Abstract
We have found previously that, in contrast to the free O initiator protein of λ phage or plasmid rapidly degraded by the Escherichia coli ClpP/ClpX protease, the λO present in the replication complex (RC) is protected from proteolysis. However, in cells growing in a complete medium, a temperature shift from 30 to 43°C resulted in the decay of the λO fraction, which indicated disassembly of RC. This process occurred due to heat shock induction of the groE operon, coding for molecular chaperones of the Hsp60 system. Here we demonstrate that an increase in the cellular concentration of GroEL and GroES proteins is not in itself sufficient to cause RC disassembly. Another requirement is a DNA gyrase-mediated negative resupercoiling of λ plasmid DNA, which counteracts DNA relaxation and starts to dominate 10 min after the temperature upshift. We presume that RC dissociates from λ DNA during the negative resupercoiling, becoming susceptible to the subsequent action of GroEL/S and ClpP/ClpX proteins. In contrast to λcro+, in λcro− plasmid-harboring cells, the RC reveals heat shock resistance. After temperature upshift of the λcrots plasmid-harboring cells, a Cro repressor-independent control of λ DNA replication and heat shock resistance of RC are established before the period of DNA gyrase-mediated negative supercoiling. We suggest that the tight binding of RC to λ DNA is due to interaction of RC with other DNA-bound proteins, and is related to the molecular basis of the λcro− plasmid replication control.
The studies on the bacteriophage λ DNA early (θ, or circle-to-circle) replication in Escherichia coli have been greatly stimulated by the availability of the phage λ-derived plasmids, called originally λdv (for a review, see reference 17). The λ plasmids may be produced from λ phage DNA by cutting out and circularization of the λ replication region oRpR-cro-cII-O-P. The origin of replication, oriλ, is situated in the middle of the O gene, and the pR promoter-initiated transcription plays a dual role: it is required for the synthesis of λ replication proteins, O and P, as well as for the transcriptional activation of oriλ (for reviews, see references 14 and 30). The binding of Cro repressor to the oR operator represents an autoregulatory loop, which is assumed to play an important role in the control of λ plasmid replication (17). The cII protein, so important for the lysis-or-lysogeny decision in the early development of phage λ, is dispensable for λ plasmid replication. The λ O protein, the initiator of λ DNA replication, is rapidly degraded in vivo by the E. coli ClpP/ClpX protease (3, 8, 46), but it becomes protected from proteolysis in the pathway of the λ replication complex (RC) assembly (21, 34, 42). This event occurs after interaction of the λP-DnaB helicase complex with the O initiator bound to oriλ at the step of the λO-λP-DnaB preprimosome formation (34). The next step, the release of DnaB helicase from λP-mediated inhibition, is performed by DnaK, DnaJ, and GrpE proteins, the molecular chaperones of the Hsp70 system (2, 6, 48). The action of DnaB helicase loaded between transiently separated (due to transcriptional activation of oriλ) complementary strands of λ DNA permits the binding of at least DnaG primase and the DNA polymerase III holoenzyme, leading to the assembly of a functional λ RC (1, 32, 43–45, 48).
The studies on the in vitro-reconstituted λ plasmid DNA replication led to an attractive model suggesting that the λ RC functional in elongation resembles that of its host, E. coli: both complexes would be devoid of the respective initiator proteins, λO or DnaA, and the respective DnaB helicase inhibitors, λP or DnaC. Moreover, it is tacitly assumed that in both systems, λ and the E. coli chromosome, the RC would be completely disassembled after one round of replication. By implication, the next round of replication should depend on the binding of the initiator, λO or DnaA, to oriλ or oriC, respectively (14). However, the present in vitro system of λ plasmid DNA replication may not exactly reflect the conditions occurring in the cell and is not suitable for studying more than one replication round.
We developed two systems for blocking the de novo assembly of the λ RC. In amino acid-starved E. coli relA cells, there was no synthesis of the rapidly destroyed λO initiator (33, 35, 36) and in E. coli wild-type (wt) cells growing in a complete medium, the λPts1 mutation (in the presence of the dnaA+ allele) blocked RC assembly at 43°C (33, 36, 44). Density shift and other experiments demonstrated that in both systems λ plasmid DNA replication does occur and occurs linearly: only one of two plasmid daughter copies was able to initiate the next round of replication (28, 33, 36, 40). This replication could occur due to RCs assembled before the onset of amino acid starvation or temperature upshift, respectively. It was dependent on λO, as well as on DnaK, DnaJ, and GrpE chaperone functions (33, 35, 36, 40, 41). We concluded that the assembled RC, containing λO and probably also λP, is not disassembled after termination of a round of replication (30, 40). This multiprotein complex (or at least the part that protects λO from proteolysis) is inherited by one of two λ plasmid daughter copies at each round of replication (29, 36). In amino acid-starved E. coli relA cells, the “old” RC-driven replication ceases after several rounds (39), but in E. coli wt cells growing in a complete medium, this replication does not seem to be time restricted (36). The above studies, together with the observation that neither the absence of nor the presence of excess λO-digesting ClpP/ClpX protease affects λ plasmid or phage replication (27), rule out the model of λO binding to oriλ as the crucial event in the control of initiation frequency.
In amino acid-starved E. coli relA cells, the Cro-mediated regulation did not work, probably due to titration out of Cro by the growing number of oR operator sequences (40, 41). In E. coli wt cells growing in complete medium, the replication of λcrotsPts1 plasmid after a temperature upshift was studied; hence, Cro regulation also did not work in this system (36). However, when the same procedure was applied to E. coli wt harboring λcro+Pts1, plasmid replication was blocked and RC disassembly (judged from proteolysis of the otherwise stable λO) was observed (36). The disassembly of RC also occurred for E. coli wt harboring λcro+P+. Examination of this phenomenon revealed that for RC disassembly observed in λcro+ plasmid-harboring wt cells, heat shock induction of the groE operon, coding for molecular chaperones of the Hsp60 system, is indispensable (37). This was the first observation that the Hsp60 chaperone proteins, known to mediate deaggregation of denatured protein aggregates, may also disassemble in vivo a highly organized protein structure (dispensable for cell survival) under stress caused by temperature upshift.
Here we present our study on the disassembly of RC, which emphasizes the role of DNA gyrase-mediated negative resupercoiling of λ plasmid DNA, which counteracts DNA relaxation that occurs immediately after the temperature upshift (20). It seems that the negative resupercoiling of λ plasmid DNA results in dissociation of RC that now becomes disassembled with the help of GroEL and GroES chaperone proteins. This finding allowed us to extend our studies on the λcrots plasmid and to formulate a hypothesis concerning heat shock resistance of RC when Cro repressor function is absent.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, and gene fusions.
