Abstract
The effects of short deletions of the C terminus of the BvgA response regulator protein of the BvgAS two-component system were examined in Bordetella pertussis. When present as a single copy in the chromosome, deletions removing as few as two amino acids conferred a completely Bvg− phenotype. When provided in trans, on the broad-host-range plasmid pRK290, under the control of the native bvgAS promoter, deletions of two or three amino acids conferred a profound growth inhibition which was dependent on the integrity and activity of the wild-type chromosomal bvgAS locus. It is proposed that this phenotype was the result of an inappropriate interaction of the mutant BvgA protein with the RNA polymerase enzyme, specifically the α subunit. Mutant strains in which this growth inhibition was relieved were isolated and characterized. Although most of the suppressor mutations affected either the mutant plasmid copy or the wild-type chromosomal bvg locus, three mutations which affected the α subunit of B. pertussis RNA polymerase were also isolated. Two of these resulted in increased levels of the α subunit, and one caused a substitution of glycine for the aspartic acid residue at position 171, in the N-terminal domain. All three mutations also resulted in a differential phenotype in that expression of fha was essentially normal, but expression of ptx was greatly reduced.
Environmentally responsive regulation of virulence gene expression in the human pathogen Bordetella pertussis is mediated by the bvgAS locus, which encodes a member of the two-component family of bacterial signal transduction systems (33). BvgS is an unorthodox sensor/transmitter protein which contains, in addition to the typical periplasmic, transmembrane, and histidine kinase domains, a response regulator domain and an additional domain termed the output domain. Through a phosphorelay mechanism, this protein is capable of transferring phosphate to the response regulator BvgA, resulting in increased affinity of BvgA for its binding sites in virulence gene promoters (1, 4, 5, 16, 37). In addition to regulating its own expression, the bvgAS locus governs the expression of all known protein virulence factors of this organism, including those encoding filamentous hemagglutinin (fha), pertussis toxin (ptx), and adenylate cyclase toxin/hemolysin (cya). Under standard culture conditions, BvgS activates BvgA to stimulate expression of these virulence genes. However, when grown in media containing MgSO4 or nicotinic acid, or at reduced temperatures, bvg-regulated expression of virulence factors is repressed—a phenomenon known as antigenic, or phenotypic, modulation (19).
In vitro transcription experiments involving the ptx and fha promoters have demonstrated a dependence on phosphorylated BvgA (6, 7, 28). At both promoters, binding of BvgA-phosphate to consensus inverted heptameric repeats upstream of the −35 promoter motif has been seen, with additional sequences between these repeats and the −35 motif also being protected in DNase I footprinting experiments (5, 6, 42). However, the repeats in the ptx promoter region are further upstream than in the fha promoter, and the inverted heptamers are separated by 10 bp (33). These differences in promoter architecture may be responsible for functional differences in the two promoters observed in vivo, in that higher concentrations of BvgA-phosphate are required to activate ptx transcription than are needed to activate fha transcription. This phenomenon has been observed in Escherichia coli as well as in B. pertussis (25, 36).
Further demonstration of functional differences between these two promoters is provided by missense mutations affecting the extreme C terminus of BvgA which abolish ptx transcription but have little or no effect on fha transcription (30). The region of the BvgA protein affected by these mutations may be involved in interaction with RNA polymerase at regulated promoters. This is suggested by studies of other members of the subfamily of response regulators to which BvgA is related by sequence similarity in its C-terminal domain. In both the LuxR protein of Vibrio fischeri and the UhpA protein of E. coli, short C-terminal deletions affected the ability of these proteins to stimulate transcription at regulated promoters (10, 38).
We report here an analysis of the phenotype conferred by short C-terminal deletions of BvgA. Unexpectedly, when provided in trans in B. pertussis, these deletions conferred a profound growth inhibition which was dependent on the presence of a functional chromosomal bvg locus. Mutations which relieved this growth inhibition included mutations which affected the level of expression or the primary structure of the B. pertussis RNA polymerase α subunit. These observations provide additional genetic evidence for an interaction between the C-terminal portion of the BvgA protein and RNA polymerase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are described in Table 1. E. coli DH5α, which was used as a transformation recipient for all cloning steps, was obtained from Bethesda Research Laboratories. E. coli strains were grown on L agar or in L broth (21) supplemented with antibiotics as appropriate. B. pertussis strains were grown on Bordet-Gengou agar (Difco) containing 1% proteose peptone (Difco) and 15% defibrinated sheep blood. Concentrations of antibiotics were 10 μg/ml for tetracycline, kanamycin, and gentamicin; 50 μg/ml for nalidixic acid; and 100 μg/ml for streptomycin. For growth of B. pertussis strains under modulating conditions, 50 mM MgSO4 was added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| E. coli K-12 | ||
| DH5α | High-efficiency transformation | BRL |
| SM10 | Conjugation-proficient donor | 26 |
| SS1840 | LE392[pSS1840] | 32 |
| B. pertussis | ||
| Tohama I | Patient isolate | 17 |
| BP338 | Tohama I, Nalr | 39 |
| BP536 | Tohama I, Nalr Smr | 29 |
| BP947 | Tohama I, Nalr Smrfha-lacZ | This study |
| BP953 | Tohama I, Nalr Smrfha-lacZ ptx-phoA | This study |
| RPV5 | Rifr derivative of a spontaneous Ptx− Cya− Fha+ mutant (↑RpoA) | 32 |
| BP1079 | Tohama I, Smr Nalr Rifrfha-lacZ ptx-phoA | This study |
| BP1187 | BP953, rpoA1187 | This study |
| BP1196 | BP953 mutant (↑RpoA) | This study |
| BP1242 | BP953 with rpoA gene replaced by using pSS2316 (↑RpoA) | This study |
