. Proc Natl Acad Sci. 2019;116:18269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo-Castillo FM, Zehr JP. Intriguing size distribution of the uncultured and globally widespread marine non-cyanobacterial diazotroph Gamma-A. ISME J. 2021;15:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo-Castillo FM. Diversity, Ecology and Evolution of Marine Diazotrophic Microorganisms. Ph.D. Thesis, Universidad Politécnica de Cataluña, 2017. [Google Scholar]
- Coyer JA, Cabellopasini A, Swift Het al. N2 fixation in marine heterotrophic bacteria - dynamics of environmental and molecular regulation. Proc Natl Acad Sci. 1996;93:3575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabundo R, Lehmann MF, Treibergs Let al. The contamination of commercial 15N2 gas stocks with 15N–labeled nitrate and ammonium and consequences for nitrogen fixation measurements. PLoS ONE. 2014;9:e110335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H, Luan X, Zhao Jet al. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Appl Environ Microbiol. 2009;75:2238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekaezemacker J, Bonnet S, Grosso Oet al. Evidence of active dinitrogen fixation in surface waters of the eastern tropical South Pacific during El Niño and La Niña events and evaluation of its potential nutrient controls. Glob Biogeochem Cycl. 2013;27:768–79. [Google Scholar]
- Dekas AE, Chadwick GL, Bowles MWet al. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environ Microbiol. 2014;16:3012–29. [DOI] [PubMed] [Google Scholar]
- Dekas AE, Connon SA, Chadwick GLet al. Activity and interactions of methane seep microorganisms assessed by parallel transcription and FISH-NanoSIMS analyses. ISME J. 2016;10:678–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekas AE, Fike DA, Chadwick GLet al. Widespread nitrogen fixation in sediments from diverse deep-sea sites of elevated carbon loading. Environ Microbiol. 2018;20:4281–96. [DOI] [PubMed] [Google Scholar]
- Dekas AE, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–6. [DOI] [PubMed] [Google Scholar]
- Delmont TO, Karlusich JJP, Veseli Iet al. Heterotrophic bacterial diazotrophs are more abundant than their cyanobacterial counterparts in metagenomes covering most of the sunlit ocean. ISME J. 2022;16:927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Quince C, Shaiber Aet al. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol. 2018;3:804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch C, Sarmiento JL, Sigman DMet al. Spatial coupling of nitrogen inputs and losses in the ocean. Nature. 2007;445:163–7. [DOI] [PubMed] [Google Scholar]
- Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–31. [DOI] [PubMed] [Google Scholar]
- Dos Santos PC, Fang Z, Mason SWet al. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics. 2012;13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte CM. Seafaring in the 21st century: the Malaspina 2010 circumnavigation expedition. Limnol and Oceanogr. 2015;24:11–14. [Google Scholar]
- Dugdale RC, Menzel DW, Ryther JH. Nitrogen fixation in the Sargasso Sea. Deep-Sea Res. 1961;7:297–300. [Google Scholar]
- Eren AM, Esen OC, Quince Cet al. Anvi'o: an advanced analysis and visualization platform for 'omics data. PeerJ. 2015;3:e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Andersson AF, Bertilsson Set al. Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PLoS ONE. 2011;6:e19223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Bentzon-Tilia M, Andersson AFet al. Active nitrogen-fixing heterotrophic bacteria at and below the chemocline of the central Baltic Sea. ISME J. 2013;7:1413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Harder J, Bentzon-Tilia Met al. Isolation of heterotrophic diazotrophic bacteria from estuarine surface waters. Environ Microbiol. 2014;16:3072–82. [DOI] [PubMed] [Google Scholar]
- Farnelid H, Oberg T, Riemann L. Identity and dynamics of putative N2-fixing picoplankton in the Baltic Sea proper suggest complex patterns of regulation. Environ Microbiol Rep. 2009;1:145–54. [DOI] [PubMed] [Google Scholar]
- Farnelid H, Tarangkoon W, Hansen Get al. Putative N2-fixing heterotrophic bacteria associated with dinoflagellate–Cyanobacteria consortia in the low-nitrogen Indian Ocean. Aquat Microb Ecol. 2010;61:105–17. [Google Scholar]
- Farnelid H, Turk-Kubo K, Ploug Het al. Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 2019;13:170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C, Farias L, Ulloa O. Nitrogen fixation in denitrified marine waters. PLoS ONE. 2011;6:e20539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Mendez M, Turk-Kubo KA, Buttigieg PLet al. Diazotroph diversity in the sea ice, melt ponds, and surface waters of the Eurasian basin of the central Arctic Ocean. Front Microbiol. 2016;7:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AA, Karl D, Lukas Ret al. Nitrogen fixation in an anticyclonic eddy in the oligotrophic North Pacific Ocean. ISME J. 2008;2:663–76. [DOI] [PubMed] [Google Scholar]