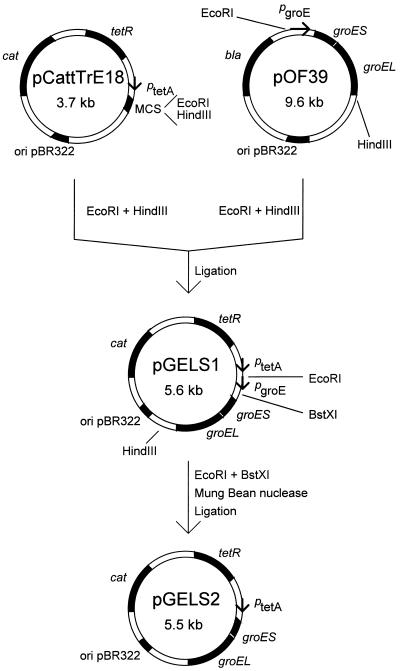
Most of the experiments (all except the gene fusion experiments) were performed in an E. coli MG1655 genetic background (12). Particular mutations were transferred by P1 transduction by the method of Silhavy et al. (25). The mutant strains used were BM270 (groEL44 linked to Tn10), obtained from C. Georgopoulos and M. Zylicz (see also reference 37); HI515 (nalA26), described by Ikeda et al. (11) and provided by K. Sekimizu and Y. Ogata; and CAG354 [rpoD800(Ts) linked to Tn10], described by Liebke et al. (16) and obtained from C. Gross. To achieve expression of the groE operon from plasmid pGELS2 (see below), it was necessary to remove a tetA gene activity from a tetracycline-resistant strain (like the groEL44 mutant). This was performed by the method of Bochner et al. (5). Plasmid pGELS1 was constructed by cloning the groE operon from pOF39 (7) into the pCattTrE18 vector (constructed in the laboratory of W. Szybalski [University of Wisconsin] by M. Koob). Plasmid pGELS2 is a derivative of pGELS1 lacking the groE promoter region; thus, transcription of groEL and groES genes is exclusively dependent on the ptetA promoter activity. Details of the construction of pGELS1 and pGELS2 are provided in Fig. 1. Plasmids derived from bacteriophage λ, pKB2 (wt), and pRLM4 (as pKB2 but crots) were described by Kur et al. (15) and Wold et al. (47), respectively. Experiments with gene fusions were performed with a Δlac strain (WAM106) described by Thomas and Glass (31). The single-copy pL-lacZ fusion carried on cryptic λcI857 prophage was provided by D. Court. The single-copy fusion of the ς32-dependent groE promoter with lacZ was described by Benvenisti et al. (4) and obtained from A. B. Oppenheim. Single-stranded DNA of phage M13mp18λI (29) was used as a template for preparation of the labeled probe used in Northern blotting experiments. Phage λclb2 (from our collection) was also used.
FIG. 1.
Construction of pGELS1 and pGELS2.
DNA techniques.
All DNA manipulations (molecular cloning and preparation of labeled probes for Northern blotting) were carried by the methods of Sambrook et al. (24).
λO protein decay.
Decay of the λO protein was measured as described previously (34, 37, 42). Briefly, bacteria harboring a λ plasmid were pulse-labeled with [35S]methionine and the label was chased by addition of excess of unlabeled l-methionine (time zero in the figures). The samples were withdrawn at the indicated times, the bacteria were lysed, and the cellular lysates were immunoprecipitated with anti-λO serum. Then the isotope-labeled proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5% polyacrylamide) and visualized by autoradiography. The bands of the λO protein remaining after the different chase times were quantitated by densitometry.
Changes in plasmid DNA linking number.
Changes in λ plasmid DNA linking number after a temperature shift were monitored by the method of Ogata et al. (20). Briefly, samples of the bacterial culture were withdrawn at the indicated times and chilled immediately by being transferred into probes containing equal volume of ice. Plasmid DNA was isolated by the alkali lysis method (24) and separated by agarose gel electrophoresis (1% agarose) in TBE buffer (24) containing 15 μg of chloroquine per ml (electrophoresis was performed at 2 V per cm of gel length). The changes in the average linking number of plasmid DNA (Δτ) were calculated by the method of Keller (13).
Analysis of the level of pR-tR2 transcripts.
Estimation of the amounts of the pR-tR2 transcripts relative to the level of rRNA (internal RNA control) and to the level of λ plasmid (template DNA) was performed as described previously (19, 29). Briefly, samples containing the same bacterial mass (estimated by measurement of the optical density at 575 nm of the culture) were withdrawn at the indicated times. Each sample was divided in two parts. RNA was isolated from the first part (see reference 29 for a detailed description of the procedure), and plasmid DNA was isolated by the alkali lysis method (24) from the second part. The relative level of rRNA was estimated densitometrically on the basis of analysis of ethidium bromide-stained agarose gels. The pR-tR2 transcripts were detected by Northern blotting and quantitated by densitometry (the template for labeling of the probe designed to detect these transcripts was single-stranded DNA isolated from phage M13mp18λI). The amount of λ plasmid DNA was estimated on the basis of densitometric analysis of DNA bands after agarose gel electrophoresis and staining with ethidium bromide, by the method of Herman et al. (9). The relative values presented in the figure were calculated by the method of Obuchowski et al. (19) by dividing the relative intensity of bands on a particular blot by the relative intensity of ethidium bromide-stained rRNA bands and then by dividing by the relative amount of λ plasmid. A value of 1 corresponds to the relative value calculated for the sample taken from the culture of bacteria harboring wild-type λ plasmid (pKB2) at 30°C (time zero), and the other values are expressed relative to this value.
Estimation of the relative amount of λ plasmid.
The relative amount of λ plasmid DNA per bacterial mass was estimated as described by Herman et al. (9) (see above). A value of 1 corresponds to the relative value calculated for the sample taken from the culture of bacteria harboring wild-type λ plasmid (pKB2) at 30°C (time zero).
Measurement of β-galactosidase activity.
The activity of β-galactosidase was measured by the method of Miller (18).
RESULTS
Fine-tuned expression of the groE operon.