| BP1246 | BP1079 mutant (↑RpoA) | This study |
| BP1247 | BP1079, bvg-1247 | This study |
| BP1248 | BP1079, bvg-1248 (bvg::IS1002) | This study |
| BP1250 | BP1079, bvg-1250 (bvg::IS481) | This study |
| BP1252 | BP1079, bvg-1251 (bvg::IS481) | This study |
| BP1253 | BP1079, bvg-1252) (bvg::IS481) | This study |
| BP1254 | BP1079, bvg-1253 (bvg::IS1002) | This study |
| Plasmids | ||
| pRK290 | Broad-host-range cloning vector, Tetr, RP4 oriV, RP4 oriT | 13 |
| pSS1129 | Allelic exchange vector, bla gen rpsL oriVColE1 oriT λ cos | 31 |
| pSS1581 | pSS1129 containing fhaB-lacZ with flanking sequences | This study |
| pSS1615 | pSS1129 containing ptx-phoA with flanking sequences | This study |
| pSS1840 | Ampr, RP4 oriT, RP4 tra, λ cos | 32 |
| pSS1853 | Ampr Genr, RP4 oriT, RP4 tra, λ cos | 32 |
| pSS1894 | Ampr Genr, RP4 oriT | 20 |
| pSS2000 | Ampr Genr, RP4 oriT, rpsL | 20 |
| pSS2141 | SalI deletion derivative of pSS2000 | This study |
| pSS2197 | bvgA allelic retrieval vector | 34 |
| pSS2306 | pSS2141 containing 7-kb B. pertussis rpoA BamHI fragment | This study |
| pSS2647 | pSS1129 containing bvgAΔC1 with flanking sequences | This study |
| pSS2650 | pSS1129 containing bvgAΔC2 with flanking sequences | This study |
| pSS2189 | pSS1129 containing bvgAΔC3 with flanking sequences | This study |
| pSS2190 | pSS1129 containing bvgAΔC6 with flanking sequences | This study |
| pSS2191 | pSS1129 containing bvgAΔC9 with flanking sequences | This study |
| pSS2192 | pSS1129 containing bvgAΔC12 with flanking sequences | This study |
| pSS2193 | pSS1129 containing bvgAΔC15 with flanking sequences | This study |
| pSS2194 | pSS1129 containing bvgAΔC18 with flanking sequences | This study |
| pSS2068 | pRK290 containing EcoRI-SphI bvgA fragment | This study |
| pSS2342 | pRK290 containing EcoRI-SphI bvgAΔC1 fragment | This study |
| pSS2344 | pRK290 containing EcoRI-SphI bvgAΔC2 fragment | This study |
| pSS2244 | pRK290 containing EcoRI-SphI bvgAΔC3 fragment | This study |
| pSS2219 | pRK290 containing EcoRI-SphI bvgAΔC6 fragment | This study |
| pSS2246 | pRK290 containing EcoRI-SphI bvgAΔC9 fragment | This study |
| pSS2248 | pRK290 containing EcoRI-SphI bvgAΔC12 fragment | This study |
| pSS2250 | pRK290 containing EcoRI-SphI bvgAΔC15 fragment | This study |
| pSS2252 | pRK290 containing EcoRI-SphI bvgAΔC18 fragment | This study |
| pSS2320 | pSS2244, bvgA1263 | This study |
| pSS2321 | pSS2244, bvgA1264 | This study |
| pSS2322 | pSS2244, bvgA1265 | This study |
| pSS2324 | pSS2244, bvgA1267 | This study |
| pSS2325 | pSS2244, bvgA1268 | This study |
| pSS2326 | pSS2244, bvgA1269 | This study |
| pSS2316 | pSS2306 containing rpoA allele from RPV5 | This study |
| pSS2318 | pSS2306 containing rpoA1187 from BP1187 | This study |
Plasmid construction.
Deletions of the bvgA gene were constructed by PCR. A fragment containing the entire bvgA gene and its promoter region was amplified by using an upstream primer including the EcoRI site at position 1 and a downstream primer including the SphI site at position 817, 6 bp downstream of the bvgA stop codon (coordinates as in GenBank accession no. M25401). Deletions removing codons immediately upstream of the stop codon were introduced by specifying the appropriate sequence in the downstream primer. In this way, genes coding for BvgA which lacked 1, 2, 3, 6, 9, 12, 15, or 18 C-terminal amino acids (bvgAΔC1-bvgAΔC18) were synthesized and subsequently cloned between the EcoRI and SphI sites of pSS1894. To permit introduction of these bvgA deletions into the B. pertussis chromosome, flanking sequences were restored and the resulting constructs were recloned into the allelic exchange vector pSS1129. In this way, plasmids pSS2189, pSS2190, pSS2191, pSS2192, pSS2193, pSS2194, pSS2647, and pSS2650 were created. To permit introduction of the mutant bvgA genes in trans, an EcoRI site was added at the SphI site by cleavage and ligation with the self-complementary oligonucleotide 5′-CATGCGAATTCG-3′. The resulting EcoRI fragments were recloned into pRK290 to create pSS2342, pSS2344, pSS2244, pSS2219, pSS2246, pSS2248, pSS2250, and pSS2252. In a similar way, pSS2068, containing the wild-type bvgA gene was created.
To create pSS2306, a BamHI fragment containing the rpoA gene of B. pertussis (9) was cloned into the allelic exchange vector pSS2141 (20).
Strain construction.
B. pertussis BP953 contains fhaB-lacZ and ptx-phoA transcriptional gene fusions to allow monitoring of virulence factor gene expression regulated by the bvgAS locus. The fhaB-lacZ fusion was created as follows. A BamHI-SalI fragment containing lacZ and derived from pRS1551 (27) was added between the most upstream BamHI site and the most downstream XhoI site in the 10.0-kb EcoRI fragment containing fhaB. The resulting EcoRI fragment containing the fusion was recloned into pSS1129 to create pSS1581. This plasmid was used to replace the chromosomal fhaB gene to create BP947, using previously described methodology (31). The ptx-phoA fusion was created as follows. Plasmid ptxA4-50-6, kindly provided by Cynthia Lee, consists of a 4.7-kb EcoRI fragment containing the pertussis toxin operon cloned into pBR325. The oligonucleotide linker 5′-pGATCTCGCGGCCGCGA-3′ was added at the unique BglII site in this plasmid, resulting in the addition of a NotI site. A fragment containing the phoA gene of E. coli was created by PCR. The upstream primer sequence was 5′-ATTCCCGGGCGAGTACATTGCGAAAATAAAGTGAAACAAAGCACTA-3′. This primer includes the first 16 bp of the phoA gene, including the native GTG start codon, 9 bp upstream of the start codon, and an XmaI site at the 5′ end. The downstream primer sequence was 5′-CGCAAGCTTGCGGCCGCATTTCAGCCCCAGAGCGGCTTTCATG-3′, which contains the last 25 bp of the phoA gene prior to the termination codon and a NotI site near the 5′ end. This PCR fragment was cloned by using its XmaI and NotI sites into the ptx operon between the naturally occurring XmaI site and the NotI site introduced at the BglII site. The resulting phoA open reading frame is predicted to code for 13 additional amino acids at its C terminus relative to wild-type phoA as a result of the lack of the native termination codon. This fusion was cloned as a BstBI fragment into the ClaI site of pSS1129 to create pSS1615, which was then used to replace the ptx locus of BP947 to create BP953 as previously described (31).