- Fulweiler RW, Brown SM, Nixon SWet al. Evidence and a conceptual model for the co–occurrence of nitrogen fixation and denitrification in heterotrophic marine sediments. Mar Ecol Progr Ser. 2013;482:57–68. [Google Scholar]
- Gaby JC, Buckley DH. A global census of nitrogenase diversity. Environ Microbiol. 2011;13:1790–9. [DOI] [PubMed] [Google Scholar]
- Gallon JR. Reconciling the incompatible: N2 fixation and O2. New Phytol. 1992;122:571–609. [Google Scholar]
- Geisler E, Bogler A, Bar-Zeev Eet al. Heterotrophic nitrogen fixation at the hyper-eutrophic Qishon River and Estuary System. Front Microbiol. 2020,1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler E, Bogler A, Rahav Eet al. Direct detection of heterotrophic diazotrophs associated with planktonic aggregates. Sci Rep. 2019;9:9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gier J, Löscher CR, Dale AWet al. Benthic dinitrogen fixation traversing the oxygen minimum zone off Mauritania (NW Africa). Front Mar Sci. 2017;4:1–16. [Google Scholar]
- Gier J, Sommer S, Löscher CRet al. Nitrogen fixation in sediments along a depth transect through the Peruvian oxygen minimum zone. Biogeosciences. 2016;13:4065–80. [Google Scholar]
- Grabarczyk DB, Berks BC. Intermediates in the Sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS ONE. 2017;12:e0173395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe R, Klopprogge K, Schmitz RA. Fnr is required for NifL-dependent oxygen control of nif gene expression in Klebsiella pneumoniae. J Bacteriol. 2001;183:1385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradoville MR, Bombar D, Crump BCet al. Diversity and activity of nitrogen-fixing communities across ocean basins. Limnol Oceanogr. 2017a;62:1895–909. [Google Scholar]
- Gradoville MR, Crump BC, Letelier RMet al. Microbiome of Trichodesmium colonies from the North Pacific subtropical gyre. Front Microbiol. 2017b;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradoville MR, Farnelid H, White AEet al. Latitudinal constraints on the abundance and activity of the cyanobacterium UCYN-A and other marine diazotrophs in the North Pacific. Limnol Oceanogr. 2020;65:1858–75. [Google Scholar]
- Gradoville MR, White AE, Böttjer Det al. Diversity trumps acidification: lack of evidence for carbon dioxide enhancement of Trichodesmium community nitrogen or carbon fixation at Station ALOHA. Limnol Oceanogr. 2014;59:645–59. [Google Scholar]
- Großkopf T, Mohr W, Baustian Tet al. Doubling of marine dinitrogen-fixation rates based on direct measurements. Nature. 2012;488:361–4. [DOI] [PubMed] [Google Scholar]
- Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–6. [DOI] [PubMed] [Google Scholar]
- Hallstrøm S, Benavides M, Salamon ERet al. Activity and distribution of diazotrophic communities across the Cape Verde Frontal Zone in the Northeast Atlantic Ocean. Biogeochem. 2022a;160:1–19. [Google Scholar]
- Hallstrøm S, Benavides M, Salamon ERet al. Pelagic N2 fixation dominated by sediment diazotrophic communities in a shallow temperate estuary. Limnol Oceanogr. 2022b;67:364–78. [Google Scholar]
- Hallstrøm S, Raina JB, Ostrowski Met al. Chemotaxis may assist marine heterotrophic bacterial diazotrophs to find microzones suitable for N2 fixation in the pelagic ocean. ISME J. 2022c;16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halm H, Lam P, Ferdelman TGet al. Heterotrophic organisms dominate nitrogen fixation in the South Pacific gyre. ISME J. 2012;6:1238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamersley MR, Sohm JA, Burns JAet al. Nitrogen fixation associated with the decomposition of the giant kelp Macrocystis pyrifera. Aquat Bot. 2015;125:57–63. [Google Scholar]
- Hamersley MR, Turk KA, Leinweber Aet al. Nitrogen fixation within the water column associated with two hypoxic basins in the Southern California Bight. Aquat Microb Ecol. 2011;63:193–205. [Google Scholar]
- Harding K, Turk-Kubo KA, Sipler REet al. Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic Ocean. Proc Natl Acad Sci. 2018;115:13371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding KJ, Turk-Kubo KA, Mak EWKet al. Cell-specific measurements show nitrogen fixation by particle-attached putative non-cyanobacterial diazotrophs in the North Pacific Subtropical Gyre. Nat Commun. 2022;13:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RL, Vetter M, van Heerden Eet al. FISH-TAMB, a fixation-free mRNA fluorescent labeling technique to target transcriptionally active members in microbial communities. Microb Ecol. 2021;84:182–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Yoshida T, Kuno Set al. The first assessment of cyanobacterial and diazotrophic diversities in the Japan Sea. Fish Sci. 2012;78:1293–300. [Google Scholar]