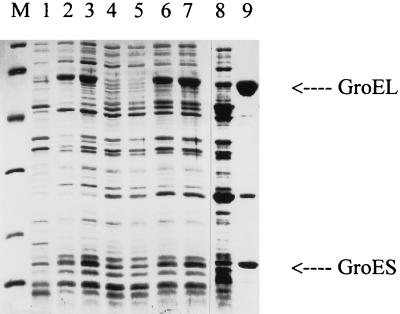
We assumed that RC disassembly may be due not only to an increased cellular concentration of GroEL/S chaperone proteins but also to some other concomitant process(es) (to be discovered), both induced by heat shock. To verify this hypothesis, it was important to raise the cellular concentration of GroEL/S proteins while keeping the temperature constant. This aim has been achieved by construction of a plasmid, pGELS2 (Fig. 1), containing the groE operon under the exclusive control of the promoter ptetA, responsible for the expression of tetracycline resistance in the Tn10 transpozon. The pGELS2 plasmid contains the tetR gene coding for the tetracycline repressor that blocks the ptetA-initiated transcription in the absence of the antibiotic. Autoclaved chlortetracycline, aCT, which has lost its antibiotic functions, inactivates TetR repressor (5, 23, 26) causing transcription of the groE operon and efficient synthesis of GroEL and GroES proteins (Fig. 2). The effect of increasing concentration of aCT on the level of GroEL/S in the pGELS2-harboring E. coli groEL44 cells was investigated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Its densitometric evaluation led to the conclusion that the level of GroEL at an aCT concentration of 0.05 μg/ml was equivalent to its level in plasmidless cells under the conditions used in our experiments before the temperature shift from 30 to 43°C; aCT at 2.0 μg/ml resulted in a three- to fourfold-higher level of GroEL (data not shown). The aCT-mediated derepression of the groE operon resulted in the production of functional GroEL/S chaperone proteins, as revealed by their activity in phage λ development (Table 1). It is worth noting that these proteins have been discovered due to their involvement in phage λ morphogenesis (for a review, see reference 22). At concentrations as low as 0.01 μg/ml, aCT caused derepression of the ptetA promoter, as revealed by the expression of the groE operon from pGELS2 (Table 1); at high aCT concentrations (e.g., 25 μg/ml), the system seems to be useful for the production of a high concentration of the desired protein in bacteria (Fig. 2, lane 9).
FIG. 2.
Overproduction of GroEL and GroES proteins from plasmids pOF39, pGELS1, and pGELS2. Bacteria were grown in Luria-Bertani medium at 30°C. When indicated, the culture was transferred to 43°C or treated with aCT (final concentration, 25 μg/ml) and incubated further for 60 min. Cell lysates were separated on 12.5% polyacrylamide gels, and protein bands were visualized by staining with Coomassie brilliant blue. Lanes: M, molecular mass standards (from top to bottom, 94, 67, 43, 30, 20.1, and 14.4 kDa), 1, MG1655; 2, MG1655/pOF39; 3, MG1655/pOF39 after a shift to 43°C; 4, MG1655/pCattTrE18; 5, MG1655/pCattTrE18 treated with aCT; 6, MG1655/pGELS1; 7, MG1655/pGELS1 treated with aCT; 8, MG1655/pGELS2; 9, MG1655/pGELS2 treated with aCT. The positions of GroEL and GroES are indicated.
TABLE 1.
Suppression of the effect of the groEL44 mutation on phage λ growth at 30°C and bacterial growth at 43°C by expression of the groE operon from plasmid pGELS2
| Strain | aCT concn (μg/ml)a | Growth of λclb2 at 30°Cb | Bacterial growth at 43°Cc |
|---|---|---|---|
| groEL44 | 0 | − | − |
| groEL44 | 25 | − | − |
| groEL44/pGELS2 | 0.010 | + | + |
| groEL44/pGELS2 | 0.025 | ++ | ++ |
| groEL44/pGELS2 | 0.050 | +++ | +++ |
| groEL44/pGELS2 | 0.100 | +++ | +++ |
| groEL44/pGELS2 | 25 | +++ | +++ |
Expression of the groE operon from pGELS2 was induced by addition of aCT to the medium.
A lysate of phage λclb2 was subjected to titer determination on the indicated host strain at 30°C. When plaques were formed, the efficiency of plating was always close to 1 relative to the wild-type (groEL+) strain. +++, formation of normal-sized (large) plaques; ++, small plaques; +, very small (tiny or pinpoint) plaques; − no plaques (efficiency of plating, <10−6).
Dilutions of bacterial culture growing at 30°C were spotted onto Luria-Bertani plates, which were then incubated overnight at 43°C. When colonies were formed, the efficiency of plating was always close to 1 relative to the same culture at 30°C. +++, formation of normal-sized (large) colonies; ++, small colonies; +, very small (tiny or pinpoint) colonies; − no colonies (efficiency of plating, <10−6).
Role of an increased GroEL/S chaperonin concentration.
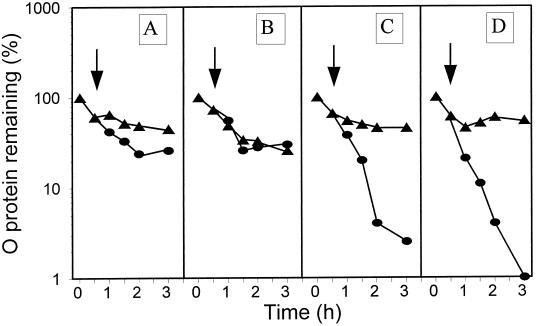
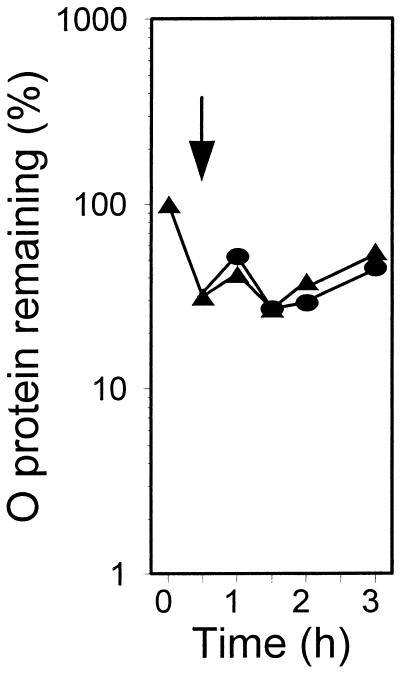
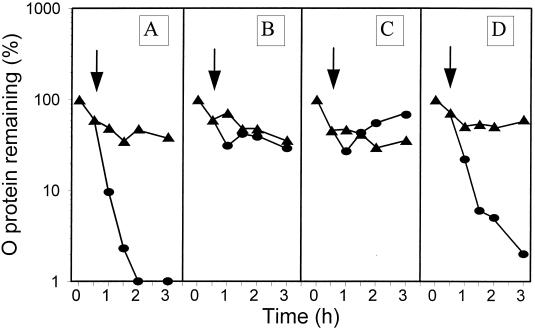
It was then possible to perform heat shock experiments with λ plasmid-harboring and pGELS2 plasmid-harboring groEL44 cells that could produce a functional GroEL protein exclusively due to aCT-mediated derepression of the groE operon of pGELS2. The aCT was added at the time of the temperature upshift. In the pulse-chase experiments in Fig. 3 (also see Fig. 4, 5, 7, and 9), the decay of λO protein occurring immediately after the start of isotope chasing at 30°C (time zero) represents ClpP/ClpX-mediated proteolysis of an excess of synthesized free λO (27, 42). The stable fraction of λO corresponds to λO protected from this protease by other components of the oriλ-bound λO-λP-DnaB preprimosome or RC (34, 37). There was no RC disassembly in the absence of aCT (Fig. 3A) or at an aCT concentration of 0.05 μg/ml (Fig. 3B), which should result in a level of functional GroEL equivalent to that present in wt cells before the heat shock (see above). However, when aCT was added to 2.0 μg/ml (see above), RC disassembly was observed (Fig. 3C). The same result was obtained at an aCT concentration of 25 μg/ml (Fig 3D). The above results support our previous finding that one of the requirements of heat shock-provoked RC disassembly is the induction of the groE operon (37). We were then ready to ask whether an increase of the concentration of GroEL/S chaperone proteins alone is sufficient to cause RC disassembly. As shown in Fig. 4, the answer was negative.