BP1079 is a spontaneous rifampin-resistant mutant of BP953 which was isolated on Bordet-Gengou agar containing 100 μg of rifampin per ml.
B. pertussis strains containing the replication-proficient derivatives of pRK290 were the result of conjugation with E. coli donor strains. Plasmids were transformed into E. coli SM10 for transfer or were transferred from DH5α in a triparental mating using SS1840.
Selection of mutants surviving growth inhibition conferred by bvgAΔC3.
Independent cultures of BP953[pS2244] or BP1079[pSS2244] were grown on Bordet-Gengou agar containing tetracycline and MgSO4. After growth for 3 to 4 days, bacteria were resuspended in phosphate-buffered saline, the bacterial density was estimated by measurement of A600, and dilutions were plated on Bordet-Gengou agar containing tetracycline but lacking MgSO4. After incubation for 3 to 4 days at 37°C, colonies surviving selection were examined for expression of alkaline phosphatase and β-galactosidase to differentiate Bvg phenotypes. Mutants were restreaked onto Bordet-Gengou medium lacking tetracycline, and colonies were screened for tetracycline sensitivity. In this way, the mutant strains were cured of plasmid pSS2244 prior to further analysis.
Visualization of alkaline phosphatase and β-galactosidase expression by B. pertussis colonies.
Colonies were allowed to adhere to nitrocellulose filters, which were then perfused by placing on 3MM filter paper (Whatman) saturated with Tris-HCl (pH 8.0) containing 200 μg of BCIP (5-bromo-4-chloro-3-indolylphosphate; Sigma) per ml and 500 μg of Magenta-Gal (Biosynth) per ml to visualize expression of alkaline phosphatase and β-galactosidase, respectively.
Enzyme assays.
For measurement of β-galactosidase, bacteria to be assayed were recovered by sterile swab from Bordet-Gengou agar and suspended in Z buffer (21). The A600 was measured, and 0.05 ml was diluted with 1 ml of Z buffer. Cells were permeabilized by the addition of 30 μl of 0.1% sodium dodecyl sulfate and 30 μl of chloroform followed by vortexing, and the assay was completed as described by Miller (21). For measurement of alkaline phosphatase, bacteria were suspended in 1.0 M Tris-HCl (pH 8.0). The A600 was measured, and 0.5 ml of cell suspension was diluted with 0.5 ml of 1.0 M Tris-HCl (pH 8.0). The cells were permeabilized as described above, and the assay was completed as described by Brickman and Beckwith (8). Units in both cases were defined by the following equation: units = [1,000 × A420 − (1.75 × A550)]/(T × V × A600), where T is the incubation time in minutes and V is the volume of permeabilized cells in milliliters.
Mapping of mutations in B. pertussis.
Donors used for Hfr mapping in this study contained selectable markers in the form of insertions of Tn2048, a derivative of the mini-Tn5 transposon delivered by pUT-Kan (11, 20). From a set of 12 such insertions spaced evenly around the B. pertussis Tohama I chromosome, three markers near the bvg locus, and three markers near the rpoA locus were used to localize mutations to these regions. The genetic background of the donor strains was Tohama I, str rif ptx-phoA. Chromosomal sequences were mobilized by a library of random B. pertussis chromosomal fragments cloned into pSS1853 by using generalized conjugation as described previously (32) into the mutant strains, with selection on Bordet-Gengou agar containing kanamycin and nalidixic acid. Linkage of a mutation to the different Tn2048 insertions was determined by visualization of the expression of ptx-phoA and fhaB-lacZ by exconjugant colonies as described above.
Mutations which behaved in these crosses in a manner consistent with a location in the bvg locus were further mapped by using suicide plasmids containing portions of the bvg locus as previously described (30) to derive an approximate location within the bvg locus. Mutations for which Hfr mapping suggested a location near rpoA were tested for rescue of their phenotype by using allelic exchange directed by pSS2306, which contains a 7-kb BamHI fragment including rpoA of B. pertussis (9).
Recovery of mutations from B. pertussis.
Plasmids pSS2320, pSS2321, pSS2322, pSS2324, pSS2325, and pSS2326 were recovered from B. pertussis mutant strains by purification of DNA by the alkaline lysis method (3) and transformation into E. coli DH5α. Plasmids containing the chromosomal bvgA gene from B. pertussis mutant strains were isolated by using the allelic recovery plasmid pSS2197 previously described for this purpose (34). Plasmids containing the rpoA gene from B. pertussis mutant strains RPV5 and BP1187 were isolated in a similar manner. Briefly, pSS2306 was transferred to the mutant strains with selection for gentamicin resistance and counterselection for nalidixic acid resistance. Strains containing pSS2306 thus integrated at the rpoA locus by a single crossover in the 7-kb BamHI fragment containing the rpoA gene were subsequently mated with SS1840 to recover the plasmid liberated by a second crossover. Plasmids so isolated were screened for the ability to confer the differential phenotype following reintroduction into BP953. In this way, pSS2316 and pSS2318 were isolated.
Western analysis.
Western blot analyses were performed as previously described (29). Samples were prepared from suspensions of B. pertussis strains grown on Bordet-Gengou agar which were adjusted to have the same optical density. The monoclonal antibody 4RA2, recognizing epitopes on the α subunit conserved between E. coli and B. pertussis, was kindly provided by Nancy Thompson and Richard Burgess (35). To detect BvgS, the murine monoclonal antibody 23/5 isolated in this laboratory was used (40).
DNA sequence analysis.
DNA sequence analysis was performed by the dideoxy-chain termination method, using a Sequenase kit with deazaGTP to reduce artifacts due to G:C compression (U.S. Biochemical). Single-stranded templates for sequencing were isolated following cloning into mp18 and mp19 (41).
RESULTS
Short C-terminal deletions of BvgA confer a bvg-dependent growth inhibition in trans.