- Heller P, Tripp HJ, Turk-Kubo Ket al. ARBitrator: a software pipeline for on-demand retrieval of auto-curated nifH sequences from GenBank. Bioinformatics. 2014;30:2883–90. [DOI] [PubMed] [Google Scholar]
- Hewson I, Moisander PH, Achilles KMet al. Characteristics of diazotrophs in surface to abyssopelagic waters of the Sargasso Sea. Aquat Microb Ecol. 2007;46:15–30. [Google Scholar]
- Hu X, Ritz T, Damjanovic Aet al. Photosynthetic apparatus of purple bacteria. Q Rev Biophys. 2002;35:1–62. [DOI] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Yet al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomura K, Bragg J, Follows MJ. A quantitative analysis of the direct and indirect costs of nitrogen fixation: a model based on Azotobacter vinelandii. ISME J. 2017;11:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir T, Jesmi Y, Vipindas PVet al. Diversity of nitrogen fixing bacterial communities in the coastal sediments of southeastern Arabian Sea (SEAS). Deep Sea Res Part II. 2018;156:51–9. [Google Scholar]
- Jayakumar A, Chang BX, Widner Bet al. Biological nitrogen fixation in the oxygen-minimum region of the eastern tropical North Pacific ocean. ISME J. 2017;11:2356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JS, Spakowicz DJ, Hong BYet al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun SR, Wassenaar TM, Nookaew Iet al. Diversity of Pseudomonas genomes, including Populus-associated isolates, as revealed by comparative genome analysis. Appl Environ Microbiol. 2016;82:375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Yet al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapili BJ, Barnett SE, Buckley DHet al. Evidence for phylogenetically and catabolically diverse active diazotrophs in deep-sea sediment. ISME J. 2020;14:971–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D, Letelier R, Tupas Let al. The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature. 1997;388:533–8. [Google Scholar]
- Karsenti E, Acinas SG, Bork Pet al. A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick JB, Fuchsman CA, Yakushev EVet al. Dark N2 fixation: nifH expression in the redoxcline of the Black Sea. Aquat Microb Ecol. 2018;82:43–58. [Google Scholar]
- Klawonn I, Lavik G, Böning Pet al. Simple approach for the preparation of 15−15N2-enriched water for nitrogen fixation assessments: evaluation, application and recommendations. Front Microbiol. 2015;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp AN, Casciotti KL, Berelson WMet al. Low rates of nitrogen fixation in eastern tropical South Pacific surface waters. Proc Natl Acad Sci. 2016;113:4398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp AN, McCabe KM, Grosso Oet al. Distribution and rates of nitrogen fixation in the western tropical South Pacific Ocean constrained by nitrogen isotope budgets. Biogeosciences. 2018;15:2619–28. [Google Scholar]
- Knapp AN. The sensitivity of marine N2 fixation to dissolved inorganic nitrogen. Front Microbiol. 2012;3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Jing H, Kataoka Tet al. Phylogenetic diversity and spatio-temporal distribution of nitrogenase genes (nifH) in the northern South China Sea. Aquat Microb Ecol. 2011;65:15–27. [Google Scholar]
- Kraft B, Strous M, Tegetmeyer HE. Microbial nitrate respiration–genes, enzymes and environmental distribution. J Biotechnol. 2011;155:104–17. [DOI] [PubMed] [Google Scholar]
- Landa M, Turk-Kubo KA, Cornejo-Castillo FMet al. Critical role of light in the growth and activity of the marine N2-fixing UCYN-A symbiosis. Front Microbiol. 2021;12:666739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolfi A, Koeve W, Dietze Het al. A new perspective on environmental controls of marine nitrogen fixation. Geophys Res Lett. 2015;42:4482–9. [Google Scholar]
- Langlois R, Grosskopf T, Mills Met al. Widespread distribution and expression of Gamma A (UMB), an uncultured, diazotrophic, gamma-Proteobacterial nifH Phylotype. PLoS ONE. 2015;10:e0128912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RJ, Hummer D, LaRoche J. Abundances and distributions of the dominant nifH phylotypes in the Northern Atlantic Ocean. Appl Environ Microbiol. 2008;74:1922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RJ, LaRoche J, Raab PA. Diazotrophic diversity and distribution in the tropical and subtropical Atlantic Ocean. Appl Environ Microbiol. 2005;71:7910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RJ, Mills MM, Ridame Cet al. Diazotrophic bacteria respond to Saharan dust additions. Mar Ecol Progr Ser. 2012;470:1–14. [Google Scholar]
- Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–77. [DOI] [PubMed] [Google Scholar]