FIG. 3.
Decay of the λO protein at 30°C (triangles) and after a shift to 43°C (circles) in the groE44 mutant harboring λcro+ (pKB2) and pGELS2 plasmids, untreated (A) or treated with aCT at 0.05 μg/ml (B), 2 μg/ml (C), or 25 μg/ml (D) at the time of the temperature shift (arrow).
FIG. 4.
Decay of the λO protein at 30°C in the groE44 mutant harboring λcro+ (pKB2) and pGELS2 plasmids, untreated (triangles) or treated with aCT at 25 μg/ml (circles) at the time indicated by the arrow.
FIG. 5.
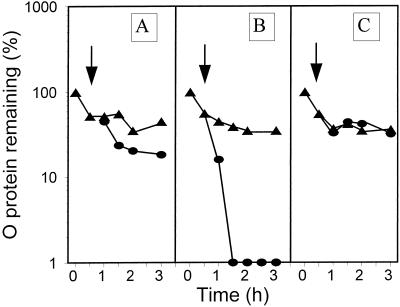
Decay of the λO protein at 30°C (triangles) and after a shift to 43°C (circles) in the wild-type strain (A to C) and the nalA26 mutant (D) harboring λcro+ (pKB2) plasmid, untreated (A), or treated with 100 μg of nalidixic acid per ml (B and D) or 50 μg of coumermycin per ml (C) at the time of the temperature shift (arrow).
FIG. 7.
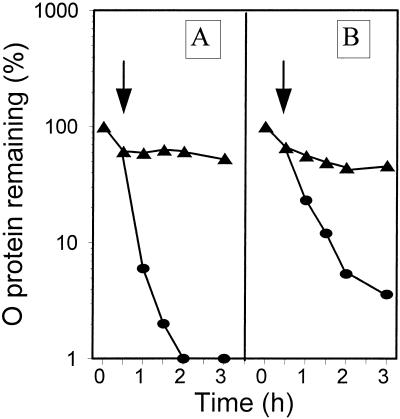
Decay of the λO protein at 30°C (triangles) and after a shift to 43°C (circles) in the wild-type strain harboring λcrots (pRLM4) plasmid, untreated (A) or treated with 25 μg of rifampin per ml (B) or 25 μg of rifampin per ml and 50 μg of coumermycin per ml (C) at the time of the temperature shift (arrow).
FIG. 9.
Decay of the λO protein at 30°C (triangles) and after a shift to 43°C (circles) in the rpoD800(Ts) mutant harboring λcro+ (pKB2) (A) or λcrots (pRLM4) (B). The temperature shift is indicated by the arrows.
Role of DNA gyrase-mediated negative supercoiling.
What else, besides induction of the groE operon, is changing after heat shock that is required for λ plasmid RC disassembly? We turned our attention to changes in supercoiling of plasmid DNA, known to occur after a temperature shift from 30 to 50°C (20), and started to study the effect of DNA gyrase inhibitors on RC disassembly under our standard experimental conditions. The disassembly of RC (Fig. 5A) was blocked by each of two DNA gyrase inhibitors, nalidixic acid (Fig. 5B) and coumermycin (Fig. 5C). The specificity of action of nalidixic acid was checked in an experiment performed with a nalidixic acid-resistant mutant, nalA26 (Fig. 5D). Gyrase may cause DNA relaxation or negative supercoiling; both processes are inhibited by nalidixic acid, in contrast to coumermycin, which specifically inhibits the ATP-dependent negative supercoiling of DNA (for a review, see reference 14).
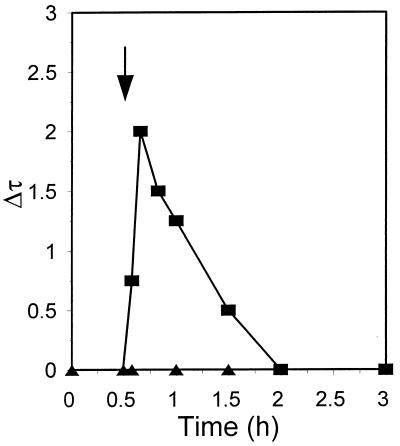
The heat shock-provoked changes in supercoiling of λ plasmid DNA are shown in Fig. 6. Measurement of changes in the average linking number of plasmid DNA revealed that immediately after the shift from 30 to 43°C there was a rapid DNA relaxation, attaining its peak value after 10 min. The maximal change that occurred in the absolute superhelical density of the plasmid population after the temperature shift was estimated to about 10 to 20% (data not shown). Then a process of negative resupercoiling began that restored the original state of supercoiling about 90 min after the temperature upshift. Each of the two DNA gyrase inhibitors nalidixic acid and coumermycin caused an even more rapid DNA relaxation, leading to a much more relaxed state of λ plasmid DNA (Δτ > 10) that persisted to the end of the experiment (results not shown). These results indicate that another enzyme, probably topoisomerase I (20), is responsible for the observed relaxation. The DNA gyrase-mediated negative supercoiling seemed to counteract DNA relaxation from the time of temperature upshift and began to dominate after 10 min. We conclude that the process which is inhibited by nalidixic acid or coumermycin is necessary for RC disassembly. The only candidate for such a process is DNA gyrase-mediated negative resupercoiling. Since this process is specifically inhibited by coumermycin, in subsequent presentations the results obtained with nalidixic acid are not shown; in all cases they matched those obtained with coumermycin.
FIG. 6.