To assess the phenotype conferred by deletion mutations affecting the C terminus of BvgA, deletions removing 1, 2, 3, 6, 9, 12, 15, and 18 amino acids were constructed and are illustrated in Fig. 1. These deleted bvgA genes were used to replace the wild-type bvgA gene of B. pertussis BP953. This strain contains transcriptional gene fusions of the fhaB gene, encoding filamentous hemagglutinin, to the lacZ gene of E. coli, encoding β-galactosidase, and of the ptx operon, encoding pertussis toxin, to the phoA gene of E. coli, which encodes alkaline phosphatase. Deletions removing more than one C-terminal amino acid conferred a Bvg− phenotype in that colonies were Lac− and Pho− as assessed by colony lifts and perfusion with colorimetric substrates BCIP and Magenta-Gal (data not shown). In addition, these colonies were nonhemolytic, indicating a lack of expression of the cya operon, encoding adenylate cyclase toxin/hemolysin (data not shown). The bvgAΔC1 mutation conferred a mild regulatory phenotype in that ptx-phoA expression was reduced approximately twofold, while fha-lacZ expression remained at wild-type levels (data not shown). To assess the effects of these mutations in trans, the deleted bvgA genes were cloned into pRK290, which is capable of replication in B. pertussis with a copy number of 5 to 7 (14). These plasmids were transferred by conjugation to BP953. As indicated in Fig. 1, deletion of one amino acid had no effect, but deletion of two or three amino acids of the bvgA gene in trans resulted in a complete inhibition of detectable growth on Bordet-Gengou agar. Deletions removing 6, 9, 12, or 15 amino acids had an intermediate effect, as revealed by a reduction in colony size but not number, and deletion of 18 residues relieved the inhibition. Expression of the growth-inhibitory phenotype conferred by the bvgAΔC3 gene of plasmid pSS2244 was dependent on the activity of the wild-type chromosomal bvgAS locus, as no inhibition was observed with a strain containing a chromosomal bvgA null mutation, or of BP953[pSS2244] grown in the presence of 50 mM MgSO4, which represses synthesis of bvgAS-activated genes, including the bvgAS locus itself (data not shown).
FIG. 1.
BvgA proteins expressed by plasmid-borne bvgA alleles in trans. The top line depicts DNA sequence features of the EcoRI-SphI fragment containing bvgA. P1 and P2 are Bvg-inducible and constitutive promoters, respectively (23, 24). BBS is a BvgA binding site (23). The coding sequence of wild-type bvgA (bvgAw.t.) is depicted by an open arrow. Nucleotide coordinates for the EcoRI and SphI sites refer to the first nucleotide of those sites with reference to GenBank accession no. M25401. The BvgA proteins are depicted as open boxes below the DNA map, with the number of residues in each protein to the right. The shaded areas correspond to the region of amino acid sequence similarity with other two-component response regulator proteins, and the stippled areas correspond to the putative helix-turn-helix (HTH) DNA binding motif. The positions of the bvgA1056 and bvgA1060 mutations which affect ptx transcription but not fha transcription (30) are depicted by arrowheads on the BvgA wild-type protein. Plasmid names and their encoded bvgA alleles are shown at the left; the growth phenotype conferred upon BP953 is shown at the right.
Selection for mutations which relieve growth inhibition conferred by BvgAΔC3.
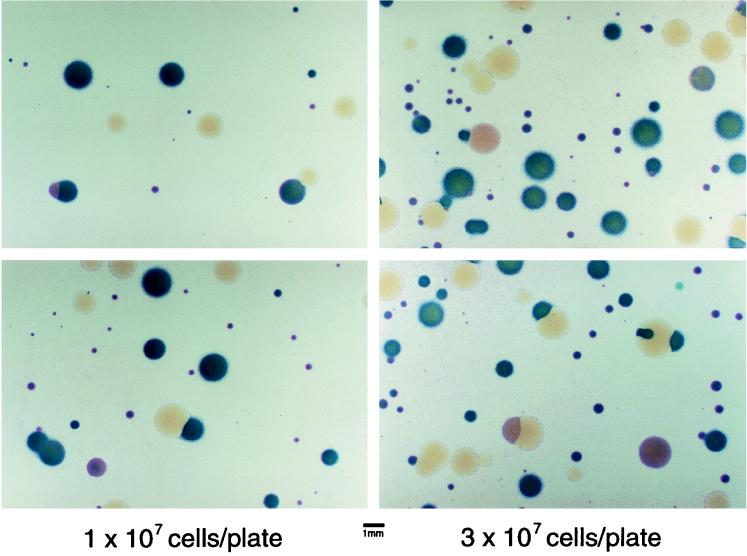
Cultures of BP953[pSS2244] or BP1079[pSS2244] grown on Bordet-Gengou agar containing MgSO4 (permissive conditions) were plated on Bordet-Gengou agar lacking MgSO4 (nonpermissive conditions). This permitted the isolation of mutants which relieved the growth-inhibitory effect of the BvgAΔC3 protein. As shown in Fig. 2, when colonies surviving this selection were examined for expression of fhaB-lacZ and ptx-phoA, a variety of colonial phenotypes were observed. Dark blue colonies expressed both fusions normally and were Cya+ by hemolysis (data not shown), a Bvg+ phenotype. Colorless colonies lacked expression of either gene fusion and were nonhemolytic, a Bvg− phenotype. Pink colonies had reduced hemolysis and expression of ptx-phoA but normal or nearly normal expression of fha-lacZ, a differential Bvg phenotype. Each of these classes were further characterized as follows.
FIG. 2.
Selection for mutations which suppress the growth-inhibitory phenotype conferred by pSS2244. BP953[pSS2244] was grown under permissive conditions and plated under nonpermissive conditions at an approximate density of 1 × 107 or 3 × 107 per plate (8.5-cm diameter). Colonies were allowed to adhere to nitrocellulose filters and developed with BCIP and Magenta-Gal as described in Materials and Methods. Dark blue colonies express ptx-phoA and fha-lacZ, pink colonies express primarily fha-lacZ, and white colonies express neither fusion.
Four independently isolated mutant strains displaying a Bvg+ phenotype were analyzed. Plasmid pSS2244, directing expression of the BvgAΔC3 protein, was extracted from these strains and reintroduced into BP953. Upon reintroduction, these plasmids failed to confer a growth-inhibitory phenotype and did not affect the Bvg+ phenotype of BP953 (data not shown). From these observations, it was concluded that mutations in this class of survivors probably represent knockouts of the bvgAΔC3 gene.