- Lema KA, Willis BL, Bourne DG. Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the Acropora millepora microbiome. Environ Microbiol. 2014;16:3345–59. [DOI] [PubMed] [Google Scholar]
- Lema KA, Willis BL, Bourne DG. Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl Environ Microbiol. 2012;78:3136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letelier RM, Bjorkman KM, Church MJet al. Climate-driven oscillation of phosphorus and iron limitation in the North Pacific subtropical gyre. Proc Natl Acad Sci. 2019;116:12720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yu K, Wang Yet al. Diazotroph diversity associated with scleractinian corals and its relationships with environmental variables in the South China Sea. Front Physiol. 2020;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R, Reyes-Ramirez F, Zhang Yet al. Signal transduction to the Azotobacter vinelandii NIFL-NIFA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 2000;19:6041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhou L, Li Jet al. Effect of mesoscale eddies on diazotroph community structure and nitrogen fixation rates in the South China Sea. Region Stud Mar Sci. 2020;35:101106. [Google Scholar]
- Löscher CR, Bourbonnais A, Dekaezemacker Jet al. N2 fixation in eddies of the eastern tropical South Pacific Ocean. Biogeosciences. 2016;13:2889–99. [Google Scholar]
- Löscher CR, Großkopf T, Desai FDet al. Facets of diazotrophy in the oxygen minimum zone waters off Peru. ISME J. 2014;8:2180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher CR, Mohr W, Bange HWet al. No nitrogen fixation in the Bay of Bengal?. Biogeosciences. 2020;17:851–64. [Google Scholar]
- Luo YW, Doney SC, Anderson LAet al. Database of diazotrophs in global ocean: abundance, biomass and nitrogen fixation rates. Earth Syst Sci Data. 2012;4:47–73. [Google Scholar]
- Mague TH, Weare MM, Holm-Hansen O. Nitrogen fixation in the north Pacific Ocean. Mar Biol. 1974;24:109–19. [Google Scholar]
- Man-Aharonovich D, Kress N, Bar Zeev Eet al. Molecular ecology of nifH genes and transcripts in the eastern Mediterranean Sea. Environ Microbiol. 2007;9:2354–63. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez C, Mohr W, Schwedt Aet al. Metabolic versatility of a novel N2-fixing Alphaproteobacterium isolated from a marine oxygen minimum zone. Environ Microbiol. 2018;20:755–68. [DOI] [PubMed] [Google Scholar]
- Masepohl B. 2017; Regulation of nitrogen fixation in photosynthetic purple nonsulfur bacteria. In: Hallenbeck PC (ed.), Modern Topics in the Phototrophic Prokaryotes: Metabolism, Bioenergetics, and Omics. New York: Springer International Publishing, 1–25. [Google Scholar]
- McGlathery KJ, Risgaard-Petersen N, Christensen PB. Temporal and spatial variation in nitrogen fixation activity in the eelgrass Zostera marina rhizosphere. Mar Ecol Progr Ser. 1998;168:245–58. [Google Scholar]
- McInnes AS, Shepard AK, Raes EJet al. Simultaneous quantification of active carbon- and nitrogen-fixing communities and estimation of fixation rates using fluorescence in situ hybridization and flow cytometry. Appl Environ Microbiol. 2014;80:6750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRose DL, Zhang X, Kraepiel AMet al. Diversity and activity of alternative nitrogenases in sequenced genomes and coastal environments. Front Microbiol. 2017;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MP, Baross JA. Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon. Science. 2006;314:1783–6. [DOI] [PubMed] [Google Scholar]
- Mehta MP, Butterfield DA, Baross JA. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol. 2003;69:960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler S, Britten GL, Dutkiewicz Set al. Constraining uncertainties of diazotroph biogeography from nifH gene abundance. Limnol Oceanogr. 2022;67:816–29. [Google Scholar]
- Messer LF, Brown MV, Van Ruth PDet al. Temperate southern Australian coastal waters are characterised by surprisingly high rates of nitrogen fixation and diversity of diazotrophs. PeerJ. 2021;9:e10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer LF, Mahaffey C, Robinson CMet al. High levels of heterogeneity in diazotroph diversity and activity within a putative hotspot for marine nitrogen fixation. ISME J. 2015,1499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer NR, Fortney JL, Dekas AE. NanoSIMS sample preparation decreases isotope enrichment: magnitude, variability and implications for single-cell rates of microbial activity. Environ Microbiol. 2021;23:81–98. [DOI] [PubMed] [Google Scholar]
- Mills MM, Ridame C, Davey Met al. Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature. 2004;429:292–4. [DOI] [PubMed] [Google Scholar]