Changes in the average linking number (Δτ) of λcro+ (pKB2) plasmid DNA in the wild-type strain at 30°C (triangles) and after a shift to 43°C (squares). The values are estimated relative to the linking number observed for the pKB2 plasmid at time zero in bacteria growing at 30°C. The temperature shift is indicated by an arrow.
λcrots plasmid: heat shock resistance of RC requires pR-initiated transcription.
The results presented in previous sections with bacteria harboring the λcro+ plasmid led us to propose a model assuming that DNA gyrase-mediated λ DNA negative resupercoiling (occurring after initial plasmid relaxation caused by a temperature upshift) results in dissociation of RC from λ DNA. We suggest that RC disassembly helped by GroEL/S chaperone proteins then occurs and that λO is no longer protected and becomes hydrolyzed by ClpP/ClpX protease. It was now possible to ask why the temperature upshift does not cause RC disassembly in λcrots plasmid-harboring cells. The Cro repressor blocks the pR-initiated transcription required for initiation of λ plasmid replication as well as for Cro synthesis (30). According to the generally accepted model (17), the transient derepression event that should occur due to dilution of Cro in the growing cell volume should produce a wave of transcription leading to a round of replication. Synthesis of Cro would restore the initial, silent state of λ plasmid. In contrast to the λcro+-harboring cells, the pR-initiated transcription should be constantly derepressed in λcrots plasmid-harboring bacteria at 43°C. Therefore, an obvious question was whether unhindered transcription from pR is indeed required for RC stability.
In a study of the heat shock resistance of RC in λcrots plasmid-harboring cells (Fig. 7A), we found that 25 μg of rifampin per ml (generally used for inhibition of transcription) added at the time of temperature upshift caused RC disassembly (Fig. 7B), which was blocked by coumermycin (Fig. 7C). Addition of rifampin 10 min before the temperature upshift did not change the results (not shown). These results were rather unexpected, since it was previously demonstrated that disassembly of RC requires the synthesis of at least GroEL and GroES proteins (37), and one might predict that the addition of rifampin should result not only in the inhibition of transcription but also in the abolition of production of new protein molecules. However, we found that under the conditions of our experiments, the amount of GroEL synthesized in the presence of rifampin (Fig. 8) was sufficient to cause RC disassembly. Upon the shift from 30 to 43°C, rifampin at 25 μg/ml completely blocked lacZ expression from a pL-lacZ fusion (repressed at 30°C by a temperature-sensitive cI857 protein whose gene was also present on the same single-copy fusion) but was allowed some β-galactosidase synthesis when lacZ was fused to the ς32-dependent groE promoter (results not shown). Therefore, it seems that rifampin at 25 μg/ml does not efficiently inhibit the initiation of transcription by RNA polymerase holoenzyme containing ς32 (Eς32), which is engaged in the groE operon transcription after heat shock.
FIG. 8.
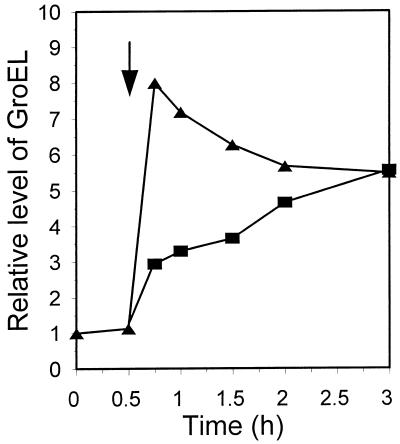
Relative level of the GroEL protein in the wt strain after a shift from 30 to 43°C (arrow). Bacteria were untreated (triangles) or treated with 25 μg of rifampin per ml (squares) at the time of the temperature shift.
The importance of unhindered transcription from the pR promoter for heat shock resistance of RC in λcrots plasmid-harboring bacteria was supported in experiments performed with an E. coli rpoDts mutant. The rpoD gene codes for the ς70 subunit of RNA polymerase that recognizes most of the host and phage promoters, including the pR promoter. After the temperature upshift, the groE operon is expressed due to the rpoH-encoded ς32 subunit, but Eς32 cannot recognize the pR promoter. As expected, in λcrots plasmid-harboring E. coli rpoDts cells, a temperature upshift caused RC disassembly (Fig. 9). This process was blocked, as usual, by DNA gyrase inhibitors (results not shown). The kinetics of the λO protein decay was somewhat slower in cells harboring λcrots plasmid than in those harboring the wild-type λ plasmid (Fig. 9). This could perhaps be explained by some residual activity of the rpoDts gene product at 43°C, since rifampin (which should block activity of Eς70 almost completely) caused the λO protein decay in the λcrots-harboring cells as efficiently as in λcro+-harboring bacteria (Fig. 7B).
λcrots plasmid: kinetics of transcription and replication.
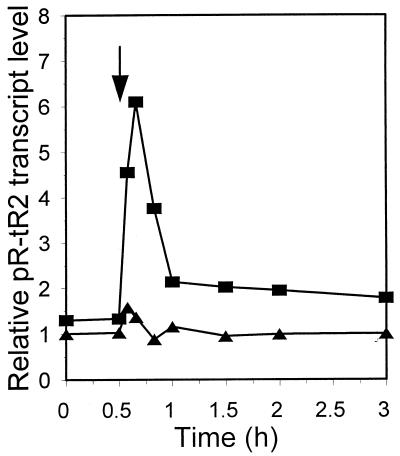
In λcrots plasmid-harboring cells, the plasmid was stably maintained after the temperature upshift (36) and the release from Cro repression did not lead to uncontrolled λ DNA replication, in contrast to the situation that would be expected on the basis of previously proposed model (17), in which the Cro repressor autoregulatory loop plays a crucial role in the regulation of replication and maintenance of λ plasmids in E. coli (see below). Therefore, transcription of the O-P region, leading to the synthesis of λ replication proteins and to activation of oriλ situated in the middle of the O gene, should be controlled by a Cro-independent system. It was interesting to check how rapidly this control is established after the temperature upshift. A convenient measure of the transcription of the O-P region is the level of the pR-tR2 transcript; the tR2 terminator is situated downstream of the P gene. The level of the pR-tR2 transcripts, calculated per plasmid copy, increased up to 10 min after the temperature upshift (Fig. 10). Since this level results from the synthesis and decay of the transcript, one may assume that its synthesis attained its peak even earlier. The rapid decrease in the level of the pR-tR2 transcripts and its stabilization observed later (Fig. 10) reveal an important element of a Cro-independent control system.
FIG. 10.
Relative level of the pR-tR2 transcripts normalized to the level of rRNA (internal RNA control) and to the amount of plasmid (i.e., template) DNA in the wt strain harboring λcro+ (pKB2) (triangles) or λcrots (pRLM4) (squares) plasmid after a shift from 30 to 43°C (arrow).