A total of 12 independent mutants displaying a Bvg− phenotype were analyzed. All retained their Bvg− phenotype when cured of pSS2244. The approximate chromosomal locations of these mutations were determined by Hfr mapping as described in Materials and Methods. All behaved in Hfr crosses in a manner indistinguishable from that of mutations in the bvg locus (data not shown). Three of these mutations were further mapped by using marker rescue by suicide plasmids, and their positions were found to be distributed in the bvgA and bvgS genes (data not shown). These mutations appear to be knockouts of the bvgAS locus which relieve growth inhibition by simply reducing expression of the bvgAΔC3, itself a bvg-regulated gene.
A total of 26 independent mutants displaying a differential Bvg phenotype were analyzed. In eight of these, the mutant genotype allowing survival and conferring a differential Bvg phenotype was demonstrated to be associated with plasmid pSS2244 by recovery of the plasmids from the mutant strains and reintroduction into BP953 by conjugation. In all eight cases, reintroduction of the plasmid resulted in the mutant phenotype (data not shown). The remaining 18 mutants were subjected to Hfr mapping in order to determine an approximate chromosomal location. Fifteen of these behaved in these crosses in a manner consistent with a location in the bvg locus, and the other three showed linkage to markers near the rpoA locus of B. pertussis (data not shown). Further characterization of these three mutant classes displaying a differential Bvg phenotype are described below.
Plasmid-borne bvgA mutations conferring a differential Bvg phenotype.
The differential phenotypes conferred by six of the plasmid-borne bvgA alleles were analyzed by β-galactosidase and alkaline phosphatase assays in order to quantitate the differential expression of ptx-phoA and fhaB-lacZ. These data are presented in Table 2 (plasmid-borne bvgA mutations) and demonstrate, in comparison to BP953 containing the pRK290 vector alone, that ptx-phoA expression is repressed upon introduction of these alleles in trans.
TABLE 2.
β-Galactosidase and alkaline phosphatase assays of mutant strains displaying a differential phenotype
| Strain | β-Galactosidase (fha expression)a | Alkaline phosphatase (ptx expression)a |
|---|---|---|
| Plasmid-borne bvgA mutations | ||
| BP953[pRK290] (vector) | 81 (9) | 91 (25) |
| BP953[pSS2320] (bvgA1263) | 82 (9) | 13 (2) |
| BP953[pSS2321] (bvgA1264) | 96 (13) | 14 (7) |
| BP953[pSS2322] (bvgA1265) | 93 (11) | 31 (5) |
| BP953[pSS2324] (bvgA1267) | 90 (9) | 46 (1) |
| BP953[pSS2325] (bvgA1268) | 85 (3) | 20 (4) |
| BP953[pSS2326] (bvgA1269) | 69 (4) | 9 (1) |
| Chromosomal bvgAS mutations | ||
| BP1247 (bvg-1247) | 40 (21) | 3 (1) |
| BP1248 (bvg-1248) | 49 (1) | 3 (1) |
| BP1250 (bvg-1250) | 66 (3) | 5 (1) |
| BP1252 (bvg-1252) | 79 (3) | 4 (0) |
| BP1253 (bvg-1253) | 90 (4) | 5 (1) |
| BP1254 (bvg-1254) | 74 (7) | 6 (1) |
| rpoA-linked mutations | ||
| BP1187 (rpoA1187) | 98 (11) | 11 (2) |
| BP1196 (↑RpoA) | 75 (5) | 11 (4) |
| BP1246 (↑RpoA) | 72 (10) | 9 (4) |
| BP1242 (RPV5 rpoA) (↑RpoA) | 94 (4) | 35 (9) |
Values are expressed as percentages of those obtained for BP953. Standard deviations calculated from the results of three separate assays performed on different cultures on different days are given in parentheses.
These six pSS2244 plasmid derivatives were subjected to DNA sequence analysis to define the nature of their mutations. As depicted in Fig. 3, one nonsense mutation, two IS481 insertions, and three +2 frameshift mutations were observed. The effect of all six mutations is to truncate the mutant BvgAΔC3 protein in the C-terminal domain upstream of the helix-turn-helix motif. This presumably relieves the growth inhibition associated with the novel C terminus of this protein by removing this apparently toxic portion of the BvgA molecule. We hypothesize that the inhibitory effect on Bvg-regulated expression of ptx is due to the interaction of truncated BvgA with wild-type BvgA or by its competition with wild-type BvgA for phosphorylation by BvgS. In either case, the overall level of BvgA activity is reduced and a differential phenotype results. That an intermediate level of BvgAS activity can lead to a differential phenotype is suggested by temporal patterns of gene expression following induction of bvgAS by temperature shift (25). This can also be demonstrated through partial modulation of the activity of BvgAS. As shown in Fig. 4, the expression of ptx-phoA in BP953 is more sensitive to the addition of MgSO4 than is fha-lacZ, being repressed at lower MgSO4 concentrations. Thus, intermediate concentrations of a modulator can result in partial BvgAS activity and reduced expression of ptx-phoA relative to fha-lacZ.
FIG. 3.
Plasmid-borne mutations in the bvgAΔC3 gene which relieve growth inhibition of pSS2244 and confer a differential phenotype. The positions and nature of the mutations are shown above the map of the EcoRI-SphI bvgAΔC3-containing fragment. Other features are as described for Fig. 1, with the exception of the hatched boxes, which represent predicted nonsense (n.s.) peptide introduced into the bvgA open reading frame as a result of frameshift (f.s.) mutation or IS insertion.
FIG. 4.
MgSO4 modulation of the bvg-regulated gene fusions fha-lacZ and ptx-phoA. BP953 was grown on Bordet-Gengou agar containing different concentrations of MgSO4. Cells were recovered and assayed as described in Materials and Methods. Each point represents the mean of three separate assays. β -Gal, β-galactosidase; Alk. PhoS., alkaline phosphatase.
Chromosomal bvg-linked mutations conferring a differential Bvg phenotype.