- Mise K, Masuda Y, Senoo Ket al. Undervalued pseudo-nifH sequences in public databases distort metagenomic insights into biological nitrogen fixers. mSphere. 2021;6:e0078521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J, Higa R, Toki Tet al. Molecular characterization of potential nitrogen fixation by anaerobic methane-oxidizing archaea in the methane seep sediments at the number 8 Kumano Knoll in the Kumano Basin, offshore of Japan. Appl Environ Microbiol. 2009;75:7153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NM, Colman AS, Tal Yet al. Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ Microbiol. 2008;10:2910–21. [DOI] [PubMed] [Google Scholar]
- Mohr W, Lehnen N, Ahmerkamp Set al. Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature. 2021;600:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisander PH, Beinart RA, Voss Met al. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J. 2008;2:954–67. [DOI] [PubMed] [Google Scholar]
- Moisander PH, Benavides M, Bonnet Set al. Chasing after non-cyanobacterial nitrogen fixation in marine pelagic environments. Front Microbiol. 2017;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisander PH, Morrison AE, Ward BBet al. Spatial-temporal variability in diazotroph assemblages in Chesapeake Bay using an oligonucleotide nifH microarray. Environ Microbiol. 2007;9:1823–35. [DOI] [PubMed] [Google Scholar]
- Moisander PH, Serros T, Paerl RWet al. Gammaproteobacterial diazotrophs and nifH gene expression in surface waters of the South Pacific Ocean. ISME J. 2014;8:1962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisander PH, Zhang R, Boyle EAet al. Analogous nutrient limitations in unicellular diazotrophs and Prochlorococcus in the South Pacific Ocean. ISME J. 2012;6:733–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Mills MM, Achterberg EPet al. Large-scale distribution of Atlantic nitrogen fixation controlled by iron availability. Nat Geosci. 2009;2:867–71. [Google Scholar]
- Moraru C, Lam P, Fuchs BMet al. GeneFISH – an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol. 2010;12:3057–73. [DOI] [PubMed] [Google Scholar]
- Moynihan MA, Goodkin NF, Morgan KMet al. Coral-associated nitrogen fixation rates and diazotrophic diversity on a nutrient-replete equatorial reef. ISME J. 2022;16:233–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland MR, Bernhardt PW, Blanco-Garcia JLet al. Rates of dinitrogen fixation and the abundance of diazotrophs in North American coastal waters between Cape Hatteras and Georges Bank. Limnol Oceanogr. 2012;57:1067–83. [Google Scholar]
- Mulholland MR, Bernhardt PW, Widner BNet al. High rates of N2 fixation in temperate, Western North Atlantic coastal waters expand the realm of marine diazotrophy. Glob Biogeochem Cycl. 2019;33:826–40. [Google Scholar]
- Nakayama T, Inagaki Y. Genomic divergence within non-photosynthetic cyanobacterial endosymbionts in rhopalodiacean diatoms. Sci Rep. 2017;7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell SE, Pritchard KR, Foster SQet al. Molecular evidence for sediment nitrogen fixation in a temperate New England estuary. PeerJ. 2016;4:e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton WE. Recent advances in understanding nitrogenases and how they work. Biol Nitrogen Fixat, 2015;1 and 2:7–20. [Google Scholar]
- Olson JB, Litaker RW, Paerl HW. Ubiquity of heterotrophic diazotrophs in marine microbial mats. Aquat Microb Ecol. 1999;19:29–36. [Google Scholar]
- Olson ND, Ainsworth TD, Gates RDet al. Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J Exp Mar Biol Ecol. 2009;371:140–6. [Google Scholar]
- Paerl HW, Prufert LE. Oxygen-poor microzones as potential sites of microbial N2 fixation in nitrogen-depleted aerobic marine waters. Appl Environ Microbiol. 1987;53:1078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl HW. Microzone formation: its role in the enhancement of aquatic N2 fixation. Limnol Oceanogr. 1985;30:1246–52. [Google Scholar]
- Paerl RW, Hansen TN, Henriksen NNet al. N-fixation and related O2 constraints on model marine diazotroph Pseudomonas stutzeri BAL361. Aquat Microb Ecol. 2018;81:125–36. [Google Scholar]
- Paulsen DM, Paerl HW, Bishop PE. Evidence that molybdenum-dependent nitrogen-fixation is not limited by high sulfate concentrations in marine environments. Limnol Oceanogr. 1991;36:1325–34. [Google Scholar]
- Pedersen JN, Bombar D, Paerl RWet al. Diazotrophs and N2-fixation associated with particles in coastal estuarine waters. Front Microbiol. 2018;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedros-Alio C. The rare bacterial biosphere. Ann Rev Mar Sci. 2012;4:449–66. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Dekas AE, Brown CTet al. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci. 2008;105:7052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierella Karlusich JJ, Pelletier E, Lombard Fet al. Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods. Nat Commun. 2021;12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Pavlekovic M, Lee NMet al. Fluorescence in situ hybridization for intracellular localization of nifH mRNA. Syst Appl Microbiol. 2009;32:186–92. [DOI] [PubMed] [Google Scholar]