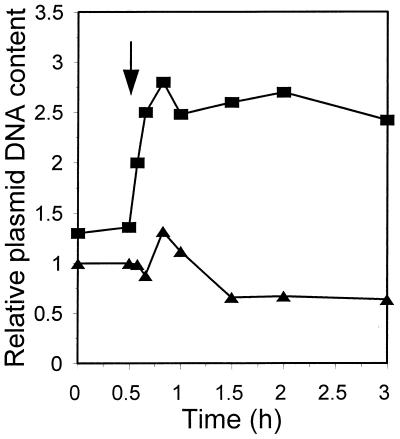
A parallel examination of the λcrots plasmid DNA revealed that after the temperature upshift from 30 to 43°C, it has already attained the level specific for 43°C (about twice as high as at 30°C) in 10 min (Fig. 11). Since the plasmid DNA content was calculated in relation to the constant bacterial mass estimated by measurement of the optical density of the bacterial culture, small disturbances occurring immediately afterward may be attributed to the change in the average cell size that follows the temperature upshift. The simplest interpretation of the results presented in Fig. 11 is that all plasmid copies, released suddenly from the Cro repression, perform one round of replication. The increased plasmid copy number, 55 to 60 per cell, persists and is characteristic for λcrots plasmids at 43°C. We checked that the kinetics of the heat shock-provoked changes in supercoiling of λcrots plasmid DNA is similar to that of λcro+; Δτ attains its highest value 10 min after the temperature upshift (results not shown).
FIG. 11.
Relative level of plasmid DNA per bacterial mass in the wt strain harboring λcro+ (pKB2) (triangles) or λcrots (pRLM4) (squares) plasmid after a shift from 30 to 43°C (arrow).
DISCUSSION
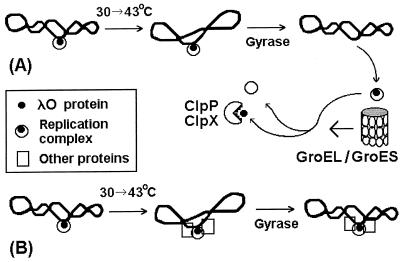
The results presented for the λcro+ plasmid demonstrate that heat shock-provoked disassembly of the λO initiator-containing structure, RC, requires both DNA gyrase-mediated negative supercoiling of plasmid DNA and induction of the groE operon. We have no data concerning the temporal sequence of action of DNA gyrase and GroEL/S chaperone proteins. However, the most probable scenario is that at first the RC dissociates from λ DNA due to gyrase-mediated negative resupercoiling, which occurs after plasmid relaxation caused by a temperature upshift (Fig. 12A). Then disassembly of the free RC, helped by the GroEL/S proteins, may occur, exposing λO to the action of ClpP/ClpX protease. We assume that, this chain of events cannot start earlier than 10 min after the temperature shift from 30 to 43°C, since the negative resupercoiling of the heat shock-relaxed plasmid DNA begins at this time (Fig. 6).
FIG. 12.
Proposed mechanism of heat shock-provoked disassembly of the λ replication complex in bacteria harboring λcro+ plasmid (A) and of heat shock resistance of the λ replication complex in bacteria harboring λcrots plasmid (B). In the case of a λcro-null plasmid, the λ replication complex should also interact with other DNA-bound proteins before temperature upshift. See the text for details.
An obvious and unanswered question is how the GroE chaperone system distinguishes between the DNA-bound and free states of RC, acting exclusively in the latter case on this viral protein structure. The exact composition of the structure which protects the λO protein from proteolysis, referred to as RC, is not yet known, but it was demonstrated that this structure must contain at least λO, λP, and DnaB proteins (34). Therefore, perhaps the crucial role in the process of distinguishing among the different states of RC by the GroE system is played by the surface of RC formed by sequential binding of RC components (λO, λP, DnaB, etc.) to oriλ and their mutual interactions, which change due to the action of DnaK, DnaJ, and GrpE chaperone proteins. After dissociation of RC from λ DNA, this part of the RC surface may appear conformationally unnatural (possibly with exposed hydrophobic groups) and become prone to GroEL/GroES attack.
The knowledge accumulated in the study of heat shock-provoked disassembly of RC in λcro+ plasmid-harboring cells allowed us to look for the cause of the heat shock resistance of RC in the case of the λcrots plasmid (36) (see above). This feature appears to be characteristic of λcro− plasmids as supported by an experiment with a recently constructed λcro-null plasmid (10). In this case, the pR-initiated transcription, unhindered by Cro repression, could occur all the time, i.e., before and after the temperature upshift. We found that the RC is heat shock resistant in λcro-null plasmid-harboring bacteria (38). We postulate that dissociation of RC from λ DNA during the negative resupercoiling does not occur, because RC is more tightly bound to DNA than in the case of the λcro+ plasmid, possibly due to interaction with other DNA-bound proteins (Fig. 12B). One of the potential candidates for such proteins may be the host DnaA protein. This protein stimulates the activity of the pR promoter (43). Several putative DnaA-binding sequences were found in the λ replication region, and one of them is located near pR (30). Therefore, one may assume a competition between Cro and DnaA in binding to the region of this promoter. If DnaA is able to interact with components of λ RC, it might stabilize this structure more efficiently in the absence of Cro function. An alternative hypothesis is that the heat shock resistance of RC under cro mutant conditions (when pR-initiated transcription is more efficient due to the lack of the Cro repressor) is based on the assumption of positive supercoiling in front of RNA polymerase, which might counteract the DNA gyrase-mediated negative resupercoiling. However, the observations that in λcrots plasmid-harboring cells the very intensive pR-initiated transcription and λ DNA replication caused by the temperature upshift terminates before the period of DNA gyrase-mediated negative resupercoiling (compare Figs. 6, 10, and 11) do not support this hypothesis. On the other hand, they suggest that the heat shock resistance of RC and the replication control specific for λcrots plasmid at 43°C are established within 10 min after the temperature upshift and before the process of negative resupercoiling of λ plasmid DNA.
The construction of the λcro-null plasmid, which is stably maintained in E. coli (10), and our observation that the copy number of the λcrots plasmid increases only a few times at 43°C relative to 30°C are in contrast to previously reported findings that Cro function is a key element in the regulation of λ plasmid replication since it is necessary for the stable maintenance of such a plasmid (17). However, the older data came from experiments with λ plasmids produced in vivo, called λdv, which usually contained relatively large fragments of phage λ genome, including the cI gene (coding for a strong repressor of pR). Currently used λ plasmids (including those used in this work) were constructed in vitro and do not contain this gene. These differences between the λ plasmids used previously and in this work may be responsible for the differences between the results obtained previously (17) and those presented in this paper.