In all but 1 of the 15 strains harboring chromosomal bvg-linked mutations, the location of the mutation conferring the differential phenotype could be unequivocally localized to the 816-bp EcoRI-SphI fragment containing the bvgA gene and upstream region. This was accomplished by using the allelic recovery plasmid pSS2197 to recover this fragment from the mutant strains and reintroduce it into the BP953 genetic background according to procedures described elsewhere (34). Except in the case of bvg-1247, which appeared to map to the region encoding the periplasmic domain of BvgS, the mutation was associated with the EcoRI-SphI bvgA fragment in all cases. Six mutants of this class were selected for further characterization. The results of enzyme assays performed to quantitate the differential expression of ptx-phoA and fhaB-lacZ in these strains are presented in Table 2 (chromosomal bvgAS mutations). The expression of the ptx locus in all six strains is reduced to very low levels, while expression of the fha locus is reduced at most twofold. In all cases, the expression of fha-lacZ was repressed by addition of MgSO4 to the growth media, demonstrating that these mutations did not affect modulation mediated by the bvgAS locus (data not shown).
The EcoRI-SphI bvgA fragments from five of these six mutant strains (the exception being BP1247) were subjected to DNA sequence analysis. In all five cases, the mutation was found to be the result of an insertion sequence. Three examples of IS481 and two examples of IS1002 were found. In all five cases, insertion occurred at exactly the same spot between the bvgA and bvgS genes such that neither open reading frame was disrupted. Restriction analysis of the remaining 8 mutants in this class suggested that a similar insertion mutation had occurred in 14 of the 15 mutants (data not shown). These mutations would be expected to affect the level of BvgA expression and/or activity by reducing expression of BvgS, possibly allowing its expression, at a lower level, from a promoter contained in IS481 or its relative IS1002. The ability of IS481 to direct transcription of a neighboring gene from a promoter within IS481 has been observed previously (12). Western blot analysis in fact demonstrated that BvgS levels were markedly reduced in these mutant strains (data not shown). Therefore, it appears that these mutations relieve inhibition of growth by reducing expression of the BvgAΔC3 protein, and the differential Bvg phenotype is a result of lower levels of BvgA activity, in a manner similar to that suggested above and illustrated in Fig. 4.
Chromosomal rpoA-linked mutations conferring a differential Bvg phenotype.
Mutations affecting RpoA have been shown previously to cause a differential Bvg phenotype (9). One of these previously described rpoA alleles was examined and was also found to relieve the growth-inhibitory phenotype conferred by pSS2244 but apparently not to the same degree, as colony size was reduced. This rpoA allele was recovered from its parental strain RPV5 and transferred by allelic exchange to the BP953 chromosome to create BP1242. A quantitative comparison of the regulatory phenotypes caused by rpoA-linked mutations isolated in this study with that from RPV5 is shown in Table 2 (rpoA-linked mutations). It can be seen from these data that the effects of these mutations were similar, although BP1187, BP1196, and BP1246 showed a somewhat more pronounced differential effect.
The rpoA allele from RPV5 directs increased expression of the protein encoded by this gene, the α subunit of RNA polymerase (9). To determine whether the same was true of the rpoA-linked mutations isolated here, Western analysis was performed. As shown in Fig. 5, both BP1196 and BP1246 show increased expression of the α subunit, while BP1187 displays wild-type levels. The magnitude of this increased expression appears comparable to that previously reported for RPV5, i.e., approximately twofold (9). Thus, although the exact locations of the mutations in these strains have not been determined, their effect appears to be due to an increase in the amount of the α subunit.
FIG. 5.
Western analysis of RNA polymerase α subunit expression. Samples were whole-cell extracts of bacterial suspensions normalized by optical density to contain the same numbers of cells. The blot was probed with monoclonal antibody 4RA2, which recognizes an epitope on the α subunit conserved between E. coli and B. pertussis, kindly provided by Nancy Thompson and Richard Burgess. Sizes are indicated in kilodaltons. Lanes: a, BP953; b, BP1187; c, BP1196; d, BP1246.
Plasmid pSS2306, which contains a 7-kb BamHI fragment bearing the wild-type rpoA gene and flanking sequences, was used to test these three strains for restoration of the wild-type phenotype by allelic exchange. By this method, only rpoA1187 was demonstrated to be within this 7-kb BamHI fragment. DNA sequence analysis of the rpoA gene from BP1187 revealed the presence of a missense mutation in the coding region of this gene whereby aspartic acid residue 171 is changed to a glycine. This mutation is shown in Fig. 6, where the B. pertussis RpoA sequence is aligned with that of E. coli. It can be seen that the residue affected is near the middle of the α subunit sequence, in the N-terminal domain, a location not typically seen for mutations in the α subunit that affect its interaction with positive regulatory factors.
FIG. 6.
Amino acid sequence alignment of the α subunits of B. pertussis and E. coli. Alignment was performed at the National Center for Biotechnology Information by using the LFASTA network service. The substitution in BP1187 is indicated above the B. pertussis RpoA (BpeRpoA) sequence, and the position of a mutation in the E. coli α subunit affecting activation of class II promoters (22) is marked by an asterisk below the E. coli RpoA (EcoRpoA) sequence.
DISCUSSION
An unexpected phenotype results from the expression, in trans, of deletions of two or three amino acid residues from the C terminus of the response regulator protein BvgA of B. pertussis. This phenotype, a profound inhibition of growth, is proposed to be the result of an inappropriate interaction of the mutant BvgA protein with the RNA polymerase enzyme, specifically the α subunit. Experimental support for this hypothesis comes from the observation that mutations which alter the amino acid sequence of the α subunit or which increase levels of the α subunit can relieve this inhibition of growth.
The portion of the BvgA molecule associated with this growth inhibition, and by inference the interaction with the α subunit, is the C terminus, downstream of the putative helix-turn-helix DNA binding motif. Experimental support for this conclusion comes from the observation that deletion of this region in BvgAΔC18 relieves the growth inhibition, as do larger truncations isolated as spontaneous mutations of the plasmid-borne bvgAΔC3 gene. It is suggested that this portion of the BvgA molecule normally interacts with the α subunit but in the BvgΔC2 and BvgΔC3 deletions does so in an uncontrolled or otherwise inappropriate way. Results from studies on other response regulators, with sequence similarity to BvgA in the C-terminal domain, are consistent with a role for the C-terminal sequence in interaction with RNA polymerase. For example, short C-terminal truncations result in an inability of LuxR to activate transcription, although it appears to be able to bind DNA normally (10). Similarly, UhpA C-terminal deletions are inactive in transcriptional activation of the uhpT promoter (38). The Bvg− phenotypes observed here with chromosomally encoded BvgA C-terminal deletions are consistent with these observations in that removal of as few as two amino acids abolishes BvgA activity. Interestingly, this region of the BvgA molecule includes the sites of amino acid substitutions in two mutants which no longer activate ptx transcription but which activate fha transcription normally (30), consistent with the hypothesis that the differential effect of those mutations involves a change in the interaction of BvgA with RNA polymerase at the ptx promoter.