- Ploug H, Bergkvist J. Oxygen diffusion limitation and ammonium production within sinking diatom aggregates under hypoxic and anoxic conditions. Mar Chem. 2015;176:142–9. [Google Scholar]
- Ploug H, Kühl M, Buchholz-Cleven Bet al. Anoxic aggregates-an ephemeral phenomenon in the pelagic environment?. Aquat Microb Ecol. 1997;13:285–94. [Google Scholar]
- Ploug H. Small-scale oxygen fluxes and remineralization in sinking aggregates. Limnol Oceanogr. 2001;46:1624–31. [Google Scholar]
- Poff KE, Leu AO, Eppley JMet al. Microbial dynamics of elevated carbon flux in the open ocean's abyss. Proc Natl Acad Sci. 2021;118:e2018269118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole RK, Hill S. Respiratory protection of nitrogenase activity in Azotobacter vinelandii--roles of the terminal oxidases. Biosci Rep. 1997;17:303–17. [DOI] [PubMed] [Google Scholar]
- Post E, Golecki JR, Oelze J. Morphological and ultrastructural variations in Azotobacter vinelandii growing in oxygen-controlled continous culture. Arch Microbiol. 1982;133:75–82. [Google Scholar]
- Postgate JR. 1982; The Fundamentals of Nitrogen Fixation. London: Cambridge University Press. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor LM. Nitrogen-fixing, photosynthetic, anaerobic bacteria associated with pelagic copepods. Aquat Microb Ecol. 1997;12:105–13. [Google Scholar]
- Raes EJ, Van de Kamp J, Bodrossy Let al. N2 fixation and new insights into nitrification from the ice-edge to the equator in the South Pacific Ocean. Front Mar Sci. 2020;7:1–20.32802822 [Google Scholar]
- Rahav E, Bar-Zeev E, Ohayon Set al. Dinitrogen fixation in aphotic oxygenated marine environments. Front Microbiol. 2013;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahav E, Giannetto M, Bar-Zeev E. Contribution of mono and polysaccharides to heterotrophic N2 fixation at the eastern Mediterranean coastline. Sci Rep. 2016;6:27858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratten J-M. 2017; Annual Cycle of Change in Bacterial Community Structure in a Temperate Coastal Marine Embayment in the North Atlantic. Thesis, Dalhousie University, Halifax, Nova Scotia. [Google Scholar]
- Raut Y, Capone DG. Macroalgal detrital systems: an overlooked ecological niche for heterotrophic nitrogen fixation. Environ Microbiol. 2021;23:4372–88. [DOI] [PubMed] [Google Scholar]
- Raut Y, Morando M, Capone DG. Diazotrophic macroalgal associations with living and decomposing sargassum. Front Microbiol. 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder CF, Löscher CR. Nitrogenases in oxygen minimum zone waters. Front Mar Sci. 2022;9:1–9.35450130 [Google Scholar]
- Rees AP, Gilbert JA, Kelly-Gerreyn BA. Nitrogen fixation in the western English Channel (NE Atlantic Ocean). Mar Ecol Progr Ser. 2009;374:7–12. [Google Scholar]
- Ribeiro CG, dos Santos AL, Marie Det al. Small eukaryotic phytoplankton communities in tropical waters off Brazil are dominated by symbioses between Haptophyta and nitrogen-fixing cyanobacteria. ISME J. 2018;12:1360–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes M, Dziallas C, Coma Ret al. Microbial diversity and putative diazotrophy in high- and low-microbial-abundance Mediterranean sponges. Appl Environ Microbiol. 2015;81:5683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridame C, Dinasquet J, Hallstrøm Set al. N2 fixation in the Mediterranean Sea related to the composition of the diazotrophic community and impact of dust under present and future environmental conditions. Biogeosciences. 2022;19:415–35. [Google Scholar]
- Riemann L, Farnelid H, Steward GF. Nitrogenase genes in non-cyanobacterial plankton: prevalence, diversity and regulation in marine waters. Aquat Microb Ecol. 2010;61:235–47. [Google Scholar]
- Riemann L, Rahav E, Passow Uet al. Planktonic aggregates as hotspots for heterotrophic diazotrophy: the plot thickens. Front Microbiol. 2022;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra W, Zeng AP, Lunsdorf Het al. Effect of oxygen on formation and structure of Azotobacter vinelandii alginate and its role in protecting nitrogenase. Appl Environ Microbiol. 2000;66:4037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino T, Hatori A. 1980; Nitrogen fixation by Trichodesmium and its significance in nitrogen cycling in the Kuroshio area and adjacent waters. In: Takenouti AY (ed.), The Kuroshio IV. Tokyo: Saikon Publishing, 697–709. [Google Scholar]
- Salazar G, Paoli L, Alberti Aet al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell. 2019;179:1068–1083.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Venceslau SS, Grein Fet al. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science. 2015;350:1541–5. [DOI] [PubMed] [Google Scholar]