As mentioned above, in λcrots plasmid-harboring cells, the heat shock resistance of RC should be established before the period of DNA gyrase-mediated negative supercoiling of plasmid DNA, which begins 10 min after the temperature shift from 30 to 43°C. In the same 10-min period, a Cro-independent control of λ plasmid replication is established. If the Cro-mediated repression of the pR promoter, and thus the Cro autoregulatory loop, were the crucial process in the control of λ plasmid replication, as suggested previously (17), one should expect a rapid and continuous increase in the λcrots plasmid copy number after a temperature upshift. However, the release of the pR promoter from Cro repression does not result in a runaway plasmid replication. On the contrary, after rapid doubling in copy number, the plasmid DNA replication proceeds in step with the growth of host cells (Fig. 11) providing a new argument in favor of the existence of a Cro-independent control of replication. We assume that the wave of the pR-initiated and tR2-terminated transcription occurring shortly after inactivation of the cro gene product, which activates oriλ, is responsible for the rapid doubling of the plasmid copy number (Fig. 10 and 11). However, this transcription soon falls off and proceeds further at a constant level (Fig. 10). This level may reflect the transcriptional activation of oriλ that should occur to ensure replication in the Cro-independent system of replication control, as described previously (36). The low level of the pR-tR2 transcripts in the absence of the functional Cro repressor may mean that the pR-initiated transcription is inhibited, prematurely terminated, or paused on the way to oriλ and finally to tR2. Since there is no reason to suspect that activation of the pR promoter by DnaA protein is abolished under these conditions, the pausing of RNA polymerase seems to serve as the best working hypothesis concerning the model of the Cro-independent control. It is tempting to speculate that, the paused RNA polymerase and other DNA-bound proteins (for example DnaA, as discussed above) can interact with the oriλ-bound RC, forming a complex which may sequester oriλ during most of the cell cycle and can be responsible for the block of reinitiation of replication which is characteristic for λcro mutant plasmids (36, 38). Although the molecular mechanism of the Cro-independent control awaits elucidation, the data presented here suggest that strong binding of RC to λ DNA (proposed in this work on the basis of the heat shock resistance of RC) is one of the elements of this control.
ACKNOWLEDGMENTS
We thank D. Court, C. Georgopoulos, C. Gross, A. B. Oppenheim, Y. Ogata, K. Sekimizu, and M. Żylicz for providing bacterial strains and M. Koob for providing plasmid pCattTrE18. We are grateful to W. Szybalski for his advice to use the TetR-ptetA system, to K. Sekimizu and Y. Ogata for discussions, and to K. Sekimizu for advice about the analysis of plasmid DNA linking number.
This work was supported by the University of Gdańsk (grant BW-1190-5-0252-7 to A.W.) and the Polish State Committee for Scientific Research (grant 6 P04A 051 08 to K.T. and G.W.).
REFERENCES
- 1.Alfano C, McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage λ replication origin during the initiation of DNA replication. J Biol Chem. 1989;264:10699–10708. [PubMed] [Google Scholar]
- 2.Alfano C, McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage λ DNA replication. J Biol Chem. 1989;264:10709–10718. [PubMed] [Google Scholar]
- 3.Bejarano I, Klemes Y, Schoulaker-Schwarz R, Engelberg-Kulka H. Energy-dependent degradation of λO protein in Escherichia coli. J Bacteriol. 1993;175:7720–7723. doi: 10.1128/jb.175.23.7720-7723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benvenisti L, Koby S, Rutman A, Giladi H, Yura T, Oppenheim A B. Cloning and primary sequence of the rpoH gene from Pseudomonas aeruginosa. Gene. 1995;155:73–76. doi: 10.1016/0378-1119(94)00829-h. [DOI] [PubMed] [Google Scholar]
- 5.Bochner B R, Huang H C, Schieven G L, Ames B. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodson M, McMacken R, Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage λ: protein association and disassociation reactions responsible for localized initiation of replication. J Biol Chem. 1989;264:10719–10725. [PubMed] [Google Scholar]
- 7.Fayet O, Louarn J-M, Georgopoulos C. Suppression of the Escherichia coli dnaA46 mutation by amplification of the groEL and groES genes. Mol Gen Genet. 1986;202:435–445. doi: 10.1007/BF00333274. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman S, Clark W P, de Crecy-Legard V, Maurizi M R. ClpX, an alternative subunit of the ATP-dependent Clp protease of Escherichia coli: sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 9.Herman A, Węgrzyn A, Węgrzyn G. Differential replication of plasmids during stringent and relaxed response of Escherichia coli. Plasmid. 1994;32:89–94. doi: 10.1006/plas.1994.1049. [DOI] [PubMed] [Google Scholar]
- 10.Herman-Antosiewicz, A., S. Śrutkowska, K. Taylor, and G. Węgrzyn. Replication and maintenance of λ plasmids devoid of the Cro repressor autoregulatory loop in Escherichia coli. Plasmid, in press. [DOI] [PubMed]
- 11.Ikeda H, Moriya K, Matsumoto T. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harbor Symp Quant Biol. 1981;46:399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- 12.Jensen K F. The Escherichia coli “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci USA. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 15.Kur J, Górska I, Taylor K. Escherichia coli dnaA initiation function is required for replication of plasmids derived from coliphage lambda. J Mol Biol. 1987;198:203–210. doi: 10.1016/0022-2836(87)90306-8. [DOI] [PubMed] [Google Scholar]
- 16.Liebke H, Gross C, Walter W, Burgess R. A new mutation rpoD800, affecting the sigma subunit of E. coli RNA polymerase is allelic to two other sigma mutants. Mol Gen Genet. 1980;177:277–282. doi: 10.1007/BF00267439. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara K. Replication control system in λ dv. Plasmid. 1981;5:32–52. doi: 10.1016/0147-619x(81)90076-7. [DOI] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 19.Obuchowski M, Węgrzyn A, Szalewska-Pałasz A, Thomas M S, Węgrzyn G. An RNA polymerase α subunit mutant impairs N-dependent transcriptional antitermination in Escherichia coli. Mol Microbiol. 1997;23:211–222. doi: 10.1046/j.1365-2958.1997.2101576.x. [DOI] [PubMed] [Google Scholar]