The growth-inhibitory phenotype observed with bvgAΔC3 in trans can also be overcome by mutations which affect the wild-type chromosomal bvg locus, either by eliminating expression (bvg knockouts) or by reducing expression and/or activity, through the insertion of IS481 or IS1002 just upstream of the bvgS gene. In the latter case, the differential phenotype observed in terms of ptx and fha transcription is likely to be due to differences in the amount of BvgA-phosphate required for maximal expression at these two loci. In a similar fashion, the truncated BvgA molecules presented in Fig. 3 may result in intermediate levels of activity, thus explaining the differential phenotype observed under these conditions. One explanation for this finding would involve the formation of mixed dimers of BvgA. Several different experimental approaches have suggested that BvgA forms dimers (2, 4, 24). Alternatively, these truncated derivatives may compete with wild-type BvgA for phosphorylation by BvgS. Genetic analysis of a similar trans-dominant phenotype conferred by truncated UhpA molecules would suggest that the latter possibility is most likely correct (38). The fact that several of the mutant strains harboring chromosomal bvg mutations have reduced transcriptional activity of fha-lacZ is consistent with their very low levels of ptx-phoA expression and the observation that the ranges of responsiveness of these two fusions to modulating signals overlap somewhat (Fig. 4). This finding suggests that these strains have even lower levels of Bvg activity than the plasmid-bearing strains shown above them in Table 2.
The dependence of the growth-inhibitory phenotype on expression of an intact bvg locus can be explained in several ways. BvgAS may simply be required for the expression of bvgAΔC3, itself a bvg-regulated gene. Alternatively, wild-type BvgA protein may play a direct role in the growth inhibition, perhaps through interaction with BvgAΔC3 to form a heteromultimer, which would then be the toxic form of the protein. To date, our efforts to distinguish between these two possibilities by expressing the bvgAΔC3 gene from a non-bvg-regulated promoter have been hampered by the unavailability of a promoter that is inducible in B. pertussis and that is of sufficient strength to achieve the growth-inhibitory phenotype, even in the presence of a wild-type chromosomal copy of bvgAS (data not shown). A third possibility is that manifestation of toxicity requires phosphorylation of BvgAΔC3 by BvgS. This hypothesis is currently being addressed by assessing the toxicity of BvgAΔC3 into which a D54N mutation has been incorporated, thus rendering it incapable of phosphorylation.
It has been previously reported that mutations affecting the level of the α subunit in B. pertussis affect bvg-regulated gene expression (9). In this case, the effect seen was a differential phenotype (Fha+ Ptx− Cya−). This has been interpreted to possibly indicate (i) an interaction between the α subunit and BvgA and (ii) titration of active BvgA-phosphate by excess α subunit. Biochemical data provide strong evidence of such an interaction as well, in that the C-terminal portion of the α subunit is required for bvg-activated transcription of the fha gene (6). The results reported here provide additional genetic data supporting such an interaction in that mutations which increase α subunit levels suppress the phenotype associated with a mutant BvgA protein.
Interestingly, a mutation affecting the primary structure of the α subunit was also found to overcome the growth inhibition conferred by BvgAΔC3. Typically, mutations in rpoA which suppress mutations in positive regulatory proteins have been found in the C-terminal third of this protein, a region believed to make up a separate conformational domain which is attached to the N-terminal portion of α by a flexible peptide linker (15). Two possible explanations for the effect of this mutation are proposed. One is that this mutation defines a site on the N-terminal domain of α which contacts the BvgA molecule in the process of transcriptional activation of the ptx promoter. However, this would not be the only contact required, because in vitro, BvgA-activated transcription of the ptx promoter is dependent on the C-terminal domain of the α subunit as well (7). A precedent for interaction of the N-terminal portion of α in transcriptional activation comes from recent reports of catabolite gene activator protein-activated transcription of the synthetic class II promoter CC(−41.5) in E. coli. In this case, a mutation affecting the glutamic acid residue at position 165 of the E. coli α subunit had a significant negative effect (22). Interestingly, as shown in Fig. 5, the aspartic acid residue that was identified in the B. pertussis α subunit, at position 171, is nine amino acids away from the B. pertussis α residue corresponding to the E. coli α glutamic acid 165 and may represent an analogous mutation. Another possibility is that the glycine 171 mutation affects the assembly of α into the RNA polymerase core enzyme, either in terms of the efficiency of assembly or in terms of the configuration adopted by α in the assembled enzyme. If efficiency of assembly was low, unincorporated α subunit could accumulate with a differential effect on transcription of the ptx and fha genes as previously suggested (9). However, inefficient assembly either would be expected to result in a reduced growth rate due to reduced levels of RNA polymerase or would require higher levels of α to maintain the same level of RNA polymerase. However, neither of these conditions was observed. Fortuitously, the contribution of the corresponding residue in the E. coli α subunit on assembly of RNA polymerase in vitro has been previously examined. It was found that a change of this aspartic acid at position 174 to an alanine negatively affected dimerization of the α subunit, the initial step in assembly, but subsequent assembly of α2β and α2ββ′ was normal (18). Inferring from this study and the observation of normal growth of the mutant B. pertussis strain, we may still ask whether the configuration and presentation of the mutant α subunit in the assembled RNA polymerase might not be altered and thus affect interaction with the BvgA protein at the ptx promoter. Clearly more study is required to distinguish between these models and to explain the behavior of this interesting mutant.
ACKNOWLEDGMENTS
I gratefully acknowledge Mei-Shin Yang for invaluable technical assistance, Nancy Thompson and Richard Burgess for providing monoclonal antibody directed against the α subunit, Richard Ebright, Phil Boucher, and Nicholas Carbonetti for useful discussions, and Joseph Devito, Michael Schmitt, and Judy Kassis for critical reading of the manuscript.