- Sato T, Shiozaki T, Taniuchi Yet al. Nitrogen fixation and diazotroph community in the subArctic Sea of Japan and Sea of Okhotsk. J Geophys Res Oceans. 2021;126:e2020JC017071. [Google Scholar]
- Scavotto RE, Dziallas C, Bentzon-Tilia Met al. Nitrogen-fixing bacteria associated with copepods in coastal waters of the North Atlantic Ocean. Environ Microbiol. 2015;17:3754–65. [DOI] [PubMed] [Google Scholar]
- Schvarcz CR, Wilson ST, Caffin Met al. Overlooked and widespread pennate diatom-diazotroph symbioses in the sea. Nat Commun. 2022;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt LC, Hoffman BM, Dean DR. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem. 2009;78:701–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selden CR, Chappell PD, Clayton Set al. A coastal N2 fixation hotspot at the Cape Hatteras front: elucidating spatial heterogeneity in diazotroph activity via supervised machine learning. Limnol Oceanogr. 2021;66:1832–49. [Google Scholar]
- Selden CR, Mulholland MR, Bernhardt PWet al. Dinitrogen fixation across physico-chemical gradients of the eastern tropical North Pacific oxygen deficient zone. Glob Biogeochem Cycl. 2019;33:1187–202. [Google Scholar]
- Shao Z, Luo YW. Controlling factors on the global distribution of a representative marine heterotrophic diazotroph phylotype (Gamma A). Biogeosciences. 2022;19:2939–52. [Google Scholar]
- Shiozaki T, Bombar D, Riemann Let al. Basin scale variability of active diazotrophs and nitrogen fixation in the North Pacific, from the tropics to the subArctic Bering Sea. Glob Biogeochem Cycl. 2017;31:996–1009. [Google Scholar]
- Shiozaki T, Bombar D, Riemann Let al. Linkage between dinitrogen fixation and primary production in the oligotrophic South Pacific Ocean. Glob Biogeochem Cycl. 2018a;32:1028–44. [Google Scholar]
- Shiozaki T, Fujiwara A, Ijichi Met al. Diazotroph community structure and the role of nitrogen fixation in the nitrogen cycle in the Chukchi Sea (western Arctic Ocean). Limnol Oceanogr. 2018b;63:2191–205. [Google Scholar]
- Shiozaki T, Fujiwara A, Inomura Ket al. Biological nitrogen fixation detected under Antarctic Sea ice. Nat Geosci. 2020;13:729–32. [Google Scholar]
- Shiozaki T, Ijichi M, Kodama Tet al. Heterotrophic bacteria as major nitrogen fixers in the euphotic zone of the Indian Ocean. Glob Biogeochem Cycl. 2014;28:1096–110. [Google Scholar]
- Shiozaki T, Nagata T, Ijichi Met al. Nitrogen fixation and the diazotroph community in the temperate coastal region of the northwestern North Pacific. Biogeosciences. 2015;12:4751–64. [Google Scholar]
- Short SM, Jenkins BD, Zehr JP. Spatial and temporal distribution of two diazotrophic bacteria in the Chesapeake Bay. Appl Environ Microbiol. 2004;70:2186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Zehr JP. Nitrogenase gene expression in the Chesapeake Bay Estuary. Environ Microbiol. 2007;9:1591–6. [DOI] [PubMed] [Google Scholar]
- Simon M, Grossart H-P, Schweitzer Bet al. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 2002;28:175–211. [Google Scholar]
- Sipler RE, Gong D, Baer SEet al. Preliminary estimates of the contribution of Arctic nitrogen fixation to the global nitrogen budget. Limnol Oceanogr Lett. 2017;2:159–66. [Google Scholar]
- Smercina DN, Evans SE, Friesen MLet al. To fix or not to fix: controls on free-living nitrogen fixation in the rhizosphere. Appl Environ Microbiol. 2019;85:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppe TF, Olson JB, Paerl HWet al. Consortial N2 fixation: a strategy for meeting nitrogen requirements of marine and terrestrial cyanobacterial mats. FEMS Microbiol Ecol. 1996;21:149–56. [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron Set al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. [DOI] [PubMed] [Google Scholar]
- Tang W, Li Z, Cassar N. Machine learning estimates of global marine nitrogen fixation. J Geophys Res Biogeosci. 2019a;124:717–30. [Google Scholar]
- Tang W, Wang S, Fonseca-Batista Det al. Revisiting the distribution of oceanic N2 fixation and estimating diazotrophic contribution to marine production. Nat Commun. 2019b;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Foster RA, Krupke Aet al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science. 2012;337:1546–50. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KAet al. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–94. [DOI] [PubMed] [Google Scholar]
- Troussellier M, Escalas A, Bouvier Tet al. Sustaining rare marine microorganisms: macroorganisms as repositories and dispersal agents of microbial diversity. Front Microbiol. 2017;8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk K, Rees AP, Zehr JPet al. Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J. 2011;5:1201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk-Kubo KA, Frank IE, Hogan MEet al. Diazotroph community succession during the VAHINE mesocosm experiment (New Caledonia lagoon). Biogeosciences. 2015;12:7435–52. [Google Scholar]