- 20.Ogata Y, Mizushima T, Kataoka K, Miki T, Sekimizu K. Identification of DNA topoisomerases involved in immediate and transient DNA relaxation induced by heat shock in Escherichia coli. Mol Gen Genet. 1994;244:451–455. doi: 10.1007/BF00583895. [DOI] [PubMed] [Google Scholar]
- 21.Pawłowicz A, Węgrzyn G, Taylor K. Effect of coliphage λ P gene mutations on the stability of the λO protein, the initiator of λ DNA replication. Acta Biochim Pol. 1993;40:29–31. [PubMed] [Google Scholar]
- 22.Polissi A, Goffin L, Georgopoulos C. The Escherichia coli heat shock response and bacteriophage λ development. FEMS Microbiol Rev. 1995;17:159–169. doi: 10.1111/j.1574-6976.1995.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 23.Posfai G, Koob M, Hradecna Z, Hasan N, Filutowicz M, Szybalski W. In vivo excision and amplification of large segments of the Escherichia coli genome. Nucleic Acids Res. 1994;22:2392–2398. doi: 10.1093/nar/22.12.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 26.Skerra A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- 27.Szalewska A, Węgrzyn G, Taylor K. Neither absence nor excess of λO initiator-digesting ClpXP protease affects λ plasmid or phage replication in Escherichia coli. Mol Microbiol. 1994;13:469–474. doi: 10.1111/j.1365-2958.1994.tb00441.x. [DOI] [PubMed] [Google Scholar]
- 28.Szalewska-Pałasz A, Węgrzyn G. An additional role of transcriptional activation of oriλ in the regulation of λ plasmid replication in Escherichia coli. Biochem Biophys Res Commun. 1994;205:802–806. doi: 10.1006/bbrc.1994.2736. [DOI] [PubMed] [Google Scholar]
- 29.Szalewska-Pałasz A, Węgrzyn A, Herman A, Węgrzyn G. The mechanism of the stringent control of λ plasmid DNA replication. EMBO J. 1994;13:5779–5785. doi: 10.1002/j.1460-2075.1994.tb06916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor K, Węgrzyn G. Replication of coliphage lambda DNA. FEMS Microbiol Rev. 1995;17:109–119. doi: 10.1111/j.1574-6976.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 31.Thomas M S, Glass R E. Escherichia coli rpoA mutation which impairs transcription of positively regulated systems. Mol Microbiol. 1991;5:2719–2725. doi: 10.1111/j.1365-2958.1991.tb01980.x. [DOI] [PubMed] [Google Scholar]
- 32.Węgrzyn A, Węgrzyn G. Transcriptional activation of oriλ regulates λ plasmid replication in amino acid-starved Escherichia coli cells. Biochem Biophys Res Commun. 1995;214:978–984. doi: 10.1006/bbrc.1995.2382. [DOI] [PubMed] [Google Scholar]
- 33.Węgrzyn A, Węgrzyn G, Taylor K. Plasmid and host functions required for λ plasmid replication carried out by the inherited replication complex. Mol Gen Genet. 1995;247:501–508. doi: 10.1007/BF00293153. [DOI] [PubMed] [Google Scholar]
- 34.Węgrzyn A, Węgrzyn G, Taylor K. Protection of coliphage λO initiator protein from proteolysis in the assembly of the replication complex in vivo. Virology. 1995;207:179–184. doi: 10.1006/viro.1995.1064. [DOI] [PubMed] [Google Scholar]
- 35.Węgrzyn A, Taylor K, Węgrzyn G. The cbpA chaperone gene function compensates for dnaJ in λ plasmid replication during amino acid starvation of Escherichia coli. J Bacteriol. 1996;178:5847–5849. doi: 10.1128/jb.178.19.5847-5849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Węgrzyn A, Węgrzyn G, Herman A, Taylor K. Protein inheritance: λ plasmid replication perpetuated by the heritable replication complex. Genes Cells. 1996;1:953–963. doi: 10.1046/j.1365-2443.1996.830283.x. [DOI] [PubMed] [Google Scholar]
- 37.Węgrzyn A, Węgrzyn G, Taylor K. Disassembly of the coliphage λ replication complex due to heat shock induction of the groE operon. Virology. 1996;217:594–597. doi: 10.1006/viro.1996.0154. [DOI] [PubMed] [Google Scholar]
- 38.Węgrzyn, A., G. Węgrzyn, and K. Taylor. Unpublished data.
- 39.Węgrzyn G. Amplification of λ plasmids in Escherichia coli relA mutants. J Biotechnol. 1995;43:139–143. doi: 10.1016/0168-1656(95)00132-5. [DOI] [PubMed] [Google Scholar]
- 40.Węgrzyn G, Taylor K. Inheritance of the replication complex by one of two daughter copies during λ plasmid replication in Escherichia coli. J Mol Biol. 1992;226:681–688. doi: 10.1016/0022-2836(92)90625-t. [DOI] [PubMed] [Google Scholar]
- 41.Węgrzyn G, Neubauer P, Krueger S, Hecker M, Taylor K. Stringent control of replication of plasmids derived from coliphage λ. Mol Gen Genet. 1991;225:94–98. doi: 10.1007/BF00282646. [DOI] [PubMed] [Google Scholar]
- 42.Węgrzyn G, Pawłowicz A, Taylor K. Stability of coliphage λ DNA replication initiator, the λO protein. J Mol Biol. 1992;226:675–680. doi: 10.1016/0022-2836(92)90624-s. [DOI] [PubMed] [Google Scholar]
- 43.Węgrzyn G, Szalewska-Pałasz A, Węgrzyn G, Obuchowski M, Taylor K. Transcriptional activation of the origin of coliphage λ DNA replication is regulated by the host DnaA initiator function. Gene. 1995;154:47–50. doi: 10.1016/0378-1119(94)00849-n. [DOI] [PubMed] [Google Scholar]
- 44.Węgrzyn G, Węgrzyn A, Konieczny I, Bielawski K, Konopa G, Obuchowski M, Helinski D R, Taylor K. Involvement of the host initiator function dnaA in the replication of coliphage λ. Genetics. 1995;139:1469–1481. doi: 10.1093/genetics/139.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Węgrzyn G, Węgrzyn A, Pankiewicz A, Taylor K. Allele specificity of the Escherichia coli dnaA gene function in the replication of plasmids derived from phage λ. Mol Gen Genet. 1996;252:580–586. doi: 10.1007/BF02172404. [DOI] [PubMed] [Google Scholar]
- 46.Wojtkowiak D, Georgopoulos C, Żylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 47.Wold M S, Mallory J B, Roberts J D, LeBowitz J N, McMacken R. Initiation of bacteriophage λ DNA replication in vitro with purified λ replication proteins. Proc Natl Acad Sci USA. 1982;79:6176–6180. doi: 10.1073/pnas.79.20.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Żylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of λ DNA replication with purified host- and bacteriophage-encoded proteins: the role of the DnaK, DnaJ and GrpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]