REFERENCES
- 1.Beier D, Deppisch H, Gross R. Conserved sequence motifs in the unorthodox BvgS two-component sensor protein of Bordetella pertussis. Mol Gen Genet. 1996;252:169–176. doi: 10.1007/BF02173217. [DOI] [PubMed] [Google Scholar]
- 2.Beier D, Schwarz B, Fuchs T M, Gross R. In vivo characterization of the unorthodox BvgS two-component sensor protein of Bordetella pertussis. J Mol Biol. 1995;248:596–610. doi: 10.1006/jmbi.1995.0245. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1520. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher P E, Menozzi F D, Locht C. The modular architecture of bacterial response regulators. Insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J Mol Biol. 1994;241:363–377. doi: 10.1006/jmbi.1994.1513. [DOI] [PubMed] [Google Scholar]
- 5.Boucher P E, Stibitz S. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J Bacteriol. 1995;177:6486–6491. doi: 10.1128/jb.177.22.6486-6491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucher P E, Murakami K, Ishihama A, Stibitz S. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J Bacteriol. 1997;179:1755–1763. doi: 10.1128/jb.179.5.1755-1763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher, P. Unpublished data.
- 8.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 9.Carbonetti N H, Fuchs T M, Patamawenu A A, Irish T J, Deppisch H, Gross R. Effect of mutations causing overexpression of RNA polymerase α subunit on regulation of virulence factors in Bordetella pertussis. J Bacteriol. 1994;176:7267–7273. doi: 10.1128/jb.176.23.7267-7273.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi S H, Greenberg E P. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci USA. 1991;88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLorenzo V, Herrero M, Jakubzik J, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeShazer D, Wood G E, Friedman R L. Molecular characterization of catalase from Bordetella pertussis: identification of the katA promoter in an upstream insertion sequence. Mol Microbiol. 1994;14:123–130. doi: 10.1111/j.1365-2958.1994.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 13.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figurski D, Meyer R, Helinski D R. Suppression of ColEI replication properties by the incP-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979;133:295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
- 15.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimova G, Bellalou J, Ullmann A. Phosphorylation-dependent binding of BvgA to the upstream region of the cyaA gene of Bordetella pertussis. Mol Microbiol. 1996;20:489–496. doi: 10.1046/j.1365-2958.1996.5231057.x. [DOI] [PubMed] [Google Scholar]
- 17.Kasuga T, Nakase Y, Ukishima K, Takatsu K. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch Exp Med. 1954;27:57–62. [PubMed] [Google Scholar]
- 18.Kimura M, Ishihama A. Functional map of the α subunit of Escherichia coli RNA polymerase: amino acid substitution within the amino-terminal assembly domain. J Mol Biol. 1995;254:342–349. doi: 10.1006/jmbi.1995.0621. [DOI] [PubMed] [Google Scholar]
- 19.Lacey B W. Antigenic modulation of Bordetella pertussis. J Hyg. 1960;31:423–434. doi: 10.1017/s0022172400038134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merkel T J, Stibitz S. Identification of a locus required for the regulation of bvg-repressed genes in Bordetella pertussis. J Bacteriol. 1995;177:2727–2736. doi: 10.1128/jb.177.10.2727-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy C, Miller J F, Falkow S. Autogenous regulation of the bvgABC operon of the bacterial pathogen Bordetella pertussis. Proc Natl Acad Sci USA. 1990;87:3763–3767. doi: 10.1073/pnas.87.10.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarlato V, Prugnola A, Arico B, Rappuoli R. Positive transcriptional feedback at the bvg locus controls expression of virulence factors in Bordetella pertussis. Proc Natl Acad Sci USA. 1990;87:6753–6757. doi: 10.1073/pnas.87.17.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarlato V, Arico B, Prugnola A, Rappuoli R. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J. 1991;10:3971–3975. doi: 10.1002/j.1460-2075.1991.tb04967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–789. [Google Scholar]
- 27.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 28.Steffen P, Goyard S, Ullmann A. Phosphorylated BvgA is sufficient for transcriptional activation of virulence-regulated genes in Bordetella pertussis. EMBO J. 1996;15:102–109. [PMC free article] [PubMed] [Google Scholar]
- 29.Stibitz S, Yang M-S. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J Bacteriol. 1991;173:4288–4296. doi: 10.1128/jb.173.14.4288-4296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stibitz S. Mutations in the bvgA gene of Bordetella pertussis that differentially affect regulation of virulence determinants. J Bacteriol. 1994;176:5615–5621. doi: 10.1128/jb.176.18.5615-5621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 32.Stibitz S, Carbonetti N H. Hfr mapping of mutations in Bordetella pertussis that define a genetic locus involved in virulence gene regulation. J Bacteriol. 1994;176:7260–7266. doi: 10.1128/jb.176.23.7260-7266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stibitz S, Miller J F. Coordinate regulation of virulence in Bordetella pertussis mediated by the vir (bvg) locus. In: Miller V L, Kaper J B, Portnoy D A, Isberg R I, editors. Molecular genetics of bacterial pathogenesis. Washington, D.C: American Society for Microbiology; 1994. [Google Scholar]
- 34.Stibitz S. Allelic retrieval: a scheme to facilitate the repeated isolation of a specific segment of the Bordetella pertussis chromosome. Gene. 1998;208:183–189. doi: 10.1016/s0378-1119(97)00643-4. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, N. E. Unpublished data.
- 36.Uhl M A, Miller J F. BvgAS is sufficient for activation of the Bordetella pertussis ptx locus in Escherichia coli. J Bacteriol. 1995;177:6477–6485. doi: 10.1128/jb.177.22.6477-6485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 38.Webber C A, Kadner R J. Action of receiver and activator modules of UhpA in transcriptional control of the Escherichia coli sugar phosphate transport system. Mol Microbiol. 1995;15:883–893. doi: 10.1111/j.1365-2958.1995.tb02358.x. [DOI] [PubMed] [Google Scholar]
- 39.Weiss A A, Hewlett E L, Meyers G A, Falkow S. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect Immun. 1983;42:33–41. doi: 10.1128/iai.42.1.33-41.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, M.-S., and S. Stibitz. Unpublished data.
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 42.Zu T, Manetti R, Rappuoli R, Scarlato V. Differential binding of BvgA to two classes of virulence genes of Bordetella pertussis directs promoter selectivity by RNA polymerase. Mol Microbiol. 1996;21:557–565. doi: 10.1111/j.1365-2958.1996.tb02564.x. [DOI] [PubMed] [Google Scholar]