- Turk-Kubo KA, Karamchandani M, Capone DGet al. The paradox of marine heterotrophic nitrogen fixation: abundances of heterotrophic diazotrophs do not account for nitrogen fixation rates in the Eastern Tropical South Pacific. Environ Microbiol. 2014;16:3095–114. [DOI] [PubMed] [Google Scholar]
- Turk-Kubo KA, Mills MM, Arrigo KRet al. UCYN-A/haptophyte symbioses dominate N2 fixation in the Southern California Current System. ISME Commun. 2021;1:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venrick EL. The distribution and significance of Richelia intracellularis Schmidt in the North Pacific Central Gyre. Limnol Oceanogr. 1974;19:437–45. [Google Scholar]
- Villareal TA. 1992; Marine nitrogen-fixing diatom - cyanobacteria symbioses. In: Carpenter EJ, Capone DJ, Rueter JG (eds), Marine Pelagic Cyanobacteria: Trichodesmium and other Diazotrophs. Dordrecht: Kluwer Academic Publishers, 163–75. [Google Scholar]
- Ward BA, Dutkiewicz S, Moore CMet al. Iron, phosphorus, and nitrogen supply ratios define the biogeography of nitrogen fixation. Limnol Oceanogr. 2013;58:2059–75. [Google Scholar]
- White AE, Granger J, Selden Cet al. A critical review of the 15N2 tracer method to measure diazotrophic production in pelagic ecosystems. Limnol Oceanogr Methods. 2020;18:129–47. [Google Scholar]
- Wu C, Kan J, Liu Het al. Heterotrophic bacteria dominate the diazotrophic community in the Eastern Indian Ocean (EIO) during pre-southwest monsoon. Microb Ecol. 2019;78:804–19. [DOI] [PubMed] [Google Scholar]
- Wu C, Sun J, Liu Het al. Evidence of the significant contribution of heterotrophic diazotrophs to nitrogen fixation in the Eastern Indian Ocean during pre-southwest monsoon period. Ecosystems. 2021;25:1066–83. [Google Scholar]
- Wyman M, Zehr J, Capone D. Temporal variability in nitrogenase gene expression in natural populations of the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1996;62:1073–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QS, Dong JD, Ahmad Met al. Analysis of nifH DNA and RNA reveals a disproportionate contribution to nitrogenase activities by rare plankton-associated diazotrophs. BMC Microbiol. 2019;19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Lee ST, Huang CRet al. Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol Oceanogr. 2016;61:1078–86. [Google Scholar]
- Yogev T, Rahav E, Bar-Zeev Eet al. Is dinitrogen fixation significant in the Levantine Basin, East Mediterranean Sea?. Environ Microbiol. 2011;13:854–71. [DOI] [PubMed] [Google Scholar]
- You M, Nishiguchi T, Saito Aet al. Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: daily rhythm during the light-dark cycle. Appl Environ Microbiol. 2005;71:8183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani S, Mellon MT, Collier JLet al. Expression of nifH genes in natural microbial assemblages in Lake George, NY detected with RT-PCR. Appl Environ Microbiol. 2000;66:3119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J, McReynolds L. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J, Mellon M, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Capone DG. 2021; Marine Nitrogen Fixation. Berlin: Springer Nature. [Google Scholar]
- Zehr JP, Capone DG. Changing perspectives in marine nitrogen fixation. Science. 2020;368:eaay9514. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Jenkins BD, Short SMet al. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol. 2003;5:539–54. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Mellon M, Braun Set al. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Montoya JP, Jenkins BDet al. Experiments linking nitrogenase gene expression to nitrogen fixation in the North Pacific subtropical gyre. Limnol Oceanogr. 2007;52:169–83. [Google Scholar]
- Zhang X, McRose DL, Darnajoux Ret al. Alternative nitrogenase activity in the environment and nitrogen cycle implications. Biogeochemistry. 2016;127:189–98. [Google Scholar]
- Zhang X, Song Y, Liu Det al. Macroalgal blooms favor heterotrophic diazotrophic bacteria in nitrogen-rich and phosphorus-limited coastal surface waters in the Yellow Sea. Estuar Coast Shelf Sci. 2015;163:75–81. [Google Scholar]
- Zhang Y, Zhao Z, Sun Jet al. Diversity and distribution of diazotrophic communities in the South China Sea deep basin with mesoscale cyclonic eddy perturbations. FEMS Microbiol Ecol. 2011;78:417–27. [DOI] [PubMed] [Google Scholar]
- Zouch H, Cabrol L, Chifflet Set al. Effect of acidic industrial effluent release on microbial diversity and trace metal dynamics during resuspension of coastal sediment. Front Microbiol. 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.